Abstract
BACKGROUND
Prostate tumors shed circulating tumor cells (CTCs) into the blood stream. Increased evidence shows that CTCs are often present in metastatic prostate cancer and can be alternative sources for disease profiling and prognostication. Here we postulate that CTCs expressing genes related to epithelial-mesenchymal transition (EMT) are strong predictors of metastatic prostate cancer.
METHODS
A microfiltration system was used to trap CTCs from peripheral blood based on size selection of large epithelial-like cells without CD45 leukocyte marker. These cells individually retrieved with a micromanipulator device were assessed for cell membrane physical properties using atomic force microscopy. Additionally, 38 CTCs from eight prostate cancer patients were used to determine expression profiles of 84 EMT-related and reference genes using a microfluidics-based PCR system.
RESULTS
Increased cell elasticity and membrane smoothness were found in CTCs compared to noncancerous cells, highlighting their potential invasiveness and mobility in the peripheral circulation. Despite heterogeneous expression patterns of individual CTCs, genes that promote mesenchymal transitioning into a more malignant state, including IGF1, IGF2, EGFR, FOXP3, and TGFB3, were commonly observed in these cells. An additional subset of EMT-related genes (e.g., PTPRN2, ALDH1, ESR2, and WNT5A) were expressed in CTCs of castration-resistant cancer, but less frequently in castration-sensitive cancer.
CONCLUSIONS
The study suggests that an incremental expression of EMT-related genes in CTCs is associated with metastatic castration-resistant cancer. Although CTCs represent a group of highly heterogeneous cells, their unique EMT-related gene signatures provide a new opportunity for personalized treatments with targeted inhibitors in advanced prostate cancer patients.
Keywords: prostate cancer, circulating tumor cells, metastasis
INTRODUCTION
During the formation and growth of a prostate tumor, malignantly transformed cells can be shed from the primary site and circulate in the bloodstream. These circulating tumor cells (CTCs) are found at very low levels, one in a billion blood cells, and most die in the circulation [1,2]. Nonetheless, a proportion of these rare cells survive and can be further preprogramed by integrins and chemokines, enabling their attachment at distant sites [3,4]. After seeding to a metastatic location, CTCs adapt to survive in inhospitable conditions, e.g., low blood oxygen perfusion or low pH for extended periods [5]. As CTCs can be obtained through routine phlebotomy, there is significant interest in their use as a measure of disease prognosis and treatment response as well as for the potential of treatment selection.
Despite the promise of CTC characterization for clinical use, detecting this rare cell population is technically challenging. The FDA-approved CellSearch® system has to date been considered the gold standard for CTC detection in the clinical setting [6,7]. This system uses antibodies against the epithelial cell adhesion molecule (EpCAM), which positively select CTCs in a magnetic field [8]. Immunocytological analysis can then be used to confirm if these enriched cells express cytokeratins or intermediate filaments of epithelial cells, but not the common leukocyte antigen CD45 [9]. Using EpCAM-based or equivalent approaches, studies have shown that the presence of high CTC counts (≥5 cells/7.5 ml of blood) is associated with shorter progression-free survival and lower overall survival in prostate cancer patients [9–11]. Furthermore, in patients with castration-resistant prostate cancer lower CTC counts detected post-treatments can be a stronger prognostic indicator for survival [6,12].
While this EpCAM-based detection technology is useful for detecting advanced prostate cancer progression, CTCs are heterogeneous and display stem cell-like properties [13]. Emerging evidence suggests that a subset of CTCs may lack EpCAM or cytokeratin expression and instead exhibit a feature of epithelial-mesenchymal transition (EMT) [14]. EMT is a gradual process, and gene markers specific for mesenchymal and stem-like cells can be detected in CTCs [15,16]. CTCs once reaching a particular site acquire an “organ-mimetic phenotype” and may lose prostate epithelial hallmarks [17,18].
In this study, we developed an approach to enrich and process CTCs based on their unique differences in sizes and deformability that are distinct from blood and non-invasive cells. Single CTCs individually retrieved using a micromanipulator system were subject to atomic force microscopy (AFM) as well as microfluidics-based PCR analyses. The results show that CTCs isolated from advanced prostate cancer patients frequently lose the typical features of epithelial prostate cancer cells. This shift was accompanied by expressing highly diverse patterns of EMT-related genes in CTCs. Furthermore, incremental increases in the expression of these genes in these circulating cells are associated with metastatic castration-resistant prostate cancer.
MATERIALS AND METHODS
Isolation of Single Circulating Tumor Cells (CTCs) Using Size-based Filtration
The University of Texas Health Science Center at San Antonio’s Institutional Review Board approved the study and consent was obtained prior to sample collection. Patient blood samples (~10 ml) were collected in K2-EDTA tubes, which were inverted five times and kept at 4°C or on ice. The patient blood was subjected to single CTC isolation. CTCs were first isolated from blood cells using ScreenCell® CC filtration kit (cat # CC 3LC-ha, ScreenCell, Paris, France) according to manufacturer’s protocol with modifications[19]. After blood filtration, the circular-filter was released onto an uncoated sterile petri dish with the cell-retained side up. From this point on, the rest of isolation process was carried out under an inverted Evos fl digital fluorescence microscope (cat # 1253460, AMG, Bothell, WA). The filter was washed 2 to 3 times with 50 μl PBS. During the washes, the residual blood cells were further carried through the filter using gentle pipetting or dragging the filter against the bottom of petri dish using sterile forceps. If blood cell clumping occurred that could interfere with single CTC isolation, clumps were dissociated by incubation with 50 μl TrypLE Express (cat # 12604-013, Invitrogen, Carlsbad, CA) for 10 min in a petri dish before PBS washes. CTCs and residual blood cells retained on the filter were stained with anti-CD45 conjugated with phycoerythrin (PE) (BD, Maryland) for 15 min and subjected to three PBS washes as described above. CTCs on the filter were incubated with 25 μl TrypLE Express for 10 min and removed and placed onto a new petri dish for CD45-negative selection and single CTC isolation using a Narishige micromanipulator and Ferty Syringe Plus Microinjector (cat # MN-153 and INJ-FS-PLUS, Origio MidAtlantic Devices, Mt. Laurel, NJ). Single CD45-negative CTCs were isolated individually, ejected in 4.5 μl PBS with 0.5 μl lysis buffer (cat #55827, Invitrogen, Carlsbad, CA) in a 0.2 ml PCR tubes and frozen on dry ice immediately and stored at −20°C until microfluidics-based PCR analysis. Some CTCs were pooled together in RPMI medium supplemented with 10% FBS and ampicillin/streptomycin for atomic force microscope analysis.
Prostate Cell Culture
Prostate cancer cell lines, LNCaP-AD (androgen-dependent), LNCaP-AI (androgen-independent) were routinely maintained in the laboratory. PC-3, and DU145 and the cell line were obtained from ATCC. The cells were cultured in RPMI medium with 10% FBS.
Analysis of CTCs and Prostate Cells Using Atomic Force Microscopy (AFM)
Individual CTCs and prostate cells suspended in ~50 μl PBS were loaded on a poly-Lys (300kD; 01% in PBS) coated glass disc glued to a steel disc. The discs were mounted in the MultiMode Nanoscope IIIa microscope (Bruker) equipped with the J type scanner and the glass chamber for in-liquid work. The SQube probes with a colloidal gold sphere with a diameter of between 1.5 to 3 micrometers as a tip, and nominal spring constant of 0.08 N/m were applied for elasticity testing and topography imaging. Probes with spherical tips were used as they produce less harsh indentation than sharp tips and are less likely to cause physical damage or trigger molecular response. The surface of the glass disc was surveyed for the presence of cells under a video camera used for probe position control, and the probe was directed above the selected cell. Subsequently, the height image of the cell for roughness analysis was collected in a contact mode followed by a cell indentation for the elasticity testing. A standard plane fit was executed on the height mode images with the Nanoscope software version 5.12. Roughness and force plots were analyzed with the SPIP v.5.11 software (Image Metrology, Denmark).
Cell elasticity
To determine the Young modulus, we performed cellular indentation mapping with the force AFM. The central area on a cell surface was probed to obtain the most consistent elasticity data. We collected a 3 x 3 array of force curves (total 9 data points) covering area of 4 μm2, with at least 5 indentations for each point. Indentation depth was restricted to 400nm. A constant pulling rate was maintained throughout all the experiments. The applied design allowed for data collection in less than 2 min per cell minimizing the cell stress response induced by the prolong instrumentation of the cell surface. For each evaluated point, the force versus indentation curve was constructed based on the force-load plots. We then applied the Hertz model to calculate the Young’s modulus using the force-indentation curves. The model describes the physical relationship between the applied force and the cantilever indentation. It assumes spherical shape of the end of a tip placed on a flat surface. The model is valid when the sphere radius is substantially larger than indentations. The elasticity for each cell was averaged, and nominal elasticity was tested against cleaned glass disks.
Cell roughness
To assess a level of morphological complexity of cell membranes, we determined their surface roughness. A contact mode image of each cell was collected using a scan size from 5 x 5 to 30 x 30 micrometers with a matrix of 512 x 512 pixels per scan at 1 Hz scan rate. We analyzed roughness values within 2 to 4 square areas of a cell surface covering from 1 to 25 μm2. When analyzing multiple patient-derived samples, we used the same spherical probe for force plots and image collecting. As a measure of a cell membrane roughness, we employed Root Mean Squared (RMS) of height calculated from heights of all image pixels included in the area of interest. Images of a glass surface surrounding the cells were used as a blank.
Single-cell Microfluidics-based RT-PCR Analysis
Single-cell microfluidics-based RT-PCR analysis was carried out using CellsDirectTM one-step qRT-PCR kit (cat # 11753-100, Invitrogen, Carlsbad, CA) and a microfluidics device, BioMark HD MX/HX system (cat # BMKHD-PKG-MH, Fluidigm, Inc., South San Francisco, CA) [20]. Single CTCs in PBS/lysis buffer were thawed, mixed well and spun down before lysed at 75°C for 10 min. To reduce contamination, genomic DNA was degraded in an 18 μl reaction volume using DNase I (5 units) with 1X DNase I buffer at RT for 5 min. PCR primers of selected genes for expression profiling were selected from the PrimerBank database. These primers were divided into two panels to fit BioMark 48x48 chips.
Reverse transcription (RT), preamplification, and PCR amplification were carried out according to the protocol of single-cell gene expression (cat # BMK-M-48.48, Fluidigm). Target genes were amplified using BioMark HB MX/HX system with 1X SsoFast EvaGreen supermix with low ROX (cat # PN172-5211, Bio-Rad, Hercules, CA) and 1X DNA binding dye sample loading reagent (cat # PN 100-3738, Fluidigm). In each chip assay, universal RNA (200 pg) from human normal tissues (cat # 4234565, BioChain, Newark, CA) and no template control (NTC) served as positive and negative controls.
Data Analysis
Expression data of genes of interest were displayed in cycles of threshold (Cts) after analysis using Real-Time PCR analysis software (Fluidigm). Relative expression values of the genes was obtained using 2−ΔΔCt method in that each gene expression is normalized to a reference gene and then normalized to lowest expressed genes that have Ct 40 as described previously [21]. Although three housekeeping genes (ACTB, GAPDH, and UBB) were initially included as reference genes, we found Ubiquitin B (UBB) to be a highly stable gene for microfluidics-based PCR analysis, as its reliability has previously been validated in a meta-analysis of over 1000 clinical samples [22]. However, expression levels of ACTB and GAPDH were less stable and weaker among different CTCs, consistent with a previous finding for single-cell CTC analysis [23]. Therefore, we only selected cells that expressed UBB at a threshold of Ct ≤30 after pre-amplification, assuming that CTCs expressing robust expression of UBB are less likely to contain degraded RNA. Log2 values of gene expression in each CTC were summed up as cumulative gene expression according to the groups of frequently expressed EMT-related genes (detected in ≥44% CTCs) and less frequently expressed EMT-related genes (present in <44% CTCs) and different oncogenic signaling pathways for comparisons. Cumulative gene expressions of CTCs from prostate cancer patients were analyzed using one-way ANOVA and unpaired Student’s t test using Prism 6 (GraphPad Software, La Jolla, CA). A p value of <0.05 is considered as statistically significant.
For in silico analysis of EMT-related gene expression in clinical samples, raw probe cel intensity (*.cel) files were obtained from Gene Expression Omnibus (GEO) series GSE6919. Expression data for samples representing Normal Prostate Tissue free of any pathological alteration (n=18), Normal Prostate Tissue Adjacent to Tumor (n=63), Primary Prostate Tumor (n=65) and Metastatic Prostate Tumor (n=25), generated using Affymetrix Human Genome U95 Version 2 Array were used for this study. RMA (Robust Multichip Average) expression measures were calculated for probes in all the samples by RMA normalization and background correction using Bioconductor Affy package in R [24]. The expression was then collapsed to gene level by averaging the measures for the probes representing a gene. These expression data were further used to compare the Metastatic samples with Normal samples by calculating the significance (using Student’s t-test along with Benjamini, Hochberg false discovery rate adjustment) and fold change.
RESULTS
Increased elasticity and smoothness of cell surface membrane in CTCs
The separation of malignant cells from the primary site via acquisition of invasive properties and transport into the bloodstream are initial steps of metastasis. To characterize CTCs, we used a microporous device to filter and select CD45-negative cells from blood samples (Table 1; see the schematic diagram in Fig. 1). Larger than blood cells, these cells showed irregular fibroblastoid morphology, suggestive of epithelial to mesenchymal transition (Fig. 1, inserts). These CTCs were individually retrieved by a micromanipulator and used to determine their surface topography and mechanical properties by AFM. The AFM-based analysis utilizes interactions between a probe (“tip”) and a cell. Raster scanning of the cell with a probe results in the image of cell surface, suitable for comparing general features of surface topography, here represented by membrane roughness, between individual single cells (Fig. 2A). On the other hand, in the AFM force mode the probe indents a cell with a controlled force load. As a result, the cantilever to which the probe is attached is deflected proportionally to the applied force [25]. Figure 2B shows an example of a force curve resulting from indentation of a single CTC at one preselected site. A blue trace represents a tip approach phase in which the tip is brought into a direct contact with a cell surface from approximately 1000 to 700nm. Next, the cantilever is progressively deflected as the tip encounters stronger cell resistance. At a preset Z position, a tip stops and then retracts (red trace) not exactly following the approach trace.
Table 1.
Clinical information of prostate cancer patients.
| Patient # | Age | Gleason score | PSA (ng/ml) | CTC isolated (# analyzed) | Metastasis | Status of treatment |
|---|---|---|---|---|---|---|
| CC01 | 72 | 9 | 331.47 | 84 (10) | Bone | Castration-resistant, chemo-resistant |
| CC02 | 61 | 9 | 88.47 | 9 (6) | Bone | Castration-resistant |
| CC03 | 71 | 9 | 96.05 | 6 (4) | Bone | Castration-resistant |
| CC04 | 53 | 9 | 79.00 | 4 (2) | Bone (small volume), lymph nodes | Castration-resistant, chemo-resistant |
| CC06 | 80 | 9 | 6.4 | 44 (5) | Bone (small volume) | Castration-resistant, immunotherapy-responsive |
| CC07 | 62 | 9 | 1191.56 | 151 (6) | Bone (small volume), lymph nodes | Castration-sensitive |
| CC08 | 60 | N/A | 13.58 | 6 (3) | Bone, lymph nodes | Castration-sensitive |
| CC09 | 64 | 8 | 0.26 | 4 (2) | None | Castration-sensitive |
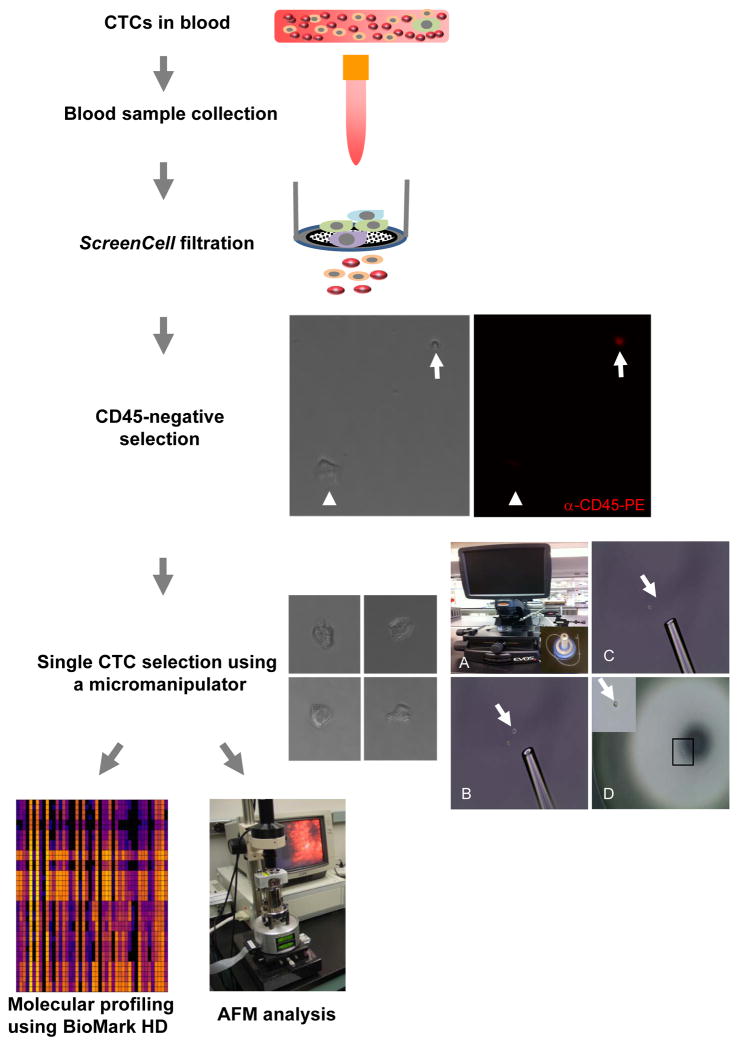
Fig. 1.
A schematic flow chart of CTC isolation and analyses. The details are described in the Materials and Methods section. In anti-CD45 negative selection, representative microscopic photos show a CTC (white arrow head) was negative for anti-CD45-PE staining, whereas a blood cell (white arrow) positive. In single CTC selection, the left panel shows four representative CTCs and the right panel illustrates the single cell isolation using a micromanipulator and an Evos fl microscope. A: An Evos fl microscope and a micromanipulator (inset). B: A pipette tip pointing to a cell (white arrow) selected using a micromanipulator. C: The single cell was aspirated into the pipette tip from the place it was previously located at (white arrow). D: The selected single cell was placed on a petri dish. A higher magnification of the single cell (black rectangle) is shown in the inset.
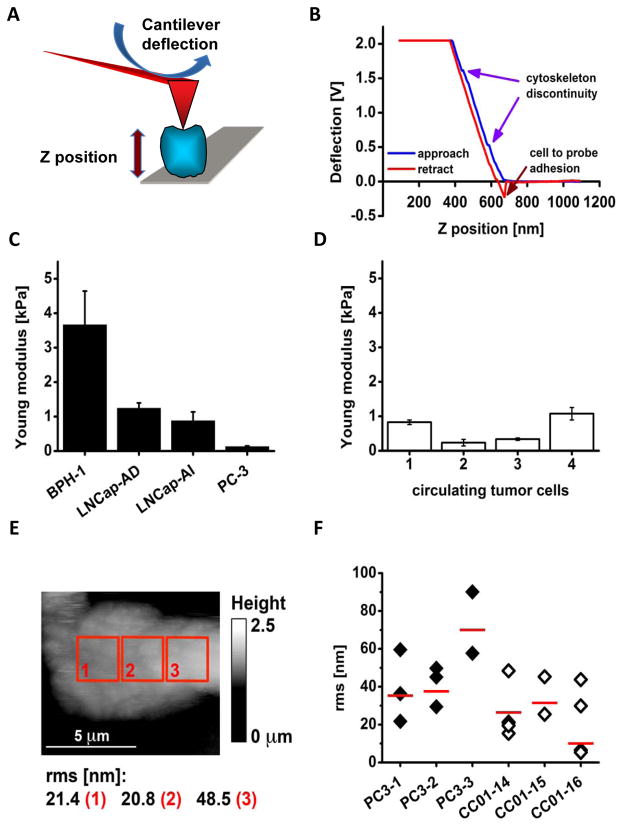
Fig. 2.
AFM probing of cell surface indicates that CTCs exhibit mechanical phenotype resembling highly metastatic cultured prostate cancer cells. A: A scheme illustrating the principle of measuring cell elasticity. A cell (blue) bound to a mica surface (grey) is indented by a tip (red triangle) mounted on a flexible cantilever (red board) proportionally to the cell elasticity. Deflection of the cantilever (blue arrow) changes a position of a laser beam reflection that measures force needed to indent the cell. The distance between a tip end and the cell is represented by the Z position (thick vertical arrow) directly measured by a piezoelectric element of the microscope. B: An example of a force plot of individual CTC (cell #4). Blue arrows point at positions of little humps at which the tip likely sensed a cytoskeleton discontinuity. Adhesion forces between the tip and the cell bent the cantilever in the opposite direction as indicated by the red arrow. C: Histogram comparing elasticity of four prostate cell lines. The elasticity is presented as the Young modulus. The benign BPH-1 cells are the stiffest (showed the largest Young modulus), whereas androgen dependent LNCap are more elastic followed by the LNCap androgen independent, and by PC-3 cells that are highly metastatic, and also the softest. Histograms represent mean values with corresponding SD. D: Histogram comparing elasticity of four individual CTCs. These cells were as soft as the cancerous cell lines presented in the panel C. Histograms represent mean values with corresponding SD. E: Height topography image of a single CTC (cell #4) recorded with a spherical tip in contact mode. Roughness (rms in nm) of the cell membrane calculated for three 2.5 by 2.5 μm. The cell is flat since it is tightly bound to a glass plate with poly-L-Lys and also may represent a strongly metastatic phenotype.
Based on a plot describing dependence of the cantilever deflection on indentation, the Young modulus constituting a measure of individual cell elasticity was derived (Fig. 2B) [26,27]. We determined the Young modulus of the cultured cells from the following established lines: the immortalized BPH-1 prostate cells and three prostate cancer cell lines, LNCap-AD, LNCap-AI, and PC-3. Noncancerous BPH-1 cells were the least elastic with the Young modulus about 3.7-kilopascal (kPa), whereas the highly metastatic PC-3 cells were almost 30X more elastic (0.13-kPa, Fig. 2C). Interestingly, androgen-independent LNCap-AD cells were more elastic then androgen-dependent LNCap-Al (0.88-kPa versus 1.2-kPa). We also measured elasticity of four CTCs isolated from blood of a patient with castrate-resistant prostate cancer and bone metastasis (Fig. 2D). Young moduli of these CTCs ranged from 0.23-kPa to 1.1-kPa, and the obtained values were similar to that of PC-3 elasticity, but much lower than those values calculated for BPH-1 cells.
To determine cell surface roughness, images of the same cells were acquired immediately after elasticity determination, by scanning the cells in contact mode with the same spherical probe. We measured roughness with a root RMS parameter, which corresponds to a variance of pixel heights included in an area of interest [28]. The RMS is measured in nm and does not depend on area size in the range of 1 to 5 μm2 (Fig. 2E). Therefore, the higher RMS value reflects a more rich relief of a cell surface and its lower value corresponds to a smoother surface. RMS values found in a single cell were quite diverse reaching from 22 to 90 nm. On average the PC-3 cells showed a rougher cell surface than CTCs, which appeared smoother. Specifically, the average RMS for all the PC-3 cells was 48.7 nm, whereas for CTCs was only 25.2 nm with the difference statistically significant at p<0.05 (Fig. 2F).
The AFM analysis presented here indicates that cell elasticity and smoothness can be considered useful parameters to distinguish between non-metastatic and metastatic cells. Differences in elasticity also reflect a histological background of a cell. The smoothness, commonly used to characterize a surface property of a variety of materials, reflects cell mobility, distribution of surface proteins, and loss of cell polarity [29,30]. These results suggest that the high deformity and high smoothness of CTC membrane surface can be the result of a morphological transitioning of these cells into mesenchymal-like cells for malignant invasion. Considering the changes in a cell membrane accompanying EMT and propensity to adhere, we expect that softer and smoother cells represent the most aggressive metastatic cells possibly indicating poor prognosis.
Loss of epithelial prostate cancer features in CTCs
Our microporous filtration-micromanipulator system was further used to isolate 308 CD45-negative CTCs from blood samples of 8 prostate cancer patients (Table 1). CTCs were not detectable in blood samples from two healthy individuals (data not shown). Sixty-two of these captured cells were subjected to single-cell microfluidics-based RT-PCR analysis of a panel of 11 known prostate epithelial markers and one negative control gene (CD45) (Table 2). Of these, 38 cells showed robust expression of UBB, and their expression data were subsequently used for normalization with the expression value of this housekeeping gene. Included in the analysis were three prostate cancer cell lines – PC-3, DU145, and LNCap-AD and universal RNA as a positive control and water as a negative control.
Table 2.
Genes (n=84) selected for single-cell microfluidics-based RT-PCR analysis.
| Gene names | Biological functions |
|---|---|
| Stem cell marker | |
| PTPRN2 | Protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| ALDH1(A1) | Aldehyde dehyfrogenase 1 family, member A1; involved in metabolism |
| CD44 | CD44 antigen; involved in cell-cell interaction, cell adhesion and migration |
| PTEN | Phosphatase and tensin homolog; tumor suppressor. Acts as a dual-specificity protein phosphatase. |
| CD133 (PROM1) | Prominin 1; binds cholesterol in cholesterol-containing plasma membrane microdomains |
| NKX3-1 | NK3 homeobox 1; transcription factor, acts as tumor suppressor controlling prostate carcinogenesis |
| MYC | V-myc avian myelocytomatosis viral oncogene homolog; activate the transcription of growth-related genes |
| ATXN1 (SCA-1) | Ataxin 1; chromatin-binding factor that repress Notch signaling |
| GATA3 | GATA binding protein 3; transcription factor contains two GATA-type zinc fingers |
| TNFSF11 (RANKL) | Tumor necrosis receptor (ligand) superfamily, member 11; ligand of cytokine |
| TNFRSF11B | Tumor necrosis receptor superfamily, member 11b; acts as decoy receptor in osteroclastogenesis. |
| TACSTD2 | Tumor-associated calcium signal transducer 2; A cell surface receptor that transduces calcium signals. |
| Other EMT-related genes | |
| CXCL13 | Chemokine (C-X-C motif) ligand 13; chemotactic for B-lymphocytes |
| ESR2 (ESRb) | Estrogen receptor 2 (ER beta); nuclear receptor transcription factors |
| ASPA | Aspartoacylase; catalyzes the conversion of N-acetyl_L-aspartic acid to aspartate and acetate |
| CDH2 | Cadherin 2; cadherin, neuronal (N-cadherin); a calcium dependent cell-cell adhesion glycoprotein; contribute to the sorting of heterogeneous cell types |
| CDH1 | Cadherin 1; E-cadherin (epithelial); a calcium dependent cell-cell adhesion glycoprotein; loss of funtion contribute to progression of several cancers. |
| COL1A2 | Collagen, type 1, alpha 2; a type-I fibril-forming collagen |
| DAB2IP | DAB2 interacting protein; functions as a Ras GTPase-activating protein. |
| FN1 | Fibronectin 1; involved in cell adhsion and migration processes |
| VIM | Vimentin; Class-III intermediate filaments found in non-epithelial cells, especially mesenchymal |
| ITGB1 (CD29) | Integrin, beta 1; membrane receptors involved in cell adhesion and recognition |
| Wnt signaling | |
| IGF1 | Insulin-like growth factor (somatomedin C); growth promoting by enhancing glucose uptake |
| IGF2 | Insulin-like growth facotr 2 (somatomedin A); growth-promoting activity |
| WNT5A | Wingless-type MMTV integration site family, member 5A |
| IGF1R | Insulin-like growth factor 1 receptor |
| FZD4 | Frizzled family receptor 4 |
| WNT11 | Wingless-type MMTV integration site family, member 11 |
| MMP14 | Matrix metallopeptidase 14 (membrane-inserted) |
| MMP9 | Matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| WNT5B | Wingless-type MMTV integration site family, member 5B |
| SNAI2 | Slug; Snail, drosophila, homolog of, 2 |
| GSK3B | Glycogen synthase kinase 3 beta |
| MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| MMP7 | Matrix metallopeptidase 7 (matrilysin, uterine) |
| NOTCH1 | Notch, drosophila, homolog of, 1; translocation-associated notch homolog (TAN1) |
| SOX9 | SRY (sex determining region Y)-box 9 |
| TCF3 | Transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) |
| CTNNB1 | Catenin, beta-1; cadherin-associated protein, beta; beta-catenin |
| FZD7 | Frizzled family receptor 7 |
| ITGA6 | Integrin, alpha 6 |
| SHH signaling | |
| PTCH1 | Patched 1 |
| GLI-3 | GLI family zinc finger 3 |
| PTCH2 | Patched 2 |
| SHH | Sonic hedgehog |
| TGFβ signaling | |
| FOXP3 | Forkhead box P3 |
| TGFB3 | Transforming growth factor, beta 3 |
| SMAD2 | SMAD family member 2 |
| TWIST1 | Twist, drosophila, homolog of 1 |
| ZEB1 | Zinc finger E-box binding homeobox 1 |
| BMP7 | Bone morphogenetic protein 7 |
| TGFB2 | Transforming growth factor, beta 2 |
| ZEB2 | Zinc finger E box-binding homeobox 2; SMAD-interacting protein 1 (SMADIP1) |
| FOXC2 | Forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| FOXA2 | Forkhead box A2 |
| TGFB1 | Transforming growth factor, beta 1 |
| EGFR signaling | |
| EGFR (Her/ERBB1) | Epidermal growth factor receptor; HER1; ERBB1 |
| ERBB2 | V-ERB-B2 avian erythroblastic leukemia viral oncogene homolog 2; NEU; HER2 |
| SRC | proto-oncogene tyrosine-protein kinase SRC |
| Clinical drug targets | |
| PIM3 | Pim-3 oncogene |
| MTOR | Mechanistic target of rapamycin (serine/threonine kinase) |
| ACP5 | Acid phosphatase 5, tartrate resistant |
| PIM1 | Pim-1 oncogene |
| PIM2 | Pim-2 oncogene |
| AXL | AXL receptor tyrosine kinase |
| ALPL (BAP) | Alkaline phosphatase, liver/bone/kidney |
| SPP1 | Secreted phosphoprotein 1 |
| ADRA2A | Adrenergic, alpha-2A-, receptor |
| HERPUD1 (MIF1) | Homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| AURKA | Aurora kinase A |
| MUC1 | Mucin 1, transmembrane |
| Prostate markers | |
| EpCAM | Epithelial cell adhesion molecule |
| ACPP (PSAP) | Acid phosphatase, prostate |
| MKI67 (Ki-67) | Antigen identified by monoclonal antibody Ki-67 |
| KLK4 | Kallikrein-related peptidase 4 |
| PCA-3 | Prostate cancer antigen 3 (non-protein coding) |
| FOLH1 (PSMA) | Folate hydrolase (prostate-specific membrane antigen) 1 |
| KLK2 | Kallikrein-related peptidase 2 |
| KRT5(CK5) | Keratin 5 |
| KRT7(CK7) | Keratin 7 |
| KRT8(CK8) | Keratin 8; cytokeratin 8 |
| PSA (KLK3) | Kallikrein-related peptidase 3 |
| Controls | |
| PTPRC (CD45) | Leukocyte-common antigen; protein-tyrosine phosphatase, receptor-type, c |
| UBB | Ubiquitin B; polyubiquitin B |
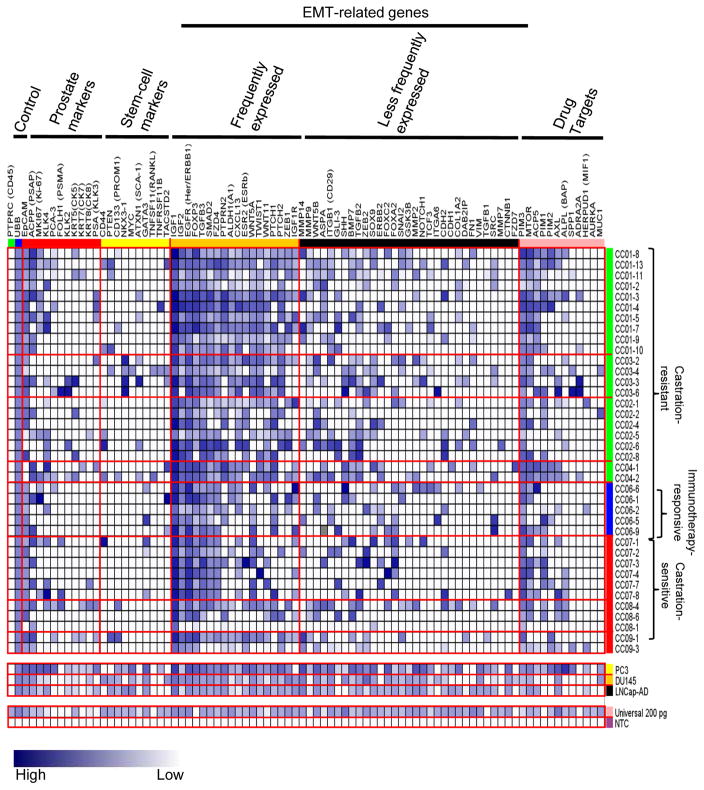
As shown in Figure 3, the heat map displays a remarkable heterogeneity of gene expression in these 38 CTCs analyzed. The majority (93%) of these cells expressed EpCAM, suggesting their epithelial origin. However, only ~20% of these CTCs showed detectable PSA and PCA-3 that are known to encode common prostate-specific antigens. Other prostate cancer markers (e.g., PSAP and PSMA) and epithelial markers (cytokeratins 5, 7, and 8) were also present in 20% of these circulating cells. Seven cells were EpCAM-negative, but expressed various prostate-related gene markers. Although we cannot rule out technical limitations of detecting some prostate cancer-related genes at the single-cell level, our initial results suggest a dramatic shift of gene expression occurring in CTCs that escaped from their primary tumor sites [16]. When seeded in metastatic locations, these cells may recirculate back into the bloodstream and progressively lose their epithelial prostate characteristics [5].
Fig. 3.
Heterogeneous expression profiles of EMT-related and other genes among CTCs. RNA from CTCs was subjected to microfluidics-based single-cell qRT-PCR analysis using a BioMark HD system. Gene expression for each gene was obtained as described in materials and methods and displayed in a blue-white gradient. Gene symbols and gene groups were labeled on the top and CTC numbers and patient groups on the right. EMT-related genes are further divided into two groups: the frequently expressed group and the less frequently expressed group.
Cumulative expression of EMT-related genes in CTCs of castration-resistant cancer
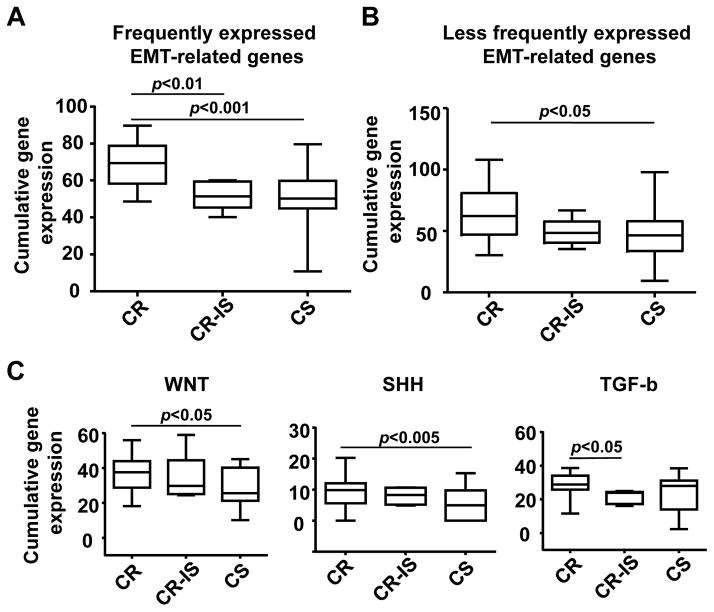
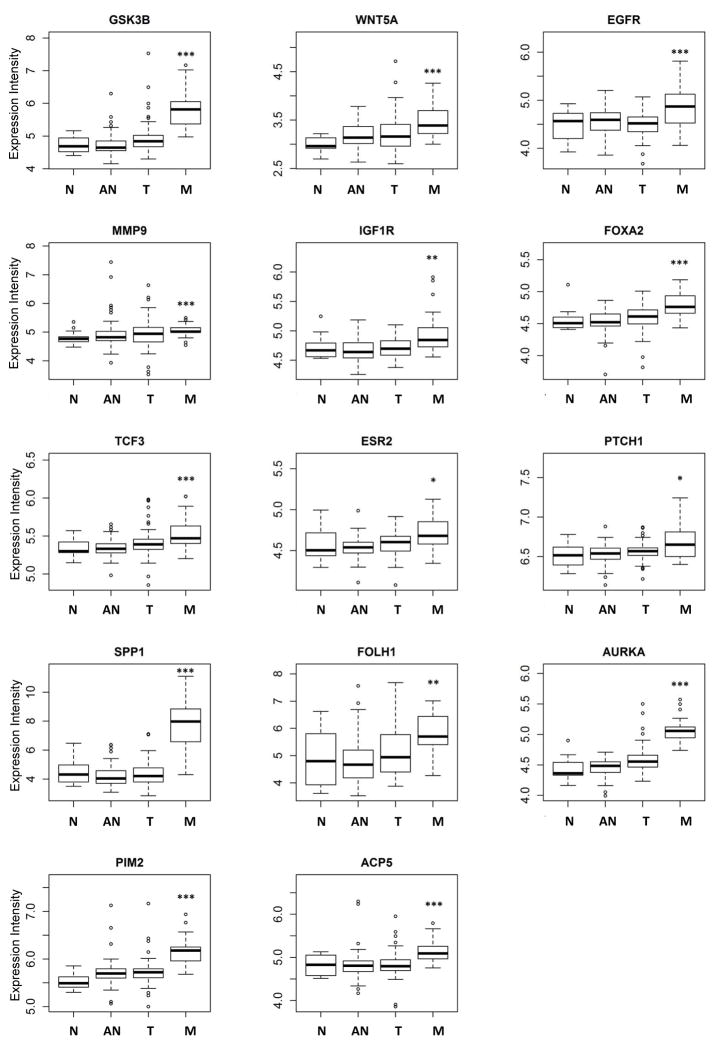
Because of the invasive nature of CTCs, we also determined expression profiles of 56 EMT-related genes in these prostate cancer patients that were categorized into castration-resistant (i.e., four patients resistant to both castration therapy and docetaxel chemotherapy), one castration-resistant/immunotherapy-responsive (in regards to patient’s serum PSA response observed following the Provenge immunotherapy), and castration-sensitive (i.e., three patients obtaining PSA response following initiation of castration therapy) groups (see Table 1). Despite high degrees of transcriptional heterogeneity, 18 of these EMT-related genes were commonly expressed in 44–100% of these CTCs analyzed (Fig. 3). Furthermore, expression levels of these genes (e.g., PTPRN2, ALDH1, ESR2, and WNT5A) were significantly higher in CTCs of castration-resistant patients than those of castration-resistant/immunotherapy-responsive (p<0.01) and castration-sensitive (p<0.001) patients (Fig. 4A). The expression of the remaining 24 EMT-related genes was less frequent (<44%) in these CTCs by the microfluidics-based PCR system. When expressed, incremental numbers and high expression values of these genes were significantly found in circulating cells isolated from castration-resistant patients (p<0.05) (Fig. 4B). When further categorizing EMT-related genes into different oncogenic signaling pathways, we found that upregulation of these genes was significantly associated with Sonic Hedgehog (p<0.005), WNT (p<0.05), and TGF-β (p<0.05), suggesting their important roles in metastatic castration-resistance and immunotherapy. In silico analysis using available expression microarray data of a published prostate cancer cohort confirmed frequent upregulation of fourteen (e.g., ESR2, WNT5A, IGF1R, PTCH1, GSK3B, MMP3, PTPRC, and EGFR) of these candidate genes in metastatic sites of prostate cancer (Fig. 5) [31].
Fig. 4.
Elevated cumulative expression of EMT-related genes and signaling pathways in CTCs from castration-resistant patients. Cumulative expression EMT-related genes in each CTC are displayed in box plots among CR, CR-IS and CS patients. A: Cumulative gene expression of frequently expressed EMT-related genes. B: Cumulative gene expression of less frequently expression EMT-related genes. C: Cumulative gene expressions of WNT, SHH and TGF-β signaling pathways. Data were analyzed using one-way ANOVA and unpaired Student’s t test. A p value of <0.05 is considered as statistically significant. CR: castration-resistant; CR-IS: castration-resistant and immunotherapy sensitive; CS: castration-sensitive.
Fig. 5.
Elevated expression of EMT-related genes and drug target genes in metastatic prostate cancer. In silico analysis of gene expression revealed that expression of nine EMT-related genes and five drug target genes are higher in clinical metastatic prostate tumors than normal prostate. Data were analyzed using Student’s t test. N: normal; AN: normal tissue adjacent to tumor; T: tumor; and M: metastatic. *, p<0.05; **, p<0.01; ***, p<0.001.
Genes encoding for the regulation and maintenance of stem-cell characteristics were detected in CTCs, but appeared as a less frequent event (~10%). However, two additional stem-cell gene markers, PTPRN2 and ALDH1, were related to EMT and were frequently expressed in CTCs of castration-resistant patients.
DISCUSSION
The current EpCAM-based technologies are largely restricted to count increased numbers of CTCs known to correlate with advanced prostate cancer [6,9,10]. Using an innovative strategy by coupling a microfiltration system with a micromanipulator device, we have developed a new system to characterize physical properties and expression patterns of individual CTCs in advanced prostate cancer patients, as well as in established prostate cancer cell lines. This novel technology has permitted us to make the unexpected discovery that the majority of EpCAM-positive CTCs show loss of epithelial characteristics. In spite of high PSA values detected in the blood of these patients, these cells may not express PSA and other frequently detectable markers in primary prostate tumors. Shedding from the primary sites, these cells become highly deformed by increasing their membranous elasticity and smoothness. It is possible that aberrant expression of EMT-related genes can completely or partially replace prostate epithelial features, instead displaying mesenchymal and stem-like characteristics [18,32]. Activation of TGF-β signaling leads to increased activities of transcription factors in the TWIST, ZEB and SNAIL gene families that repress epithelial cell adhesion and induce other mesenchymal proteins [16]. Overexpression of WNT agonists, FZD7 and FZD4, results in increased expression of MMP gene families that promote metastatic dissemination [33].
Clinically relevant to this discussion is the difference observed in the EMT-related gene profiles between the patients with advanced castrate-sensitive prostate cancer (i.e., responding to castration with PSA response) and patients who are castrate- and chemo-resistant (i.e., progressed on both castration therapy and docetaxel chemotherapy). Patients with newly diagnosed advanced prostate cancer are almost always treated with medical castration therapy, the majority of which will respond favorably to therapy with improvement in PSA response, defined as a PSA≤4 ng/ml at 7 months after therapy, with those achieving a PSA≤0.2 ng/ml having a much better median overall survival of 75 months. About a third of men however fail to achieve a PSA≤4 ng/ml, develop early castrate-resistant disease, and have a median OS of just 13 months. Identifying this subset of patients early in their course of castration therapy based on expression patterns of EMT-related genes in CTCs would have prognostic value.
A further interesting finding relates to the EMT-related gene profile for one patient with castrate-resistant disease treated with the Provenge immunotherapy instead of docetaxel chemotherapy. Provenge is now being used to treat men with asymptomatic or advanced metastatic castrate-resistant prostate cancer. Despite improvement in median overall survival, most patients did not achieve PSA response to therapy [34]. This patient, however, had an improvement of PSA response from 9.29 ng/ml to 6.4 ng/ml following immunotherapy. Interestingly, his EMT-related gene profile in CTCs most closely resembles that of patients with castrate-sensitive disease. One of the limitations of our study is small sample size, however, it is possible that this type of single-cell analysis may have a predictive role in a subset of patients with castrate-resistant disease who would benefit from immunotherapy.
In this regard, we additionally conducted a microfluidics-based PCR analysis of 12 oncogenes for which targeted inhibitors are readily available in early phase clinical studies at our institution. The CTC analysis on these patients will allow their clinicians to consider targeted treatments, such as PIM kinase inhibitors, mTOR inhibitors, G-202 (a PSMA targeting pro-drug), Axl and MUC-1 inhibitors, as therapeutic options for these men with castrate and chemo-resistant disease who have exhausted all FDA-approved agents available to them. For example, three genes - PIM3, MTOR, and ACP5 were frequently found in CTCs of both castration-resistant and -sensitive patients (see Fig. 3). This finding suggests that metastatic potential of CTCs may depend on the oncogenic addiction of related signal transduction. Consideration should be given to this type of assessment for patients with advanced prostate who have failed hormone ablation and second-line therapies. It is noteworthy to mention that recently Darshan et al reported a significant correlation between cytoplasmic sequestration of AR and clinical response to chemotherapy using CTCs from patients [35]. In addition this correlation was observed in EpCAM-positive, PSMA positive and CD45-negative CTCs. Further mutations in AR were also detected using CTCs from CRPCA patients [36]. However we did not include AR in our panel of genes as we solely focused our efforts on EMT processes not AR signaling. Although EMT has been demonstrated to play a critical role in tumorigenesis, whether AR plays a significant role in EMT is relatively unexplored. Nevertheless recent reports show that androgen deprivation induces EMT in both normal prostate and prostate cancer [37,38]. Given these emerging data showing relationship between EMT and AR, identification of changes in expression of AR in CTCs would be interesting and useful. Therefore future studies will be conducted by placing these CTCs in short-term cell culture for testing specific inhibitors that target EMT-related signaling and exploring the role of AR signaling in EMT.
Acknowledgments
Grant Sponsors: U54CA113001 (Integrative Cancer Biology Program), U01CA086402 (Early Detection Research Network), and P30CA054174 (Cancer Center Support Grant) of the National Institutes of Health, the Cancer Therapy and Research Center Foundation, and the University of Texas STARS program
The authors would like to thank the staff at the Core for Advanced Translational Technologies for providing their expertise in single-cell microfluidics-based PCR. This work is supported by grants U54CA113001 (Integrative Cancer Biology Program), U01CA086402 (Early Detection Research Network), and P30CA054174 (Cancer Center Support Grant) of the National Institutes of Health, the University of Texas STARS award, and gifts from the Cancer Therapy and Research Center Fund and the Voelcker Fund.
References
- 1.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, Beitsch PD, Leitch M, Hoover S, Euhus D, Haley B, Morrison L, Fleming TP, Herlyn D, Terstappen LW, Fehm T, Tucker TF, Lane N, Wang J, Uhr JW. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 3.Dittmar T, Heyder C, Gloria-Maercker E, Hatzmann W, Zanker KS. Adhesion molecules and chemokines: the navigation system for circulating tumor (stem) cells to metastasize in an organ-specific manner. Clin Exp Metastasis. 2008;25:11–32. doi: 10.1007/s10585-007-9095-5. [DOI] [PubMed] [Google Scholar]
- 4.Sun YX, Schneider A, Jung Y, Wang J, Dai J, Cook K, Osman NI, Koh-Paige AJ, Shim H, Pienta KJ, Keller ET, McCauley LK, Taichman RS. Skeletal localization and neutralization of the SDF-1(CXCL12)/CXCR4 axis blocks prostate cancer metastasis and growth in osseous sites in vivo. J Bone Miner Res. 2005;20:318–329. doi: 10.1359/JBMR.041109. [DOI] [PubMed] [Google Scholar]
- 5.Alix-Panabieres C, Schwarzenbach H, Pantel K. Circulating tumor cells and circulating tumor DNA. Annu Rev Med. 2012;63:199–215. doi: 10.1146/annurev-med-062310-094219. [DOI] [PubMed] [Google Scholar]
- 6.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, Scher HI. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 7.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 8.Helo P, Cronin AM, Danila DC, Wenske S, Gonzalez-Espinoza R, Anand A, Koscuiszka M, Vaananen RM, Pettersson K, Chun FK, Steuber T, Huland H, Guillonneau BD, Eastham JA, Scardino PT, Fleisher M, Scher HI, Lilja H. Circulating prostate tumor cells detected by reverse transcription-PCR in men with localized or castration-refractory prostate cancer: concordance with CellSearch assay and association with bone metastases and with survival. Clin Chem. 2009;55:765–773. doi: 10.1373/clinchem.2008.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, Heller G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 11.Goodman OB, Jr, Fink LM, Symanowski JT, Wong B, Grobaski B, Pomerantz D, Ma Y, Ward DC, Vogelzang NJ. Circulating tumor cells in patients with castration-resistant prostate cancer baseline values and correlation with prognostic factors. Cancer Epidemiol Biomarkers Prev. 2009;18:1904–1913. doi: 10.1158/1055-9965.EPI-08-1173. [DOI] [PubMed] [Google Scholar]
- 12.Coumans FA, Doggen CJ, Attard G, de Bono JS, Terstappen LW. All circulating EpCAM+CK+CD45- objects predict overall survival in castration-resistant prostate cancer. Ann Oncol. 2010;21:1851–1857. doi: 10.1093/annonc/mdq030. [DOI] [PubMed] [Google Scholar]
- 13.Danila DC, Pantel K, Fleisher M, Scher HI. Circulating tumors cells as biomarkers: progress toward biomarker qualification. Cancer J. 2011;17:438–450. doi: 10.1097/PPO.0b013e31823e69ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor RA, Toivanen R, Risbridger GP. Stem cells in prostate cancer: treating the root of the problem. Endocr Relat Cancer. 2010;17:R273–285. doi: 10.1677/ERC-10-0145. [DOI] [PubMed] [Google Scholar]
- 16.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyen J, Alix-Panabieres C, Hofman P, Parks SK, Chamorey E, Naman H, Hannoun-Levi JM. Circulating tumor cells in prostate cancer: a potential surrogate marker of survival. Crit Rev Oncol Hematol. 2012;81:241–256. doi: 10.1016/j.critrevonc.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Danila DC, Fleisher M, Scher HI. Circulating tumor cells as biomarkers in prostate cancer. Clin Cancer Res. 2011;17:3903–3912. doi: 10.1158/1078-0432.CCR-10-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desitter I, Guerrouahen BS, Benali-Furet N, Wechsler J, Janne PA, Kuang Y, Yanagita M, Wang L, Berkowitz JA, Distel RJ, Cayre YE. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011;31:427–441. [PubMed] [Google Scholar]
- 20.Citri A, Pang ZP, Sudhof TC, Wernig M, Malenka RC. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat Protoc. 2012;7:118–127. doi: 10.1038/nprot.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Popovici V, Goldstein DR, Antonov J, Jaggi R, Delorenzi M, Wirapati P. Selecting control genes for RT-QPCR using public microarray data. BMC bioinformatics. 2009;10:42. doi: 10.1186/1471-2105-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, Sheth S, Kurian AW, Ford JM, Stockdale FE, Quake SR, Pease RF, Mindrinos MN, Bhanot G, Dairkee SH, Davis RW, Jeffrey SS. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 25.Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 26.Faria EC, Ma N, Gazi E, Gardner P, Brown M, Clarke NW, Snook RD. Measurement of elastic properties of prostate cancer cells using AFM. Analyst. 2008;133:1498–1500. doi: 10.1039/b803355b. [DOI] [PubMed] [Google Scholar]
- 27.Lekka M, Pogoda K, Gostek J, Klymenko O, Prauzner-Bechcicki S, Wiltowska-Zuber J, Jaczewska J, Lekki J, Stachura Z. Cancer cell recognition - Mechanical phenotype. Micron. 2012;43:1259–1266. doi: 10.1016/j.micron.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 28.Girasole M, Pompeo G, Cricenti A, Congiu-Castellano A, Andreola F, Serafino A, Frazer BH, Boumis G, Amiconi G. Roughness of the plasma membrane as an independent morphological parameter to study RBCs: a quantitative atomic force microscopy investigation. Biochim Biophys Acta. 2007;1768:1268–1276. doi: 10.1016/j.bbamem.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Chen DT, Weeks ER, Crocker JC, Islam MF, Verma R, Gruber J, Levine AJ, Lubensky TC, Yodh AG. Rheological microscopy: local mechanical properties from microrheology. Phys Rev Lett. 2003;90:108301. doi: 10.1103/PhysRevLett.90.108301. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Wan Z, Liu W, Li L, Ren L, Wang X, Sun P, Zhao H, Tu Q, Zhang Z, Song N, Zhang L. Atomic force microscope study of tumor cell membranes following treatment with anti-cancer drugs. Biosens Bioelectron. 2009;25:721–727. doi: 10.1016/j.bios.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 32.Nauseef JT, Henry MD. Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle? Nat Rev Urol. 2011;8:428–439. doi: 10.1038/nrurol.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, Rantala J, Alanen K, Nees M, Kallioniemi O. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- 34.Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, Redfern CH, Ferrari AC, Dreicer R, Sims RB, Xu Y, Frohlich MW, Schellhammer PF. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 35.Darshan MS, Loftus MS, Thadani-Mulero M, Levy BP, Escuin D, Zhou XK, Gjyrezi A, Chanel-Vos C, Shen R, Tagawa ST, Bander NH, Nanus DM, Giannakakou P. Taxane-induced blockade to nuclear accumulation of the androgen receptor predicts clinical responses in metastatic prostate cancer. Cancer Res. 2011;71:6019–6029. doi: 10.1158/0008-5472.CAN-11-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y, Palma JF, Agus DB, Wang Y, Gross ME. Detection of androgen receptor mutations in circulating tumor cells in castration-resistant prostate cancer. Clin Chem. 2010;56:1492–1495. doi: 10.1373/clinchem.2010.143297. [DOI] [PubMed] [Google Scholar]
- 37.Zhu ML, Kyprianou N. Role of androgens and the androgen receptor in epithelial-mesenchymal transition and invasion of prostate cancer cells. FASEB J. 2010;24:769–777. doi: 10.1096/fj.09-136994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y, Wang BE, Leong KG, Yue P, Li L, Jhunjhunwala S, Chen D, Seo K, Modrusan Z, Gao WQ, Settleman J, Johnson L. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 2012;72:527–536. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]