Abstract
The mechanisms whereby immune therapies affect progression of Type 1 diabetes (T1D) are not well understood. Teplizumab, an FcR non-binding anti-CD3 mAb, has shown efficacy in multiple randomized clinical trials. We previously reported an increase in the frequency of circulating CD8+ central memory (CD8CM) T cells in clinical responders, but the generalizability of this finding and the molecular effects of teplizumab on these T cells have not been evaluated. We analyzed data from 2 randomized clinical studies of teplizumab in patients with new and recent onset T1D. At the conclusion of therapy clinical responders showed a significant reduction in circulating CD4+ effector memory (CD4EM) T cells. Afterwards, there was an increase in the frequency and absolute number of CD8CM T cells. In vitro, teplizumab expanded CD8CM T cells by proliferation and conversion of non-CM T cells. Nanostring analysis of gene expression of CD8CM T cells from responders and non-responders vs placebo-treated control subjects identified decreases in expression of genes associated with immune activation and increases in expression of genes associated with T cell differentiation and regulation. We conclude that CD8CM T cells with decreased activation and regulatory gene expression are associated with clinical responses to teplizumab in patients with T1D.
Keywords: anti-CD3 mAb, Type 1 diabetes, immune therapy, CD8 T cells, tolerance
Introduction
Type 1 diabetes is a progressive autoimmune disease resulting from T cell-mediated, targeted destruction of β cells that leads to the loss of insulin production and dependence on exogenous insulin [1]. Metabolic control with insulin therapy does not achieve the same metabolic control as β cells in the islets of Langerhans. Studies of insulitis in humans have highlighted the role of islet-infiltrating CD8+ T cells in the disease process, including those with specificities for known diabetes antigens [2].
Over the past 15 years, clinical trials with Fc receptor-nonbinding humanized anti-CD3 mAbs, have shown slowed progression of disease [3–10]. Even the Protégé trial, that did not meet its clinical endpoint showed improvement in C-peptide treated subjects.
However, not all patients respond to treatment. Those who do respond may have a remarkably robust preservation of insulin production: In the AbATE trial the responders had < 10% loss of C-peptide responses two years from diagnosis [5]. Studies to date have not identified the immunologic basis for responses in these subjects. Identifying biomarkers of responsiveness are important objectives for understanding the mechanisms of the treatment and the disease, maximizing efficacy, and avoiding treatment of those who are not likely to respond to teplizumab.
Several metabolic and immunologic features have been found to distinguish responders to teplizumab. In the AbATE trial of teplizumab, we found that differences in glycemic control, insulin use, and changes in subsets of both CD4+ and CD8+ T cells at baseline predicted response; in other trials younger age was a predictor [5]. However, these markers are not related to the actions of the anti-CD3 mAb and may identify differences in β cells, insulin sensitivity, or other parameters unrelated to the pathogenesis or drug response. Previously, we tracked the frequency of diabetes and other antigen-specific CD8+ T cells, and found that treatment with teplizumab did not eliminate antigen-specific or other effector T cells [11]. We also reported that responders to teplizumab could be distinguished from non-responders, surprisingly, by an increase in the number of circulating CD8+ central memory (CD8CM) T cells in the former [12]. This is surprising because other successful immune therapies have been associated with a decrease in CD4+ and CD8+ memory T cells [13, 14].
A number of mechanisms of anti-CD3 mAbs have been suggested. Previous studies from our group and others showed induction of subpopulations of regulatory T cells [15–18]. Recent work has suggested that adaptive regulatory T cells that produce IL10 and/or TGF-β may be induced following migration of T cells to the gut following treatment with teplizumab [19, 20]. Belghith et al found that anti-CD3 mAb induced TGFβ-dependent CD4+ Tregs in the pancreatic draining lymph nodes in NOD mice [17]. Finally, CD8+ T cells isolated directly from drug-treated patients have regulatory function in ex vivo assays [15, 16, 18]. These cells were distinguished by low levels of expression of NKG2A (KLRC1). Collectively, the findings suggest that regulatory mechanisms are involved, either by direct induction of regulatory T cells or inactivation of subpopulations, such as memory T cells, that are involved in disease progression.
In this analysis, we determined the effects of teplizumab treatment on T cell subsets in vitro and in vivo using cells and data from two randomized clinical trials of patients with T1D in order to identify cellular correlates of clinical responses [5, 12]. We identified changes in memory T cells immediately after drug treatment but clinical responses were associated with an increase in the frequency of CD8CM T cells. We analyzed gene expression in these cells and, in clinical responders, found reduced expression of genes associated with cell activation and changes in genes associated with differentiation and regulation.
Results
Teplizumab slows the rate of C-peptide loss in patients with T1D
Data and samples were collected from subjects with T1D enrolled in two randomized clinical trials of teplizumab [5, 7]. The AbATE trial enrolled subjects with new-onset disease and the Delay trial enrolled patients with T1D of 4–12 months duration. The patient demographics have been published and were similar in the two trials. In both trials, patients with T1D, age range 8–35, were randomized to a control group (placebo in Delay, open label in AbATE) or teplizumab. The dosing regimen of teplizumab was the same in both trials and was daily IV doses of 51 μg/m2, 103 μg/m2, 207 μg/m2, and 413 μg/m2 on Study Days 0–3, respectively, and 826 μg/m2 on each of Study Days 4–13. The total dose for a 14-day course was 9,034 μg/m2.
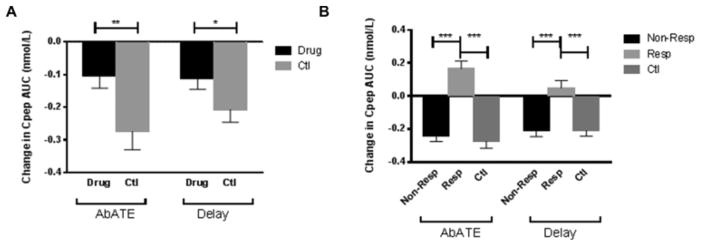
The primary clinical outcomes from these trials were reported [5, 7]. The C-peptide responses (AUC) to a 4-hr MMTT were measured at study entry and at 6 and 12 months after treatment. The 12-month change in C-peptide was significantly improved in drug-treated individuals in both studies (AbATE: −0.104±0.037 nmol/L vs −0.274±0.056 nmol/L, p=0.002, and Delay: −0.110±0.035 nmol/L vs −0.207±0.039 nmol/L, p=0.03) (Figure 1A).
Figure 1. C-peptide changes in patients treated with teplizumab.
(A) Comparison of 12-month changes in C-peptide between drug-treated and control subjects in Delay and AbATE. In AbATE (p=0.002) and Delay (p=0.03) (Student’s t-test) teplizumab treatment decreased the rate of C-peptide loss in the first year after treatment. (B) Comparison of 12-month change in C-peptide of responders, non-responders and controls subjects in Delay and AbATE. Responders were defined as having lost < 7.5% of C-peptide in the first year after treatment [8]. In both studies, responders had a positive increase in C-peptide in the first year of treatment and C-peptide change in non-responders was indistinguishable from controls (*p<0.05, ***p<0.001) (repeated measures mixed model). In A and B, the changes in C-peptide in the Delay study were corrected for imbalance in the baseline HbA1c levels in the mixed linear model [12].
Changes in T cell subsets distinguishes clinical responders to treatment
Not all patients receiving teplizumab therapy showed the same response. To identify the changes in T cells that distinguished responders and non-responders and to allow direct comparison between these two and previous trials [8], we designated drug-treated patients as responders or non-responders, based on a previously used definition of responders as having ≤ 7.5% loss of baseline levels of C-peptide after 12 months [6] (Table 1). The C-peptide responses at study entry were not significantly different in the responders and non-responders in AbATE or Delay. The percentage of responders to therapy was similar in the two trials (AbATE: 38.8%, Delay: 41.9%, p=0.82). Responders, on average, had an improvement in C-peptide response at 12 months compared to baseline (0.166±0.044 nmol/L and 0.048±0.045 nmol/L in AbATE and Delay, respectively), while non-responders showed losses that were similar to untreated or placebo-treated control subjects (−0.24±0.035 nmol/L and −0.207±0.038 nmol/L, AbATE and Delay, respectively) (Figure 1B).
Table 1.
Demographics at entry of responders and non-responders in AbATE and Delay
| Trial | Group | N | Age | Duration of diabetes (days) | Baseline C-peptide (nmol/L) |
|---|---|---|---|---|---|
| AbATE | Responders | 19 | 13.5±1.33 | 40.7±1.97 | 0.76±.071 |
| AbATE | Non-responders | 30 | 12.2±0.84 | 41.0±1.43 | 0.70±.053 |
| AbATE | Controls | 25 | 12.3±0.82 | 37.6±7.58 | 0.67±0.056 |
| Delay | Responders | 13 | 12.15±1.09 | 210±20.6 | 0.588±0.07 |
| Delay | Non-responders | 18 | 13±1.06 | 219±18.7 | 0.639±0.08 |
| Delay | Placebo | 28 | 12.0±0.76 | 216±13.9 | 0.6±0.08 |
Responders were defined as having lost < 7.5% of their baseline C-peptide response at month 12 (6).
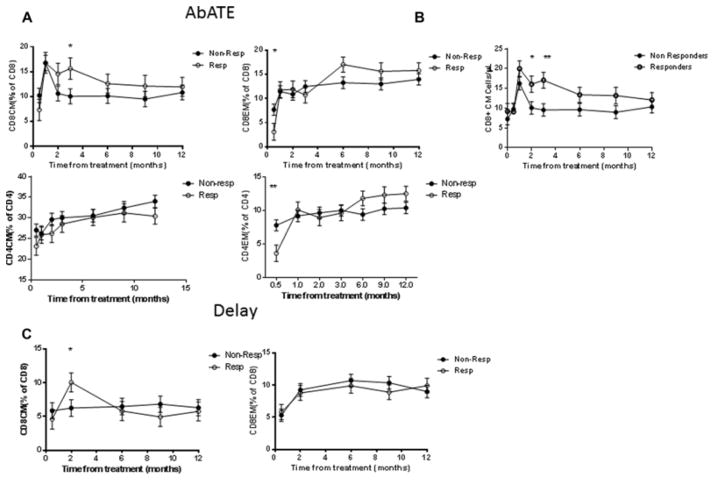
The percentages and absolute numbers of T cell subsets in the peripheral blood were evaluated in both trials in real time with freshly isolated PBMC before and during the year following treatment with anti-CD3 mAb by flow cytometry. The laboratories used for analysis were different for the two trials but the same laboratories were used for all participants within each trial. The first analysis of T cells after treatment was performed on day 14 (2 weeks: after the completion of treatment) in the AbATE trial, but at month 2 in the Delay trial. In the AbATE trial, the relative proportion of CD4EM and CD8EM T cells were significantly lower in the responders vs non-responders at that time (p=0.006, p=0.03 respectively) but these cells recovered by the next analysis at 1 month (Figure 2A).
Figure 2. Changes in CD4+ and CD8+ T cell subsets in patients with T1D following treated with teplizumab.
(A) CD4+ and CD8+ CM and EM (percent of total) T cells in responders and non-responders in the first year after treatment in AbATE. The data shown represent the geometric mean±SEM from the repeated measures mixed model corrected for the baseline values. (*p<0.05, **p<0.01). (B) The changes in the CD8CM T cells were due to an increase in the absolute number of the cells in the responders in the AbATE trial (* p<0.05, **p<0.01). (C) In the Delay trial there was also a significant increase in the proportion of CD8CM T cells at month 2 (*p<0.05), corrected for the baseline values. There were not significant differences in the proportion of CD8EM T cells.
The relative proportions of CD8CM T cells were significantly increased in responders compared to the non-responders following drug treatment in both trials (Figure 2). In the AbATE trial, there was a trend for an increase in the frequency of CD8CM T cells by 2 mos after the last dose of teplizumab (resp: 14.6±2.16% vs non-resp: 10.6±1.5%, p=0.13) and a significant increase at 3 mos (resp: 15.6±2.03% vs non-resp: 10.1±1.5%, p=0.029) (Figure 2A). The differences in the proportion of CD8CM T cells was due to an increase in the absolute number of cells (Figure 2B, p=0.05) after 2 mos (16.1±2.10 cells/μl vs 10.1±1.6 cells/μl, p=0.032), and 3 mos (17.1±2.00 cells/μl vs 9.56±1.59 cells/μl, p=0.004). In the Delay trial we found a significant increase in the proportion of CD8CM T cells at 2 mos (Figure 2C, 10.0±1.41% vs 6.25±1.23%, p=0.046). The findings from the AbATE trial show that both responders and non-responders had an initial increase in CD8CM T cells with treatment, but the subpopulation of CD8CM T cells remained elevated in the responders for a longer period of time.
Source of CD8CM T cells
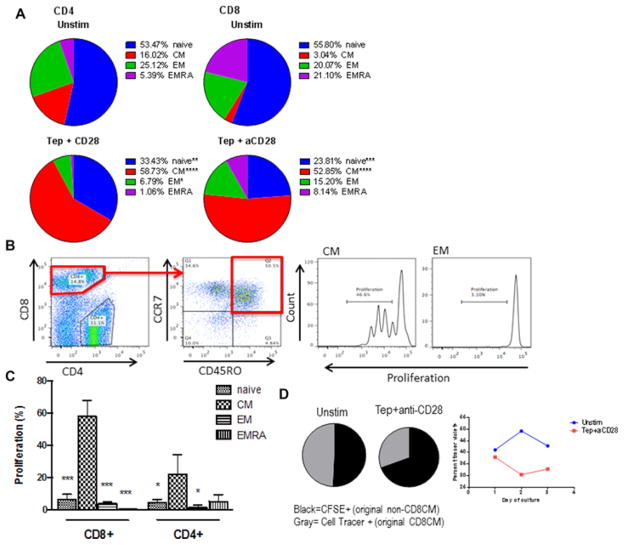
We cultured cells in vitro with teplizumab to determine how it affected CD8CM T cells. There was an increase in the proportion of CD4CM and CD8CM T cells after culture with teplizumab (Chi-squared p<0.0001 for both) (Figure 3A). The changes in the proportions could reflect proliferation of CD8CM or killing of the non-CM populations, but the CD8CM T cell subset showed increased proliferation (Figure 3B and C).
Figure 3. Proliferative responses of CD8+ T cell subpopulations to teplizumab.
(A) Changes in the subpopulations of cells after culture with teplizumab and anti-CD28 mAb. PBMC from 3 healthy control subjects were cultured for 3 days in the presence or absence of teplizumab and anti-CD28 mAbs as described in Materials and Methods. The changes in the proportion of cells was determined by flow cytometry at the conclusion of the cultures. The proportion of subgroups of CD4 and CD8 T cells changed with culture (p<0.0001) and there was an increase in the proportion of CD4CM and CD8CM T cells. (*p<0.05, **p<0.01, **** p<0.0001, ANOVA) (B) To determine whether the changes in proportions of cells reflected proliferation of the CM subsets, we studied the proliferation of cell subgroups by dilution of CellTrace dye. An example of the gating strategy used to analyze proliferation of T cell subsets in vitro is shown on the left. (C) Proliferation was assessed as percent of cells and was greatest in the CD8CM T cells (*p<0.05, ***p<0.001). (D) To determine the source of the CD8CM T cells in the cultures, we sorted CD8CM T cells from PBMC and labeled them with CellTrace dye. These labeled cells were placed into culture with PBMC that had been stained with CFSE and we measured the proportion of CellTrace dye+ CD8CM T cells after 1, 2, and 3 days in culture with or without teplizumab+anti-CD28 mAb. The pie charts show the proportion of CellTrace Dye+ and CFSE+ cells after 2 days in culture. (gray=CD8CM T cells labeled at the start of culture, black=CD8CM T cells that had differentiated from other subsets.) The graph shows the changes in the proportion of the CellTrace dye+ cells during cultures. The data is from a single experiment representative of 3 independent experiments. There was an increase proportion of CD8CM T cells derived from other subpopulations in cultured PBMC.
To determine the source of the CD8CM T cells, we sorted CD8CM T cells, labeled them with CellTrace dye, and seeded them into CFSE-labeled PBMC from which they had been sorted. The cells were then cultured for 2 – 3 days with teplizumab and anti-CD28 mAb. The proportion of CellTrace dye+ CD8CM T cells declined with time in culture while the frequency of CFSE+CD8CM T cells increased with time (4.6% (median) CD8CM cells in control wells vs 17.9% in teplizumab+anti-CD28 wells at 3 days) suggesting that some of the CD8CM T cells came from non-CD8CM T cells (Figure 3D). These studies show that the CD8CM cells at the end of the cultures contained previously naïve cells that had differentiated into CM T cells. When we labeled naïve cells with CellTrace dye, between 3–9% of the CD8CM cells were positive for the dye at the end of the 3 day culture period (not shown).
Transcriptional analysis of CD8CM T cells from trial participants
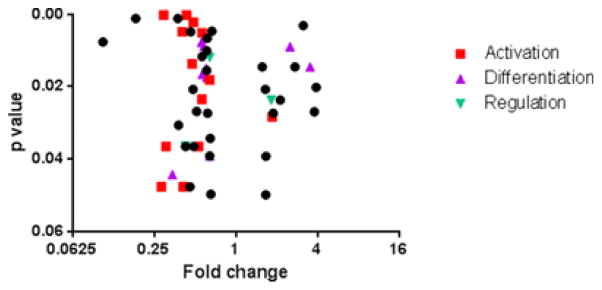
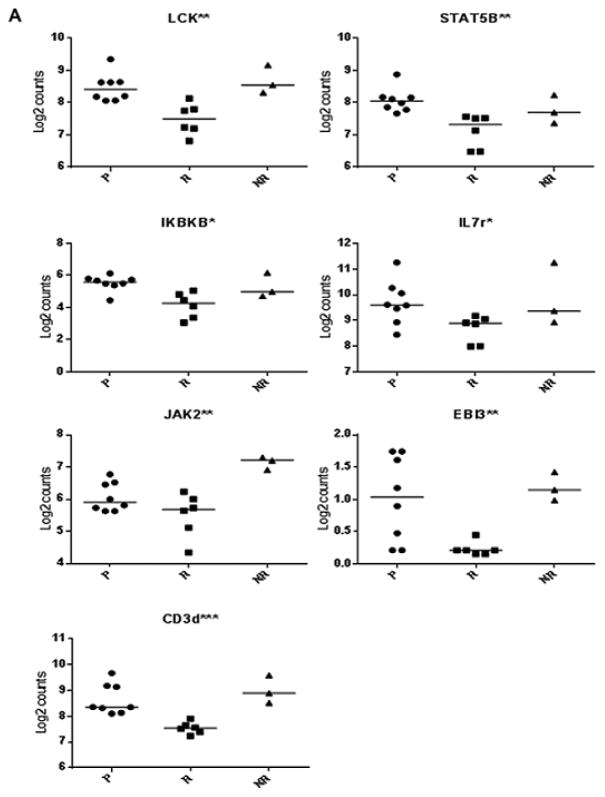
We sorted CD8CM T cells from PBMC collected at the 2 month visit in the Delay trial in responders and non-responders, and from placebo treated subjects and measured gene expression by nanostring, using the human immunology codeset covering 594 genes (Figure 4). Since drug treated non-responders and placebo treated subjects showed a similar response, in our first analysis we pooled data from the placebo treated and drug treated non-responders to identify differences in gene signatures that were associated with preservation of C-peptide responses in the drug treated responders. There were 53 genes that showed at least a 1.5 fold difference in expression between these groups with a p value < 0.05 by Wilcoxon test (Table 2, Figure 4). Thirteen of the differentially expressed genes encode molecules associated with cell activation such as LCK, EBI3, JAK1, and IKBKB as well as decreased expression of 2 components of the CD3 complex. The expression of STAT5B was also modestly reduced in the responder group (1.4 fold, p=0.001). There were differences in expression of twelve genes differentially regulated that were associated with cell differentiation. These included decreased expression of IL17A, and increased expression of 2 IL10 family members (IL20 and IL26). The latter may enhance IL10 secretion [21]. There was decreased expression of IL22 which is coproduced with IL17 [22]. Expression of IL10 itself was also increased 2.1 fold (p=0.06). There was also decreased expression of IKZF1 (ikaros family zinc finger protein 1) which is a T1D susceptibility gene [23].
Figure 4. Effects of teplizumab on gene expression in CD8CM T cells, measured by nanostring, from teplizumab-treated responders, non-responders, and placebo-treated subjects after treatment.
Volcano plot of a comparison of gene expression in CD8CM T cells in drug-treated responders (n=6) vs drug-treated non-responders (n=3) and placebo treated (n=6) subjects. The log2 of the fold change (X-axis) and p value are plotted. Differences in gene expression of at least 1.5-fold with a p<0.05 (Wilcoxon Rank Sum test) are shown. Genes that are associated with T cell activation, differentiation, and regulation are shown. A listing of data of the individual genes is shown in Table 2.
Table 2.
Comparison of gene expression in CD8CM T cells from responders vs non-responders+placebo
| Activation | Differentiation | Regulation | Others | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Fold R/NR+P | p | Name | Fold R/NR+P | p | Name | Fold R/NR+P | p | Name | Fold R/NR+P | p |
| IRF3 | 0.281 | 0.048 | IKZF1 | 0.293 | 0.000 | KLRC1 | 0.424 | 0.037 | ATG10 | 0.104 | 0.008 |
| CTLA4-TM | 0.305 | 0.037 | IL15 | 0.339 | 0.044 | IDO1 | 0.641 | 0.012 | KLRC2 | 0.183 | 0.001 |
| IKBKB | 0.402 | 0.005 | GZMA | 0.409 | 0.048 | LIF | 1.820 | 0.024 | ITGA6 | 0.372 | 0.001 |
| IL7R | 0.475 | 0.014 | ETS1 | 0.557 | 0.008 | CFH | 0.377 | 0.031 | |||
| LCK | 0.486 | 0.002 | IL17A | 0.565 | 0.017 | GBP1 | 0.425 | 0.037 | |||
| CD3EAP | 0.531 | 0.037 | IL22 | 0.580 | 0.009 | CD3D | 0.432 | 0.000 | |||
| CD3D | 0.432 | 0.000 | IL20 | 2.507 | 0.009 | BCL2 | 0.458 | 0.048 | |||
| KIT | 0.562 | 0.023 | IL26 | 3.535 | 0.015 | CCND3 | 0.464 | 0.005 | |||
| EBI3 | 0.564 | 0.005 | IL9 | 0.605 | 0.015 | CD8A | 0.483 | 0.021 | |||
| JAK1 | 0.641 | 0.018 | IL7 | 0.608 | 0.017 | KLRD1 | 0.489 | 0.037 | |||
| CXCR2 | 0.653 | 0.050 | BATF3 | 2.122 | 0.024 | NOD1 | 0.513 | 0.027 | |||
| IL32 | 1.855 | 0.028 | CXCL13 | 0.641 | 0.039 | CD34 | 0.561 | 0.012 | |||
| IRF5 | 3.140 | 0.003 | CEACAM6 | 0.607 | 0.015 | ||||||
| RAG1 | 0.610 | 0.010 | |||||||||
| BAX | 0.614 | 0.007 | |||||||||
| ADA | 0.618 | 0.027 | |||||||||
| ARG1 | 0.646 | 0.034 | |||||||||
| HLA-B | 0.665 | 0.005 | |||||||||
| SELPLG | 1.566 | 0.015 | |||||||||
| ATG10 | 0.104 | 0.008 | |||||||||
| KLRC2 | 0.183 | 0.001 | |||||||||
| ITGA6 | 0.372 | 0.001 | |||||||||
| CFH | 0.377 | 0.031 | |||||||||
| GBP1 | 0.425 | 0.037 | |||||||||
| CD3D | 0.432 | 0.000 | |||||||||
| BCL2 | 0.458 | 0.048 | |||||||||
| CCND3 | 0.464 | 0.005 | |||||||||
We compared the expression of these genes in the 3 groups (i.e. drug treated responder and non-responders and placebo treated) (Figure 5). We found decreased expression of genes associated with T cell activation: LCK (p=0.002), STAT5B (p=0.002), IKBKB (p=0.01), JAK2 (p=0.007), IL7R (p=0.03), EBI3 (p=0.007), and CD3d (p=0.0003; all by Kruskal-Wallis test, Figure 5A). We confirmed the differential expression of IL7R by FACS on CD8CM cells from the patients (p=0.02, Kruskal-Wallis test) (Supplemental Figure 1), but did not find a difference in the expression of CD3 on the CD8CM T cell surfaces (not shown).
Figure 5. Gene expression among responder subsets.
We compared gene expression by nanostring in the 3 subgroups. (*p<0.05, **p<0.01, ***p<0.001 by Kruskal-Wallis ANOVA). (A) Genes associated with T cell activation, (B) Genes associated with T cell differentiation and regulation.
We likewise found differences between these subgroups in genes associated with cell differentiation including the IL10 family members (IL10 (p=0.009), IL20 (p=0.006), IL22 (p=0.02), IL26 (p=0.01)), IL17A (p=0.03), and IKZF1 (p=0.0004) (Figure 5B).
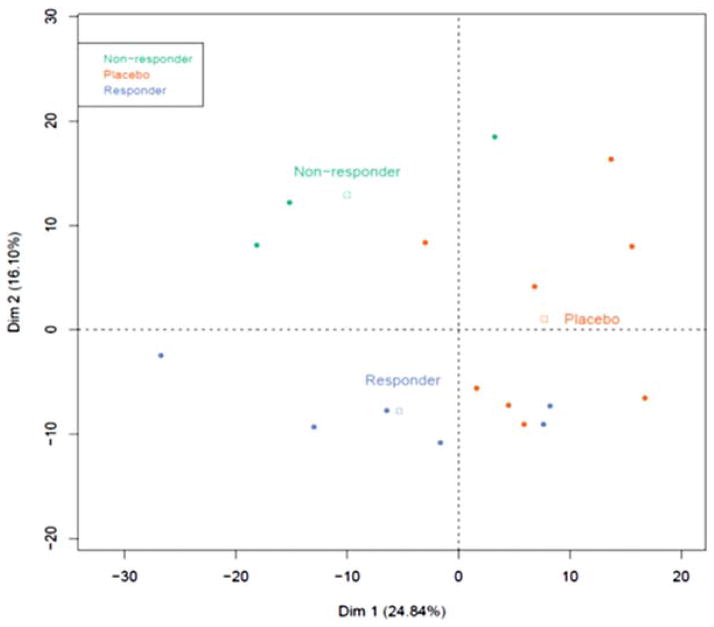
We performed a principal components analysis (PCA, Figure 6) of all genes in the 3 groups. We identified 2 dimensions that define the subgroups. The first dimension explained 24.8% of the overall variance and was correlated to placebo status (p=0.009); 241/579 individual genes were in this dimension (p<0.05 by Spearman’s correlation). The second dimension explained 16.1% of the overall variance and was correlated to non-responder and responder status (p=0.008, 0.011 respectively), with 160/579 individual genes loading to this dimension (p<0.05) (Table 3). Thirty-three genes overlapped between the two dimensions (Table 3). In addition genes identified above, this analysis also showed a correlation with the expression of FOXP3 (r=−0.75, p=0.0005), GATA3 (r=−0.725, p=0.001), and CTLA4_all (r=−0.83, p=0.3.82E-5).
Figure 6. Principal component analysis of the 3 groups.
We performed a PCA analysis with nanostring data. Each circular symbol represents an individual subject who was a drug treated responder (blue), non-responder (green), or placebo treated subject (orange). The squares indicate the median values of the two dimensions of each group. The components of Dim2, which differentiated responders and non-responders are listed in Table 3.
Table 3.
Listing of genes in Dim2 and correlation (Bold indicates genes that overlap between Dim1 and Dim2)
| Name | Correlation | p | Name | Correlation | p | Name | Correlation | p | Name | Correlation | p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AIRE | −0.546 | 0.023 | CFH | 0.588 | 0.013 | IL18RAP | 0.681 | 0.003 | NFATC1 | −0.503 | 0.040 |
| APP | −0.553 | 0.021 | CFI | −0.891 | 0.000 | IL1R2 | −0.592 | 0.012 | NFKB1 | −0.552 | 0.022 |
| ARG1 | 0.529 | 0.029 | CFP | −0.593 | 0.012 | IL1RL1 | 0.521 | 0.032 | NFKB2 | −0.635 | 0.006 |
| ATG10 | 0.942 | 0.000 | CIITA | 0.775 | 0.000 | IL20 | −0.723 | 0.001 | NFKBIZ | −0.503 | 0.040 |
| ATG5 | −0.518 | 0.033 | CMKLR1 | 0.685 | 0.002 | IL22 | 0.708 | 0.001 | PDGFB | −0.626 | 0.007 |
| B3GAT1 | −0.574 | 0.016 | CSF1 | −0.773 | 0.000 | IL26 | −0.592 | 0.012 | PIGR | 0.559 | 0.020 |
| BATF3 | −0.497 | 0.042 | CSF2 | 0.580 | 0.015 | IL28A | −0.711 | 0.001 | PRF1 | 0.849 | 0.000 |
| BAX | 0.629 | 0.007 | CTLA4_all | −0.830 | 0.000 | IL28A/B | −0.568 | 0.017 | PSMB7 | −0.534 | 0.027 |
| BCAP31 | 0.533 | 0.028 | CTLA4-TM | 0.602 | 0.011 | IL29 | 0.679 | 0.003 | PSMB8 | 0.747 | 0.001 |
| BCL10 | −0.570 | 0.017 | CTNNB1 | −0.751 | 0.001 | IL32 | −0.646 | 0.005 | PSMC2 | −0.651 | 0.005 |
| BCL3 | −0.605 | 0.010 | CTSC | 0.529 | 0.029 | IL4 | 0.765 | 0.000 | PSMD7 | −0.551 | 0.022 |
| C14orf166 | 0.571 | 0.017 | CTSG | 0.526 | 0.030 | IL5 | 0.491 | 0.046 | PTGER4 | −0.680 | 0.003 |
| C1QB | 0.508 | 0.037 | CXCR1 | 0.636 | 0.006 | IL6ST | −0.672 | 0.003 | RAG1 | 0.577 | 0.015 |
| C4BPA | 0.849 | 0.000 | CXCR6 | 0.519 | 0.033 | IL7 | 0.596 | 0.012 | RAG2 | −0.523 | 0.031 |
| C6 | −0.716 | 0.001 | DEFB103B | 0.697 | 0.002 | IL7R | 0.507 | 0.038 | SELPLG | 0.534 | 0.027 |
| C7 | 0.483 | 0.049 | DEFB4A | 0.611 | 0.009 | IL9 | 0.803 | 0.000 | SIGIRR | −0.510 | 0.036 |
| C9 | 0.678 | 0.003 | EBI3 | 0.848 | 0.000 | ILF3 | −0.547 | 0.023 | SLAMF1 | 0.630 | 0.007 |
| CASP1 | 0.523 | 0.031 | ETS1 | 0.538 | 0.026 | IRF4 | −0.658 | 0.004 | SLAMF6 | 0.633 | 0.006 |
| CCBP2 | −0.536 | 0.027 | FCAR | −0.497 | 0.042 | IRGM | −0.514 | 0.035 | SMAD3 | 0.519 | 0.033 |
| CCL18 | 0.643 | 0.005 | FCGR1A/B | 0.662 | 0.004 | ITGB1 | 0.711 | 0.001 | SRC | −0.573 | 0.016 |
| CCL19 | 0.523 | 0.031 | FN1 | −0.522 | 0.032 | JAK1 | 0.557 | 0.020 | STAT2 | 0.482 | 0.050 |
| CCL23 | −0.681 | 0.003 | FOXP3 | −0.751 | 0.001 | JAK2 | 0.849 | 0.000 | STAT3 | −0.530 | 0.029 |
| CCL4 | 0.493 | 0.044 | GATA3 | −0.725 | 0.001 | KIR3DL1 | 0.511 | 0.036 | STAT5A | −0.734 | 0.001 |
| CCL8 | 0.722 | 0.001 | GBP1 | 0.494 | 0.044 | KIR3DL2 | −0.639 | 0.006 | STAT5B | 0.523 | 0.031 |
| CCND3 | 0.567 | 0.018 | GZMA | 0.812 | 0.000 | KLRB1 | 0.662 | 0.004 | TBX21 | 0.483 | 0.050 |
| CCR5 | 0.498 | 0.042 | HFE | 0.748 | 0.001 | KLRC2 | 0.590 | 0.013 | TCF7 | −0.546 | 0.023 |
| CCR7 | −0.561 | 0.019 | HLA-B | 0.554 | 0.021 | KLRD1 | 0.690 | 0.002 | TFRC | −0.759 | 0.000 |
| CCRL1 | −0.652 | 0.005 | HLA-DOB | −0.528 | 0.029 | KLRG1 | 0.598 | 0.011 | TGFBR2 | −0.615 | 0.009 |
| CD160 | 0.553 | 0.021 | ICOS | −0.520 | 0.032 | LCK | 0.643 | 0.005 | TIRAP | −0.675 | 0.003 |
| CD164 | −0.699 | 0.002 | IDO1 | 0.707 | 0.002 | LIF | −0.502 | 0.040 | TLR1 | −0.586 | 0.013 |
| CD1A | 0.683 | 0.003 | IFNA2 | 0.572 | 0.016 | LITAF | −0.613 | 0.009 | TLR3 | −0.509 | 0.037 |
| CD247 | −0.639 | 0.006 | IFNAR2 | 0.693 | 0.002 | LY96 | −0.539 | 0.026 | TLR7 | −0.696 | 0.002 |
| CD34 | 0.662 | 0.004 | IFNB1 | 0.746 | 0.001 | MAF | 0.599 | 0.011 | TLR9 | −0.788 | 0.000 |
| CD3D | 0.850 | 0.000 | IKBKB | 0.582 | 0.014 | MALT1 | −0.800 | 0.000 | TNF | 0.482 | 0.050 |
| CD45R0 | 0.535 | 0.027 | IKZF1 | 0.723 | 0.001 | MAP4K4 | −0.780 | 0.000 | TNFAIP6 | −0.571 | 0.017 |
| CD5 | −0.572 | 0.016 | IL10 | −0.731 | 0.001 | MAPK11 | −0.571 | 0.017 | TRAF5 | 0.668 | 0.003 |
| CD81 | 0.823 | 0.000 | IL11RA | −0.506 | 0.038 | MAPK14 | −0.499 | 0.042 | VCAM1 | 0.667 | 0.003 |
| CD8A | 0.540 | 0.025 | IL15 | 0.864 | 0.000 | MBL2 | 0.553 | 0.021 | VTN | 0.846 | 0.000 |
| CEACAM6 | 0.542 | 0.025 | IL17A | 0.731 | 0.001 | MUC1 | −0.564 | 0.018 | XCR1 | −0.495 | 0.043 |
| CEACAM8 | −0.490 | 0.046 | IL17F | −0.548 | 0.023 | NCAM1 | −0.912 | 0.000 | ZBTB16 | 0.669 | 0.003 |
Discussion
In 4 randomized clinical trials of patients with new and recent onset T1D, teplizumab has caused improvement in C-peptide responses for a year after treatment, compared to untreated or placebo treated patients. Previous clinical and preclinical studies have suggested that anti-CD3 mAb may affect immune responses by induction of regulatory cells, release of inhibitory cytokines, or even by depleting effector T cells but these studies did not distinguish the immunologic effects of the drug in general from those that were associated with clinical responses. In this analysis we have focused on the immunologic differences that lead to preservation of insulin secretion. At the conclusion of 14-day drug treatment, clinical responders had a decline in CD8 and CD4 CM and EM subsets that was significantly greater in the CD8EM and CD4EM subsets. However, these acute changes alone are unlikely to account for the lasting effects of the drug because these cell subpopulations recovered quickly and the changes in these subpopulations were not lasting. In both trials, an increase in CD8CM T cells identified the responders compared to drug-treated non-responders and controls. The analysis of cell counts in the AbATE trial which showed that there was a similar expansion of the CD8CM T cells in both responders and non-responders initially but a decline in the proportion and absolute number of circulating CD8CM T cells by 2 mos after treatment in the non-responders. This observation may not have been appreciated in the Delay or other trials due to less frequent sampling of PBMC between the 0 and 2 mo time-points.
The differences in the CD8CM T cells are not likely to be due to differences in the drug pharmacokinetics since drug is not detectable on the surfaces of T cells after 3–4 weeks ([6, 8] and personal observation), and the levels of coating and modulation of CD3 cannot identify clinical responders.
Our studies in vitro showed that the anti-CD3 mAb preferentially induced proliferation of CD8CM T cells. The source of the CD8CM cells involved both conversion of naïve or other non-CD8CM cells that had been activated by the anti-CD3 mAb, as well as proliferation of CD8CM T cells, because there were CD8+ cells that became CM T cells during the cultures. We are unable to perform similar lineage tracing studies in vivo but the acute changes that were seen in the effector population would suggest that there has been differentiation of the CD8+ cell population following teplizumab treatment, and our previous studies also suggest changes in CD8 subpopulations [11]. The effects of the anti-CD3 mAb on the original CM T cells and the newly differentiated CM cells may not be the same. In this regard, the timing of the anti-CD3 mAb treatment has been shown to be an important determinant of responses – drug efficacy appears to be greater after initiation of an immune response [5, 24–26].
Since our objective was to identify gene signatures associated with clinical responses, we first combined samples from placebo and drug treated non-responders and compared them to the drug treated responders. We identified 53 genes that differentiated the responders from the two other groups. These genes suggest that modulation of cell signaling, induction of regulatory cells or cytokines, or other mechanisms may be involved. Several of the genes are involved in T cell signaling (LCK, EBI3, JAK1, IKBK, KIR), These observations suggest that the anti-CD3 mAb may reduce signaling of CD8CM T cells. Since these cells may represent a source of diabetogenic effector T cells, this mechanism may account for the long term action of the drug in preventing disease progression.
We also found differences in the expression of genes associated with T cell differentiation including pathways involved in the development of pathogenic or regulatory T cells. The role of IL17A in T1D has been controversial but diabetes antigen specific T cells have been shown to produce IL17 and IL22 in response to their antigens [27]. In other studies, IL10 production in response to islet antigens was seen in healthy subjects [28]. We found lower expression of IL17A and IL22 in the responders compared to the non-responders and increased expression of other IL10 family cytokines such as IL20 and IL26. One of the differences we identified was in the expression of IKZF1, ikaros family zinc finger 1, which is also a T1D susceptibility gene. This gene is a regulator of lymphopoiesis and immune homeostasis. Its link to T1D was shown in genome-wide associated studies – the minor allele (rs10272724) is protective for the disease but the susceptibility allele is not correlated with the levels of transcripts in peripheral blood cells [23].
The changes in the expression of genes associated with immune regulation were variable: The expression of IDO1 was decreased but we found increased transcripts of LIF, which has been shown to induce Tregs [29]. Interestingly, we also found lower levels of expression of KLRC1 (NKG2A). In previous studies, we had shown that low expression of NKG2A identified CD8+ T cells with regulatory function from teplizumab treated subjects [16].
When we separately compared gene expression in the 3 subgroups, we also found differential expression of genes associated with T cell activation, differentiation, and regulation in the responders and non-responders. Our PCA analysis allowed us to identify a gene signature that could distinguish responders and non-responders and also to identify genes that were associated with anti-CD3 mAb treatment but not necessarily with clinical response. For example, consistent with our previous findings in humanized mice we found increased expression of CCL20 (MIP3A) by the CD8CM T cells that was correlated with the first principal component distinguishing placebo from treatment samples (Spearman’s ρ = −0.49, p=0.047), but this correlation is not found with the second principal component which distinguished responders and non-responders. We also found increased intracellular CCL20 production by CD8CM T cells from responders+non-responders from month 2 compared to CD8CM T cells from placebo treated subjects when they were stimulated with PMA/ionomycin (1.02±0.22% vs 0.37±0.05%, p=0.008, Mann-Whitney) but CCL20 production did not distinguish responders from non-responders (not shown).
The results of these studies should be interpreted cautiously. First, the sample numbers that we have studied were relatively small. In addition, we did not correct for multiple comparisons in our statistical analysis of gene expression. Although our studies of T cell proliferation in vitro suggest that the anti-CD3 mAb itself caused the expansion of the CD8CM T cells, it is possible that some cells within the expanded CD8CM population represent viral antigen reactive T cells. In a previous analysis of CD8+ T cells following treatment with otelixizumab, Keymeulen et al reported that there was transient EBV reactivation that was associated with an EBV-specific T cell response [30]. In participants in the AbATE trial, we found that the frequency of CD8CM T cells was higher in the EBV sero+ individuals (@ month 3: 86±24 × 103/ml (n=7) vs 294±106 × 103/ml (n=6), p=0.14, Student’s t-test with Welch’s correction) but together, these values were greater than the CD8CM T cells in EBV sero- non-responders (182±57 × 103/ml vs 41±12 × 103/ml, (n= 13), p=0.003, Mann-Whitney). We cannot exclude the possibility that EBV sero+ subjects are more likely to respond to the anti-CD3 mAb but the experience from that trial of otelixizumab, in which all subjects were EBV sero+ at entry would suggest that other factors determine whether individuals will respond to the treatment.
In summary, this study of participants in two clinical studies has identified a gene signature that is associated with clinical responses and can differentiate drug treated responders and non-responders. Reduced expression of genes associated with activation of T cells was found in the CD8CM T cell subpopulation that distinguishes the responders. In addition, there was enhanced expression of genes associated with differentiation of regulatory cells such as IL10 and KLRC1, and reduced transcripts of pathogenic cytokines such as IL17A. These findings may be used to identify individual factors that can predict clinical responses to anti-CD3 mAb which will help to improve the safety and efficacy of this form of immune modulation.
Materials and Methods
Human Subjects
We studied subjects with T1D between the ages of 8–35 enrolled in the AbATE and Delay studies (NCT00129259 and NCT00378508). The AbATE study was a randomized open-label study of patients with new-onset (< 3 months) diabetes, and the Delay study was a randomized placebo-controlled study that enrolled subjects with 4–12 months duration of disease. The drug-treated subjects in both trials received the same dose of teplizumab for 14 days (total dose = 9.033 mg/m2). The descriptions of the study groups and results of the primary endpoint analysis for both trials have previously been published [5, 7]. The data presented herein are limited to the first year of each study to permit direct comparison of studies. The study protocols and use of samples for the mechanistic studies were approved at each study site. Written informed consent was obtained from the participants.
Analysis of C-peptide responses/designation of responders
The subjects in both trials underwent a 4-hr mixed meal tolerance test (MMTT) at the time of study entry and after 6 and 12 months. C-peptide levels were measured at Northwest Lipid Research Laboratory (Seattle, WA), and the area under the curve (AUC) was calculated using the trapezoidal rule. We defined responders as subjects who lost < 7.5% of their baseline C-peptide response at month 12 [6]. This criterion was more stringent than criteria used to designate responders in the AbATE trial based on year-2 C-peptide results but had been used in previous analyses of treatment responses [6].
Flow cytometry
Fresh whole blood samples from AbATE were analyzed at the Immune Tolerance Network Flow Cytometry Core and fresh whole blood samples from the Delay trial were analyzed at the Yale Flow Cytometry Core. Fresh samples from study participants were stained with mAbs to CD8, CD4, CD45RA, CD45RO, and CD62L (BD Pharmingen, CA) to identify the following subsets of CD4+ and CD8+ T cells: CM: CD45RA−CD45RO+CD62L+; EM: CD45RA−CD45RO+CD62L−; EMRA: CD45RA+CD45RO−CD62L−; naïve: CD45RA+CD45RO−CD62L+. Gating strategies were kept consistent throughout each study and percentages of cell types or absolute cell counts (using the number of circulating lymphocytes from the CBC) were compared to identify changes with treatment. For gene expression analysis and cell culture studies, frozen cells were thawed prior to study. Because CD62L expression is affected by freezing and thawing [31], CCR7 was used to identify CD4+ or CD8+ T cell subsets: CM: CD45RO+CCR7+; EM: CD45RO+CCR7−; EMRA: CD45RO−CCR7−; naïve: CD45RO+CCR7+. Cells were acquired with an LSRII or a FACSAria cytometer and analyzed with FlowJo software. Samples were analyzed without knowledge of the treatment assignment.
Cell cultures
To analyze the effects of teplizumab on T cell proliferation, PBMCs from 3 different healthy controls were intracellularly labeled with CellTrace Violet dye (Life Technologies, Grand Island, NY). Thawed PBMCs were cultured with 0.5 μg/ml teplizumab and 1.0 μg/ml of anti-CD28 mAb for 3 days. Proliferation of different T cell subsets was determined based on dilution of CellTrace Violet dye, staining with the above mAbs and flow cytometry. For some experiments, CD8CM or naïve cells were sorted from PBMC and labeled with CellTrace Violet dye. The remaining cells were labeled with CFSE. The two populations were combined and cultured with teplizumab and anti-CD28 mAb as above. The frequency of CellViolet+ CD8CM T cells was determined after 1, 2, and 3 days by flow cytometry.
Gene expression analysis
To identify differences in gene expression among CD8CM T cells, PBMC from the 2 month time point from drug treated responders (n=6), non-responders (n=4), and placebo-treated control subjects (n=8) from the 2 mo time point in the Delay trial were labeled with mAbs to CD2, CD8, CD45RO, and CCR7 for cell sorting. CD8CM T cells (CD2+CD8+CD45RO+CCR7+) were sorted and RNA was prepared. The nanostring nCounter® GX Human Immunology gene expression panel was used for gene expression analysis of 594 immunology-related genes.
Pathway and network analyses
The Nanostring data were normalized by (Nalini transformation) followed by quantile normalization across all samples [32, 33] as implemented in the R BioConductor Project library preprocessCore (Bioconductor: Open software development for computational biology and bioinformatics R. (Gentleman, V. J. Carey, et al Genome Biology, Vol. 5, R80, 2004). For technical reasons, the results from one of the non-responders could not be normalized to the other 17 samples and the results from this sample was not used in the analyses. Principle Component Analysis was performed as described [34].
Statistical analyses
We compared the change in C-peptide AUC over 1 year in the drug-treated responders and non-responders vs control or placebo-treated subjects in each trial. Due to a baseline imbalance in HbA1c between placebo and drug-treated subjects in the Delay trial [7], C-peptide analysis was corrected for HbA1c levels at study entry in a mixed linear model. Changes in the proportion of T cell subsets from flow studies were compared by repeated measures ANOVA and corrected for the baseline levels. Absolute cell counts (Figure 2B) were calculated from the proportion of cells multiplied by the lymphocyte counts at the same blood draw. The presented flow cytometry data were corrected for the baseline unless otherwise indicated. Unless otherwise indicated, the data shown in the figures represent the LSMEANS values from the statistical model performed with SAS. Grouped data were compared either by t-tests, Wilcoxon test, Chi-squared test, ANOVA, or Kruskal-Wallis test (for data without normal distribution) as indicated. Calculations were done with GraphPad Prism (version 6.0) and SAS (version 9.3).
Supplementary Material
Expression of the IL7r on T cells by flow cytometry from each of the indicated groups. (p=0.014 by Kruskal-Wallis ANOVA).
Acknowledgments
Supported by grants R01 DK057846, U01AI1U01-02011-01, 2UL1RR024139 from the National Institutes of Health. This research was performed as a project of the Immune Tolerance Network (NIH Contract #N01 AI15416), an international clinical research consortium headquartered at the Benaroya Research Institute and supported by the National Institute of Allergy and Infectious Diseases and the Juvenile Diabetes Research Foundation.
Footnotes
Conflicts of interest: The authors have no conflicts of interest to declare.
References
- 1.Herold K, Vignali DA, Cooke A, Bluestone J. Type 1 diabetes: Translating mechanistic observations into effective clinical outcomes. Nat Rev Immunol. 2013:13. doi: 10.1038/nri3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coppieters KT, Dotta F, Amirian N, Campbell PD, Kay TW, Atkinson MA, Roep BO, von Herrath MG. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J Exp Med. 2012;209:51–60. doi: 10.1084/jem.20111187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagopian W, Ferry RJ, Jr, Sherry N, Carlin D, Bonvini E, Johnson S, Stein KE, Koenig S, Daifotis AG, Herold KC, Ludvigsson J. Teplizumab preserves C-peptide in recent-onset type 1 diabetes: two-year results from the randomized placebo-controlled Protege trial. Diabetes. 2013;62:3901–3908. doi: 10.2337/db13-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herold KC, Gitelman S, Greenbaum C, Puck J, Hagopian W, Gottlieb P, Sayre P, Bianchine P, Wong E, Seyfert-Margolis V, Bourcier K, Bluestone JA. Treatment of patients with new onset Type 1 diabetes with a single course of anti-CD3 mAb teplizumab preserves insulin production for up to 5 years. Clin Immunol. 2009 doi: 10.1016/j.clim.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herold KC, Gitelman SE, Ehlers MR, Gottlieb PA, Greenbaum CJ, Hagopian W, Boyle KD, Keyes-Elstein L, Aggarwal S, Phippard D, Sayre PH, McNamara J, Bluestone JA. Teplizumab (anti-CD3 mAb) treatment preserves C-peptide responses in patients with new-onset type 1 diabetes in a randomized controlled trial: Metabolic and immunologic features at baseline identify a subgroup of responders. Diabetes. 2013 doi: 10.2337/db13-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herold KC, Gitelman SE, Masharani U, Hagopian W, Bisikirska B, Donaldson D, Rother K, Diamond B, Harlan DM, Bluestone JA. A Single Course of Anti-CD3 Monoclonal Antibody hOKT3{gamma}1(Ala-Ala) Results in Improvement in C-Peptide Responses and Clinical Parameters for at Least 2 Years after Onset of Type 1 Diabetes. Diabetes. 2005;54:1763–1769. doi: 10.2337/diabetes.54.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herold KC, Gitelman SE, Willi SM, Gottlieb PA, Waldron-Lynch F, Devine L, Sherr J, Rosenthal SM, Adi S, Jalaludin MY, Michels AW, Dziura J, Bluestone JA. Teplizumab treatment may improve C-peptide responses in participants with type 1 diabetes after the new-onset period: a randomised controlled trial. Diabetologia. 2012 doi: 10.1007/s00125-012-2753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herold KC, Hagopian W, Auger JA, Poumian-Ruiz E, Taylor L, Donaldson D, Gitelman SE, Harlan DM, Xu D, Zivin RA, Bluestone JA. Anti-CD3 monoclonal antibody in new-onset type 1 diabetes mellitus. N Engl J Med. 2002;346:1692–1698. doi: 10.1056/NEJMoa012864. [DOI] [PubMed] [Google Scholar]
- 9.Keymeulen B, Vandemeulebroucke E, Ziegler AG, Mathieu C, Kaufman L, Hale G, Gorus F, Goldman M, Walter M, Candon S, Schandene L, Crenier L, De Block C, Seigneurin JM, De Pauw P, Pierard D, Weets I, Rebello P, Bird P, Berrie E, Frewin M, Waldmann H, Bach JF, Pipeleers D, Chatenoud L. Insulin needs after CD3-antibody therapy in new-onset type 1 diabetes. N Engl J Med. 2005;352:2598–2608. doi: 10.1056/NEJMoa043980. [DOI] [PubMed] [Google Scholar]
- 10.Sherry N, Hagopian W, Ludvigsson J, Jain SM, Wahlen J, Ferry RJ, Jr, Bode B, Aronoff S, Holland C, Carlin D, King KL, Wilder RL, Pillemer S, Bonvini E, Johnson S, Stein KE, Koenig S, Herold KC, Daifotis AG. Teplizumab for treatment of type 1 diabetes (Protege study): 1-year results from a randomised, placebo-controlled trial. Lancet. 2011 doi: 10.1016/S0140-6736(11)60931-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cernea S, Herold KC. Monitoring of antigen-specific CD8 T cells in patients with type 1 diabetes treated with antiCD3 monoclonal antibodies. Clin Immunol. 2010;134:121–129. doi: 10.1016/j.clim.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herold KC, Gitelman SE, Willi SM, Gottlieb PA, Waldron-Lynch F, Devine L, Sherr J, Rosenthal SM, Adi S, Jalaludin MY, Michels AW, Dziura J, Bluestone JA. Teplizumab treatment may improve C-peptide responses in participants with type 1 diabetes after the new-onset period: a randomised controlled trial. Diabetologia. 2013;56:391–400. doi: 10.1007/s00125-012-2753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orban T, Beam CA, Xu P, Moore K, Jiang Q, Deng J, Muller S, Gottlieb P, Spain L, Peakman M Type 1 Diabetes TrialNet Abatacept Study G. Reduction in CD4 central memory T-cell subset in costimulation modulator abatacept-treated patients with recent-onset type 1 diabetes is associated with slower C-peptide decline. Diabetes. 2014;63:3449–3457. doi: 10.2337/db14-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigby M, DiMeglio L, Rendell M, Felner E, Dostou J, Gitelman S, Patel C, Griffin K, Tsalikian E, Gottlieb P, Greenbaum C, Sherry N, Moore W, Roshanak M, Willi S, Raskin P, Moran A, Russell W, Pinckney A, Keyes-Elstein L, Howell M, Aggarwal S, Lim N, Phippard D, Nepom G, McNamara J, Ehlers MR. Targeting effector memory T cells with alefacept in new onset type 1 diabetes: 12 month results from the TIDAL study. Lancet Endocrinology and Metabolism. 2013 doi: 10.1016/S2213-8587(13)70111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ablamunits V, Bisikirska B, Herold KC. Acquisition of regulatory function by human CD8(+) T cells treated with anti-CD3 antibody requires TNF. Eur J Immunol. 2010;40:2891–2901. doi: 10.1002/eji.201040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ablamunits V, Henegariu O, Preston-Hurlburt P, Herold KC. NKG2A is a marker for acquisition of regulatory function by human CD8+ T cells activated with anti-CD3 antibody. Eur J Immunol. 2011;41:1832–1842. doi: 10.1002/eji.201041258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belghith M, Bluestone JA, Barriot S, Megret J, Bach JF, Chatenoud L. TGF-beta-dependent mechanisms mediate restoration of self-tolerance induced by antibodies to CD3 in overt autoimmune diabetes. Nat Med. 2003;9:1202–1208. doi: 10.1038/nm924. [DOI] [PubMed] [Google Scholar]
- 18.Bisikirska B, Colgan J, Luban J, Bluestone JA, Herold KC. TCR stimulation with modified anti-CD3 mAb expands CD8 T cell population and induces CD8CD25 Tregs. J Clin Invest. 2005;115:2904–2913. doi: 10.1172/JCI23961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldron-Lynch F, Henegariu O, Deng S, Preston-Hurlburt P, Tooley J, Flavell R, Herold KC. Teplizumab induces human gut-tropic regulatory cells in humanized mice and patients. Sci Transl Med. 2012;4:118ra112. doi: 10.1126/scitranslmed.3003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O’Connor W, Jr, Rongvaux A, Van Rooijen N, Haberman AM, Iwakura Y, Kuchroo VK, Kolls JK, Bluestone JA, Herold KC, Flavell RA. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hor S, Pirzer H, Dumoutier L, Bauer F, Wittmann S, Sticht H, Renauld JC, de Waal Malefyt R, Fickenscher H. The T-cell lymphokine interleukin-26 targets epithelial cells through the interleukin-20 receptor 1 and interleukin-10 receptor 2 chains. J Biol Chem. 2004;279:33343–33351. doi: 10.1074/jbc.M405000200. [DOI] [PubMed] [Google Scholar]
- 22.Nikoopour E, Bellemore SM, Singh B. IL-22, cell regeneration and autoimmunity. Cytokine. 2015;74:35–42. doi: 10.1016/j.cyto.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Swafford AD, Howson JM, Davison LJ, Wallace C, Smyth DJ, Schuilenburg H, Maisuria-Armer M, Mistry T, Lenardo MJ, Todd JA. An allele of IKZF1 (Ikaros) conferring susceptibility to childhood acute lymphoblastic leukemia protects against type 1 diabetes. Diabetes. 2011;60:1041–1044. doi: 10.2337/db10-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You S, Zuber J, Kuhn C, Baas M, Valette F, Sauvaget V, Sarnacki S, Sawitzki B, Bach JF, Volk HD, Chatenoud L. Induction of allograft tolerance by monoclonal CD3 antibodies: a matter of timing. Am J Transplant. 2012;12:2909–2919. doi: 10.1111/j.1600-6143.2012.04213.x. [DOI] [PubMed] [Google Scholar]
- 25.Chatenoud L, Primo J, Bach JF. CD3 antibody-induced dominant self tolerance in overtly diabetic NOD mice. J Immunol. 1997;158:2947–2954. [PubMed] [Google Scholar]
- 26.Goto R, You S, Zaitsu M, Chatenoud L, Wood KJ. Delayed anti-CD3 therapy results in depletion of alloreactive T cells and the dominance of Foxp3+ CD4+ graft infiltrating cells. Am J Transplant. 2013;13:1655–1664. doi: 10.1111/ajt.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arif S, Moore F, Marks K, Bouckenooghe T, Dayan CM, Planas R, Vives-Pi M, Powrie J, Tree T, Marchetti P, Huang GC, Gurzov EN, Pujol-Borrell R, Eizirik DL, Peakman M. Peripheral and islet interleukin-17 pathway activation characterizes human autoimmune diabetes and promotes cytokine-mediated beta-cell death. Diabetes. 2011;60:2112–2119. doi: 10.2337/db10-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arif S, Tree TI, Astill TP, Tremble JM, Bishop AJ, Dayan CM, Roep BO, Peakman M. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J Clin Invest. 2004;113:451–463. doi: 10.1172/JCI19585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao W, Thompson L, Zhou Q, Putheti P, Fahmy TM, Strom TB, Metcalfe SM. Treg versus Th17 lymphocyte lineages are cross-regulated by LIF versus IL-6. Cell Cycle. 2009;8:1444–1450. doi: 10.4161/cc.8.9.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keymeulen B, Candon S, Fafi-Kremer S, Ziegler A, Leruez-Ville M, Mathieu C, Vandemeulebroucke E, Walter M, Crenier L, Thervet E, Legendre C, Pierard D, Hale G, Waldmann H, Bach JF, Seigneurin JM, Pipeleers D, Chatenoud L. Transient Epstein-Barr virus reactivation in CD3 monoclonal antibody-treated patients. Blood. 2010;115:1145–1155. doi: 10.1182/blood-2009-02-204875. [DOI] [PubMed] [Google Scholar]
- 31.Reimann KA, Chernoff M, Wilkening CL, Nickerson CE, Landay AL. Preservation of lymphocyte immunophenotype and proliferative responses in cryopreserved peripheral blood mononuclear cells from human immunodeficiency virus type 1-infected donors: implications for multicenter clinical trials. The ACTG Immunology Advanced Technology Laboratories. Clin Diagn Lab Immunol. 2000;7:352–359. doi: 10.1128/cdli.7.3.352-359.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolstad BM, Collin F, Simpson KM, Irizarry RA, Speed TP. Experimental design and low-level analysis of microarray data. Int Rev Neurobiol. 2004;60:25–58. doi: 10.1016/S0074-7742(04)60002-X. [DOI] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Le S, Josse J, Husson F, Factomine R. Journal of Statistical Software. 2008:25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of the IL7r on T cells by flow cytometry from each of the indicated groups. (p=0.014 by Kruskal-Wallis ANOVA).