Abstract
Translated regions distinct from annotated coding sequences have emerged as essential elements of the proteome. This includes upstream open reading frames (uORFs) present in mRNAs controlled by the integrated stress response (ISR) that show “privileged” translation despite inhibited eukaryotic initiation factor 2–guanosine triphosphate–initiator methionyl transfer RNA (eIF2·GTP·Met-tRNAiMet). We developed tracing translation by T cells to directly measure the translation products of uORFs during the ISR. We identified signature translation events from uORFs in the 5′ untranslated region of binding immunoglobulin protein (BiP) mRNA (also called heat shock 70-kilodalton protein 5mRNA) that were not initiated at the start codon AUG. BiP expression during the ISR required both the alternative initiation factor eIF2A and non–AUG-initiated uORFs. We propose that persistent uORF translation, for a variety of chaperones, shelters select mRNAs from the ISR, while simultaneously generating peptides that could serve as major histocompatibility complex class I ligands, marking cells for recognition by the adaptive immune system.
Homeostatic mechanisms facilitate adaptation to a variety of environmental conditions and cellular dysfunction. The integrated stress response (ISR) is one such mechanism, triggered when cells encounter an array of stress stimuli. These stimuli include misfolded proteins, which elicit the unfolded protein response (UPR) and thereby activate the endoplasmic reticulum (ER)–resident kinase (PERK) (1–3). In addition, three related kinases are activated by other stimuli, such as the interferon-induced double-stranded RNA (dsRNA)–dependent eIF2α kinase (PKR) (by viral infection) (4, 5); the general control nonderepressible 2 (GCN2) (by amino acid deprivation) (6); and the hemeregulated inhibitor kinase (HRI) (by heme deficiency, oxidative stress, heat shock, or osmotic shock) (7). Each of these conserved kinases initiate the ISR by phosphorylating the same single residue (Ser51) on the α subunit of eukaryotic initiation factor 2α (eIF2α) and down-regulate translation initiation at AUG start codons by the eukaryotic initiation factor 2–guanosine triphosphate (GTP)–initiator methionyl transfer RNA (tRNA) (eIF2·GTP·Met-tRNAiMet) ternary complex. Phosphorylation of eIF2α (eIF2α-P) inhibits exchange of guanosine diphosphate for GTP by eIF2B, the dedicated eIF2 guanine nucleotide exchange factor, which causes inhibition of total protein synthesis (8). The blockade in translation is important for cell survival and the eventual switch into apoptosis if homeostasis cannot be reestablished.
Although eIF2α-P limits global translation, it is required for the regulated expression of several proteins, such as activating transcription factor 4 (ATF4 or CREB-2) (9–11) and C/EBP homologous protein (12, 13), that finely tune cell survival (14). These ISR-induced proteins are translated from mRNAs and harbor a series of upstream open reading frames (uORFs) in the 5′ untranslated region (5′ UTR) that limit ribosome access to the main coding sequence (CDS), as first characterized in the budding yeast Saccharomyces cerevisiae (15). According to the prevailing model, under normal growth conditions, ribosome initiation occurs predominantly at uORFs, which prevents access to the downstream CDS. By contrast, when the ISR is induced and eIF2α-P levels rise, stochastic ribosome bypass of the uORFs allows access to the downstream CDS AUG start codon.
Another subset of mRNAs remains efficiently translated during the ISR. These include mRNAs encoding heat shock and UPR proteins (1, 16–18) and a variety of inflammatory cytokines in response to viral (19, 20) and bacterial (21) pathogens. In the context of the UPR, for example, translation of mRNAs encoding ER chaperones is imperative to alleviate ER stress. BiP [immunoglobulin heavy chain–binding protein, also known as heat shock 70 kD protein (HSP70), heat shock protein family A member 5 (HSPA5), or glucose-regulated protein 78] is an essential HSP70-type chaperone in the ER and is expressed persistently during ER stress (22–24). It plays a role in cancer progression (25) and is a therapeutic target for a variety of diseases (26, 27). Yet, it has remained a mystery how BiP and other stress-response mRNAs escape translational down-regulation imposed by the ISR. Elements in the 5′ UTRs, including internal ribosome entry sites (IRESs), uORFs, and nucleotide modifications, have all been suggested to confer translational privilege to these mRNAs (28, 29).
Recent genome-wide approaches predict that nearly half of all mammalian mRNAs harbor uORFs in their 5′ UTRs, and many are initiated with non-AUG start codons (30–34). The presence of uORFs in 5′ UTRs may reflect a general mechanism to regulate downstream CDS expression, such as proto-oncogenes and growth factors (30), as well as other disease-causing proteins (35), including hereditary thrombocythemia (36–38). Given the abundance of uORFs and their potential for regulatory roles, as well as the emerging plethora of short open reading frames (sORFs) (39–41) with bioactive properties (42), we developed a method to measure translation from RNA regions outside of annotated CDSs systematically.
Development of tracing translation by T cells (3T) to measure translation outside of annotated coding sequences
Ribosome-profiling experiments reveal that mRNAs encoding stress-response proteins harbor a particularly high abundance of uORFs (43, 44). Yet, despite the thousands of peptides predicted by ribosome profiling to be translated from uORFs, very few uORF peptides have been identified by mass spectrometry (45). Currently, there is considerable effort to improve proteomic approaches for the detection of peptides from uORFs and other sORFs (39). Here, we exploited the exquisite sensitivity and specificity of T cells to detect such translation products.
We developed an approach, termed 3T, where cells are supplied with DNA vectors containing noncoding RNA elements, such as 5′ UTRs, harboring sequences that encode tracer peptides (Fig. 1A). If the resulting RNA is translated, cells proteolytically process the translated polypeptides. The resulting peptides are then transported by the transporter associated with antigen processing (TAP) into the ER. In the ER, the peptides (also called antigens) are loaded onto major histocompatibility complex class I (MHC I) molecules and displayed on the cell surface. We assessed presentation of the translated tracer peptide by MHC I by addition of a peptide–MHC I–cognate T cell hybridoma, engineered to express LacZ from the interleukin 2 (IL-2) promoter under control of the N-FAT enhancer (46). Engagement with tracer peptide–loaded MHC I cells triggered LacZ expression, which resulted in production of its translation product, β-galactosidase. We detected β-galactosidase using its substrate, chlorophenol red–β-d-galactopyranoside (CPRG), which yielded a red cleavage product. In the absence of tracer peptide, the T cell hybridoma did not engage cells, and LacZ was transcriptionally inactive (Fig. 1A) (47).
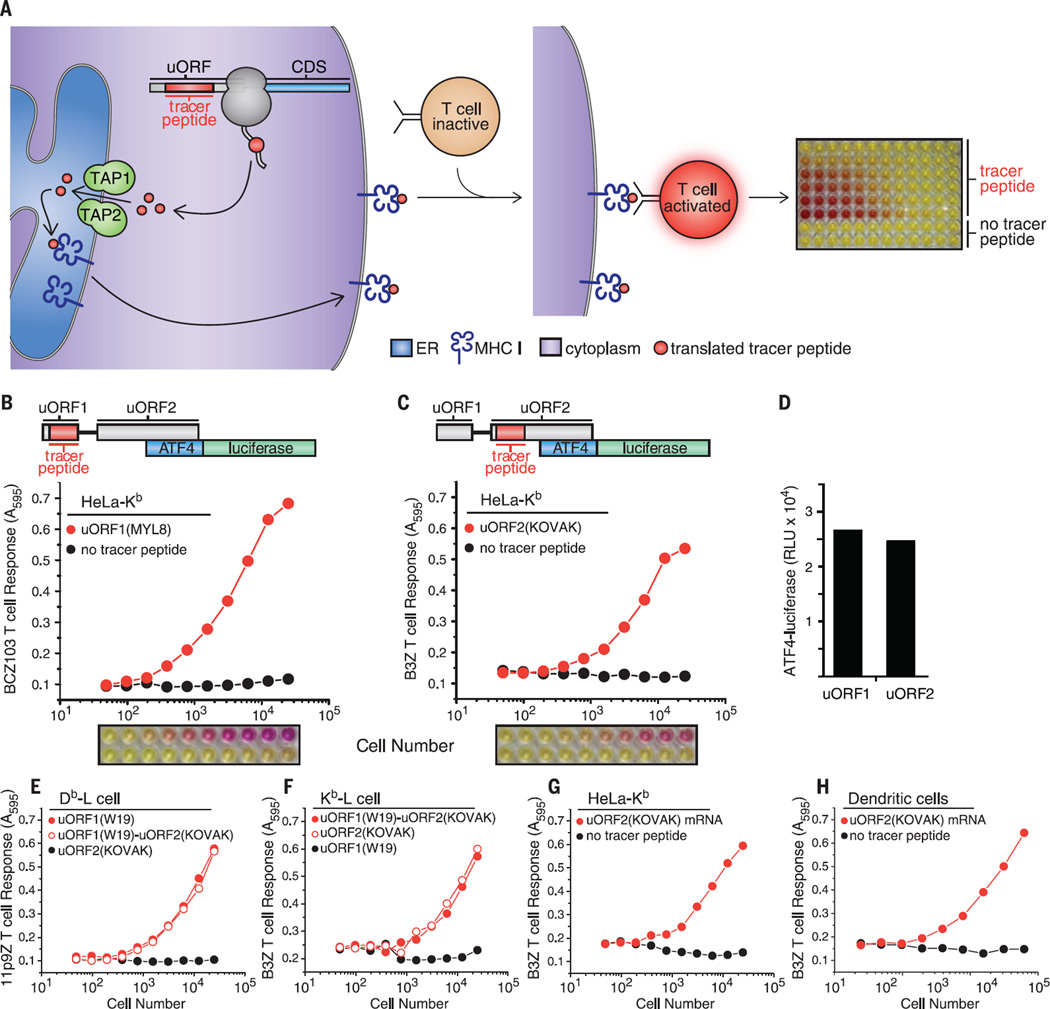
Fig. 1. 3T reveals the translational capacity of uORFs in 5′ UTRs.
(A) Schematic of 3T: A tracer peptide inserted into noncoding regions of the 5′ UTR, such as uORFs, followed by translation, peptide processing, and transport by TAP into the ER for loading onto MHC I molecules. The tracer peptide–MHC I complex transits to the cell surface and is recognized by a T cell hybridoma. Recognition of the tracer peptide–MHC I complex on the cell surface activates the T cell hybridoma, which is measured by a colorimetric assay and indicates the presence of a uORF translation product. Schematics of the ATF4 5′ UTR with nested tracer peptides. Plotted T cell responses and colorimetric readout of the 3Tassay from tracer peptides generated from transfection of HeLa-Kb cells with (B) uORF1(MYL8)–ATF4-luciferase detected with the BCZ103 T cell hybridoma and (C) uORF2(KOVAK)–ATF4-luciferase detected with the B3Z T cell hybridoma with simultaneous detection of ATF4-luciferase expression (D). T cell responses from tracer peptides generated from uORF1(WI9)–ATF4-luciferase, uORF1 (WI9)-uORF2(KOVAK)–ATF4-luciferase, or uORF2(KOVAK)–ATF4-luciferase transfected (E) Db-L cells detected with the 11p9Z T cell hybridoma or (F) Kb-L cells detected with the B3Z T cell hybridoma. 3T responses from uORF2(KOVAK)–ATF4-luciferase (detected with the B3Z T cell hybridoma) generated from mRNA transfection of HeLa-Kb cells (G) and primary bone marrow–derived dendritic cells (H). T cell responses are representative of n ≥ 3. A595, absorption at 595 nm.
To validate 3T, we tested the coding capacity of the activating transcription factor 4 (ATF4) 5′ UTR, which contains well-characterized uORFs (fig. S1). To this end, we inserted tracer peptides into two highly conserved uORFs that were previously shown to regulate ATF4 expression in response to eIF2α phosphorylation (11, 48) (Fig. 1, B and C, and fig. S1). To assess the levels of uORF translation, we transfected cells harboring the appropriate MHC I with uORF (tracer peptide)–ATF4–luciferase constructs. We first nested the coding sequence for the peptide MTFNYRNL (MYL8) (fig. S1) into uORF1 of ATF4-luciferase and the peptide KSIINFEHLK (KOVAK) (fig. S1) into uORF2. Both tracer peptides are presented by MHC I H-2Kb (Kb MHC I). We then added, as a titration of increasing cell numbers, an equal number of T cell hybridomas specific for the tracer peptides displayed [BCZ103 for uORF1(MYL8) or B3Z for uORF2(KOVAK)], followed by CPRG. As expected, T cell responses were only observed when the tracer peptide was present in either uORF (Fig. 1, B and C).
The tracer peptide constructs allowed robust detection of translation from multiple different regions of the mRNA. For example, we measured ATF4-luciferase activity resulting from CDS translation (Fig. 1D) from cells expressing tracer peptides from either uORF1 or uORF2 (Fig. 1, B and C, and fig. S2). Similarly, using the tracer peptide WMHHNMDLI (WI9), presented by MHC I H-2Db (Db MHC I) and detected by a different T cell hybridoma, we measured uORF1 expression from either the uORF1(WI9) or the uORF1(WI9)-uORF2(KOVAK) constructs (Fig. 1E). As expected, uORF2(KOVAK) generated a peptide that was only detected by the appropriate T cell hybridoma (compare Fig. 1, E and F). Similarly, the KOVAK tracer peptide was detected independently of its placement in uORF1 or uORF2 (Fig. 1F).
3T can be adapted for detection of tracer peptides from mRNAs directly transfected into cells. In particular, we observed uORF2 tracer peptide expression from uORF2(KOVAK)–ATF4-luciferase mRNA (Fig. 1G). Furthermore, we readily observed tracer peptide expression from uORF2 from primary bone marrow–derived dendritic cells after only a 3-hour mRNA transfection (Fig. 1H). These observations underscore the possibility that any mRNA and a diverse range of cell types can be used to measure uORF expression, as long as the MHC I peptide presentation pathway is constitutively expressed in these cells.
For 3T to reflect expression from distinct regions of the mRNA reliably, tracer peptide placement should not deregulate the expression of the main CDS during steady-state and stress conditions. Therefore, we tested CDS expression (ATF4-luciferase activity) from constructs bearing various uORF tracer peptide insertions. Indeed, insertion of tracer peptides into either uORF1 or uORF2 or simultaneously into both uORFs did not substantially impair the inhibitory function of the uORFs, as assessed by luciferase activity compared with wild type–ATF4-luciferase and a constitutively active variant of ATF4-luciferase (49) (Fig. 2A and fig. S3).
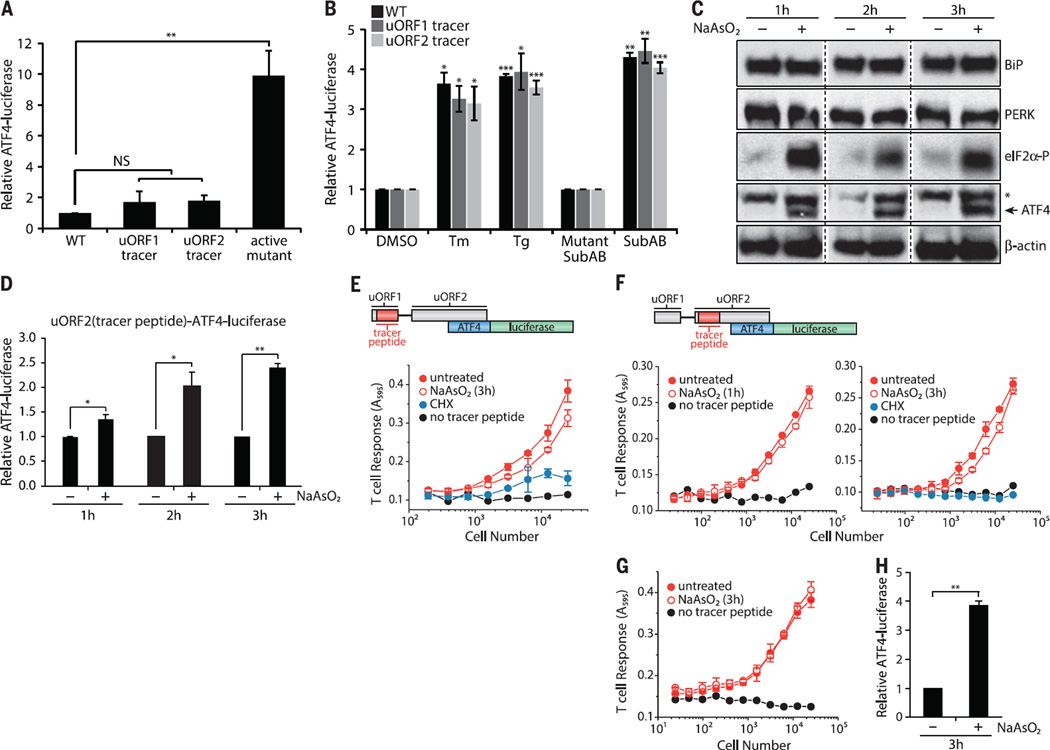
Fig. 2. uORFs are expressed during stress-induced expression of ATF4.
(A) ATF4-luciferase levels during normal growth conditions were measured from HeLa-Kb cells transfected with wild type–ATF4-luciferase, ATF4-luciferase constructs with tracer peptide insertions [uORF1(MYL8) or uORF2(KOVAK) tracer peptides], or a constitutively active ATF4-luciferase variant (means ± SEM; n = 3 to 4). (B) Stress-induced ATF4-luciferase levels were measured from HeLa-Kb cells transfected with wild type–ATF4-luciferase and ATF4-luciferase constructs with tracer peptide insertions [uORF1(MYL8) or uORF2(KOVAK) tracer peptides] and after treatments with tunicamycin (Tm) (1 µg/ml), Tg (1 µM), or Mutant SubAB or SubAB (0.2 µg/ml) for 6 hours (means ± SEM; n = 3 to4). HeLa-Kb cells from independent tracer peptide DNA transfections were treated with NaAsO2 (10 µM) for 1 to 3 hours and analyzed by immunoblot (n = 3; *nonspecific ATF4-specific antibody signal) (C) or for luciferase expression from uORF2(KOVAK)–ATF4-luciferase–transfected cells (means ± SD; n = 3 to 4) (D). Tracer peptide expression was measured from HeLa-Kb cells transfected with either uORF1(MYL8)–ATF4 luciferase (E) or uORF2(KOVAK)–ATF4 luciferase (F) after treatment with NaAsO2 (10 µM) (means ± SD from two biological replicates are representative of n = 3). Stable uORF2(KOVAK)–ATF4-luciferase HeLa-Kb cells were treated for 3 hours with NaAsO2 (10 µM) and assayed for uORF2(KOVAK) tracer peptide expression (G) (means ± SD from two biological replicates are representative of n = 3) or for ATF4-luciferase expression (H) (means ± SEM; n = 3). Statistical significance was evaluated with the unpaired t test (NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001).
To test whether control of ATF4 induction by the ISR was likewise intact in the reporter cell lines bearing the tracer peptide insertions, we exposed cells to subtilase cytotoxin (SubAB), a bacterial AB toxin that is endocytosed by cells and retrotransported to the ER lumen, where it destroys BiP by proteolysis (50). BiP destruction induced protein misfolding in the ER and PERK-catalyzed phosphorylation of eIF2α, which inhibited cellular translation (fig. S4A). The translation block was readily reversed by ISRIB, a small molecule that overcomes the effects of eIF2α phosphorylation (51–53) (fig. S4A). SubAB triggered PERK activation, expression of endogenous ATF4, eIF2α phosphorylation (fig. S4B), and up-regulation of the plasmid-borne ATF4-luciferase reporter (fig. S4C). ISRIB inhibited endogenous ATF4 expression and ATF4-luciferase expression, which indicated that the ATF4 transgene behaved like endogenous ATF4 (fig. S4). Furthermore, multiple UPR inducers stimulated ATF4-luciferase expression equivalently from constructs harboring either a uORF1 or uORF2 tracer peptide (Fig. 2B). These results indicate that tracer peptide placement does not compromise the control of ATF4 expression during ER stress.
3T relies on proper processing of tracer peptides, including import into the ER and loading onto MHC I molecules. To induce eIF2α phosphorylation without disturbing the normal processes in the ER lumen, such as protein folding (compromised by SubAB cleavage of BiP), or N-linked glycosylation (inhibited by tunicamycin), or directly activating the T cell hybridomas [calcium fluxes with thapsigargin (Tg)], we treated cells with NaAsO2, an inducer of oxidative stress, which rapidly induces eIF2α phosphorylation and ATF4 induction by activating cytosolic HRI (54). We observed rapid and robust expression of endogenous ATF4 without activation of PERK or altered expression of BiP (Fig. 2C and fig. S5). Pertinent to the utility of 3T, ATF4-luciferase harboring a nested uORF2 tracer peptide showed the expected induction with NaAsO2 treatment (Fig. 2D). Peptide expression from uORF1 (Fig. 2E) and uORF2 (Fig. 2, F to H, and fig. S6) largely persisted when ATF4-luciferase was induced, a finding that differs from studies with yeast GCN4 (55). However, the pervasive uORF2 peptide expression measured here is consistent with recent ribosome-profiling studies on ATF4 mRNA upon ISR induction (43, 44). These results validate 3T as a sensitive and robust indicator of uORF expression. As such, we next applied this method to ask whether uORF translation is a general strategy that cells use to ensure privileged protein expression during the ISR.
3T reveals a novel regulatory element in the 5′ UTR of BiP mRNA
BiP synthesis, which is initiated at a standard AUG start codon (fig. S7), persists during ER stress, when the ISR is induced (56). To explore this phenomenon, we treated cells with SubAB, resulting in PERK activation, eIF2α phosphorylation, and a massive reduction in protein synthesis (Fig. 3, A and B). As expected, treatment with a catalytically inactive form of SubAB (Mut SubAB) did not result in ISR induction (Fig. 3, A and B, and fig. S8). To assess directly whether BiP is synthesized after onset of the ISR, we transfected cells with DNA encoding FLAG-tagged BiP [or BiP(L416D)-FLAG, a SubAB-resistant mutant] and treated them with Mut SubAB or SubAB. We next pulse-labeled cells with [35S] Met-Cys to label newly synthesized proteins, followed by immunoprecipitation of BiP. In agreement with previous results, BiP from both constructs was readily synthesized despite ISR induction (Fig. 3C). Thus, elements other than conventional initiation at the annotated BiP AUG start codon or its CDS may be required to ensure privileged expression during the UPR.
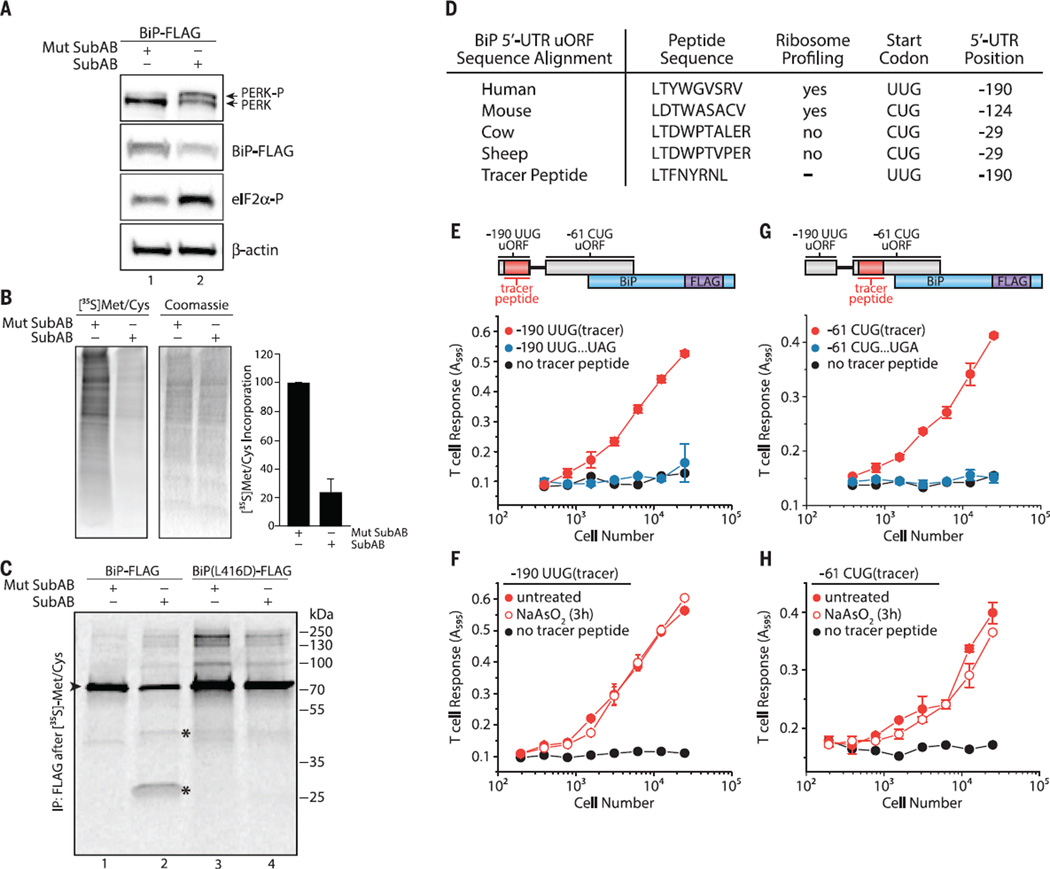
Fig. 3. The BiP 5′ UTR harbors non–AUG-initiated uORFs constitutively expressed during the ISR.
Cells treated with Mutant SubAB or SubAB (0.2 µg/ml) for 2 hours in HeLa-Kb cells were analyzed by (A) immunoblot or (B) [35S]Met-Cys pulse-labeled to measure total protein synthesis (means ± SEM; n = 4). (C) SubAB-treated HeLa-Kb cells (1 hour) followed by [35S]Met-Cys pulse-labeling (1 hour) and BiP-FLAG immunoprecipitation (►is full-length BiP and * indicates BiP cleavage products; data representative of n=2). (D) Amino acid sequence alignment of the BiP 5′ UTR −190 uORF with non-AUG start codons. (E) Schematic of the BiP 5′ UTR with the tracer peptide LYL8 at the −190 UUG uORF. Translation of the −190 UUG uORF was measured from−190 UUG uORF tracer peptide BiP-FLAG–transfected HeLa-Kb cells detected with the BCZ103 T cell hybridoma and compared with cells transfected with an identical construct containing an in-frame UAG stop codon inserted in the middle of the tracer peptide (see fig. S9) or transfected with a no-tracer peptide construct (BiP-FLAG) and (F) after treatment with NaAsO2 (10 µM) for 3 hours (means ± SD from two biological replicates and are representative of n = 3). (G) Schematic of the BiP 5′ UTR with the nested tracer peptide KOVAK in the −61 CUG uORF. Translation of the −61 CUG uORF was measured from −61 CUG uORF tracer peptide BiP-FLAG–transfected HeLa-Kb cells detected with the B3Z T cell hybridoma and compared with cells transfected with an identical construct containing an in-frame UGA stop codon inserted after the −61 CUG uORF start codon but before the tracer peptide (see fig. S9) or transfected with a no-tracer peptide construct (BiP-FLAG) and (H) after treatment with NaAsO2 (10 µM) for 3 hours (data are presented as means ± SD of two biological replicates and are representative of n = 3).
Recent ribosome profiling measurements indicate that BiP mRNA shows substantial levels of ribosome occupancy in the 5′ UTR (33). The most prominent ribosome initiation signal was detected at an UUG codon at position −190 (relative to BiP coding sequence +1 AUG), encoding a putative nine–amino acid peptide (Fig. 3D and fig. S9A). To test whether this predicted uORF is translated, we nested a tracer peptide [LTFNYRNL (LYL8)] in the −190 UUG uORF (Fig. 3D), which did not alter the expression of BiP during basal conditions or its targeting to the ER as assayed by SubAB sensitivity (fig. S10A). When assessed by 3T, we readily detected the −190 UUG leucine codon–initiated expression of the tracer peptide (Fig. 3E). Translation of the −190 UUG uORF persisted upon eIF2α phosphorylation induced by NaAsO2 treatment (Fig. 3F). Given that an UUG codon codes for leucine and that leucine initiation is resistant to NaAsO2 treatment and reduced eIF2·GTP·Met-tRNAiMet levels (57, 58), a noncanonical UUG or CUG initiation mechanism may function in the BiP 5′ UTR.
We similarly tested expression of the nested KOVAK tracer peptide in the predicted −61 leucine CUG-initiated uORF in the BiP 5′ UTR (33) (Fig. 3G and fig. S9B). This uORF is out of frame with the BiP AUG-initiated CDS. Indeed, the −61 CUG-initiated uORF supports expression of a peptide in cells when present out of frame (Fig. 3G) or in frame with the BiP coding sequence (fig. S10, A and B). Expression of the −61 CUG uORF was not markedly reduced in the presence of NaAsO2 (Fig. 3H), which suggests that multiple noncanonical initiation events are involved in sustaining BiP expression during stress.
BiP uORF expression is regulated by eIF2A
The alternative initiation factor eIF2A, a monomeric protein structurally and functionally distinct from the trimeric eIF2, coordinates noncanonical leucine (CUG) initiation (58) and expression of a CUG leucine–initiated phosphatase and tensin homolog deleted on chromosome 10 isoform (59). Given these findings, we examined the consequences of eIF2A depletion on −190 UUG uORF and BiP expression. Although when eIF2A is depleted (eIF2A knockdown) with small interfering RNA (siRNA), this did not measurably impair global protein synthesis (fig. S11), and it substantially impaired expression of the −190 UUG uORF (Fig. 4A and fig. S12), which required nearly 40% more tracer-expressing cells to stimulate T cells to a half-maximal response (Fig. 4B). This observation is consistent with a requirement for eIF2A-dependent initiation of the BiP −190 UUG uORF, which may regulate the levels of BiP.
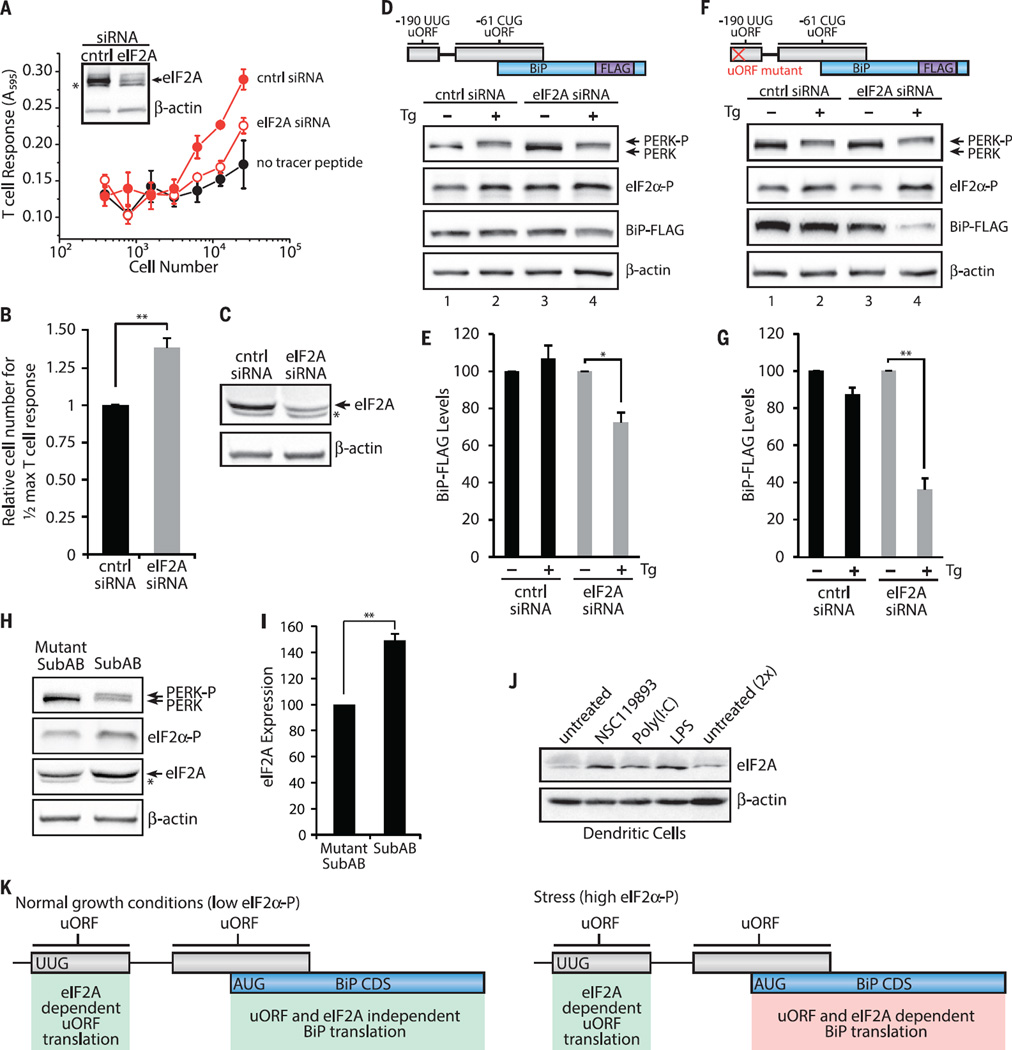
Fig. 4. eIF2A and 5′ UTR uORFs are required for sustained BiP expression during the ISR.
(A) Translation of the −190 UUG uORF from −190 UUG uORF tracer peptide (LYL8) BiP-FLAG–transfected HeLa-Kb cells after 48 hours siRNA knockdown detected with the BCZ103 T cell hybridoma and compared with cells transfected with a no-tracer peptide construct (data are presented as means ± SD of two biological replicates and are representative of n = 4). Inset shows an immunoblot for eIF2A knockdown and a β-actin loading control. (B) Relative number of cells required to achieve half-maximal T cell response for −190 UUG uORF expression with eIF2A siRNA knockdown (mean ± SEM; n = 4). Expression of BiP-FLAG analyzed by immunoblot from BiP-FLAG (D and E) or uORF mutant BiP-FLAG (F and G) transfected HeLa-Kb cells after 48 hours of siRNA knockdown (C) and treatment with DMSO or Tg (1 µM) for 3 hours (n = 3). BiP levels are presented relative to untreated for each siRNA. (H and I) HeLa-Kb cells treated with Mut SubAB or SubAB (0.2 µg/ml) for 2 hours and analyzed by immunoblot for eIF2A expression (*nonspecific anti-eIF2A antibody signal) (means ± SEM; n = 3). (J) Primary bone marrow–derived dendritic cells analyzed by immunoblot for eIF2A expression after treatment with NSC119893 (50 µM), poly(I:C) (1 µg/ml), or LPS (1 µg/ml) for 3 hours (n=2). (K) Working model for uORF translation regulation of BiP CDS expression. During normal growth conditions, BiP CDS expression is not dependent on uORF translation and eIF2A. Stress up-regulates eIF2α-P levels, leads to elevated eIF2A levels, and leads to constitutive uORF translation, which positively regulates BiP CDS expression. Statistical significance was evaluated with the unpaired t test (*P < 0.05; **P < 0.01)
Indeed, siRNA knockdown of eIF2A (Fig. 4C) measurably compromised expression of BiP-FLAG, during induction of the ISR with Tg, an ER stress inducer (Fig. 4D, lane 4, and 4E); deregulated the steady-state levels of endogenous BiP; and compromised its expression during the ISR (fig. S13). Notably, deletion of the −190 UUG uORF concurrently with eIF2A siRNA knockdown dramatically impaired the expression of BiP in ER-stressed cells (Fig. 4F, lane 4, and 4G). Moreover, the expression of eIF2A was induced by SubAB in our cell model (Fig. 4, H and I) and in primary mouse dendritic cells in response to a variety of other stresses (Fig. 4J). In these cells, eIF2A was induced by poly(I:C) and lipopolysaccharide (LPS) (which are activators of PKR) and the small molecule NSC119893 that blocks Met-tRNAiMet binding to eIF2 and compromises canonical AUG-dependent, but not noncanonical, initiation (58, 60). eIF2A up-regulation during acute loss of eIF2·GTP·Met-tRNAiMet ternary complex caused by NSC119893 and dependence on eIF2A for −190 UUG uORF expression are consistent with a requirement for eIF2A in order for BiP to be expressed during cellular stress (Fig. 4K).
uORFs harbor predicted HLA epitopes
In addition to regulating BiP levels during stress, the peptide translated from the −190 UUG uORF is predicted (61) to be a potent human leukocyte antigen (HLA)–presented epitope (human MHC I–peptide) recognized by human T cells (Fig. 5A and fig. S14), as is a peptide potentially generated by translation and processing of the −61 CUG uORF (fig. S14). Peptide expression from these uORFs, during basal and/or ISR conditions, suggested that there may be widespread peptide expression that could serve a variety of extracellular regulatory roles, such as HLA presentation, yet escape detection by currently available techniques, as these peptides are smaller than 10 kD.
Fig. 5. Peptides translated from uORFs and other RNAs during the ISR are predicted to be MHC I peptides.
(A) HLA epitope (human MHC I) prediction from translation of the BiP 5′ UTR −190 UUG uORF using the immune epitope database (IEDB) analysis resource consensus tool (94). (B) Schematic for TAP diffusion [TAP1(GFP)-TAP2 complex] in the ER in the presence or absence of peptides. (C) Meljuso cells stably expressing TAP1-GFP (62) treated for 1 hour with NSC119893 (25 µM) or CHX (50 µg/ml) and labeled with 35S (n = 3) or (D) HeLa-Kb cells treated for 8 hours with bruceantin (100 nM); NSC119893 (50 µM, replenished every 2 hours); or CHX 100 µg/ml and analyzed by immunoblot (n = 3). (E) Meljuso cells stably expressing TAP1-GFP were treated with either NSC119893 or CHX for 1 hour and imaged using fluorescence confocal microscopy for lateral mobility of TAP with treatment (means ± SEM; n = 4 to 7). (F) uORFs predicted from ribosome profiling of chaperone mRNAs (33) generate peptides predicted to be HLA epitopes using the IEDB resource consensus tool (94). Statistical significance was evaluated with the unpaired t test (***P < 0.001).
To begin to assess the prevalence of these translation events, we measured the presence of peptides under conditions of limiting ternary complex using a global readout of their abundance. To this end, we measured diffusion of green fluorescent protein (GFP)–labeled TAP after photobleaching (TAP-FRAP) (62) (Fig. 5B). TAP is an ER-resident adenosine triphosphate– binding cassette transporter that consists of two subunits, TAP1 and TAP2. Antigenic precursor peptides are transported from the cytosol into the ER lumen by TAP (63) and loaded onto MHC I molecules with the aid of ER-resident chaperones (64). Note that the mobility of TAP in the ER membrane depends on peptide abundance. When peptides are lacking, TAP mobility in the ER membrane is measurably enhanced, which is indicative of peptide engagement and, hence, provides a robust readout for peptide abundance (62). We treated cells with NSC119893 to induce the ISR and block eIF2-mediated initiation or with cycloheximide (CHX) to globally inhibit translation (Fig. 5C). As expected, BiP expression was not inhibited by NSC119893 but was by other translation inhibitors (Fig. 5D and fig. S15). In the TAP-FRAP assay, untreated cells showed baseline levels of TAP diffusion, indicative of high peptide availability and TAP engagement, as shown by dimethyl sulfoxide (DMSO) (Fig. 5E). As previously shown (62), CHX inhibited peptide supply and, therefore, enhanced TAP diffusion (Fig. 5E). By contrast, treatment with NSC119893 showed only a modest increase in TAP diffusion (Fig. 5E), which was substantially less than that observed with CHX, despite an equivalent reduction of full-length protein synthesis as assayed by [35S]Met-Cys incorporation (Fig. 5C). These results provide independent evidence for the ubiquitous presence of newly synthesized peptides during normal growth and ISR conditions.
Stress-resistant expression is a common feature of chaperones and heat shock proteins (65), with the 5′ UTR often necessary and sufficient to reconstitute expression during stress (66, 67). We examined ribosome profiling data (33) for a variety of chaperones to identify non-AUG uORFs in their 5′ UTRs. This analysis revealed that UUG- and CUG-initiated uORFs are present in the 5′ UTR of a variety of human chaperones (Fig. 5F). Furthermore, several of these uORFs encode peptides, which, after cellular processing, are predicted to be bound by HLA with strong affinity (Fig. 5F). Taken together, our results suggest that the HLA peptide repertoire is larger than previously predicted and includes peptides derived from 5′ UTR uORFs that may be selectively translated under stress conditions.
Discussion
Here, we exploited the sensitivity and specificity of T cells to assess whether translation occurs outside of the standard protein CDS using 3T. The basis for 3T is the insertion of a tracer peptide coding sequence, known to be presented by MHC I to T cells, into a predefined DNA sequence, which is transcribed into mRNA and translated by ribosomes. The translated tracer peptides, whether nested in a polypeptide or translated from their own ORFs, are efficiently processed and loaded onto MHC I molecules in the ER and travel to the cell surface, where the peptide–MHC I complex is detected by exquisitely specific and sensitive T cell hybridomas. This candidate approach allows customized insertion of tracer peptides into any region of the genome and is amenable to genome tiling of tracer peptides for large-scale discovery and validation of translated uORFs, sORFs, protein isoforms, and “noncoding” RNAs, such as introns, long noncoding RNAs, and 3′ UTRs.
We validated 3T by detecting translated peptides from the uORFs of ATF4, a transcription factor only expressed during the ISR. We measured a high degree of peptide expression from both ATF4 uORF1 and uORF2 during both normal growth and stress conditions, a finding recently predicted from ribosome profiling measurements (43, 44). The high degree of uORF expression during stress, however, was not predicted on the basis of the current reinitiation model for ATF4 expression (11) or the related yeast homolog GCN4 during stress (55). In this model, the small ribosomal subunit is expected to remain mRNA bound and scans past uORF2 and acquires eIF2·GTP·Met-tRNAiMet and the large ribosomal subunit in time for initiation at the conventional AUG start codon of the CDS. Our measurements indicate either that reinitiation commences after only a few ribosomes skip uORF2 or that translation of the ATF4 CDS is a combination of reinitiation and scanning past the uORFs (“leaky scanning”), two events that are technically challenging to distinguish (45). Our finding that uORFs are constitutively expressed during stress and limiting eIF2·GTP· Met-tRNAiMet levels prompts us to speculate that uORF translation might serve an additional role during the ISR, which provides an explanation for the high level of ribosome occupancy within the 5′ UTRs of mRNAs expressed during stress (43). Constitutive uORF expression could thus protect mRNAs from translational down-regulation during stress and thereby serve to control gene expression.
Using 3T, we showed that BiP harbors uORFs that are constitutively translated during stress. By contrast to the AUG-initiated uORFs of ATF4, BiP 5′ UTR uORFs are exclusively initiated by the non-AUG leucine codons UUG and CUG. Expression of the −190 UUG uORF from the BiP 5′ UTR requires the alternative initiation factor eIF2A. By contrast to initiation with the conventional eIF2·GTP·Met-tRNAiMet ternary complex, eIF2A is a monomer and does not bind GTP (68). In addition to initiation at CUG start codons (58, 59), eIF2A is required for initiation at the AUG start codon of a hepatitis C virus internal ribosome entry site (IRES) reporter (69) and viral proteins from the Sindbis alphavirus (70), during stress conditions and high levels of eIF2α phosphorylation. Stress-induced eIF2A could substitute for eIF2 during initiation at the BiP AUG start codon. Our data support a model where eIF2A and a combination of uORFs maintain the levels of BiP synergistically during translational attenuation accompanying cellular stress. We propose that elevated levels of eIF2A, as we observed upon ISR induction, protect BiP mRNA from translational shutdown upon eIF2α phosphorylation.
It was suggested previously that BiP mRNA harbors an IRES in its 5′ UTR, required for BiP expression during continuous heat shock (71, 72). Our data reveal an added level of regulation within the BiP 5′ UTR wherein persistent, noncanonical uORF translation and eIF2A function as a cis-acting regulatory element necessary for privileged BiP expression during stress. The role for eIF2A and noncanonical initiation in the 5′ UTR during ribosome scanning of cellular mRNAs is an emerging property of translational regulation (59), which highlights a new translational mechanism used during the integrated stress response. These findings add to a growing list of alternative initiation events, such as initiation from structured 5′ UTRs with DEAD/DExH-box proteins (73, 74) or eIF2-independent recruitment of Met-tRNAiMet on viral mRNAs by Ligatin and MCT-1 or DENR (75). The observation that other chaperones, also stably expressed during stress, harbor non–AUG-initiated uORFs in the 5′ UTR indicates that uORF expression may be a general mechanism used to regulate the synthesis of proteins necessary during the ISR. Thus, substantial evidence now suggests that uORFs are required for regulation of CDSs and that uORF mutations are linked to a number of diseases [reviewed in (45)].
The notion that some uORF peptides fall outside the detectable range of mass spectrometry analysis because of their short length or selective proteolysis (76) presents an advantage for 3T. The TAP channel readily transports short peptides into the ER for the peptide-loading complex, which is poised to efficiently facilitate loading onto MHC I molecules. Therefore, the detection of translated products from very short uORFs, such as ATF4 uORF1 (three amino acids), is ideal for 3T, because the entirety of the translated uORF serves to generate the mature peptide for MHC I loading and subsequent detection by T cells. Additionally, the uORF products with short half-lives that are efficiently processed serve to enhance the sensitivity of 3T, because proteolysis is required before TAP transport and MHC I loading. Last, the 3T assay is exquisitely sensitive because of the inherent sensitivity of T cells that can detect even a few copies of the peptide–MHC I and trigger the LacZ response (46, 77).
The apparent disconnect between the number of translated uORFs predicted by ribosome profiling (32, 33) and the number of translated uORFs identified by mass spectrometry is likely explained by the challenges encountered by extensive mass spectrometry analysis of HLA-bound peptides over the years (78, 79). These HLA-associated peptide analyses have revealed that mass spectrometry analysis is unlikely to capture all the different peptides presented by MHC I molecules for a variety of reasons. Early analyses predicted that 10,000 to 20,000 different peptides are presented on the cell surface by both MHC I and MHC II molecules, although recent estimates suggest that MHC I alone could display between 30,000 and 120,000 peptides on each cell surface (80). An analogous disconnect is observed between the 42,271 human sORFs (40 to 100 amino acids long) predicted by ribosome profiling (41) and the 1259 alternative proteins detected by mass spectrometry (81). Absolute peptide abundance may limit the recovery of some peptides if they are present at low copy numbers, such as 1 to 1000 molecules per cell (39, 78, 82). Furthermore, assignment of peptides from low-molecular-weight precursors (less than 100 amino acids) by mass spectrometry is limited by the presence of many unidentified ion peaks and the lack of comprehensive low-molecular-weight reference databases (83), which is currently being addressed (81, 83). In addition, standard assignment of recovered HLA-bound peptides is typically performed by alignment with the CDS of the mRNA (78, 79) not to regions outside the annotated open reading frames. Examples of MHC I peptides generated from translation of noncoding RNA—such as 5′ UTRs, introns, and intron-exon junctions (84)—highlight the need to explore regions outside of the CDS for sources of MHC I peptides and other bioactive peptides. The recent reports of non-AUG start codon initiation (58, 59, 67) suggest that standard peptide alignments should also include ORFs initiated with CUG, UUG, and other non-AUG start codons, whether inside or outside of the annotated CDS (81).
Discovery of uORF translation from the 5′ UTR of BiP and prediction of uORF translation in other stress-related chaperones could generate a pool of peptides that serve a variety of biological functions. For example, uORF peptides may shape the immune response as self-antigens during cancer progression and autoimmune disease (80). Indeed, nonmutant tumor antigens generate physiologically relevant immune responses for cancer immunotherapy (85). Therefore, a reexamination of HLA-associated peptide libraries (78) promises to uncover peptides that are generated from translation outside of annotated CDS, such as uORFs initiated with non- AUG start codons. Translated uORFs could also generate bioactive peptides that directly or indirectly regulate expression of the main CDS, as seen with the peptide translated from the uORF in S-adenosylmethionine decarboxylase mRNA (86), or function in signaling pathways, such as Toddler—a short, secreted peptide essential for embryogenesis (42).
3T offers a robust approach for characterization of the numerous predicted translation events in 5′ UTRs and other noncoding RNA (32, 33). These results emphasize the importance of translated regulatory features in 5′ UTRs, which regulate translational control of the downstream CDS, as indicated by a variety of disease-causing 5′ UTR uORF mutations (35). We propose that this phenomenon presents an extracellular signature during the ISR to modulate T cell immune responses (84).
Supplementary Material
Acknowledgments
We are grateful for technical help from K. Banta and S. J. Yang (University of California, Berkeley) and K. Crotty (University of California, San Francisco). We thank M. Elvekrog, E. Costa, M. Lam, and W. Merrick for critical reading of the manuscript and C. Sidrauski, S. Ramundo, H. Tran, N. Ingolia, and J. Weissman for helpful discussions and advice. The human retinal pigment epithelial cell line (RPE-19) was a gift from X. Gong (School of Optometry, University of California, Berkeley). This research was supported by grants from the NIH to N.S. and P.W. P.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/351/6272/aad3867/suppl/DC1
Material and Methods
Figs. S1 to S15
References (87– 98)
REFERENCES AND NOTES
- 1.Walter P, Ron D. The unfolded protein response: From stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. pmid: 22116877. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, et al. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. pmid: 9819435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. pmid: 9930704. [DOI] [PubMed] [Google Scholar]
- 4.Galabru J, Katze MG, Robert N, Hovanessian AG. The binding of double-stranded RNA and adenovirus VAI RNA to the interferon-induced protein kinase. Eur. J. Biochem. 1989;178:581–589. doi: 10.1111/j.1432-1033.1989.tb14485.x. pmid: 2912723. [DOI] [PubMed] [Google Scholar]
- 5.Meurs E, et al. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. pmid: 1695551. [DOI] [PubMed] [Google Scholar]
- 6.Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur. J. Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. pmid: 10504407. [DOI] [PubMed] [Google Scholar]
- 7.Lu L, Han AP, Chen JJ. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 2001;21:7971–7980. doi: 10.1128/MCB.21.23.7971-7980.2001. pmid: 11689689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. pmid: 20094052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hai TW, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: An extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3(12B):2083–2090. doi: 10.1101/gad.3.12b.2083. pmid: 2516827. [DOI] [PubMed] [Google Scholar]
- 10.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. pmid: 15479734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. pmid: 15277680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fornace AJ, Jr, Alamo I, Jr, Hollander MC. DNA damage-inducible transcripts in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. pmid: 3194391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. pmid: 1547942. [DOI] [PubMed] [Google Scholar]
- 14.Han J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. pmid: 23624402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. pmid: 24499181. [DOI] [PubMed] [Google Scholar]
- 16.Joslin G, Hafeez W, Perlmutter DH. Expression of stress proteins in human mononuclear phagocytes. J. Immunol. 1991;147:1614–1620. pmid: 1880418. [PubMed] [Google Scholar]
- 17.Panniers R. Translational control during heat shock. Biochimie. 1994;76:737–747. doi: 10.1016/0300-9084(94)90078-7. pmid: 7893824. [DOI] [PubMed] [Google Scholar]
- 18.Richter K, Haslbeck M, Buchner J. The heat shock response: Life on the verge of death. Mol. Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. pmid: 20965420. [DOI] [PubMed] [Google Scholar]
- 19.Der SD, Lau AS. Involvement of the double-stranded-RNA-dependent kinase PKR in interferon expression and interferon-mediated antiviral activity. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8841–8845. doi: 10.1073/pnas.92.19.8841. pmid: 7568028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilfoy FD, Mason PW. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J. Virol. 2007;81:11148–11158. doi: 10.1128/JVI.00446-07. pmid: 17686861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu L-C, et al. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature. 2004;428:341–345. doi: 10.1038/nature02405. pmid: 15029200. [DOI] [PubMed] [Google Scholar]
- 22.Shiu RP, Pouyssegur J, Pastan I. Glucose depletion accounts for the induction of two transformation-sensitive membrane proteins[ ]in Rous sarcoma virus-transformed chick embryo fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 1977;74:3840–3844. doi: 10.1073/pnas.74.9.3840. pmid: 198809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. pmid: 6417546. [DOI] [PubMed] [Google Scholar]
- 24.Munro S, Pelham HR. An Hsp70-like protein in the ER: Identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. pmid: 3087629. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Lee AS. Stress induction of GRP78/BiP and its role in cancer. Curr. Mol. Med. 2006;6:45–54. doi: 10.2174/156652406775574523. pmid: 16472112. [DOI] [PubMed] [Google Scholar]
- 26.Gorbatyuk MS, Gorbatyuk OS. The molecular chaperone GRP78/BiP as a therapeutic target for neurodegenerative disorders: A mini review. J. Genet. Syndr. Gene Ther. 2013;4:128. doi: 10.4172/2157-7412.1000128. pmid: 23750325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Booth L, et al. GRP78/BiP/HSPA5/Dna K is a universal therapeutic target for human disease. J. Cell. Physiol. 2015;230:1661–1676. doi: 10.1002/jcp.24919. pmid: 25546329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. pmid: 11445534. [DOI] [PubMed] [Google Scholar]
- 29.Zhou J, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. pmid: 26458103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. pmid: 19372376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resch AM, Ogurtsov AY, Rogozin IB, Shabalina SA, Koonin EV. Evolution of alternative and constitutive regions of mammalian 5′UTRs. BMC Genomics. 2009;10:162. doi: 10.1186/1471-2164-10-162. pmid: 19371439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. pmid: 22056041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, et al. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. pmid: 22927429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingolia NT, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Reports. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. pmid: 25159147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cazzola M, Skoda RC. Translational pathophysiology: A novel molecular mechanism of human disease. Blood. 2000;95:3280–3288. pmid: 10828006. [PubMed] [Google Scholar]
- 36.Kondo T, et al. Familial essential thrombocythemia associated with one-base deletion in the 5′-untranslated region of the thrombopoietin gene. Blood. 1998;92:1091–1096. pmid: 9694695. [PubMed] [Google Scholar]
- 37.Wiestner A, Schlemper RJ, van der Maas AP, Skoda RC. An activating splice donor mutation in the thrombopoietin gene causes hereditary thrombocythaemia. Nat. Genet. 1998;18:49–52. doi: 10.1038/ng0198-49. pmid: 9425899. [DOI] [PubMed] [Google Scholar]
- 38.Ghilardi N, Skoda RC. A single-base deletion in the thrombopoietin (TPO) gene causes familial essential thrombocythemia through a mechanism of more efficient translation of TPO mRNA. Blood. 1999;94:1480–1482. pmid: 10484635. [PubMed] [Google Scholar]
- 39.Slavoff SA, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. pmid: 23160002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014;15:193–204. doi: 10.1038/nrg3520. pmid: 24514441. [DOI] [PubMed] [Google Scholar]
- 41.Olexiouk V, et al. sORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2015:gkv1175. doi: 10.1093/nar/gkv1175. pmid: 26527729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pauli A, et al. Toddler: An embryonic signal that promotes cell movement via Apelin receptors. Science. 2014;343:1248636. doi: 10.1126/science.1248636. pmid: 24407481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreev DE, et al. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2015;4:e03971. doi: 10.7554/eLife.03971. pmid: 25621764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife. 2015;4 doi: 10.7554/eLife.05033. pmid: 25719440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Somers J, Pöyry T, Willis AE. A perspective on mammalian upstream open reading frame function. Int. J. Biochem. Cell Biol. 2013;45:1690–1700. doi: 10.1016/j.biocel.2013.04.020. pmid: 23624144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karttunen J, Sanderson S, Shastri N. Detection of rare antigen-presenting cells by the lacZ T-cell activation assay suggests an expression cloning strategy for T-cell antigens. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6020–6024. doi: 10.1073/pnas.89.13.6020. pmid: 1378619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Materials and methods are available as supplementary materials on Science Online
- 48.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. pmid: 12667446. [DOI] [PubMed] [Google Scholar]
- 49.Mielnicki LM, Hughes RG, Chevray PM, Pruitt SC. Mutated Atf4 suppresses c-Ha-ras oncogene transcript levels and cellular transformation in NIH3T3 fibroblasts. Biochem. Biophys. Res. Commun. 1996;228:586–595. doi: 10.1006/bbrc.1996.1702. pmid: 8920955. [DOI] [PubMed] [Google Scholar]
- 50.Paton AW, et al. AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature. 2006;443:548–552. doi: 10.1038/nature05124. pmid: 17024087. [DOI] [PubMed] [Google Scholar]
- 51.Sidrauski C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife. 2013;2:e00498. doi: 10.7554/eLife.00498. pmid: 23741617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sidrauski C, et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife. 2015;4:e07314. doi: 10.7554/eLife.07314. pmid: 25875391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sekine Y, et al. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science. 2015;348:1027–1030. doi: 10.1126/science.aaa6986. pmid: 25858979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhan K, et al. Phosphorylation of eukaryotic initiation factor 2 by heme-regulated inhibitor kinase-related protein kinases in Schizosaccharomyces pombe is important for resistance to environmental stresses. Mol. Cell. Biol. 2002;22:7134–7146. doi: 10.1128/MCB.22.20.7134-7146.2002. pmid: 12242291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abastado JP, Miller PF, Jackson BM, Hinnebusch AG. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol. 1991;11:486–496. doi: 10.1128/mcb.11.1.486. pmid: 1986242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gülow K, Bienert D, Haas IG. BiP is feed-back regulated by control of protein translation efficiency. J. Cell Sci. 2002;115:2443–2452. doi: 10.1242/jcs.115.11.2443. pmid: 12006628. [DOI] [PubMed] [Google Scholar]
- 57.Schwab SR, Shugart JA, Horng T, Malarkannan S, Shastri N. Unanticipated antigens: Translation initiation at CUG with leucine. PLOS Biol. 2004;2:e366. doi: 10.1371/journal.pbio.0020366. pmid: 15510226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starck SR, et al. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science. 2012;336:1719–1723. doi: 10.1126/science.1220270. pmid: 22745432. [DOI] [PubMed] [Google Scholar]
- 59.Liang H, et al. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 2014;19:836–848. doi: 10.1016/j.cmet.2014.03.023. pmid: 24768297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robert F, et al. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2·GTP·Met-tRNAi Met ternary complex availability. Mol. Biol. Cell. 2006;17:4632–4644. doi: 10.1091/mbc.E06-06-0478. pmid: 16928960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. pmid: 10602881. [DOI] [PubMed] [Google Scholar]
- 62.Reits EA, Vos JC, Grommé M, Neefjes J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature. 2000;404:774–778. doi: 10.1038/35008103. pmid: 10783892. [DOI] [PubMed] [Google Scholar]
- 63.Androlewicz MJ, Cresswell P. How selective is the transporter associated with antigen processing? Immunity. 1996;5:1–5. doi: 10.1016/s1074-7613(00)80304-0. pmid: 8758889. [DOI] [PubMed] [Google Scholar]
- 64.Hammer GE, Kanaseki T, Shastri N. The final touches make perfect the peptide-MHC class I repertoire. Immunity. 2007;26:397–406. doi: 10.1016/j.immuni.2007.04.003. pmid: 17459809. [DOI] [PubMed] [Google Scholar]
- 65.Brandvold KR, Morimoto RI. The chemical biology of molecular chaperones—Implications for modulation of proteostasis. J. Mol. Biol. 2015;427:2931–2947. doi: 10.1016/j.jmb.2015.05.010. pmid: 26003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu B, Hunt C, Morimoto R. Structure and expression of the human gene encoding major heat shock protein HSP70. Mol. Cell. Biol. 1985;5:330–341. doi: 10.1128/mcb.5.2.330. pmid: 2858050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, et al. Translational control of the cytosolic stress response by mitochondrial ribosomal protein L18. Nat. Struct. Mol. Biol. 2015;22:404–410. doi: 10.1038/nsmb.3010. pmid: 25866880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zoll WL, Horton LE, Komar AA, Hensold JO, Merrick WC. Characterization of mammalian eIF2A and identification of the yeast homolog. J. Biol. Chem. 2002;277:37079–37087. doi: 10.1074/jbc.M207109200. pmid: 12133843. [DOI] [PubMed] [Google Scholar]
- 69.Kim JH, Park SM, Park JH, Keum SJ, Jang SK. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454–2464. doi: 10.1038/emboj.2011.146. pmid: 21556050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ventoso I, et al. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. pmid: 16391235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. pmid: 1652694. [DOI] [PubMed] [Google Scholar]
- 72.Kim YK, Jang SK. Continuous heat shock enhances translational initiation directed by internal ribosomal entry site. Biochem. Biophys. Res. Commun. 2002;297:224–231. doi: 10.1016/s0006-291x(02)02154-x. pmid: 12237106. [DOI] [PubMed] [Google Scholar]
- 73.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. pmid: 18596238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. pmid: 19109895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skabkin MA, et al. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. pmid: 20713520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oyama M, et al. Analysis of small human proteins reveals the translation of upstream open reading frames of mRNAs. Genome Res. 2004;14(10B):2048–2052. doi: 10.1101/gr.2384604. pmid: 15489325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat. Immunol. 2004;5:524–530. doi: 10.1038/ni1058. pmid: 15048111. [DOI] [PubMed] [Google Scholar]
- 78.Hunt DF, et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. pmid: 1546328. [DOI] [PubMed] [Google Scholar]
- 79.Escobar H, et al. Large scale mass spectrometric profiling of peptides eluted from HLA molecules reveals N-terminal-extended peptide motifs. J. Immunol. 2008;181:4874–4882. doi: 10.4049/jimmunol.181.7.4874. pmid: 18802091. [DOI] [PubMed] [Google Scholar]
- 80.Engelhard VH. The contributions of mass spectrometry to understanding of immune recognition by T lymphocytes. Int. J. Mass Spectrom. 2007;259:32–39. doi: 10.1016/j.ijms.2006.08.009. pmid: 18167512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vanderperre B, et al. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLOS ONE. 2013;8:e70698. doi: 10.1371/journal.pone.0070698. pmid: 23950983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crotzer VL, et al. Immunodominance among EBV-derived epitopes restricted by HLA-B27 does not correlate with epitope abundance in EBV-transformed B-lymphoblastoid cell lines. J. Immunol. 2000;164:6120–6129. doi: 10.4049/jimmunol.164.12.6120. pmid: 10843661. [DOI] [PubMed] [Google Scholar]
- 83.Lubec G, Afjehi-Sadat L. Limitations and pitfalls in protein identification by mass spectrometry. Chem. Rev. 2007;107:3568–3584. doi: 10.1021/cr068213f. pmid: 17645314. [DOI] [PubMed] [Google Scholar]
- 84.Starck SR, Shastri N. Non-conventional sources of peptides presented by MHC class I. Cell. Mol. Life Sci. 2011;68:1471–1479. doi: 10.1007/s00018-011-0655-0. pmid: 21390547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kowalewski DJ, et al. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL) Proc. Natl. Acad. Sci. U.S.A. 2015;112:E166–E175. doi: 10.1073/pnas.1416389112. pmid: 25548167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Law GL, Raney A, Heusner C, Morris DR. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 2001;276:38036–38043. doi: 10.1074/jbc.M105944200. pmid: 11489903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.