Abstract
The digestive tracts of many animals are epithelial tubes with specialized compartments to break down food, remove wastes, combat infection, and signal nutrient availability. C. elegans possesses a linear, epithelial gut tube with foregut, midgut, and hindgut sections. The simple anatomy belies the developmental complexity that is involved in forming the gut from a pool of heterogeneous precursor cells. Here, I focus on the processes that specify cell fates and control morphogenesis within the embryonic foregut (pharynx) and the developmental roles of the pharynx after birth. Maternally donated factors in the pregastrula embryo converge on pha-4, a FoxA transcription factor that specifies organ identity for pharyngeal precursors. Positive feedback loops between PHA-4 and other transcription factors ensure commitment to pharyngeal fate. Binding-site affinity of PHA-4 for its target promoters contributes to the progression of the pharyngeal precursors towards differentiation. During morphogenesis, the pharyngeal precursors form an epithelial tube in a process that is independent of cadherins, catenins, and integrins but requires the kinesin zen-4/MKLP1. After birth, the pharynx and/or pha-4 are involved in repelling pathogens and controlling aging.
Keywords: foxa2, endoderm, morphogenesis, epithelia, dietary restriction, aging
INTRODUCTION
To form the pharynx, C. elegans faces developmental challenges that are similar to organ formation in more complex animals. With the ability to visualize individual cells during organogenesis and the development of powerful genetic and genomic tools, scientists have begun to dissect the pathways that control cell fate specification, morphogenesis, and postembryonic roles of the pharynx.
ANATOMY OF THE MATURE PHARYNX
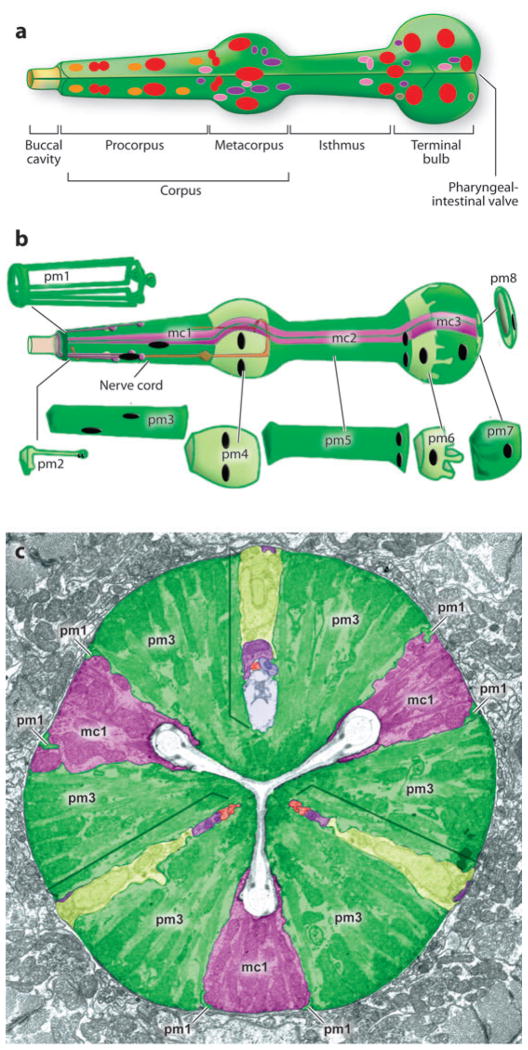
Ultrastructural studies have provided an in-depth view of pharynx architecture (Albertson & Thomson 1976). The pharynx is a bilobed, linear tube that is organized into three sections: At the anterior, the corpus pumps food (bacteria) into the pharynx and concentrates it by expelling excess water. The bacteria then pass through the isthmus by peristalsis and are ground up by chitinous projections in the terminal bulb (Figure 1) (Avery & Horvitz 1989). The pharynx is made up of 95 cells grouped into seven cell types: arcade cells, muscle or myoepithelial cells, epithelia, neurons, glands, marginal cells, and valves (Albertson & Thomson 1976, Horner et al. 1998). The bulk of the pharynx is composed of eight sections of muscles, pm1-pm8, that are positioned as rings along the longitudinal axis (Figure 1). The muscles and interdigitating marginal cells are arranged with threefold symmetry. The muscle fibers are organized radially so that contraction widens the pharyngeal lumen from a Y-shape to a triangle (Figure 1), whereas the marginal cells sit at the vertices of the Y and anchor the lumen during muscle contraction and relaxation (Figure 1). Both the muscles and marginal cells have characteristics of epithelia with adherens junctions and an apical surface that faces the lumen.
Figure 1.
Pharynx anatomy. (a) Nuclei within the pharynx are shown as muscles (red ), neurons ( purple), epithelia (orange), marginal cells ( pink), and glands (brown). Arcade cells and pharyngeal intestinal valves are not shown. (b) The bulk of the pharynx is composed of eight layers of muscles (pm1–8) ( green) and three groups of structural marginal cells (mc1–3) ( purple). (c) Muscles and marginal cells are arranged with threefold rotational symmetry, as shown in the cross section. Adapted from Mango (2007) and Altun & Hall [Wormatlas (http://www.wormatlas.org)].
The pharyngeal lumen is lined with cuticle, which is continuous with the cuticle of the epidermis, although structurally distinct. Specialized cuticular fingers project into the lumen of the corpus and terminal bulb and may function as a sieve and teeth, respectively. These additions help propel bacteria down the pharynx and crush them to initiate the digestion process.
Five glands and twenty neurons are embedded within the pharyngeal myoepithelium (Figure 1) (Albertson & Thomson 1976). The neurons are organized bilaterally with processes that extend along the left and right subventral surfaces or along the dorsal surface. They synapse on pharyngeal muscles or other pharyngeal neurons and control the rate of feeding (Avery & Horvitz 1987, 1989). The gland cells send processes into the pharyngeal lumen and secrete vesicles prior to hatching, at each larval molt, and during feeding. The glands produce mucins that may line the lumen and lubricate the passage of food (Smit et al. 2008). At the anterior, the pharynx attaches to the buccal cavity and epidermis with three rings of arcade cells and epithelial cells. At the posterior, the last pharyngeal muscle, pm8, links to the intestine via a toroid of six valve cells. For additional discussion of pharyngeal anatomy, see the Atlas of C. elegans anatomy (http://www.wormatlas.org/htm).
CELL FATE SPECIFICATION DURING EMBRYOGENESIS
Overview
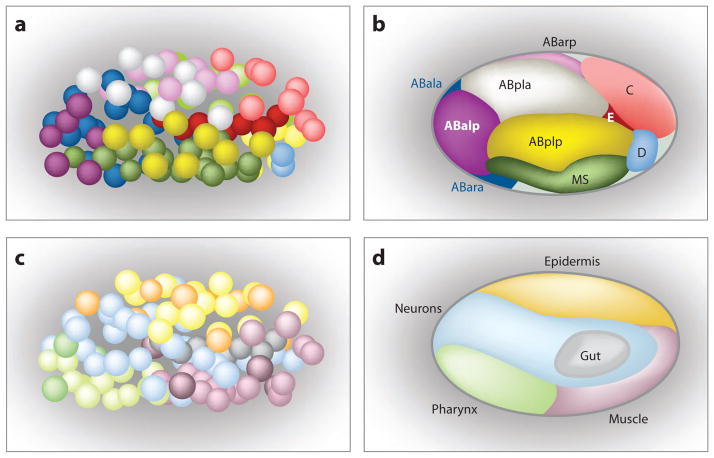
To build the mature pharynx, C. elegans embryos first establish a pool of pharyngeal precursors and subsequently specify different cell-type identities such as pharyngeal muscle or pharyngeal gland. Three patterning systems govern these events. During the first 100 minutes of embryogenesis, maternally deposited factors establish six founder cells, two of which give rise to pharyngeal cells. The maternal regulatory circuits segue into two zygotic patterning systems (Figure 2). One system distinguishes antero-posterior fates in daughter cells that divide along the A-P axis. This system functions reiteratively and helps specify the founder cells in the pregastrula embryo, the pharyngeal precursors at the onset of gastrulation, and cell types within the pharynx during the terminal cleavage stages. A second patterning system generates a cluster of organ precursors in the early gastrula. Rather than lineage, cells within this domain are united to each other by their pharyngeal identity and the homogeneous expression of the FoxA transcription factor PHA-4. Subsequently, additional tissue and lineage-restricted factors establish different cell types within the pharynx in combination with PHA-4. The three pathways—founder cells, A-P patterning and tissue/organ patterning—illustrate how a cohesive organ is built progressively from a heterogeneous pool of cells.
Figure 2.
Two zygotic patterning systems. (a, b) For lineage-based regulation, cells that derive from a single progenitor blastomere cluster together. With each antero-posterior division, anterior versus posterior fates are established between pairs of daughter cells. Cell nuclei are shown in (a) and fields of cells are shown in (b). (c, d ) During organ/tissue regulation, cells from different lineages that are destined to produce common cell types cluster together, revealing a shared organ or tissue identity. Nuclei are shown in (c) and fields of cells are shown in (d ). Adapted from Labousse & Mango (1999).
Founder Cells Are Established by Maternally Donated Factors
Two founder cells called AB and MS produce the pharyngeal cells and also generate other nonpharyngeal cell types. Independent pathways specify AB and MS and control their ability to produce pharynx (reviewed in Mango 2007). Ultimately, however, the pharyngeal cells produced by AB and MS are close neighbors, and some even fuse with each other (Figure 1) (Albertson & Thomson 1976). AB is formed by an asymmetric cell division at the one-cell stage (reviewed by Cowan & Hyman 2007). The Notch receptor glp-1 is maternally supplied and selectively translated in early AB descendants (see Table 1 for a list of genes with roles in pharyngeal development) (Evans & Hunter 2005). Two rounds of GLP-1/Notch-mediated signaling ensure that a subset of AB descendants generate pharyngeal cells (Evans & Hunter 2005, Priess 2005). At the 4-cell stage, AB gives rise to an anterior daughter called ABa, which produces pharyngeal cells, and a posterior ABp daughter, which does not. The distinction in pharynx production depends on the ref-1 family of bHLH transcription factors, which are induced only in ABp by GLP-1/Notch signaling (Figure 3) (Neves & Priess 2005). The ref-1 family is comprised of six genes distantly related to the E(spl) family of transcriptional repressors (Neves & Priess 2005). These REF proteins repress transcription of the closely related T-box factors, tbx-37 and tbx-38, most likely directly (Neves & Priess 2005). tbx-37 and tbx-38 constitute a pair of redundant factors that are critical to produce pharynx from the AB lineage; their absence ensures that no ABp descendants generate pharyngeal cells in wild-type embryos (Good et al. 2004). This regulatory hierarchy explains why the inactivation of GLP-1 signaling at the 4-cell stage leads to an overabundance of pharyngeal cells, which are derived from ABp (Good et al. 2004).
Table 1.
Summary of genes involved in pharynx development
| Gene | Homology | M/Z | Expression (initiation) | Pha phenotype | Targets | Binding sequence | Upstream genes | References |
|---|---|---|---|---|---|---|---|---|
| ceh-2 | Homeobox ems, EMX | Z | I3, NSM, M3, pm2, e2 | Poor feeding, M3 defective | ? | ? | Aspock et al. 2003 | |
| ceh-22 | Nkx 2.2 Homeobox TF | Z | pm3–5, pm7 | Indistinct BM around pharynx | myo-2 | CACTTAT |
pha-4 ceh-22 ceh-2 pha-2 |
Kalb et al. 1998, Mango et al. 1994, Okkema & Fire 1994, Okkema et al. 1997 |
| ceh-24 | Nkx 2.3 TF | Z | pm8 | None | ? | ? | ? | Harfe et al. 1998 |
| ceh-28 | Nkx 2 TF | Z | M4 | Peristalsis defective, M4 outgrowth | Ray et al. 2008 | |||
| ceh-43 | Distal-less homeobox | Z | Non-pha neurons, epidermis | Pun, anterior leakage | Aspock & Burglin 2001, Burglin & Aspock 1999 | |||
| daf-12 | NHR Zn TF | Z | Broadly | Daf |
myo-2 ceh-22 |
AGTGCA | daf-9 | Ao et al. 2004 |
| eya-1 | Eyes absent | Z | Broadly ≥ bean | Thin, asymmetric | ? | NA | ? | Furuya et al. 2005 |
| gei-17 | E3 sumo ligase | ? | ? | Thick isthmus | ? | NA | ? | Roy Chowdhuri et al. 2006 |
| glp-1 | Notch Rcp | M | ABa@4 ABa@12 |
Aph |
ref-1 pha-4 |
NA | ? | Kalb et al. 1998, Mango et al. 1994, Priess et al. 1987 |
| hlh-6 | Sage-like | Z | Glands | Loss of some g2 glands, misformed g1 glands, PGM1 activation | phat |
lag-1 pha-4 |
Raharjo & Gaudet 2007, Smit et al. 2008 | |
| htz-1 | H2A.Z histone variant | Z | Broadly from the 28-cell stage | Delayed activation |
myo-2, R07B1.9 many |
NA | ? | Updike & Mango 2006, Whittle et al. 2008 |
| lag-1 | Su(H) TF | M | Broadly | Aph |
ref-1 pha-4 |
RTGGGAA | glp-1 | Christensen et al. 1996, Smith & Mango 2007 |
| lin-12 | Notch receptor | Z | pm8 | pm8 tubulogenesis |
lag-1 aff-1 eff-1 |
NA | lag-2/Delta | Rasmussen et al. 2008 |
| med-1/2 | GATA-like Zn TF | M, Z | EMS@4 and maternal | Ppa | tbx-35 | RRRAGTATAC | skn-1 | Broitman-Maduro et al. 2005, Maduro |
| myo-2 | Myosin heavy chain | Z | pm1–pm8 | NA |
pha-4 ceh-22 peb-1 daf-12 daf-3 |
Ao et al.2004, Gaudet & Mango 2002, Mango et al. 1994, Okkema & Fire 1994, Thatcher et al. 2001 | ||
| peb-1 | FLYWCH Zn TF | Z | Broadly | Glands distended | myo-2 | YDTGCCRW | ? | Beaster-Jones & Okkema 2004, Fernandez et al. 2004, Thatcher et al. 2001 |
| pha-1 | Novel | Z | Broadly | Arrested diffn, loss of TF expression |
ceh-22 pha-4 |
NA or ? | ? | Fay et al. 2004, Granato et al. 1994, Schnabel & Schnabel 1990 |
| pha-2 | Homeobox | Z | pm5, I4, epi | pm4, pm5, morph |
ceh-22 ceh-2 |
Avery 1993, Morck et al. 2004 | ||
| pha-4 | FoxA | Z | ABa@44 MS@28 |
Pha, adult longevity | many | TRTTKRY |
lag-1 tbx-37/38 med-1/2 glp-1 tbx-35, tbx-2 |
Broitman-Maduro et al. 2006, Gaudet & Mango 2002, Good et al. 2004, Maduro et al. 2005b, Mango et al. 1994, Smith & Mango 2007 |
| ref-1 family | bHLH TF | Z | ABa@26 ABp@4 EMS@24 |
None |
tbx-37 tbx-2 |
CANNTG | lag-1, glp-1 | Neves & Priess 2005, Smith & Mango 2007 |
| skn-1 | Zipper TF | M | EMS@4 | Pha |
ref-1 end-1 med-1, med-2 |
G/ATCAT + A/T | ? | Bowerman et al. 1992, Maduro et al. 2005b, Mango et al. 1994, Neves & Priess 2005 |
| smk-1 | PPH phosphatase regulatory subunit | ? | Broadly | Adult aging, DNA damage | sod-1, sod-2, sod-3, sod-4, sod-5, ctl-1, lys-8 | ? | ? | Wolff et al. 2006, Panowski et al. 2007 |
| tbx-2 | T-box | Z | ABa@8E | ABa muscles absent |
pha-4 ceh-22? |
? |
ref-1 tbx-37/38? |
Roy Chowdhuri et al. 2006, Smith & Mango 2007 |
| tbx-37/38 | T-box | Z | ABa@24 | Aph | pha-4? | ? | ref-1 in some cell types | Good et al. 2004 |
| tmy-1 | Tropomyosin | Z | Pha muscles pm1, pm3, pm4, pm5, pm7, intestine, gonad | Lethal | NA |
pha-4 ceh-22 |
Anokye-Danso et al. 2008 | |
| ubc-9 | E2 sumo ligase | ? | ? | ABa muscles absent | ? | NA | ? | Roy Chowdhuri et al. 2006 |
| unc-39 | Six4/5 | Z | Mesoderm arc | Pun metacorpus | ? | ? | ? | Yanowitz et al. 2004 |
Genes that are implicated in pharynx development are listed from earliest stages of specification to later events. Aph: anterior pharynx absent, Daf: dauer defective, M/Z: maternal or zygotic contribution of RNA or protein, Pha: pharynx absent, pm: pharyngeal muscle, PPa: posterior pharynx absent, Pun: pharynx unattached, Rcp: receptor, TF: transcription factor, Zn: zinc, ?: unknown. Adapted and extended from Mango (2007).
Figure 3.
Early developmental pathways. Features of pharyngeal development from the 4-cell stage to the 28-cell stage. This period is under the control of maternal factors, but transitions to zygotic control with the activation of tbx-35, tbx-37, tbx-38, and pha-4. (Left panels) show cells and the cell types they produce. (Right panels) illustrate genetic regulatory relationships. The pharynx is generated from a subset of blastomeres ( green) (i.e., ABa and EMS at the 4-cell stage). Descendents of green cells that do not produce pharyngeal cells are ( gray). Reprinted from Mango (2007).
All ABa descendants activate tbx-37 and tbx-38 expression, rendering these blastomeres competent to produce pharynx in response to inductive cues. At the 12-cell stage, GLP-1-mediated signaling from MS to two ABa granddaughters activates the LAG-1 transcription factor (Mango et al. 1994, Moskowitz et al. 1994, Priess et al. 1987). The combination of active LAG-1 and TBX-37/TBX38 induces the organ selector gene pha-4, either directly or indirectly, by the 44-cell stage (or 4E stage) in a subset of ABa descendants (Good et al. 2004, Horner et al. 1998, Smith & Mango 2007). A selector gene is a transcription factor that establishes positional, cell type, or in this case organ identity for groups of cells (Garcia-Bellido 1975, Mann & Carroll 2002). PHA-4 is the central selector regulator for the pharynx and its activity is essential for all pharyngeal development (Mango et al. 1994). The appearance of tbx-37/tbx-38 and pha-4 marks the transition to zygotic control of pharyngeal development within the AB lineage.
The MS founder cell is born at the 7-cell stage and generates pharyngeal cells by a pathway distinct from that of AB (Good et al. 2004, Priess & Thomson 1987). MS fate depends on SKN-1, a bZIP-related transcription factor that functions at the 4–8 cell stage to specify MS, its sister cell E, and their mother cell (Figure 3) (Bowerman et al. 1992, 1993). In the absence of skn-1, no pharynx is produced because MS and E are transformed into their cousin the C blastomere, which neither generates pharynx nor produces Notch ligands to signal to AB descendants. Conversely, mutants with ectopic SKN-1 activity produce extra MS-like cells and ectopic pharynx, revealing an instructive role for the SKN-1 protein (Mello et al. 1992). SKN-1 activates the transcription of a pair of GATA factors med-1 and med-2 in the 7-cell stage embryo (Maduro et al. 2001, 2007). The loss of both med genes leads to the loss of MS blastomere fate (Maduro et al. 2001, 2006), with controversial effects on E development (Captan et al. 2007; Goszczynski & McGhee 2005; Maduro et al. 2001, 2007). The phenotype of med genes is similar to that of skn-1 for the MS lineage, suggesting the med genes are the major target of SKN-1 within the MS blastomere.
The MED factors recognize the consensus sequence RAGTATAC (R = A/G), a derivative of the canonical GATA site HGATAR (H = A/T/C) (Broitman-Maduro et al. 2005). A genome-wide search for pairs of MED consensus sites uncovered MED target genes, including the transcription factor tbx-35 (Broitman-Maduro et al. 2005, 2006). tbx-35 is expressed within MS and its descendants and is necessary and sufficient to produce MS-derived body wall muscles and pharyngeal cells. However, differences between tbx-35 and med loss-of-function phenotypes suggest there are additional important MED targets within the MS lineage (Broitman-Maduro et al. 2005, 2006; Robertson et al. 2004). Intriguingly, tbx-35 is closely related to tbx-37 and tbx-38, revealing a similar requirement for T-box factors during AB and MS pharyngeal development. The heavy reliance on T-box factors is reminiscent of vertebrate endoderm development, including VegT in Xenopus and eomesodermin in zebrafish and mice (Arnold et al. 2008, Bjornson et al. 2005, Heasman 2006). The vertebrate T-box proteins differ in their timing and the target genes they regulate (Grapin-Botton 2008), raising the question of whether T-box usage in endoderm specification represents evolutionary conservation or convergence. For example, Xenopus VegT is maternally contributed and initiates endoderm formation, whereas eomesodermin is zygotically activated and functions downstream of other initiating cues similar to C. elegans (Grapin-Botton 2008). Both the upstream activators of eomesodermin and its downstream effectors appear distinct from those of TBX-35, TBX-37, and TBX-38.
These studies reveal the interplay between heterogeneous pools of precursor cells and the regulators that drive them toward pharyngeal fate. Similar processes function in vertebrate organs. For example, the pancreas is formed from separate pools of dorsal and ventral foregut endoderm. Like C. elegans, different molecular pathways are important to generate pancreatic cell types from these different populations of cells (reviewed by Zaret 2008). The different requirements of different precursor cells may have important consequences for our ability to manipulate developmental pathways, a goal of regenerative medicine. For example, GLP-1/Notch signaling represses pharynx development in AB descendants at the 4-cell stage but induces pharynx development in these same cells at the 12-cell stage, which depends on the presence or absence of TBX-37 and TBX-38. These observations underscore the importance of understanding the cellular context of regulatory pathways during development.
Transition to Zygotic Control: A-P Patterning
Most divisions in the C. elegans embryo align along the A-P axis. A global system of A-P patterning distinguishes anterior blastomeres from their posterior sisters (Kaletta et al. 1997, Labouesse & Mango 1999, Lin et al. 1995). Anterior versus posterior identity, in combination with maternally contributed factors, generates a code that determines cell fates. For the pharynx, the best-described role of the A-P patterning system is to distinguish the anterior MS founder cell from its posterior sister E. This system depends on the Lef transcription factor POP-1 and its cofactor SYS-1/b-catenin (Huang et al. 2007; Kidd et al. 2005; Lin et al. 1995, 1998; Phillips et al. 2007) (reviewed in Eisenmann 2005, Mizumoto & Sawa 2007). In MS, high POP-1 and low SYS-1 ensure POP-1 functions as a repressor, whereas in E, low POP-1 and high SYS-1 switch POP-1 into an activator. The levels of nuclear POP-1 and SYS-1 are set by the wnt and MAPK signaling pathways (Calvo et al. 2001; Kaletta et al. 1997; Kidd et al. 2005; Lin et al. 1995, 1998; Lo et al. 2004; Maduro et al. 2001; Siegfried et al. 2004).
Two important POP-1 targets in MS and E are end-1 and end-3, which encode a pair of redundant GATA factors that specify E (intestinal) fate (Broitman-Maduro et al. 2005, Calvo et al. 2001, Maduro et al. 2002, Maduro & Rothman 2002, Shetty et al. 2005). POP-1 repression of end-1 and end-3 transcription in MS quenches intestinal development. Thus, MS founder identity is established by SKN-1, MED-1/2, high POP-1, and low SYS-1. This regulatory hierarchy explains why reduced POP-1 activity leads to end-1 derepression in MS, conversion of MS into E, and therefore loss of MS-derived pharyngeal cells (Lin et al. 1995, Maduro et al. 2005b). The direct transcriptional regulation of end-1 and end-3 by POP-1 is the only known example of how the A-P patterning system links to tissue/organ specification. POP-1 continues to be expressed at a high level in anterior daughters and a low level of posterior daughters throughout the embryo, but additional targets of POP-1 are unknown.
Specification of mesendoderm or endoderm by β-catenin/Lef signaling is conserved in diverse species ranging from vertebrates to cnidaria, echinoderms, and some lophotrochozoa, suggesting this may be an ancestral function (Grapin-Botton 2008, Henry et al. 2008, Schneider & Bowerman 2007, Wikramanayake et al. 2003). However, two aspects of this pathway are very different in C. elegans from most other animals. First, the Lef/β-catenin system has expanded in C. elegans to control multiple A-P decisions throughout embryogenesis (Kaletta et al. 1997, Lin et al. 1998). For example, POP-1 contributes to pharyngeal development by distinguishing A-P identities at the 12-cell stage (Lin et al. 1998). This event establishes ABalp and ABara, which produce pharyngeal cells in response to GLP-1/Notch signaling and TBX-37/38 at later stages. A second difference is that vertebrate Lef/β-catenin activates Nodal signaling, which is a key inducer of endoderm in mammals (Grapin-Botton 2008). C. elegans lacks Nodal and does not use TGFβ signaling during embryogenesis (Savage-Dunn 2005). Perhaps the small cell number and invariant lineage have enabled nematodes to discard signaling pathways, such as TGFβ and hedgehog, and evolve a more autonomous A-P patterning system instead.
Transition to Zygotic Control: pha-4 and Tissue/Organ Patterning
LAG-1, TBX-35, TBX-37/38, POP-1, and the MED factors initiate the zygotic phase of pharyngeal development. As this phase gets under way, cells destined to produce the pharynx and that derive from different cell lineages coordinate their development to form an integrated organ (Figure 2). One of the first signs of integration is that cells from different lineages but similar fates (e.g., pharynx) cluster together within the gastrulating embryo. Mutations that alter cell fate also alter cell positioning, suggesting that the cell clusters reflect developmental identities (Horner et al. 1998, Labouesse & Mango 1999, Schnabel et al. 2006). Another indication of altered embryonic organization is that many genes that function during the zygotic phase have phenotypes that affect a specific tissue or organ rather than an entire cell lineage (Mango et al. 1994, Okkema & Fire 1994, Okkema et al. 1997, Roy Chowdhuri et al. 2006, Smith & Mango 2007). LAG-1, TBX-35, TBX-37/38, and the MED factors contribute to the transition—their mutant phenotypes affect cell lineages (e.g., MS)—but their downstream targets, described below, are geared towards tissues and organs such as MS-derived pharynx or muscle (Broitman-Maduro et al. 2006, Good et al. 2004, Maduro et al. 2005a, Smith & Mango 2007).
The maternal patterning genes converge on the FoxA transcription factor pha-4. pha-4 is the central regulator of pharynx development and the only zygotic gene that deletes the entire pharynx when mutated (Figure 3) (Horner et al. 1998, Kalb et al. 1998, Mango et al. 1994). This dramatic phenotype reflects the direct involvement of PHA-4 in transcribing many, and perhaps all, genes selectively transcribed in the pharynx, including early acting developmental regulators and terminal differentiation genes that encode structural proteins and digestive enzymes (Anokye-Danso et al. 2008, Gaudet & Mango 2002, Kalb et al. 1998, Mango 2007, Morck et al. 2004, Raharjo & Gaudet 2007, Vilimas et al. 2004). The global expression and requirement for pha-4 in all pharyngeal cells suggests that the function of pha-4 is to specify organ identity within the pharynx (Horner et al. 1998).
Organ identity genes may exist in other animals. In the mammalian pancreas, both dorsal and ventral pancreatic progenitors activate pdx1, which is required for the outgrowth and maintenance of the pancreatic buds; pdx1 mutants selectively lack a pancreas ( Jonsson et al. 1994, Offield et al. 1996). Similarly, the combined action of FoxA1 and FoxA2 establishes the liver, and double mutants fail to form even a liver bud in response to inductive cues (Lee et al. 2005). Global organ regulators like pha-4, FoxA1/2, and pdx1 may help coordinate the development of disparate populations of precursor cells into a cohesive organ.
Transcription of pha-4 is activated at the 2E–4E stage in MS-derived pharyngeal precursors and the 4E stage in AB-derived pharyngeal precursors (Baugh et al. 2003, Good et al. 2004, Horner et al. 1998, Smith & Mango 2007) and maintained in all pharyngeal cells throughout life (Alder et al. 2003, Azzaria et al. 1996, Horner et al. 1998, Kalb et al. 1998). This expression pattern fits well with the earliest defect that is associated with pha-4 mutations (Horner et al. 1998). Whereas wild-type pharyngeal precursors cluster together and ingress during gastrulation (4E–8E stages), pha-4 mutant cells remain dispersed at the embryo surface (Horner et al. 1998). The timing of pha-4 expression and activity suggests that pha-4 may be regulated directly by LAG-1 and TBX-37/38 within AB descendents at the 24-cell stage (Good et al. 2004) and TBX-35 in the MS lineage (Broitman-Maduro et al. 2006). pha-4 is also expressed in midgut and hindgut cells, some neurons, and the somatic gonad, although its functions in these other tissues are not well understood (Azzaria et al. 1996, Chen & Riddle 2008, Horner et al. 1998, Updike & Mango 2007).
The mechanism by which PHA-4 modulates transcription is unknown, but it may involve changes in the chromatin environment. In vertebrates, FoxA factors can alter the compaction of chromatin that surrounds target genes, which improves access for additional transcription factors (Cirillo et al. 2002, Lupien et al. 2008). In C. elegans, a subset of pharyngeal promoters recruit the histone variant HTZ-1/H2A.Z, and the loss of htz-1 activity is associated with delayed transcriptional onset (Updike & Mango 2006). Genome-wide, the presence of HTZ-1 correlates well with the presence of RNA polymerase II; but surprisingly, it does not correlate with transcriptional activity as monitored by microarray (Whittle et al. 2008). These observations suggest that PHA-4 may modulate the chromatin environment and prime genes for activation but that PHA-4 itself is probably not the trigger for transcriptional onset.
Animals from cnidaria to humans have FoxA transcription factors, and these are always associated with the digestive tract (Lai et al. 1990, Lee et al. 2005, Duncan et al. 1998, Friedman & Kaestner 2006, Fritzenwanker et al. 2004, Olsen & Jeffry 1997, Weigel et al. 1989b). In pha-4 mutants, some pharyngeal cells convert into non-neuronal ectoderm (Chanal & Labouesse 1997, Horner et al. 1998), and a fraction of embryos lack hindgut cells (Mango et al. 1994). These phenotypes are reminiscent of Drosophila fork head mutants, which lack foregut and hindgut, and instead form ectopic ectodermal head structures ( Jurgens & Weigel 1988, Weigel et al. 1989a). Vertebrates also require FoxA factors to form the foregut and some of its derivatives, such as liver (Ang & Rossant 1994, Dufort et al. 1998, Lee et al. 2005, Weinstein et al. 1994). In the liver, FoxA proteins activate a wide array of targets, analogous to C. elegans PHA-4 (Barthel et al. 1999, Duncan et al. 1998, Gualdi et al. 1996, Lee et al. 2002, Lehmann & Korge 1996, Tuteja et al. 2008, Wederell et al. 2008). Thus, the involvement of FoxA factors for foregut specification, differentiation, and function is evolutionarily ancient.
Developmental Plasticity and Commitment to Pharyngeal Fate
When do embryonic cells commit to a pharyngeal fate? Despite the stereotyped cell lineage, four observations suggest that early C. elegans blastomeres are born developmentally plastic and not yet restricted in their cell-fate choices. First, prior to gastrulation (≤28 cell stage, ≤2E), the cell lineage reveals that most blastomeres will generate many different types of cells. For example, MS produces pharyngeal cells, body wall muscles, the somatic gonad, scavenger cells, and even some neurons (Sulston et al. 1983). By approximately the 200-cell stage (8E-12E), most blastomeres are destined to produce cells of a single tissue or organ, such as pharyngeal cells from MSaaaap. Second, embryonic blastomeres adopt alternative fates when they are challenged with a heterologous regulator (Fukushige & Krause 2005, Gilleard & McGhee 2001, Horner et al. 1998, Kiefer et al. 2007, Smith & Mango 2007, Zhu et al. 1998). A blastomere fated to produce pharynx, for example, can instead give rise to epidermis or muscle (Fukushige & Krause 2005, Gilleard & McGhee 2001, Kiefer et al. 2007). Third, embryo manipulations have revealed that AB-derived blastomeres undergo regulative development and can respond to repositioning within the embryo by changing fate (Priess & Thomson 1987, Wood 1991). Signaling by the Notch and wnt pathways may redirect the development of repositioned cells (Eisenmann 2005, Priess 2005). Fourth, many genes that specify cell identity are expressed at or before the 8E stage (Bowerman et al. 1992, Good et al. 2004, Horner et al. 1998, Priess et al. 1987, Roy Chowdhuri et al. 2006, Smith & Mango 2007). Loss of function mutations in these regulators produce cell-fate transformations, whereas genes expressed later are often associated with subtler pharyngeal phenotypes that involve morphology or differentiation (e.g., ceh-22, ceh-24, ceh-28, pha-1, pha-2, peb-1) (Fay et al. 2004, Fernandez et al. 2004, Harfe et al. 1998, Morck et al. 2004, Okkema & Fire 1994, Okkema et al. 1997, Ray et al. 2008, Schnabel & Schnabel 1990). Together, these observations suggest that C. elegans embryonic blastomeres are born developmentally plastic, and they acquire pharyngeal identity as plasticity is lost during gastrulation.
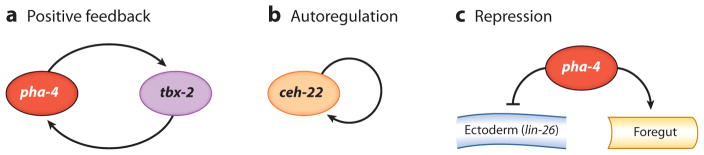
Three strategies ensure that embryonic blastomeres commit to pharyngeal fate (Figure 4). The first is positive-feedback loops between pha-4 and regulators of pharyngeal cell types. For example, a positive regulatory loop between PHA-4 and the helix-loop-helix protein HLH-6 is required to establish the pharyngeal glands (Raharjo & Gaudet 2007, Smit et al. 2008). In the absence of hlh-6 activity, g2 glands are often missing and g1 glands fail to differentiate properly (Smit et al. 2008). PHA-4 directly activates hlh-6 transcription at the bean stage, but HLH-6 is required to maintain pha-4 expression in g2 glands (Raharjo & Gaudet 2007, Smit et al. 2008). A similar positive regulatory loop between PHA-4 and TBX-2 contributes to the production of pharyngeal muscles (Roy Chowdhuri et al. 2006, Smith & Mango 2007). The interdependence of PHA-4 and either HLH-6 or TBX-2 suggests that commitment to pharyngeal identity is intimately linked to cell type specification within the pharynx.
Figure 4.
Strategies for cell fate commitment. Commitment to pharyngeal fate depends on (a) positive feedback loops between pairs of transcription factors, (b) positive autoregulation, and (c) repression of alternative fates. Adapted from Mango (2007).
All of the pharyngeal glands derive from the MS blastomere, whereas all muscles affected by tbx-2 derive from AB. This observation suggests that hlh-6 and tbx-2 are constrained by cell lineage in addition to tissue type. How lineage and tissue type converge on these factors is unknown but could theoretically involve the A-P patterning system or founder cell identities.
The second strategy for cell fate commitment is positive autoregulation (Figure 4). HLH-6, CEH-22 (an NK2.2 factor for pharyngeal muscle), and PHA-2 (a Hex factor in pharyngeal muscles and epithelia) all activate their own transcription, most likely directly (Kuchenthal et al. 2001, Morck et al. 2004, Raharjo & Gaudet 2007). For example, the ceh-22 locus contains two enhancer elements that initiate or maintain ceh-22 expression (Kuchenthal et al. 2001, Vilimas et al. 2004). The maintenance enhancer carries a CEH-22 binding site that is necessary and sufficient for activity. Thus, positive autoregulation contributes to robust, stable ceh-22 expression and a commitment to pharyngeal muscle fate. A similar scenario may exist for HLH-6 and PHA-2 in glands and epithelia, respectively.
The third strategy for cell fate commitment is transcriptional repression, which is important to inhibit alternative cell fates (Figure 4). pha-4 is necessary and sufficient to inhibit ectodermal fate and to repress ectodermal genes, such as lin-26 and elt-3 (Horner et al. 1998, Kiefer et al. 2007). FoxA in sea urchin, mammals, and Drosophila may also function as a repressor (Haberman et al. 2003, Oliveri et al. 2006, Sekiya & Zaret 2007). How does PHA-4 distinguish between activation and repression activities? Repression of ectodermal fate requires the repressive Nucleosome Remodeling and Deacetylase (NuRD) complex and the Tripartite Motif TRIM factor TAM-1, which associates with PHA-4 in yeast two-hybrid assays (Kiefer et al. 2007, Li et al. 2004). In other animals, TRIM proteins physically interact with NuRD, suggesting a possible link between these proteins (Schultz et al. 2001). In vertebrates and Drosophila, FoxA factors interact with Groucho repressors to alter chromatin organization and histone acetylation (Sekiya & Zaret 2007, Wang et al. 2000). These observations suggest that PHA-4 and other FoxA factors may function as either repressors or activators, depending on the cofactors and promoter context, an idea to be tested in future biochemical experiments.
Downstream of PHA-4: Spatial and Temporal Control During Organogenesis
As development advances from gastrulation to differentiation, a developing organ passes through a series of transient states that are characterized by successive waves of transcription. What mechanisms ensure the progression of development while also providing a stable commitment to pharyngeal fate? One key input is combinatorial control and feed-forward regulation. During pharyngeal muscle formation, for example, PHA-4 activates ceh-22 transcription during mid-embryogenesis (bean stage); and PHA-4 and CEH-22 together activate terminal muscle genes, such as the myosins myo-1 and myo-2 (Gaudet & Mango 2002, Kalb et al. 1998, Mango et al. 1994, Okkema & Fire 1994, Okkema et al. 1997). CEH-22 provides temporal precision and cell type specificity, because it is expressed later than PHA-4 and only in pharyngeal muscles (Okkema & Fire 1994). However, CEH-22 is not the only input, because myo-2 is expressed normally in ceh-22 mutants (Okkema et al. 1997). A good candidate to compensate for ceh-22 is the Hex homeobox factor pha-2, which is expressed in a subset of pharyngeal muscles (Avery 1993, Morck et al. 2004). PEB-1, a FLYWCH zinc finger factor related to Drosophila Mod(mdg4) (Beaster-Jones & Okkema 2004, Kalb et al. 2002, Thatcher et al. 2001); and DAF-12, a nuclear hormone receptor (Antebi et al. 2000, Ao et al. 2004); also target the myo-2 promoter and contribute to activity. The novel protein pha-1 is required to maintain ceh-22 and myo-2 expression by an unknown mechanism (Okkema et al. 1997). Thus, multiple inputs ensure that the pharyngeal muscles are made and myo-2 is expressed. Layers of regulators probably also guarantee that other pharyngeal cell types are produced, similar to the pharyngeal muscles (Raharjo & Gaudet 2007, Smit et al. 2008).
Combinatorial control relies on weak transcription factors, none of which are sufficient for transcriptional activation. PHA-4, for example, is a feeble transactivator both in C. elegans (Horner et al. 1998) and in yeast (Kalb et al. 2002). The architecture of pharyngeal promoters also dampens the effect of any one factor. Binding sites for a particular transcription factor are often suboptimal and exist in only 1–2 copies per regulatory region (Gaudet & Mango 2002). The number of sites for a given factor can alter the impact of that factor. For example, three PHA-4-binding sites are sufficient for activation, but most pharyngeal regulatory regions have only one PHA-4 site, which is not sufficient (Gaudet et al. 2004). Instead, pharyngeal promoters are targeted by several transcription factors to ensure that promoter firing depends on multiple weak inputs. The exact number of factors per promoter/enhancer region is unclear. Using a yeast one-hybrid approach, Deplancke and colleagues discovered an average of four factors per promoter (Deplancke et al. 2006). The factors discovered in this screen were presumably those that could bind as monomers or homomers in yeast, suggesting this is just the monomeric tip of a largely heteromeric iceberg. Additional bioinformatic screens have identified potential new regulatory sequences (Ao et al. 2004, Beer & Tavazoie 2004, GuhaThakurta et al. 2002), while forward and reverse genetic screens will continue to discover trans-acting factors (e.g., recently (Axang et al. 2008, Ray et al. 2008, Schmitz et al. 2008, Smit et al. 2008, Trzebiatowska et al. 2008, Updike & Mango 2007).
Surprisingly, the transcriptional network that guides pharyngeal development does not resemble the core endodermal network in mammals or sea urchins, at least not yet. Remnants of the vertebrate endodermal network exist in C. elegans: the previously discussed pha-4/FoxA and T-box gene, ceh-22, which has homologs in flies and vertebrates that are involved in visceral endoderm and pancreas development, respectively (Okkema et al. 1997); pha-2, whose vertebrate homologs pattern the foregut (Avery 1993, Morck et al. 2004), and hlh-6, which resembles the Drosophila salivary gland regulator Sage (Smit et al. 2008). Homologs of Sox17, FoxH1, or Mix genes have not yet been implicated in C. elegans gut development. Otx is critical for endoderm development in echinoderms (Hinman et al. 2003), but no gut role is known for Otx factors in C. elegans despite its expression in the pharynx and intestine (Hobert 2005, Lanjuin et al. 2003). The spotty conservation between C. elegans and vertebrates has led some to suggest the C. elegans pharynx is the cognate of the vertebrate heart, but parallels between the heart and pharynx likely reflect convergent evolution and not conservation (discussed in-depth in Mango 2007). Instead, the absence of many endodermal patterning transcription factors in C. elegans may reflect the simple anatomy of the foregut and the small number of cells.
Combinatorial control is important, but it is not the only input for pharyngeal patterning. A second important contributor toward expression timing is the affinity of PHA-4 for its binding sites within promoters (Figure 5) (Gaudet & Mango 2002, Gaudet et al. 2004). Mutations that raise the affinity of PHA-4-binding sites shift the onset of target gene expression earlier, whereas lower-affinity sites delay activation (Gaudet & Mango 2002). These temporal shifts occur in the context of the promoter, and they are not an absolute predictor of transcriptional activation of target genes. For example, the myo-2 locus has high-affinity PHA-4-binding sites, but it is activated late, after the addition of CEH-22 and other transcription factors (Gaudet & Mango 2002, Okkema & Fire 1994). The temporal response of pharyngeal genes to PHA-4-binding site affinity reflects the increasing levels of PHA-4 during embryogenesis: low PHA-4 levels at the 2E and 4E stages, in which PHA-4 is first expressed, rising to maximum levels around the bean stage (Horner et al. 1998, Smith & Mango 2007). Partner proteins, and/or modifications that alter PHA-4 binding or activity, may also contribute to the differential responses of PHA-4 target genes, but these are currently unknown.
Figure 5.
Strategies for temporal control. (a) As embryogenesis proceeds, PHA-4 protein accumulates. Affinity of PHA-4 protein for its DNA-binding site contributes to early (high affinity) versus late (lower affinity) onset of expression. (b) Feed-forward regulation contributes to late onset expression of target genes, including those with high-affinity PHA-4-binding (e.g., myo-2). (c) Pharyngeal genes are regulated by additional factors (A and B) in addition to PHA-4. These can include both activators and repressors. Adapted from Mango (2007).
Binding-site affinity has emerged as an important contributor to transcriptional control in multiple ways. In some instances, affinity contributes to temporal control, as it does for the pharynx. In these cases, the onset of expression reflects the concentration of active transcription factor in response to developmental progression (Ballas et al. 2005, Pearson & Doe 2003) or environmental cues (Chechik & Koller 2009, Lam et al. 2008). The role of affinity for expression timing has not yet been examined with regard to vertebrate FoxA proteins, but a survey of FoxA2 target genes suggested that affinity impacts the cellular focus. Genes with low-affinity FoxA2 sites are more likely to be expressed in the liver, which has high levels of FoxA2 and additional cooperative transcription factors, than genes with high-affinity sites (Tuteja et al. 2008). This regulation is reminiscent of Bicoid and Dorsal within the syncytial Drosophila embryo, in which DNA binding affinity contributes to spatial control (Ashe & Briscoe 2006). Target genes with low-affinity binding sites are active in embryonic regions that contain high levels of nuclear Bicoid or Dorsal, whereas targets with high-affinity binding sites can respond to lower levels in regions distal to the source of transcription factor. In addition to time and space, DNA-binding affinity can impact the strength of expression, as has been observed for the Pho5 locus in yeast (Lam et al. 2008). In these cases, higher affinity translates into stronger expression. In the pharynx, the strength of expression depends on combinatorial regulation with other transcription factors, and not PHA-4-binding-site affinity (Ao et al. 2004, Gaudet & Mango 2002, Gaudet et al. 2004). This result implies that a high-affinity, PHA-4-binding-site is not sufficient for robust transcriptional activity. The weakness of PHA-4 as a transcription factor (Gaudet et al. 2004, Kalb et al. 2002) provides a means to separate strength of expression from onset. In other situations, low-affinity sites may contribute to selectivity of target genes. For example, binding site affinity for the ets family distinguishes whether a target is recognized by ets alone (high-affinity site) or by ets in conjunction with binding partners (low-affinity sites) (Hollenhorst et al. 2007). In invertebrates, the binding of the dosage compensation complex relies on multiple low-affinity sites that ensure selective recognition of appropriate regions within the genome (Straub & Becker 2008). Thus, DNA-binding affinity can impact promoter selectivity and dynamics in multiple ways.
The transcriptional strategies of binding site affinity and combinatorial control are distinct from those of other organs such as the C. elegans intestine. Unlike the pharynx, the intestine is a simple organ composed of one cell type that derives from a single blastomere called E (Sulston et al. 1983). The anatomical simplicity is mirrored at the level of promoter architecture. The intestine is specified by tiers of GATA transcription factors that function for 1–2 cell divisions and elicit a relatively homogeneous transcriptional output (Maduro 2008, McGhee 2007). These regulatory GATA factors are more potent activators than PHA-4 and therefore less dependent on combinatorial inputs. For example, widespread expression of the GATA factor END-1 (but not PHA-4) converts all somatic cells to intestinal fate (Horner et al. 1998, Zhu et al. 1998), and the ELT-2 GATA factor is a potent activator in yeast one-hybrid assays (Kalb et al. 2002). Targets of the MED-1/2 GATA factors carry two copies of the sequence RRRAGTATAC in a 100 bp stretch within 2 kb of the ATG start codon, and these features are sufficient to identify novel MED target genes (Broitman-Maduro et al. 2005). Once specified, intestinal differentiation and function is guided by ELT-2 (Fukushige et al. 1998). ELT-2 is the only known factor for intestinal differentiation and probably acts in combination with additional transcription factors to respond to diverse stimuli, such as Notch signaling during morphogenesis (McGhee et al. 2009, Neves et al. 2007, Rasmussen et al. 2008). Thus, the simplicity of the intestine is mirrored with a simpler transcriptional strategy compared to the pharynx: potent transcription factors, less consensus sequence heterogeneity, and simpler promoter architecture. Additional analysis of promoter structures will reveal if these distinctions continue to hold true for the pharynx and intestine.
MORPHOGENESIS
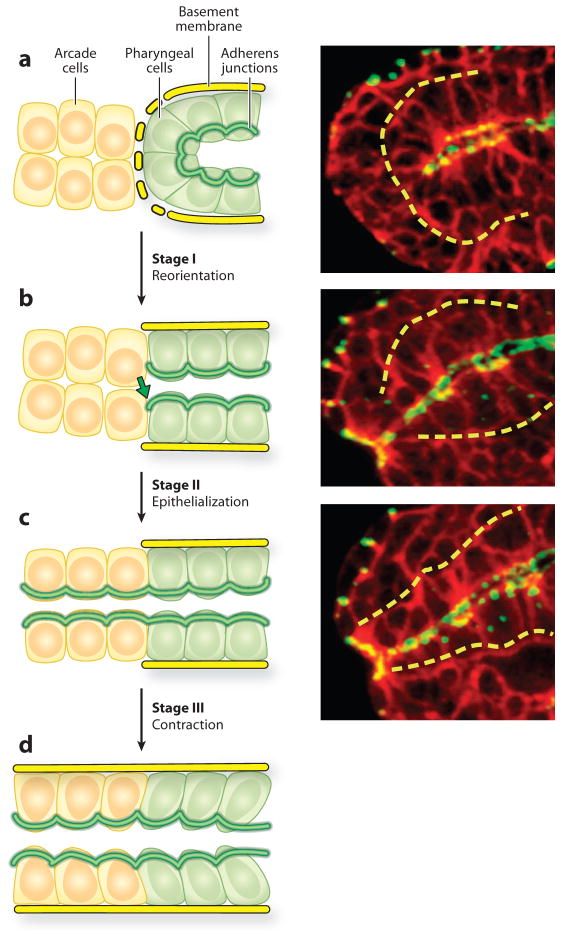
Gastrulation and cell division are virtually complete by mid-embryogenesis, and the pharyngeal primordium is visible as a single-cell layered cyst bordering the nascent intestine (Portereiko & Mango 2001, Sulston et al. 1983). Adherens junctions link the pharyngeal cells to each other and to the intestine in the interior of the embryo (Leung et al. 1999). Over the next 60 minutes, the pharyngeal cells become reorganized to form a linear gut tube that connects the gut to the external epidermis (Portereiko & Mango 2001). This process is initiated by the anterior pharyngeal epithelial cells, which reorient their apicobasal polarity to convert the pharyngeal cyst into a short tube that borders the anteriorly located arcade cells (Figure 6). In the second step the arcade cells undergo a mesenchymal to epithelial transition, which establishes a continuous epithelium between the epidermis and the pharynx (Figure 6). Finally, in the third step, the cells of the pharynx, buccal cavity, and epidermis appear to contract their apical surfaces, which pulls the cells more closely together. These events produce a continuous pharyngeal epithelium; subsequent events shape the pharynx into a bilobed tube.
Figure 6.
Pharyngeal morphogenesis. Left panels depict stages of reorientation (a to b, stage 1), epithelialization (b to c, stage 2), and contraction (c to d, stage 3). Yellow cells denote arcade cells, which are initially mesenchymal (a, b), but later become epithelialized (c, d ). Green cells represent cells in the pharyngeal primordium. Right panels show midstage embryos stained for cell periphery (red ) (αUNC-70/β spectrin) and adherens junctions ( green), (MH27/α-AJM-1) merge is yellow. The basement membrane surrounding the pharynx is denoted by a dotted yellow line in both sets of panels. Reprinted from Mango (2007).
Epithelium Formation of the Pharyngeal Arcade Cells
Like their vertebrate and Drosophila counterparts, C. elegans epithelia possess apical and basolateral domains separated by junctions, which is the apical junction or CeAJ in C. elegans (Labouesse 2006, Lockwood et al. 2008). The CeAJ resembles a combination of the zonula adherens and septate junctions of other species. Adherens proteins, such as HMR-1/cadherin, HMP-1/alpha-catenin, HMP-2/beta-catenin, JAC-1/p120-catenin, ZOO-1/ZO, and VAB-9/claudin-like, populate the upper half of the of the CeAJ toward the apical surface (Costa et al. 1998; Lockwood et al. 2008; Pettitt et al. 1996, 2003; Simske et al. 2003). These components anchor actin cables within the CeAJ but, surprisingly, they are not needed to establish epithelial polarity or intercellular adhesion (Costa et al. 1998, Pettitt et al. 2003, Simske et al. 2003). The lower half of the CeAJ contains the MAGUK protein DLG-1/discs large and its binding partner, the coiled-coil protein AJM-1 (Bossinger et al. 2001, Koppen et al. 2001). This complex contributes to the integrity of the junction, judging by ultrastructural studies, but does not localize cadherin/catenin to the CeAJ (Koppen et al. 2001, McMahon et al. 2001). Inactivation of components from both the upper and lower domains of the CeAJ leads to defects in adhesion and rupturing of the epithelium (Koppen et al. 2001, McMahon et al. 2001, Simske et al. 2003). LET-413/scribble is confined to the basolateral domain, in which it restricts the localization of CeAJ and apical domain components (Chanal & Labouesse 1997, Koppen et al. 2001, Legouis et al. 2000, McMahon et al. 2001). In the absence of let-413, junctional components spread basally. The apical domain shares components with the vertebrate tight junction and subapical domain of Drosophila such as the PAR-3/PAR-6/aPKC complex (Izumi et al. 1998, Leung et al. 1999, Simske & Hardin 2001, Tabuse et al. 1998, Totong et al. 2007). The inactivation of par-6 interferes with the localization of other apical proteins and junctional integrity. However, epithelial cells are still polarized along the apicobasolateral axis in par-6 mutants (Aono et al. 2004, Nance 2005, Totong et al. 2007).
These studies and others have demonstrated that many proteins that are required to establish or maintain epithelia in other animals are not essential to form the pharyngeal epithelium. Examples include homologs of Crumbs, cadherins, catenins discs-large, ZO-1, Scribble, and α- or β-integrins (Baum & Garriga 1997, Chanal & Labouesse 1997, Costa et al. 1998, Drubin & Nelson 1996, Legouis et al. 2000, Pettitt et al. 1996, Simske & Hardin 2001, Williams & Waterston 1994). The data suggest that additional molecules are needed in C. elegans to build epithelia. Studies with other organisms suggest that alternative routes for epithelium formation exist in these animals also (Baas et al. 2004, Bilder et al. 2003, Harris & Peifer 2004). For example, in the absence of armadillo/β-catenin, Drosophila gut cells retain some epithelial characteristics (Harris & Peifer 2004, 2005).
The kinesin-like protein ZEN-4/MKLP and its partner CYK-4/RhoGAP are required to polarize the arcade cells (Portereiko et al. 2004). Apical and adherens junction proteins fail to accumulate at the cell surface of arcade cells from zen-4 mutants, even though these proteins are synthesized in the cell. Thus, zen-4 and perhaps also cyk-4 are important for targeting proteins to the apical surface and CeAJ. Recent studies suggest that CYK-4 modulates cell polarity in other contexts by controlling Rho family GTPase activity and the contractile actomyosin cytoskeleton (D’Avino & Glover 2009, Jenkins et al. 2006). Future studies will determine if it fulfills the same role in epithelia.
Epithelial Remodeling to Build a Single-Celled Tube
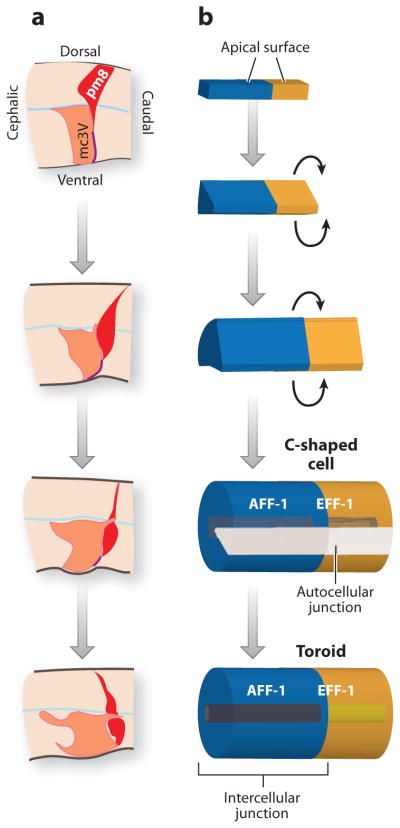
Once the epithelium is formed, individual cells acquire stereotyped shapes during the process of morphogenesis. For example, pm5 muscles are long and thin and form the isthmus, whereas pm6 and pm7 are squat discs within the terminal bulb. Little is known about the processes that dictate pharyngeal cell shapes with the exception of pm8. pm8 is a toroid-shaped muscle that links the pharynx to the pharyngeal intestinal valve and intestine. Prior to morphogenesis, pm8 sits at the dorsal surface of the pharyngeal primordium. It detaches from the basal lamina and migrates ventrally along a tract of LAM-3 and EPI-1 laminins to span the dorsoventral axis (Figure 7) (Rasmussen et al. 2008). To form a tube, pm8 wraps around finger-like projections from nearby marginal cells and forms a C-shaped tube. This tube fuses into a toroid in response to the fusogen AFF-1 (Rasmussen et al. 2008).
Figure 7.
Building a single-celled tube. (a) Prior to morphogenesis, pharyngeal muscle pm8 (red ) sits at the dorsal surface of the pharyngeal primordium. pm8 delaminates from the dorsal surface and migrates along a track of laminin laid down by an adjacent pharyngeal cell (mc3V) to span the DV axis. (b) To form a tube, pm8 forms a C-shaped cell and subsequently fuses with itself to generate a toroid. Adapted from Rasmussen et al. (2008).
Notch is involved in at least two of these morphogenetic steps. Notch is required for pm8 to delaminate from the dorsal surface. Notch signaling also activates AFF-1 expression and represses EFF-1 in pm8, which ensures pm8 fuses with the right partner (itself) and not an alternative cell. Although Notch proteins play a key role in pm8 morphogenesis, neither LIN-12 nor GLP-1 are expressed in other pharyngeal cells. Thus, the dramatic remodeling of most pharyngeal cells depends on other undescribed pathways.
In addition to these well-characterized examples, there are additional factors that affect pharynx morphogenesis by unknown processes such as cell fate, differentiation, or morphogenesis (Table 2). The predicted transcription factors ast-1/ETS TF, die-1/ZnF, unc-39/SixTF, ceh-43/distal-less, and elt-5/GATA are associated with Pun phenotypes, but their targets are unknown (Aspock & Burglin 2001, Heid et al. 2001, Koh et al. 2002, Schmid et al. 2006, Yanowitz et al. 2004). Double mutant combinations between lin-35/Rb and the ubiquitin conjugating enzyme ubc-18 (Fay et al. 2003), or between pha-1/DUF1114 and either ubc-18 or ari-1/Ariadne, lead to a Pun phenotype (Fay et al. 2004, Qiu & Fay 2006). One model to explain these interactions is that inappropriate expression of an unknown factor X leads to a Pun phenotype. Normally, X is kept in check by transcription repression (e.g., LIN-35) and protein degradation (e.g., UBC-18 or ARI-1).
Table 2.
Genes involved in pharyngeal morphogenesis
| Gene | Homology | M/Z | Cellular expression | Localization | Function | References |
|---|---|---|---|---|---|---|
| aff-1 | Novel fusogen, eff-1 | Z | pm8 | Cell surface | pm8 self-fusion | Rasmussen et al. 2008 |
| agr-1 | Agrin | Z | Buccal cavity, arcade cells, pharynx | Basement membrane | No phenotype | Hrus et al. 2007 |
| ari-1 | Ariadne RING finger | ? | Broadly | ? | pha attachment (Pun with pha-1) | Qiu & Fay 2006 |
| ast-1 | ETS | Z | Head | N/C | pha attachment (Pun at 1.5 fold) | Schmid et al. 2006 |
| ceh-43 | Distal-less | Z? | Non-Pha, epidermis | N/C | Pun | Aspock 2001 |
| crp-1 | cdc-42 related | Z | Muscles and nonphx epithelia | Endosomes, trans-Golgi network | Apical trafficking | Jenna et al. 2005 |
| cyk-4 | MgcRacGAP RhoGAP |
M,Z | Broadly | ? | Apicobasal polarity | Portereiko et al. 2004 |
| die-1 | Zinc finger TF | Z | Epithelia, pharynx | Nuclear | pha attachment, Pun or intestinal detachment | Heid et al. 2001 |
| eff-1 | Novel | Z | Muscles, nonphx fusing epithelia, vpi | Contact points, homotypic | Epithelial fusion | Mohler et al. 2002, Shemer et al. 2004, Rasmussen et al. 2008 |
| elt-5/egl-18 | GATA TF | Z | Nonpha epidermis, neurons | Nuclear | pha attachment, | Koh et al. 2002, Koh & Rothman 2001 |
| fbl-1 | Fibulin | Z | Nonphx | Basement membrane | pha morphology | Muriel et al. 2005 |
| ham-2 | Zinc finger | Z | Embryo | Nuclear | pha attachment | Baum et al. 1999 |
| Inx-3 | Innexin | Z | Broadly, epithelia | Basal surface | pha attachment, pha morphology | Starich et al. 2003 |
| let-413 | Scribble | Z | Epithelia | Basolateral | Confinement of CeAJ | Legouis et al. 2000, McMahon et al. 2001 |
| lin-35 | Rb repressor | ? | Broadly | Nuclear | pha attachment (Pun with ubc-18) | Fay et al. 2003, 2004 |
| nid-1 | Nidogen | Basement membrane, under dense bodies | No phenotype | Kang & Kramer 2000, Trzebiatowska et al. 2008 | ||
| pha-1 | DUF1114 | Z | Ubiquitous | Cytoplasmic | pha attachment (Pun with ubc-18 or ari-1) pha diffn. |
Fay et al. 2004, Granato et al. 1994, Qiu & Fay 2006, Schnabel & Schnabel 1990 |
| sma-1 | βH-spectrin | Z | Epithelia | Apical | pha elongation | McKeown et al. 1998 |
| spc-1 | α-spectrin | Z | Epithelia | Apical | pha elongation | Norman & Moerman 2002 |
| spon-1 | F-spondin | Z | Body wall muscles | Pha basement membrane | No pha phenotype; lethal | Woo et al. 2008 |
| ten-1 | Teneurin | Z | Pharynx | Basement membrane | Trzebiatowska et al. 2008 | |
| ubc-18 | Ubiquitin conj ez | ? | ? | ? | pha attachment (Pun with pha-1 or lin-35) | Fay et al. 2003 |
| unc-39 | Six TF | ? | Pharyngeal arcade cells | ? | Pha attachment or morphology | Yanowitz et al. 2004 |
| zen-4 | MKLP1 kinesin | M,Z | Broadly | ? | Apicobasal polarity | Portereiko et al. 2004 |
Genes that lead to a morphogenesis defect when mutated. M/Z: maternal or zygotic contribution, phx: pharyngeal, Pun: pharynx unattached, ?: unknown. Adapted and extended from Mango (2007).
Mutations in cdh-4 generate some arrested animals with unattached pharynges (Schmitz et al. 2008). cdh-4 and its paralog cdh-3 encode Fat-like cadherins with cadherin, laminin G, and EGF domains; and both are expressed in the pharynx (Pettitt et al. 1996, Schmitz et al. 2008). In other animals, Fat-like cadherins have been implicated in polar cell polarity (PCP) (Simons & Mlodzik 2008). C. elegans may possess a PCP-like pathway (Eisenmann 2005, Park et al. 2004, Wu & Herman 2006), but its role in pharyngeal development is unclear.
The shape of the pharyngeal isthmus depends on the homeobox factor pha-2 (Morck et al. 2004) and the βH(heavy)-spectrin sma-1 (McKeown et al. 1998). Animals that lack pha-2 have an overly thick pharyngeal isthmus and behave like non-isthmus cells, which suggests that pha-2 establishes isthmus (pm5) fate or morphology (Morck et al. 2004). Conversely, mutations in eya-1, which is similar to the phosphatase component eyes absent (Rebay et al. 2005), result in thin pharynges and reduced pumping rates (Furuya et al. 2005). eya-1 is partially redundant with vab-3/pax-6, suggesting that the regulatory circuit that controls eye development in other animals may have adopted a new function for anterior development in C. elegans, which lacks eyes (Furuya et al. 2005).
Mutations in ten-1 teneurin and nid-1 nidogen disrupt the basement membrane surrounding the pharynx (Trzebiatowska et al. 2008). ceh-22 mutants also have defects in basement membrane biogenesis (Okkema et al. 1997), but whether CEH-22 regulates basement membrane components is unknown.
PHA-4 AND THE PHARYNX AFTER BIRTH: AGING
Most studies of pha-4 and the pharynx have focused on embryonic roles, but recent data implicate this pair for postembryonic life. pha-4 is required after birth for the development of both the pharynx and gonad (Ao et al. 2004, Gaudet & Mango 2002, Updike & Mango 2007). In adults, pha-4 functions downstream of let-363/Target of Rapamycin (TOR) kinase and its partner daf-15/Raptor to mediate lifespan extension (Hansen et al. 2007, Jia et al. 2004, Panowski et al. 2007, Sheaffer et al. 2008, Vellai et al. 2003). The inactivation of TOR, or components of the translation machinery, prolongs life (Chen et al. 2007; Curran & Ruvkun 2007; Hansen et al. 2005, 2007; Kaeberlein & Kennedy 2008; Pan et al. 2007; Syntichaki et al. 2007; Vellai et al. 2003). Although TOR controls protein homeostasis (Hansen et al. 2007, Meissner et al. 2004, Wullschleger et al. 2006), genetic epistasis experiments suggest that TOR and the protein translation machinery extend lifespans with different mechanisms. For example, the loss of the translation initiation factor ife-2/eIF4E reduces protein biosynthesis and extends lifespan in a pathway that may require daf-16/FoxO but not pha-4/FoxA (Hansen et al. 2007, Sheaffer et al. 2008, Syntichaki et al. 2007). On the other hand, reduced rsks-1/S6 kinase or TOR signaling lowers protein biosynthesis and prolongs life in a pathway that depends on pha-4 but not daf-16 (Hansen et al. 2007, Pan et al. 2007, Sheaffer et al. 2008, Vellai et al. 2003). A simple model is that TOR activates S6 kinase in C. elegans, as it does in other organisms, and these kinases regulate pha-4. Given that PHA-4 protein levels and localization do not change in response to TOR or S6 kinase (Sheaffer et al. 2008), kinases probably modulate the activity of PHA-4, either directly or indirectly. The activity of PHA-4 has not been monitored, in part because there are no known targets of PHA-4 that mediate longevity. Previous studies implicated PHA-4 for transcriptional activation of super-oxide dismutases sod-1, sod-2, sod-4, and sod-5 in adults (Panowski et al. 2007). However, it is unclear whether these genes are direct PHA-4 targets and whether they modulate longevity. The inactivation of the sod genes alone or in combination does not shorten C. elegans lifespan (Doonan et al. 2008, Van Raamsdonk & Hekimi 2009). Conversely, SOD mimetics do not prolong life (Keaney et al. 2004). Genes involved in autophagy have emerged as another possible set of targets, based on genetic and cell biological analyses (Hansen et al. 2008).
Where do TOR and PHA-4 function to promote longevity? The pharynx ages over time, which could contribute to the health and longevity of animals. The rate of pharyngeal pumping declines with age (Bolanowski et al. 1981, Huang et al. 2004), and the pharynx exhibits sarcopenia with loss and damage of muscles (Chow et al. 2006, Garigan et al. 2002, Herndon et al. 2002). However, PHA-4 is expressed in other organs as well as the pharynx, namely the gonad, the intestine, and parts of the nervous system (Azzaria et al. 1996, Chen & Riddle 2008, Horner et al. 1998), all of which are involved in lifespan (Mukhopadhyay & Tissenbaum 2007).
Reduced food intake without malnutrition or dietary restriction (DR) is a potent means to extend life for C. elegans and other species. DR can be induced by different approaches in nematodes, and these have different genetic requirements (Bishop & Guarente 2007, Greer et al. 2007, Honjoh et al. 2009, Hosono et al. 1989, Houthoofd et al. 2002, Kaeberlein et al. 2006, Klass 1977, Lee et al. 2006, Mair et al. 2009, Smith et al. 2008). For example, DR on petri plates (sDR) requires daf-16/FoxO, AMP kinase, and insulin signaling (Greer & Brunet 2009, Greer et al. 2007), whereas DR in liquid (Houthoofd et al. 2002, Klass 1977) or by genetic manipulation (Lakowski & Hekimi 1998, Meissner et al. 2004) is independent of daf-16 but requires pha-4/FoxA and other factors (Bishop & Guarente 2007, Hansen et al. 2005, Houthoofd et al. 2002, Kaeberlein et al. 2006, Lee et al. 2006, Panowski et al. 2007). Phenotypic and genetic epistasis analyses suggest that TOR also functions in the DR pathway (Hansen et al. 2007, Meissner et al. 2004, Pan et al. 2007, Sheaffer et al. 2008). In organisms in which it has been tested, both the TOR pathway (e.g., yeast, Drosophila) and DR (e.g., yeast, Drosophila, rodents) extend lifespan, suggesting that this is an ancient pathway to control longevity. For additional details on DR, protein translation, and longevity, see the recent review by Kaeberlein & Kennedy (2008).
In addition to aging, the pharynx is susceptible to pathogens such as Salmonella enterica (Millet & Ewbank 2004). The pharynx participates in the immune response by physically crushing bacteria with the pharygneal grinder and secreting antimicrobial peptides (Millet & Ewbank 2004). Resistence of the pharynx to infection depends on the CED-1 and TOL-1 receptors and a noncanonical unfolded protein response (UPR) (Haskins et al. 2008, Tenor & Aballay 2008, Troemel et al. 2006). The inactivation of any of these factors allows S. enterica to invade the pharynx and leads to death. CED-1 is required to induce the UPR response, whereas TOL-1 induces defensin-like factor ABF-2 and the heat-shock protein HSP 16.41 (Haskins et al. 2008, Tenor & Aballay 2008). The pathway from receptors to activated genes is currently mysterious.
SUMMARY POINTS.
The C. elegans pharynx offers a powerful system to track organogenesis from the earliest stages of embryonic cell fate specification to the final stages of aging and death.
Maternally-contributed factors specify founder blastomeres in the pregastrula embryo. Subsequently, two zygotic patterning systems—one centered on cellular ancestry and one on organ identity—establish the pharyngeal precursor cells.
Commitment to pharyngeal fate depends on positive feedback loops between the central regulator of the pharynx, pha-4/FoxA, and additional transcriptional regulators on positive autoregulation by transcription factors and on negative repression of factors that dictate alternative fates.
DNA binding affinity contributes towards the selection of target genes by FoxA transcription factors. In C. elegans, PHA-4 binding affinity modulates the onset of expression of targets in combination with additional transcription factors.
During mid-embryogenesis the pharynx precursors undergo a mesenchymal to epithelial transition (MET), which is the first step of tube formation. MET depends on pathways independent of cadherins, catenins, and integrins.
Tube formation is only understood for the pm8 muscle precursor, which undergoes an epithelial-mesenchymal transition and cell reorientation along the dorsoventral axis. These events are guided by glp-1/Notch signaling and the extracellular matrix. pm8 wraps around its neighboring cells to form a C-shaped cell, which self-seals in response to the AFF-1 fusogen to form a single-celled tube.
More recently, the pharynx and pha-4 have emerged as regulators of post-embryonic processes, notably aging and the immune response, both in adults.
FUTURE ISSUES.
Although we have a broad outline of how patterning events occur, many mechanistic details are still lacking. For example, in the early embryo, we do not understand how the embryo transits from maternal regulation, which specifies founder cells and their lineages, to zygotic control of organs and tissues. Nor do we understand how the A-P patterning system dictates cell identities at a mechanistic level. This will require identifying targets of POP-1 and elucidating their roles in cell-fate specification.
We have only a rudimentary understanding of how different cell types are established during the terminal stages of pharynx development. Thus, a challenge for the future will be to determine if the examples we understand can be generalized to all cell types and all regulatory circuitry.
During morphogenesis, a future goal is to determine how to form epithelia by pathways that do not require cadherins or integrins and how to shape those epithelia into the distinctive morphologies of the mature pharynx.
Finally, the involvement of the pharynx and pharyngeal factors in immunity and aging is a recent discovery, and there are a host of unanswered questions. Future analyses are needed to identify new players and flesh out their regulatory relationships.
Acknowledgments
Many thanks to B. Bowerman, K. Kaestner, J. McGhee, J. Priess, and A. Schier for discussion; B. Bowerman, A. Schier, S. von Stetina, and K. Sheaffer for comments on the manuscript; and D. Lim for the illustrations. S.E.M. is supported by R01 DK070184 and R01 GM056264 from the NIH. She receives institutional support from the Huntsman Cancer Institute, Department of Oncological Sciences, and the MacArthur Foundation.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc London Ser B. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Alder MN, Dames S, Gaudet J, Mango SE. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA. 2003;9:25–32. doi: 10.1261/rna.2650903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang S-L, Rossant J. HNF-3beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–74. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F, Anyanful A, Sakube Y, Kagawa H. Transcription factors GATA/ELT-2 and forkhead/HNF-3/PHA-4 regulate the tropomyosin gene expression in the pharynx and intestine of Caenorhabditis elegans. J Mol Biol. 2008;379:201–11. doi: 10.1016/j.jmb.2007.11.103. [DOI] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–27. [PMC free article] [PubMed] [Google Scholar]
- Ao W, Gaudet J, Kent WJ, Muttumu S, Mango SE. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science. 2004;305:1743–46. doi: 10.1126/science.1102216. [DOI] [PubMed] [Google Scholar]
- Aono S, Legouis R, Hoose WA, Kemphues KJ. PAR-3 is required for epithelial cell polarity in the distal spermatheca of C. elegans. Development. 2004;131:2865–74. doi: 10.1242/dev.01146. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development. 2008;135:501–11. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe HL, Briscoe J. The interpretation of morphogen gradients. Development. 2006;133:385–94. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- Aspock G, Burglin TR. The Caenorhabditis elegans distal-less ortholog ceh-43 is required for development of the anterior hypodermis. Dev Dyn. 2001;222:403–9. doi: 10.1002/dvdy.1201. [DOI] [PubMed] [Google Scholar]
- Aspock G, Ruvkun G, Burglin TR. The Caenorhabditis elegans ems class homeobox gene ceh-2 is required for M3 pharynx motoneuron function. Development. 2003;130:3369–78. doi: 10.1242/dev.00551. [DOI] [PubMed] [Google Scholar]
- Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell. 1987;51:1071–78. doi: 10.1016/0092-8674(87)90593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron. 1989;3:473–85. doi: 10.1016/0896-6273(89)90206-7. [DOI] [PubMed] [Google Scholar]
- Axang C, Rauthan M, Hall DH, Pilon M. Developmental genetics of the C. elegans pharyngeal neurons NSML and NSMR. BMC Dev Biol. 2008;8:38. doi: 10.1186/1471-213X-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzaria M, Goszczynski B, Chung MA, Kalb JM, McGhee JD. A fork head/HNF-3 homolog expressed in the pharynx and intestine of the Caenorhabditis elegans embryo. Dev Biol. 1996;178:289–303. doi: 10.1006/dbio.1996.0219. [DOI] [PubMed] [Google Scholar]
- Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, et al. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell. 2004;116:457–66. doi: 10.1016/s0092-8674(04)00114-x. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–57. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Barthel A, Okino ST, Liao J, Nakatani K, Li J, et al. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem. 1999;274:20281–86. doi: 10.1074/jbc.274.29.20281. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Baum PD, Garriga G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron. 1997;19:51–62. doi: 10.1016/s0896-6273(00)80347-5. [DOI] [PubMed] [Google Scholar]
- Baum PD, Guenther C, Frank CA, Pham BV, Garriga G. The Caenorhabditis elegans gene ham-2 links Hox patterning to migration of the HSN motor neuron. Genes Dev. 1999;13:472–83. doi: 10.1101/gad.13.4.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaster-Jones L, Okkema PG. DNA binding and in vivo function of C. elegans PEB-1 require a conserved FLYWCH motif. J Mol Biol. 2004;339:695–706. doi: 10.1016/j.jmb.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–98. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Bilder D, Schober M, Perrimon N. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat Cell Biol. 2003;5:53–58. doi: 10.1038/ncb897. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–49. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bjornson CR, Griffin KJ, Farr GH, 3rd, Terashima A, Himeda C, et al. Eomesodermin is a localized maternal determinant required for endoderm induction in zebrafish. Dev Cell. 2005;9:523–33. doi: 10.1016/j.devcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Bolanowski MA, Russell RL, Jacobson LA. Quantitative measures of aging in the nematode Caenorhabditis elegans. I Population and longitudinal studies of two behavioral parameters. Mech Ageing Dev. 1981;15:279–95. doi: 10.1016/0047-6374(81)90136-6. [DOI] [PubMed] [Google Scholar]
- Bossinger O, Klebes A, Segbert C, Theres C, Knust E. Zonula adherens formation in Caenorhabditis elegans requires dlg-1, the homologue of the Drosophila gene discs large. Dev Biol. 2001;230:29–42. doi: 10.1006/dbio.2000.0113. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Draper BW, Mello CC, Priess JF. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74:443–52. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–75. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G, Lin KT, Hung WW, Maduro MF. Specification of the C. elegans MS blastomere by the T-box factor TBX-35. Development. 2006;133:3097–106. doi: 10.1242/dev.02475. [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G, Maduro MF, Rothman JH. The noncanonical binding site of the MED-1 GATA factor defines differentially regulated target genes in the C. elegans mesendoderm. Dev Cell. 2005;8:427–33. doi: 10.1016/j.devcel.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Burglin TR, Aspock G. Exon duplication from a fork head to a homeodomain protein. Dev Genes Evol. 1999;209:629–33. doi: 10.1007/s004270050298. [DOI] [PubMed] [Google Scholar]
- Calvo D, Victor M, Gay F, Sui G, Luke MP, et al. A POP-1 repressor complex restricts inappropriate cell type-specific gene transcription during Caenorhabditis elegans embryogenesis. EMBO J. 2001;20:7197–208. doi: 10.1093/emboj/20.24.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Captan VV, Goszczynski B, McGhee JD. Neither maternal nor zygotic med-1/med-2 genes play a major role in specifying the Caenorhabditis elegans endoderm. Genetics. 2007;175:969–74. doi: 10.1534/genetics.106.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanal P, Labouesse M. A screen for genetic loci required for hypodermal cell and glial-like cell development during Caenorhabditis elegans embryogenesis. Genetics. 1997;146:207–26. doi: 10.1093/genetics/146.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechik G, Koller D. Timing of gene expression responses to environmental changes. J Comput Biol. 2009;16:279–90. doi: 10.1089/cmb.2008.13TT. [DOI] [PubMed] [Google Scholar]
- Chen D, Pan KZ, Palter JE, Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6:525–33. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Riddle DL. Function of the PHA-4/FOXA transcription factor during C. elegans post-embryonic development. BMC Dev Biol. 2008;8:26. doi: 10.1186/1471-213X-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow DK, Glenn CF, Johnston JL, Goldberg IG, Wolkow CA. Sarcopenia in the Caenorhabditis elegans pharynx correlates with muscle contraction rate over lifespan. Exp Gerontol. 2006;41:252–60. doi: 10.1016/j.exger.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H) Development. 1996;122:1373–83. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Acto-myosin reorganization and PAR polarity in C. elegans. Development. 2007;134:1035–43. doi: 10.1242/dev.000513. [DOI] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Glover DM. Cytokinesis: mind the GAP. Nat Cell Biol. 2009;11:112–14. doi: 10.1038/ncb0209-112. [DOI] [PubMed] [Google Scholar]
- Deplancke BA, Mukhopadhyay W, Ao A, Elewa CA, Grove NJ, Martinez R, et al. A gene-centered protein-DNA interaction network of C. elegans digestive tract genes provides insights into metazoan differential gene expression at a systems level. Cell. 2006;125(6):1193–205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, et al. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–41. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–44. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Dufort D, Schwartz L, Harpal K, Rossant J. The transcription factor HNF3beta is required in visceral endoderm for normal primitive streak morphogenesis. Development. 1998;125:3015–25. doi: 10.1242/dev.125.16.3015. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Navas MA, Dufort D, Rossant J, Stoffel M. Regulation of a transcription factor network required for differentiation and metabolism. Science. 1998;281:692–95. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM. Wnt signaling. Worm Book. 2005 Jun;25:1–17. doi: 10.1895/wormbook.1.7.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TC, Hunter CP. Translational control of maternal RNAs. Worm Book. 2005 Nov;10:1–11. doi: 10.1895/wormbook.1.34.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Large E, Han M, Darland M. lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development. 2003;130:3319–30. doi: 10.1242/dev.00561. [DOI] [PubMed] [Google Scholar]
- Fay DS, Qiu X, Large E, Smith CP, Mango S, Johanson BL. The coordinate regulation of pharyngeal development in C. elegans by lin-35/Rb, pha-1, and ubc-18. Dev Biol. 2004;271:11–25. doi: 10.1016/j.ydbio.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Fernandez AP, Gibbons J, Okkema PG. C. elegans peb-1 mutants exhibit pleiotropic defects in molting, feeding, and morphology. Dev Biol. 2004;276:352–66. doi: 10.1016/j.ydbio.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–28. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzenwanker JH, Saina M, Technau U. Analysis of forkhead and snail expression reveals epithelial-mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev Biol. 2004;275:389–402. doi: 10.1016/j.ydbio.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- Fukushige T, Krause M. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development. 2005;132:1795–805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- Furuya M, Qadota H, Chisholm AD, Sugimoto A. The C. elegans eyes absent ortholog EYA-1 is required for tissue differentiation and plays partially redundant roles with PAX-6. Dev Biol. 2005;286:452–63. doi: 10.1016/j.ydbio.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A. Genetic control of wing disc development in Drosophila. Ciba Found Symp. 1975;29:161–82. doi: 10.1002/9780470720110.ch8. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–12. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–25. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Muttumu S, Horner M, Mango SE. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2004;2:e352. doi: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, McGhee JD. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol. 2001;21:2533–44. doi: 10.1128/MCB.21.7.2533-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good K, Ciosk R, Nance J, Neves A, Hill RJ, Priess JR. The T-box transcription factors TBX-37 and TBX-38 link GLP-1/Notch signaling to mesoderm induction in C. elegans embryos. Development. 2004;131:1967–78. doi: 10.1242/dev.01088. [DOI] [PubMed] [Google Scholar]
- Goszczynski B, McGhee JD. Reevaluation of the role of the med-1 and med-2 genes in specifying the Caenorhabditis elegans endoderm. Genetics. 2005;171:545–55. doi: 10.1534/genetics.105.044909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, Schnabel H, Schnabel R. Genesis of an organ: molecular analysis of the pha-1 gene. Development. 1994;120:3005–7. doi: 10.1242/dev.120.10.3005. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A. Endoderm specification. Stem Book, editor. The Stem Cell Res Community Harv Stem Cell Inst (HSCI) 2008 http://www.stembook.org. [PubMed]
- Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–27. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–56. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–82. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- GuhaThakurta D, Palomar L, Stormo GD, Tedesco P, Johnson TE, et al. Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 2002;12:701–12. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman AS, Isaac DD, Andrew DJ. Specification of cell fates within the salivary gland primordium. Dev Biol. 2003;258:443–53. doi: 10.1016/s0012-1606(03)00140-4. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–28. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Gomes AV, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998;12:2623–35. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J Cell Biol. 2004;167:135–47. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol. 2005;170:813–23. doi: 10.1083/jcb.200505127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins KA, Russell JF, Gaddis N, Dressman HK, Aballay A. Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev Cell. 2008;15:87–97. doi: 10.1016/j.devcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J. Patterning the early Xenopus embryo. Development. 2006;133:1205–17. doi: 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- Heid PJ, Raich WB, Smith R, Mohler WA, Simokat K, et al. The zinc finger protein DIE-1 is required for late events during epithelial cell rearrangement in C. elegans. Dev Biol. 2001;236:165–80. doi: 10.1006/dbio.2001.0315. [DOI] [PubMed] [Google Scholar]
- Henry JQ, Perry KJ, Wever J, Seaver E, Martindale MQ. Beta-catenin is required for the establishment of vegetal embryonic fates in the nemertean, Cerebratulus lacteus. Dev Biol. 2008;317:368–79. doi: 10.1016/j.ydbio.2008.02.042. [DOI] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–14. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci USA. 2003;100:13356–61. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Specification of the nervous system. Worm Book. 2005 Aug;8:1–19. doi: 10.1895/wormbook.1.12.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–94. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–30. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homologue, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–52. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono R, Nishimoto S, Kuno S. Alterations of life span in the nematode Caenorhabditis elegans under monoxenic culture conditions. Exp Gerontol. 1989;24:251–64. doi: 10.1016/0531-5565(89)90016-8. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, et al. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp Gerontol. 2002;37:1371–78. doi: 10.1016/s0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Hrus A, Lau G, Hutter H, Schenk S, Ferralli J, et al. C. elegans agrin is expressed in pharynx, IL1 neurons and distal tip cells and does not genetically interact with genes involved in synaptogenesis or muscle function. PLoS ONE. 2007;2:e731. doi: 10.1371/journal.pone.0000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci USA. 2004;101:8084–89. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Shetty P, Robertson SM, Lin R. Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and its coactivator beta-catenin SYS-1. Development. 2007;134:2685–95. doi: 10.1242/dev.008268. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Hirose T, Tamai Y, Hirai S, Nagashima Y, et al. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N, Saam JR, Mango SE. CYK-4/GAP Provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- Jenna S, Caruso ME, Emadali A, Nguyen DT, Dominguez M, et al. Regulation of membrane trafficking by a novel Cdc42-related protein in Caenorhabditis elegans epithelial cells. Mol Biol Cell. 2005;16:1629–39. doi: 10.1091/mbc.E04-08-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–9. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Jurgens G, Weigel D. Terminal versus segmental development in the Drosophila embryo: the role of the homeotic gene fork head. Roux’s Arch Dev Biol. 1988;197:345–54. doi: 10.1007/BF00375954. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Kennedy BK. Protein translation, 2008. Aging Cell. 2008;7:777–82. doi: 10.1111/j.1474-9726.2008.00439.x. [DOI] [PubMed] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–94. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Kalb JM, Beaster-Jones L, Fernandez AP, Okkema PG, Goszczynski B, McGhee JD. Interference between the PHA-4 and PEB-1 transcription factors in formation of the Caenorhabditis elegans pharynx. J Mol Biol. 2002;320:697–704. doi: 10.1016/s0022-2836(02)00555-7. [DOI] [PubMed] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, et al. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–80. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- Kaletta T, Schnabel H, Schnabel R. Binary specification of the embryonic lineage in Caenorhabditis elegans. Nature. 1997;390:294–99. doi: 10.1038/36869. [DOI] [PubMed] [Google Scholar]
- Kang SH, Kramer JM. Nidogen is nonessential and not required for normal type IV collagen localization in Caenorhabditis elegans. Mol Biol Cell. 2000;11:3911–23. doi: 10.1091/mbc.11.11.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaney M, Matthijssens F, Sharpe M, Vanfleteren J, Gems D. Superoxide dismutase mimetics elevate superoxide dismutase activity in vivo but do not retard aging in the nematode Caenorhabditis elegans. Free Radic Biol Med. 2004;37:239–50. doi: 10.1016/j.freeradbiomed.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Kidd AR, 3rd, Miskowski JA, Siegfried KR, Sawa H, Kimble J. A beta-catenin identified by functional rather than sequence criteria and its role in Wnt/MAPK signaling. Cell. 2005;121:761–72. doi: 10.1016/j.cell.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Kiefer JC, Smith PA, Mango SE. PHA-4/FoxA cooperates with TAM-1/TRIM to regulate cell fate restriction in the C. elegans foregut. Dev Biol. 2007;303:611–24. doi: 10.1016/j.ydbio.2006.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors infuencing life span. Mech Aging Dev. 1977;6:413–29. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Koh K, Peyrot SM, Wood CG, Wagmaister JA, Maduro MF, et al. Cell fates and fusion in the C. elegans vulval primordium are regulated by the EGL-18 and ELT-6 GATA factors–apparent direct targets of the LIN-39 Hox protein. Development. 2002;129:5171–80. doi: 10.1242/dev.129.22.5171. [DOI] [PubMed] [Google Scholar]
- Koh K, Rothman JH. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128:2867–80. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, et al. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–91. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- Kuchenthal CA, Chen W, Okkema PG. Multiple enhancers contribute to expression of the NK-2 homeobox gene ceh-22 in C. elegans pharyngeal muscle. Genesis. 2001;31:156–66. doi: 10.1002/gene.10018. [DOI] [PubMed] [Google Scholar]
- Labouesse M. Epithelial junctions and attachments. Worm Book. 2006 Jan;13:1–21. doi: 10.1895/wormbook.1.56.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M, Mango SE. Patterning the C. elegans embryo: moving beyond the cell lineage. Trends Genet. 1999;15:307–13. doi: 10.1016/s0168-9525(99)01750-3. [DOI] [PubMed] [Google Scholar]
- Lai E, Prezioso VR, Smith E, Litvin O, Costa RH, Darnell JE. HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990;4:1427–36. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:13091–96. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, O’Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–50. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev Cell. 2003;5:621–33. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–47. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Lee CS, Sund NJ, Vatamaniuk MZ, Matschinsky FM, Stoffers DA, Kaestner KH. Foxa2 controls Pdx1 gene expression in pancreatic beta-cells in vivo. Diabetes. 2002;51:2546–51. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, et al. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–24. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legouis R, Gansmuller A, Sookhareea S, Bosher JM, Baillie DL, Labouesse M. LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat Cell Biol. 2000;2:415–22. doi: 10.1038/35017046. [DOI] [PubMed] [Google Scholar]
- Lehmann M, Korge G. The fork head product directly specifies the tissue-specfic hormone responsiveness of the Drosophila Sgs-4 gene. EMBO J. 1996;15:4825–34. [PMC free article] [PubMed] [Google Scholar]
- Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–34. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–43. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 1998;92:229–39. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Lo MC, Gay F, Odom R, Shi Y, Lin R. Phosphorylation by the beta-catenin/MAPK complex promotes 14–3–3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell. 2004;117:95–106. doi: 10.1016/s0092-8674(04)00203-x. [DOI] [PubMed] [Google Scholar]
- Lockwood CA, Lynch AM, Hardin J. Dynamic analysis identifies novel roles for DLG-1 subdomains in AJM-1 recruitment and LET-413-dependent apical focusing. J Cell Sci. 2008;121:1477–87. doi: 10.1242/jcs.017137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF. Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim Biophys Acta. 2008;1789:250–60. doi: 10.1016/j.bbagrm.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH. Maternal deployment of the embryonic SKN-1–>MED-1,2 cell specification pathway in C. elegans. Dev Biol. 2007;301:590–601. doi: 10.1016/j.ydbio.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Hill RJ, Heid PJ, Newman-Smith ED, Zhu J, et al. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev Biol. 2005a;284:509–22. doi: 10.1016/j.ydbio.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Kasmir JJ, Zhu J, Rothman JH. The Wnt effector POP-1 and the PAL-1/Caudal home-oprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev Biol. 2005b;285:510–23. doi: 10.1016/j.ydbio.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Lin R, Rothman JH. Dynamics of a developmental switch: recursive intracellular and intranuclear redistribution of Caenorhabditis elegans POP-1 parallels Wnt-inhibited transcriptional repression. Dev Biol. 2002;248:128–42. doi: 10.1006/dbio.2002.0721. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3 beta homolog is mediated by MED-1 and -2 in C. elegans. Mol Cell. 2001;7:475–85. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Rothman JH. Making worm guts: the gene regulatory network of the Caenorhabditis elegans endoderm. Dev Biol. 2002;246:68–85. doi: 10.1006/dbio.2002.0655. [DOI] [PubMed] [Google Scholar]
- Mair W, Panowski SH, Shaw RJ, Dillin A. Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS ONE. 2009;4:e4535. doi: 10.1371/journal.pone.0004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango SE. The C. elegans pharynx: a model for organogenesis. Worm Book. 2007 Jan;22:1–26. doi: 10.1895/wormbook.1.129.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994;120:3019–31. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- McGhee JD. The C. elegans intestine. Worm Book. 2007 Mar;27:1–36. doi: 10.1895/wormbook.1.133.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Fukushige T, Krause MW, Minnema SE, Goszczynski B, et al. ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev Biol. 2009;327:551–65. doi: 10.1016/j.ydbio.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown C, Praitis V, Austin J. sma-1 encodes a betaH-spectrin homolog required for Caenorhabditis elegans morphogenesis. Development. 1998;125:2087–98. doi: 10.1242/dev.125.11.2087. [DOI] [PubMed] [Google Scholar]
- McMahon L, Legouis R, Vonesch JL, Labouesse M. Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J Cell Sci. 2001;114:2265–77. doi: 10.1242/jcs.114.12.2265. [DOI] [PubMed] [Google Scholar]
- Meissner B, Boll M, Daniel H, Baumeister R. Deletion of the intestinal peptide transporter affects insulin and TOR signaling in Caenorhabditis elegans. J Biol Chem. 2004;279:36739–45. doi: 10.1074/jbc.M403415200. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Krause M, Weintraub H, Preiss JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–76. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- Millet AC, Ewbank JJ. Immunity in Caenorhabditis elegans. Curr Opin Immunol. 2004;16:4–9. doi: 10.1016/j.coi.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Sawa H. Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol. 2007;17:465–73. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, et al. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–62. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Morck C, Rauthan M, Wagberg F, Pilon M. pha-2 encodes the C. elegans ortholog of the homeodomain protein HEX and is required for the formation of the pharyngeal isthmus. Dev Biol. 2004;272:403–18. doi: 10.1016/j.ydbio.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Moskowitz IPG, Gendreau SB, Rothman JH. Combinatorial specification of blastomere identity by glp-1 dependent cellular interactions in the nematode Caenorhabditis elegans. Development. 1994;120:3325–38. doi: 10.1242/dev.120.11.3325. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Tissenbaum HA. Reproduction and longevity: secrets revealed by C. elegans. Trends Cell Biol. 2007;17:65–71. doi: 10.1016/j.tcb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Muriel JM, Dong C, Hutter H, Vogel BE. Fibulin-1C and Fibulin-1D splice variants have distinct functions and assemble in a hemicentin-dependent manner. Development. 2005;132:4223–34. doi: 10.1242/dev.02007. [DOI] [PubMed] [Google Scholar]
- Nance J. PAR proteins and the establishment of cell polarity during C. elegans development. Bio Essays. 2005;27:126–35. doi: 10.1002/bies.20175. [DOI] [PubMed] [Google Scholar]
- Neves A, English K, Priess JR. Notch-GATA synergy promotes endoderm-specific expression of ref-1 in C. elegans. Development. 2007;134:4459–68. doi: 10.1242/dev.008680. [DOI] [PubMed] [Google Scholar]
- Neves A, Priess JR. The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Dev Cell. 2005;8:867–79. doi: 10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Norman KR, Moerman DG. Alpha spectrin is essential for morphogenesis and body wall muscle formation in Caenorhabditis elegans. J Cell Biol. 2002;157:665–77. doi: 10.1083/jcb.200111051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–95. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Fire A. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development. 1994;120:2175–86. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Ha E, Haun C, Chen W, Fire A. The Caenorhabditis elegans NK-2 homeobox gene ceh-22 activates pharyngeal muscle gene expression in combination with pha-1 and is required for normal pharyngeal development. Development. 1997;124:3965–73. doi: 10.1242/dev.124.20.3965. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–81. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- Olsen CL, Jeffry WR. A forkhead gene related to HNF-3beta is required for gastrulation and axis formation in the ascidian embryo. Development. 1997;124:3609–19. doi: 10.1242/dev.124.18.3609. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–19. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–55. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Park FD, Tenlen JR, Priess JR. C. elegans MOM-5/frizzled functions in MOM-2/Wnt-independent cell polarity and is localized asymmetrically prior to cell division. Curr Biol. 2004;14:2252–58. doi: 10.1016/j.cub.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425:624–28. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- Pettitt J, Cox EA, Broadbent ID, Flett A, Hardin J. The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J Cell Biol. 2003;162:15–22. doi: 10.1083/jcb.200212136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt J, Wood WB, Plasterk RH. cdh-3, a gene encoding a member of the cadherin superfamily, functions in epithelial cell morphogenesis in Caenorhabditis elegans. Development. 1996;122:4149–57. doi: 10.1242/dev.122.12.4149. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Kidd AR, 3rd, King R, Hardin J, Kimble J. Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:3231–36. doi: 10.1073/pnas.0611507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko MF, Mango SE. Early morphogenesis of the Caenorhabditis elegans pharynx. Dev Biol. 2001;233:482–94. doi: 10.1006/dbio.2001.0235. [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Saam J, Mango SE. ZEN-4/MKLP1 is required to polarize the foregut epithelium. Curr Biol. 2004;14:932–41. doi: 10.1016/j.cub.2004.05.052. [DOI] [PubMed] [Google Scholar]
- Priess JR. Notch signaling in the C. elegans embryo. Worm Book. 2005 Jun;25:1–16. doi: 10.1895/wormbook.1.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–11. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Priess JR, Thomson JN. Cellular interactions in early C. elegans embryos. Cell. 1987;48:241–50. doi: 10.1016/0092-8674(87)90427-2. [DOI] [PubMed] [Google Scholar]
- Qiu X, Fay DS. ARI-1, an RBR family ubiquitin-ligase, functions with UBC-18 to regulate pharyngeal development in C. elegans. Dev Biol. 2006;291:239–52. doi: 10.1016/j.ydbio.2005.11.045. [DOI] [PubMed] [Google Scholar]
- Raharjo I, Gaudet J. Gland-specific expression of C. elegans hlh-6 requires the combinatorial action of three distinct promoter elements. Dev Biol. 2007;302:295–308. doi: 10.1016/j.ydbio.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Rasmussen JP, English K, Tenlen JR, Priess JR. Notch signaling and morphogenesis of single-cell tubes in the C. elegans digestive tract. Dev Cell. 2008;14:559–69. doi: 10.1016/j.devcel.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Schnabel R, Okkema PG. Behavioral and synaptic defects in C. elegans lacking the NK-2 homeobox gene ceh-28. Dev Neurobiol. 2008;68:421–33. doi: 10.1002/dneu.20599. [DOI] [PubMed] [Google Scholar]
- Rebay I, Silver SJ, Tootle TL. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 2005;21:163–71. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Robertson SM, Shetty P, Lin R. Identification of lineage-specific zygotic transcripts in early Caenorhabditis elegans embryos. Dev Biol. 2004;276:493–507. doi: 10.1016/j.ydbio.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhuri S, Crum T, Woollard A, Aslam S, Okkema PG. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev Biol. 2006;295:664–77. doi: 10.1016/j.ydbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Savage-Dunn C. TGF-beta signaling. Worm Book. 2005 Sep;9:1–12. doi: 10.1895/wormbook.1.22.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C, Schwarz V, Hutter H. AST-1, a novel ETS-box transcription factor, controls axon guidance and pharynx development in C. elegans. Dev Biol. 2006;293:403–13. doi: 10.1016/j.ydbio.2006.02.042. [DOI] [PubMed] [Google Scholar]
- Schmitz C, Wacker I, Hutter H. The Fat-like cadherin CDH-4 controls axon fasciculation, cell migration and hypodermis and pharynx development in Caenorhabditis elegans. Dev Biol. 2008;316:249–59. doi: 10.1016/j.ydbio.2008.01.024. [DOI] [PubMed] [Google Scholar]
- Schnabel H, Schnabel R. An organ-specific differentiation gene, pha-1, from Caenorhabditis elegans. Science. 1990;250:686–88. doi: 10.1126/science.250.4981.686. [DOI] [PubMed] [Google Scholar]
- Schnabel R, Bischoff M, Hintze A, Schulz AK, Hejnol A, et al. Global cell sorting in the C. elegans embryo defines a new mechanism for pattern formation. Dev Biol. 2006;294:418–31. doi: 10.1016/j.ydbio.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Schneider SQ, Bowerman B. beta-Catenin asymmetries after all animal/vegetal-oriented cell divisions in Platynereis dumerilii embryos mediate binary cell-fate specification. Dev Cell. 2007;13:73–86. doi: 10.1016/j.devcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–43. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Updike DL, Mango SE. The target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–64. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer G, Suissa M, Kolotuev I, Nguyen KC, Hall DH, Podbilewicz B. EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr Biol. 2004;14:1587–91. doi: 10.1016/j.cub.2004.07.059. [DOI] [PubMed] [Google Scholar]
- Shetty P, Lo MC, Robertson SM, Lin R. C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Dev Biol. 2005;285:584–92. doi: 10.1016/j.ydbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Kidd AR, 3rd, Chesney MA, Kimble J. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics. 2004;166:171–86. doi: 10.1534/genetics.166.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Mlodzik M. Planar cell polarity signaling: from fly development to human disease. Annu Rev Genet. 2008;42:517–40. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simske JS, Hardin J. Getting into shape: epidermal morphogenesis in Caenorhabditis elegans embryos. Bio Essays. 2001;23:12–23. doi: 10.1002/1521-1878(200101)23:1<12::AID-BIES1003>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Simske JS, Koppen M, Sims P, Hodgkin J, Yonkof A, Hardin J. The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans. Nat Cell Biol. 2003;5:619–25. doi: 10.1038/ncb1002. [DOI] [PubMed] [Google Scholar]
- Smit RB, Schnabel R, Gaudet J. The HLH-6 transcription factor regulates C. elegans pharyngeal gland development and function. PLoS Genet. 2008;4:e1000222. doi: 10.1371/journal.pgen.1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ED, Kaeberlein TL, Lydum BT, Sager J, Welton KL, et al. Age- and calorie-independent life span extension from dietary restriction by bacterial deprivation in Caenorhabditis elegans. BMC Dev Biol. 2008;8:49. doi: 10.1186/1471-213X-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PA, Mango SE. Role of T-box gene tbx-2 for anterior foregut muscle development in C. elegans. Dev Biol. 2007;302:25–39. doi: 10.1016/j.ydbio.2006.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich TA, Miller A, Nguyen RL, Hall DH, Shaw JE. The Caenorhabditis elegans innexin INX-3 is localized to gap junctions and is essential for embryonic development. Dev Biol. 2003;256:403–17. doi: 10.1016/s0012-1606(02)00116-1. [DOI] [PubMed] [Google Scholar]
- Straub T, Becker PB. DNA sequence and the organization of chromosomal domains. Curr Opin Genet Dev. 2008;18:175–80. doi: 10.1016/j.gde.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–26. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998;125:3607–14. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008;9:103–9. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher JD, Fernandez AP, Beaster-Jones L, Haun C, Okkema PG. The Caenorhabditis elegans peb-1 gene encodes a novel DNA-binding protein involved in morphogenesis of the pharynx, vulva, and hindgut. Dev Biol. 2001;229:480–93. doi: 10.1006/dbio.2000.9978. [DOI] [PubMed] [Google Scholar]
- Totong R, Achilleos A, Nance J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 2007;134:1259–68. doi: 10.1242/dev.02833. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzebiatowska A, Topf U, Sauder U, Drabikowski K, Chiquet-Ehrismann R. Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol Biol Cell. 2008;19:3898–908. doi: 10.1091/mbc.E08-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G, Jensen ST, White P, Kaestner KH. Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 2008;36:4149–57. doi: 10.1093/nar/gkn366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Mango SE. Temporal regulation of foregut development by HTZ-1/H2A.Z and PHA-4/FoxA. PLoS Genet. 2006;2:e161. doi: 10.1371/journal.pgen.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Mango SE. Genetic suppressors of C. elegans pha-4/FoxA identify the predicted AAA helicase ruvb-1/RuvB. Genetics. 2007;177:819–33. doi: 10.1534/genetics.107.076653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Vilimas T, Abraham A, Okkema PG. An early pharyngeal muscle enhancer from the Caenorhabditis elegans ceh-22 gene is targeted by the Forkhead factor PHA-4. Dev Biol. 2004;266:388–98. doi: 10.1016/j.ydbio.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Wang JC, Waltner-Law M, Yamada K, Osawa H, Stifani S, Granner DK. Transducin-like enhancer of split proteins, the human homologs of Drosophila groucho, interact with hepatic nuclear factor 3beta. J Biol Chem. 2000;275:18418–23. doi: 10.1074/jbc.M910211199. [DOI] [PubMed] [Google Scholar]
- Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–64. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM, Crothers DM. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991;66:577–88. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- Weigel D, Bellen JJ, Jurgens G, Jackle H. Primordium specific requirement of the homeotic gene fork head in the developing gut of the Drosophila embryo. Roux’s Arch Dev Biol. 1989a;198:201–10. doi: 10.1007/BF00375906. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jurgens G, Kuttner F, Seifert E, Jackle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989b;57:645–58. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, et al. The winged-helix transcription factor HNF-3beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–88. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, et al. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 2008;4:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, et al. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–50. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–90. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–53. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Woo WM, Berry EC, Hudson ML, Swale RE, Goncharov A, Chisholm AD. The C. elegans F-spondin family protein SPON-1 maintains cell adhesion in neural and non-neural tissues. Development. 2008;135:2747–56. doi: 10.1242/dev.015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB. Evidence from reversal of handedness in C. elegans embryos for early cell interactions determining cell fates. Nature. 1991;349:536–38. doi: 10.1038/349536a0. [DOI] [PubMed] [Google Scholar]
- Wu M, Herman MA. A novel noncanonical Wnt pathway is involved in the regulation of the asymmetric B cell division in C. elegans. Dev Biol. 2006;293:316–29. doi: 10.1016/j.ydbio.2005.12.024. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yanowitz JL, Shakir MA, Hedgecock E, Hutter H, Fire AZ, Lundquist EA. UNC-39, the C. elegans homolog of the human myotonic dystrophy-associated homeodomain protein Six5, regulates cell motility and differentiation. Dev Biol. 2004;272:389–402. doi: 10.1016/j.ydbio.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Zaret KS. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet. 2008;9:329–40. doi: 10.1038/nrg2318. [DOI] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee JD, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 1998;12:3809–14. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]