Abstract
The Clostridium difficile toxin B is one of the main virulence factors and plays an important role in the pathogenesis of C. difficile infection (CDI). We recently revealed crucial residues in the translocation domain of TcdB for the pore formation and toxin translocation. In this study, we investigated the effects of mutating a critical site involved in pore formation, Leu-1106, to residues that differ in size and polarity (Phe, Ala, Cys, Asp). We observed a broad range of effects on TcdB function in vitro consistent with the role of this site in pore formation and translocation. We show that mice challenged systemically with a lethal dose (LD100) of the most defective mutant (L1106K) showed no symptoms of disease highlighting the importance of this residue and the translocation domain in disease pathogenesis. These findings offer insights into the structure function of the toxin translocation pore, and inform novel therapeutic strategies against CDI.
Keywords: Clostridium difficile, toxin, TcdB, translocation domain
Defection of pore formation in translocation domain of Clostridium difficile toxin B significantly reduces its in vitro and in vivo toxicity.
Graphical Abstract Figure.
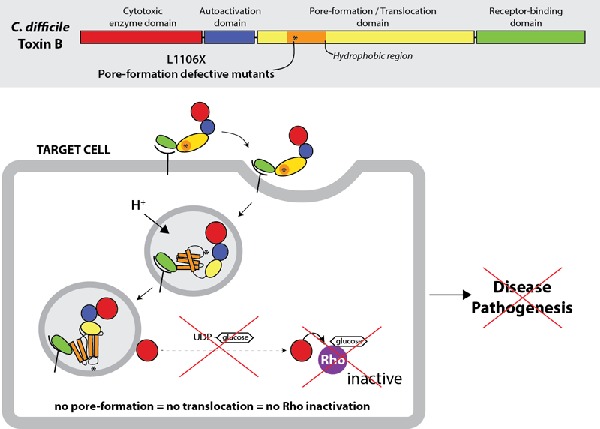
Defection of pore formation in translocation domain of Clostridium difficile toxin B significantly reduces its in vitro and in vivo toxicity.
INTRODUCTION
Clostridium difficile (C. difficile) is the major cause of antibiotic-associated diarrhea and pseudomembranous colitis and is associated with significant morbidity and mortality as well as financial costs. The incidence of C. difficile infections (CDI) has dramatically increased in the past two decades partially due to the emergence of hypervirulent strains that are resistant to antimicrobial therapy (McDonald et al. 2005; Kelly and LaMont 2008).
CDI pathogenesis is mainly mediated by the disruption of colonic microflora due to antibiotic usage leading to C. difficile colonization and the production of two large toxins, toxin A (TcdA) and toxin B (TcdB) (Lyerly et al. 1985; Voth and Ballard 2005). Both toxins consist of at least four key domains, the N-terminal glucosyltransferase domain (GTD), cysteine protease domain (CPD), translocation domain (TD) and C-terminal receptor-binding domain (Jank and Aktories 2008). Toxins may enter cells by clathrin-mediated endocytosis (Papatheodorou et al. 2010) where acidification causes conformational changes that result in the ion-conductive pore formation (Qa'dan et al. 2005; Zhang et al. 2014). Both GTD and CPD are translocated into cytosol (Li et al. 2013) where inositol hexakisphosphate activates CPD leading to the autoprocessing and the release of GTD (Egerer et al. 2007; Reineke et al. 2007). In the cytosol, the GTD inactivates host Rho-GTPases regulatory proteins leading to disorganization of the actin cytoskeleton and cell death (Jank and Aktories 2008).
Our previous work focused on identifying the elements in pore formation of TcdB. We have identified several single-point mutations within the hydrophobic region of TD that result in major defects in pore formation and translocation. Among those point mutations, position L1106K displayed the most defective in toxicity with over 3 logs relative to wild-type TcdB (Zhang et al. 2014). L1106K mutant showed a defect of pore formation both in an in vitro system (planar lipid bilayer electrophysiology) and on biological membranes (86rubidium release of ions from CHO-K1 cells upon binding to the cell membrane). In this study, we further investigated the role of defective pore formation mutations within translocation of TcdB in CDI pathogenesis.
MATERIALS AND METHODS
Generation of recombinant wild-type TcdB and TD mutants
Recombinant TcdB wild type was made from a Bacillus megaterium expression vector pHis1522 encoding strain VPI10463 (Yang et al. 2008). Proteins were expressed and purified as previously described (Zhang et al. 2014). Single-point mutations at 1106 site with residues differing in size and/or polarity were made in the TcdB sequence using QuikChange lightning multimutagenesis kit (Agilent Technologies). Sequenced plasmids with confirmed mutations were transformed and expressed as wild type. The purity of toxins was tested on Coomassie blue stained Novex® 4–20% Tris-glycine protein gel (Invitrogen).
Cell viability assay
Chinese hamster ovary cells CHO-K1 cells were cultured in Ham's F-12 medium supplemented with 10% FBS and 1% penicillin and streptomycin (all from Wisent) and seeded in 96-well CellBind plates (Corning). The next day, medium was exchanged with serum-free medium and cells were intoxicated with either wild-type TcdB or TD mutant toxins at a serial dilution of one-third starting at 1 nM for 48 h. The cell viability was assessed by PrestoBlue Cell Viability Reagent (Life Technologies). Fluorescence was read on a Spectramax M5 plate reader (Molecular Devices).
Mouse systemic toxin challenge
Six- to eight-week-old female CD1 mice were purchased from Harlan Laboratory (IN, USA). Mice were housed in pathogen-free facilities. The animal study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the University of Maryland Dental School (protocol #06-14-007). Mice were euthanized when became moribund after toxin challenge, and all efforts were made to minimize suffering. To assess in vivo toxicity of the toxins (TcdB and L1106K), groups of mice were challenged intraperitoneally (IP) with either lethal dose (LD100) of wild-type TcdB (5 μg kg−1 mouse) or (0.5 g kg−1 mouse) of L1106K (100× LD100 of TcdB). Mouse survival was monitored and data were analyzed by Kaplan–Meier survival analysis with Log rank test of significance.
Rubidium release assay
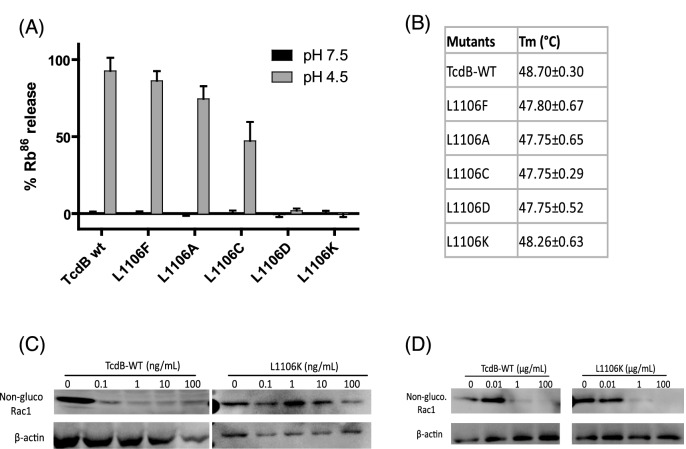
86Rb+ release assay was performed as previously reported (Genisyuerek et al. 2011) with slight modifications. Briefly, CHO-K1 cells were seeded in 96-well plates in the medium (Ham's F-12 with 10% FBS), supplemented with 1 μCi mL−1 86Rb+ (PerkinElmer) at a density of 1 × 104 cells per well. Cells were incubated at 37°C, 5% CO2 overnight. Medium was exchanged with fresh growth medium with 100 nM bafilomycin A1 (Sigma) and continued to incubate for another 20 min. Then, cells were chilled on ice and ice-cold medium containing TcdB mutants (10 nM) was added. Cells were kept on ice for toxin binding for 1 h at 4°C before they were washed with ice-cold PBS twice to remove unbound toxins. pH-dependent insertion into the plasma membrane was induced by warm, acidified growth medium (37°C, pH 4.5 or pH 7.5) for 5 min at 37°C. After 1 h of further incubation on ice, medium containing released 86Rb+ was removed from the cell plate and the amount of 86Rb+ released was determined by liquid scintillation counting with TopCount NXT (PerkinElmer).
Differential scanning fluorometry
Differential scanning fluorometry was performed in a similar manner as described previously (Tam et al. 2015). TcdB protein was diluted in phosphate buffer (100 mM KPO4, 150 mM NaCl, pH 7) containing 5× SYPRO Orange (Invitrogen). A Bio-Rad CFX96 qRT-PCR thermocycler was used to establish a temperature gradient from 15°C to 95°C in 30 s increments, while simultaneously recording the increase in SYPRO Orange fluorescence as a consequence of binding to hydrophobic regions exposed on unfolded proteins. The Bio-Rad CFX Manager 3.1 software was used to integrate the fluorescence curves to calculate the melting point.
Glucosyltransferase activity of the toxins
The wild-type and mutant toxins were assessed their glucosyltransferase activity by assaying glucosylation of the Rho GTPase Rac1, both intracellularly and in cell-free assays (Zhang et al. 2013). In intracellular assay, Vero cells in 12-well plates were exposed to different concentrations of toxins for 4 h at 37°C. Cells were lysed with SDS sampling buffer. In cell-free assays, Vero cell pellets were resuspended in glucosylation buffer (50 mM HEPES, pH 7.5, 100 mM KCl, 1 mM MnCl2 and 2 mM MgCl2) and lysed with a syringe (25G, 40 passes through the needle). After centrifugation (15 000 rpm, 15 min), the cell lysate was incubated with different doses of TcdB, or L1106K at 37°C for 1 h. The reaction was terminated by heating at 100°C for 5 min in SDS sample buffer. In both cell-free and intracellular assays, Rac1 glucosylation was detected using antibody that specifically recognizes the non-glucosylated form of Rac1 (clone 102, BD Bioscience). Anti-β-actin (clone AC-40, Sigma) was used to detect β-actin and ensure that samples were loaded evenly on the SDS polyacrylamide gels.
RESULTS
Generation of the functional residues at amino acid 1106
We investigated the role of residues of different size and/or polarity at 1106 on function.
CHO-K1 cells were treated with TcdB wild type or mutants for 48 h, and cell viability was assessed by measuring the fluorescence of cell treated with PrestoBlue cell viability reagent. Substitutions to hydrophobic residues of any size (Leu, Ala, Phe) had the least impact on TcdB translocation, followed by polar/uncharged (Cys), then acidic (Asp) and thenbasic substitutions (Lys) (Fig. 1A). This is consistent with the idea that this residue is a key site for membrane insertion; the more polar the residue at this position, the greater the impact on pore formation/translocation and thus intoxication.
Figure 1.
In vitro and in vivo toxicity of mutant TcdB. (A) CHO-K1 cells were treated with TcdB wild-type or mutants and cell viability was measured by fluorescence of cells treated with cell viability reagent (PrestoBlue). (B) CD1 mice were challenged IP with either LD100 of toxins (5 μg kg−1 mouse) or 100× LD100 of L1106K (0.5 g kg−1 mouse). Mouse survival was monitored and data were analyzed by Kaplan–Meier survival curve with Log-rank test of significance. (n = 5, P < 0.0001, between wild-type and mutant toxin groups).
L1106K mutation diminishes its activity in vivo
Reduced cellular in vitro toxicity of L1106K mutant led us to assess toxicity of wild-type TcdB and the most defective mutant-L1106K by challenging the mice systemically (IP). Mice challenged with LD100 (5 μg kg−1 mouse) of TcdB developed signs of systemic disease rapidly and all became moribund and died within six hours. In contrast, none of mice challenged with L1106K mutant at the same dose showed any systemic disease symptoms, while mice challenged with 100× LD100 (0.5g kg−1 mouse) of L1106K mutant showed signs of disease 48 h post-challenge (Fig. 1B). This data clearly demonstrate that mutation at 1106 residue is critical for TcdB toxicity and L1106K mutant is substantially attenuated in in vivo toxicity.
Characterization of L1106X activities in vitro
In an attempt to ascribe a molecular mechanism to functional consequences observed on toxin function, we conducted further in vitro studies on the various L1106X mutants. Wild-type toxin and mutant were tested for the ability to release 86Rubidium ions from CHO-K1 cells upon binding to the cell surface and acidification of the medium to trigger insertion into the plasma membrane. As shown in Fig. 2A, a remarkable range of effects on pore formation are seen upon substituting residues position 1106. To show that these effects were not due to toxin misfolding, the protein thermal stability of each toxin was tested (Fig. 2B). To check for the translocation ability of TcdB and L1106K mutant, we performed for Rac1 glucosylation intracellularly and in cell-free assays. Glucosylated Rac1 levels in Vero cells did not change with increasing concentration of TcdB mutant (Fig. 2C). On the other hand, L1106K showed similar activity in Rac1 glucosylation in cell-free assay (Fig. 2D). These data suggest that L1106K mutant has an intact glucosyltransferase activity and structure as wild type but is defective in pore formation that is essential for cytosolic translocation of glucosyltransferase domain of TcdB.
Figure 2.
Characterization of mutant TcdB. (A) Pore formation on biological membranes. Pore formation of purified mutant toxins was tested on CHO cells preloaded with 86Rb+. Pore formation was induced by acidification of the external medium (control pH 7.5; black bars, pH 4.5; gray bars)—see the section ‘Materials and Methods’ for details of assay (n = 2). (B) Temperature-dependent fluorescence measurements of TcdB melting temperature (Tm). Values represent the mean ± SD from three independent experiments. (C) Glucosyltransferase activity of the mutant toxins. Vero cells were exposed to different concentrations of TcdB-WT or L1106K mutant for 4 h. (D) Vero cell cytosolic fraction was collected and exposed to wild-type and mutant toxins at the indicated concentrations for 1 h. In both C and D, western blot was performed using monoclonal antibody (Clone 102) that only binds to non-glucosylated Rac1. β-actin was used as an equal loading control.
DISCUSSION
TcdB is one of the essential virulence factors of C. difficile as a number of pathogenic, clinically isolated strains express functional TcdB alone (Johnson et al. 2001; Drudy, Fanning and Kyne 2007). The role of enzymatic domains of TcdB in disease pathogenesis has been studied recently (Li et al. 2015; Yang et al. 2015); however, information regarding the TD and pore formation in pathogenesis has not yet been reported.
In this study, we demonstrate that defective variants of TcdB are defined by impaired pore formation, which results in reducing toxicity to cells and animals. We have identified several residues located between amino acids 1035 and 1107 that when individually mutated, markedly reduced the toxic activity by over 1000-fold. Therefore, this sensitive segment appears to be a crucial structure for the function of TcdB translocation and pore formation (Zhang et al. 2014). The observation that L1106K toxicity is substantially reduced in vitro and in vivo yet maintains its cellular binding, internalization and glucosyltransferase activities. Zhang et al. (2014) highlights the importance of this domain in translocating TcdB across the endosomal membrane into the cytosol and CDI pathogenesis.
We have previously demonstrated that pore-defective mutants have reduced toxicity in cell culture (Zhang et al. 2014). In this study, we further assessed the role of residues that differs in size and/or polarity at 1106 on function. Reduced cellular toxicity was seen in cells treated with hydrophobic residues (Leu, Ala, Phe), followed by Cys, then Asp and then basic substitutions (Lys) (Fig. 1A). This is consistent with the idea that this residue is a key site for membrane insertion; more polar, more defective in pore formation resulting in an inability to intoxicate cells.
CDI is the leading cause of healthcare-associated diarrhea in Europe and North America (Poutanen and Simor 2004; Elliott et al. 2007). Colonization of C. difficile in the intestinal mucosa leads to toxins production and leakage to the systemic circulation (Steele et al. 2012; Yu et al. 2015) causing adverse effects which includes disruption of cytoskeleton, diarrhea, colitis and death (Voth and Ballard 2005). In this study, we observed typical signs of systemic disease in mice challenged with LD100 (5 μg kg−1 mouse) of wild-type TcdB while a similar dose of L1106K exhibited no disease symptoms. Only at a dose of 100× LD100 (0.5 g kg−1 mouse), L1106K caused 40% death rate. Thus, mutations in TD not only hold a great promise to understand the mechanism of toxin translocation through pore formation, but also provide insights in the development of putative vaccines against hypervirulent multidrug-resistant bacteria such as C. difficile.
Conflict of interest. None declared.
REFERENCES
- Drudy D, Fanning S, Kyne L. Toxin A-negative, toxin B-positive Clostridium difficile. Int J Infect Dis. 2007;11:5–10. doi: 10.1016/j.ijid.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Egerer M, Giesemann T, Jank T, et al. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J Biol Chem. 2007;282:25314–21. doi: 10.1074/jbc.M703062200. [DOI] [PubMed] [Google Scholar]
- Elliott B, Chang BJ, Golledge CL, et al. Clostridium difficile-associated diarrhoea. Intern Med J. 2007;37:561–8. doi: 10.1111/j.1445-5994.2007.01403.x. [DOI] [PubMed] [Google Scholar]
- Genisyuerek S, Papatheodorou P, Guttenberg G, et al. Structural determinants for membrane insertion, pore formation and translocation of Clostridium difficile toxin B. Mol Microbiol. 2011;79:1643–54. doi: 10.1111/j.1365-2958.2011.07549.x. [DOI] [PubMed] [Google Scholar]
- Jank T, Aktories K. Structure and mode of action of clostridial glucosylating toxins: the ABCD model. Trends Microbiol. 2008;16:222–9. doi: 10.1016/j.tim.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Johnson S, Kent SA, O'Leary KJ, et al. Fatal pseudomembranous colitis associated with a variant Clostridium difficile strain not detected by toxin A immunoassay. Ann Intern Med. 2001;135:434–8. doi: 10.7326/0003-4819-135-6-200109180-00012. [DOI] [PubMed] [Google Scholar]
- Kelly CP, LaMont JT. Clostridium difficile—more difficult than ever. N Engl J Med. 2008;359:1932–40. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- Li S, Shi L, Yang Z, et al. Cytotoxicity of Clostridium difficile toxin B does not require cysteine protease-mediated autocleavage and release of the glucosyltransferase domain into the host cell cytosol. Pathog Dis. 2013;67:11–8. doi: 10.1111/2049-632X.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shi L, Yang Z, et al. Critical roles of Clostridium difficile toxin B enzymatic activities in pathogenesis. Infect Immun. 2015;83:502–13. doi: 10.1128/IAI.02316-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyerly DM, Saum KE, MacDonald DK, et al. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun. 1985;47:349–52. doi: 10.1128/iai.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. New Engl J Med. 2005;353:2433–41. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- Papatheodorou P, Zamboglou C, Genisyuerek S, et al. Clostridial glucosylating toxins enter cells via clathrin-mediated endocytosis. PLoS One. 2010;5:e10673. doi: 10.1371/journal.pone.0010673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutanen SM, Simor AE. Clostridium difficile-associated diarrhea in adults. CMAJ. 2004;171:51–8. doi: 10.1503/cmaj.1031189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qa'dan M, Christensen KA, Zhang L, et al. Membrane insertion by anthrax protective antigen in cultured cells. Mol Cell Biol. 2005;25:5492–8. doi: 10.1128/MCB.25.13.5492-5498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke J, Tenzer S, Rupnik M, et al. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446:415–9. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- Steele J, Chen K, Sun X, et al. Systemic dissemination of Clostridium difficile toxins A and B is associated with severe, fatal disease in animal models. J Infect Dis. 2012;205:384–91. doi: 10.1093/infdis/jir748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J, Beilhartz GL, Auger A, et al. Small molecule inhibitors of Clostridium difficile toxin B-induced cellular damage. Chem Biol. 2015;22:175–85. doi: 10.1016/j.chembiol.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18:247–63. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Zhou B, Wang J, et al. Expression of recombinant Clostridium difficile toxin A and B in Bacillus megaterium. BMC Microbiol. 2008;8:192. doi: 10.1186/1471-2180-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhang Y, Huang T, et al. Glucosyltransferase activity of Clostridium difficile toxin B is essential for disease pathogenesis. Gut Microbes. 2015:221–4. doi: 10.1080/19490976.2015.1062965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen K, Wu J, et al. Identification of toxemia in patients with Clostridium difficile infection. PLoS One. 2015;10:e0124235. doi: 10.1371/journal.pone.0124235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shi L, Li S, et al. A segment of 97 amino acids within the translocation domain of clostridium difficile toxin B is essential for toxicity. PLoS One. 2013;8:e58634. doi: 10.1371/journal.pone.0058634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Park M, Tam J, et al. Translocation domain mutations affecting cellular toxicity identify the Clostridium difficile toxin B pore. P Natl Acad Sci USA. 2014;111:3721–6. doi: 10.1073/pnas.1400680111. [DOI] [PMC free article] [PubMed] [Google Scholar]