SUMMARY
While alternative splicing is known to diversify the functional characteristics of some genes, the extent to which protein isoforms globally contribute to functional complexity on a proteomic scale remains unknown. To address this systematically, we cloned full-length open reading frames of alternatively spliced transcripts for a large number of human genes and used protein-protein interaction profiling to functionally compare hundreds of protein isoform pairs. The majority of isoform pairs share less than 50% of their interactions. In the global context of interactome network maps, alternative isoforms tend to behave like distinct proteins rather than minor variants of each other. Interaction partners specific to alternative isoforms tend to be expressed in a highly tissue-specific manner and belong to distinct functional modules. Our strategy, applicable to other functional characteristics, reveals a widespread expansion of protein interaction capabilities through alternative splicing and suggests that many alternative ‘isoforms’ are functionally divergent (i.e., ‘functional alloforms’).
Graphical Abstract
eTOC Blurb
Alternatively-spliced isoforms of proteins exhibit strikingly different interaction profiles and thus in the context of global interactome networks appear to behave as if encoded by distinct genes, rather than as minor variants of each other.
INTRODUCTION
Humans are more complex than worms or fruit flies, yet they appear to have roughly the same number of protein-coding genes (Blencowe, 2006). One way to address this apparent paradox is to investigate the extent to which functionally different polypeptides can be encoded by individual genes in various species.
Eukaryotic genes can encode multiple protein ‘forms’ via alternative transcription, splicing, 3′ end formation, translation, and post-translational modification. Alternative splicing produces transcript ‘isoforms’ for most human genes (Pan et al., 2008; Wang et al., 2008), providing functional diversity at the level of enzymatic activities, subcellular localizations, as well as protein-protein, protein-DNA, and protein-ligand physical interactions (Kelemen et al., 2013). An isoform may exhibit dominant negative effects over other isoforms encoded by the same gene, be up- or down-regulated instead of constitutively active, or even have opposing cellular functions. For example, two isoforms encoded by the BCL2L1 gene have opposite functions in apoptosis, the longer isoform inhibits the process while the shorter one promotes it (Schwerk and Schulze-Osthoff, 2005). In another example, ubiquitous alternative splicing of D. melanogaster Dscam1 generates thousands of different polypeptides, each with different binding specificities to enable self-recognition of neurons (Wojtowicz et al., 2007). Altogether, several hundred human genes are known to encode alternatively spliced isoforms with distinct functional characteristics (Kelemen et al., 2013).
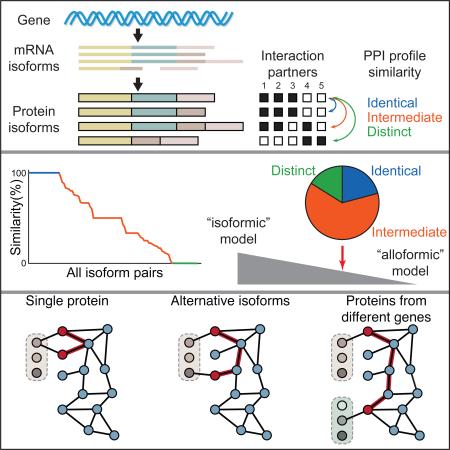
What remains unclear is how widespread this phenomenon is at the scale of the whole proteome, which is of much higher complexity than originally anticipated (Tran et al., 2011). As many as 100,000 distinct isoform transcripts could be produced from the ~20,000 human protein-coding genes (Pan et al., 2008), collectively leading to perhaps over a million distinct polypeptides obtained by post-translational modification of products of all possible transcript isoforms (Smith and Kelleher, 2013). How such proteomic complexity relates to global cellular processes is essentially unknown. To what extent are pairs of isoforms encoded by a common gene functionally different from each other? How widespread is isoform-specific functional diversity in any given species? How might such functional diversity vary between species? What role does this diversity play in evolution? Altogether, the central challenge is to determine the extent to which two distinct, yet non-mutually exclusive, models might apply: 1) alternative isoforms tend to mediate similar functions, i.e., they mostly behave as ‘functional isoforms’; and 2) alternative isoforms tend to display distinct functions, i.e., they should mostly be considered as ‘functional alloforms’ (Figure 1A).
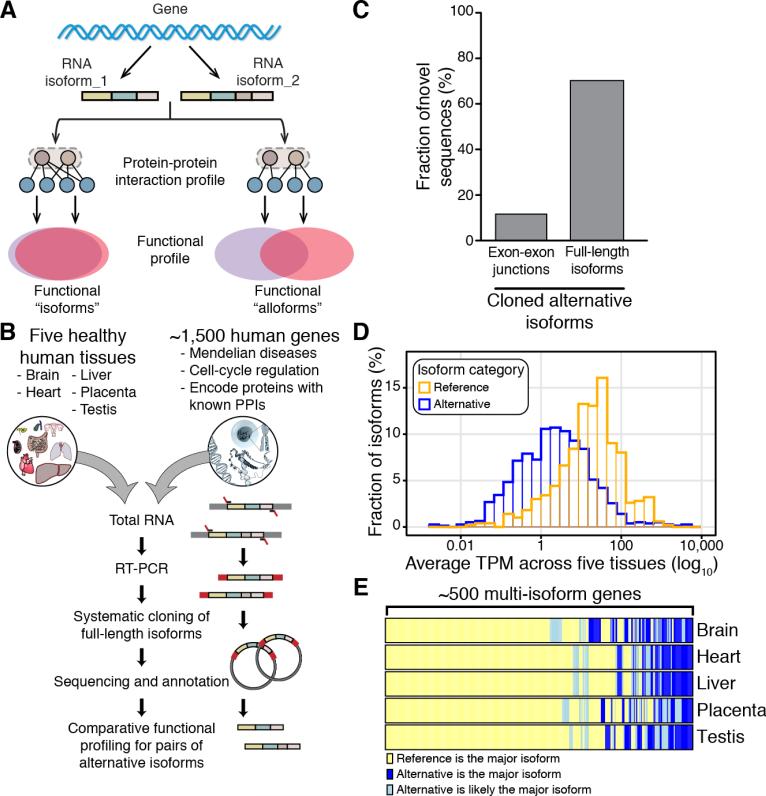
Figure 1. Cloning of Novel Alternatively Spliced Isoforms Using ORF-Seq.
(A) Comparative functional profiling of alternative isoforms.
(B) Pipeline for systematic cloning of alternatively spliced ORFs, or “altORFs”.
(C) Fraction of novel exon-exon junctions vs. novel full-length isoforms among cloned altORFs.
(D) Distribution of endogenous transcript abundance for reference and alternatively spliced isoform clones.
(E) Heatmap distinguishing cases where the reference isoform (yellow) or an alternatively spliced isoform (blue and light blue) was the major isoform detected. See also Figure S1 and Table S1.
So far, investigations into the role of alternative splicing have focused on the functions alternative protein isoforms can or cannot perform, relative to their so-called ‘reference’ counterpart (Buljan et al., 2012; Ellis et al., 2012). To begin addressing the questions outlined above in a systematic and unbiased manner, large-scale functional profiling approaches are needed to quantify the extent to which all isoforms encoded by large numbers of genes are functionally similar or different from each other, taking all pairwise combinations of isoforms encoded by the same gene into consideration. This, in turn, requires novel methodologies to identify, clone, and exogenously express full-length open reading frames (ORFs) for all isoforms across a wide range of genes.
Contemporary attempts at systematically discovering alternatively spliced isoforms genome-wide have been based on next-generation sequencing (NGS) methods. For example, RNA-Seq provides relatively deep sampling (Pan et al., 2008; Wang et al., 2008). However, the short length of RNA-Seq reads has hampered the discovery of contiguous exon connectivity for full-length alternatively spliced isoforms. Full-length sequencing of single cDNA molecules, or “Iso-seq” (Eid et al., 2009), has proven successful in generating improved models of full-length transcript isoforms (Sharon et al., 2013). Another strategy captures co-association of distant alternatively spliced exons by limiting the number of RNA molecules in the pools used to generate sequencing libraries (Tilgner et al., 2015). However, none of the above strategies provide the large-scale physical clone collections needed to systematically express and study the function of alternative isoforms.
Here, we apply a new strategy, “ORF-Seq”, to discover, characterize, exogenously express, and functionally investigate large numbers of alternatively spliced full-length ORFs. We have applied this strategy to the study of binary protein-protein interactions (PPIs) and identified widespread interaction differences due to alternative splicing (Figure 1A). Alternatively spliced protein isoforms tend to behave like completely distinct genes in interactome networks rather than minor variants of each other. Thus, a sizable proportion of alternative isoforms in the human proteome are “functional alloforms” (Figure 1A).
RESULTS
Comparative Functional Profiling of Alternative Isoforms
To characterize functional diversity between pairs of alternatively spliced isoforms encoded by common genes, or to simplify: “alternative isoforms”, across the whole genome, we designed the following strategy (Figure 1B). First, full-length ORFs corresponding to known and novel isoforms are amplified by reverse transcription followed by PCR (RT-PCR) using gene-specific primers. Pools of resulting RT-PCR products are Gateway-cloned (Walhout et al., 2000) and individual ORFs are sequenced using an NGS-based deep-well approach (Salehi-Ashtiani et al., 2008). Second, Gateway-cloned full-length isoform ORFs are transferred into various expression vectors to allow systematic functional analyses such as binary protein-protein and protein-DNA interaction assays or measurement of enzymatic activities. Large numbers of pairs of alternative isoforms can thus be functionally profiled to evaluate the extent to which their activities might be identical (“functional isoforms”), similar, or completely distinct from each other (“functional alloforms”).
Systematic Discovery of Full-Length Alternatively Spliced ORFs Using ORF-Seq
We concentrated on ~10% of all human protein-coding genes, including genes implicated in Mendelian diseases, involved in cell-cycle regulation, or encoding proteins with well-characterized PPIs (Venkatesan et al., 2009) (Figure 1B), while making sure that protein families were roughly equally represented (Figure S1A).
We carried out targeted isoform cloning of 1,492 human genes (Table S1A) for which pairs of PCR primers, one at the start codon and the other at the stop codon, had been previously validated in the context of our human “ORFeome” cloning pipeline (Lamesch et al., 2007). The ORFs in our human ORFeome collection (hORFeome) were initially obtained by PCR amplification of full-length cDNAs with GenBank accessions and RefSeq annotations from the Mammalian Gene Collection (Temple et al., 2009) and were considered to be “reference ORFs”. Our gene-specific reference ORF primers (Table S1A) were used to amplify ORF sequences from pooled reverse transcribed RNA obtained from brain, heart, liver, placenta, and testis (Figure 1B; See Experimental Procedures).
We successfully recovered at least one unique ORF clone for ~85% of the tested genes (1,266 out of 1,492), leading to the identification of 1,423 different ORF clones, of which 917 exhibited sequence differences relative to their corresponding reference ORF due to alternative splicing events and thus were defined as alternatively spliced ORFs or “altORFs”. Our human isoform ORF collection used for all subsequent analyses (Table S1B) contains one reference ORF along with one or more unique altORF(s) for a total of 1,423 isoforms (506 reference ORFs and 917 altORFs) for 506 genes (Figures S1B and S1C). GO-slim term analysis showed no significant differences between the genes with one or multiple cloned alternative isoforms (Figures S1D-S1F).
To structurally annotate novel alternatively spliced isoforms, the sequences of our 917 altORFs were compared to transcripts and coding region sequences from seven publicly available databases (Aceview, CCDS, Gencode, MGC, Human ORFeome, RefSeq, and UCSC). The majority (89%) of the individual exon-exon junctions identified within altORFs correspond to junctions already curated in at least one of the databases, suggesting that most clones in our collection are derived from genuine splicing events (Figure 1C). More importantly, ~70% of altORFs represent novel exon-exon full-length cis-connectivities and thus potentially novel polypeptides (Figure 1C; Table S1B).
A substantial proportion of splicing events are known to be associated with tissue-specific expression patterns (Barbosa-Morais et al., 2012; Buljan et al., 2012; Ellis et al., 2012; Merkin et al., 2012). Although RNA-seq does not provide unambiguous counts of full-length transcripts, expression levels of alternative isoforms can be estimated. To compare the abundance of all 506 reference and 917 alternatively spliced transcripts in the five human tissues used here, we applied RNA-Seq Expectation Maximization (RSEM) to estimate abundance in transcripts per million (TPM) (Li and Dewey, 2011) (Table S1C). On average, the abundance of the reference transcripts (average TPM = 73.2, median TPM = 15.1) was higher than that of the alternatively spliced transcripts (average TPM = 28.2, median TPM = 2.4) (Figure 1D), likely explaining why these particular forms were enriched in previous collections. Despite this, we found for 46% of genes (235/506), an alternative transcript is more abundant than its cognate reference transcript in at least one tissue (Figure 1E). Thus, depending on the tissue or cell-type, alternatively spliced transcripts can be the predominant product of a gene, thus making the notion of a reference isoform somewhat arbitrary.
Interaction Profiling of Alternative Isoforms
Because PPIs are inherent to most cellular processes, we initiated our functional studies by comparing interaction profiles of isoform pairs for 1,035 isoforms consisting of 398 reference ORFs and 637 altORFs using a stringent binary interaction platform validated by an empirical framework (Dreze et al., 2010; Venkatesan et al., 2009) (Figure 2A; Table S2A).
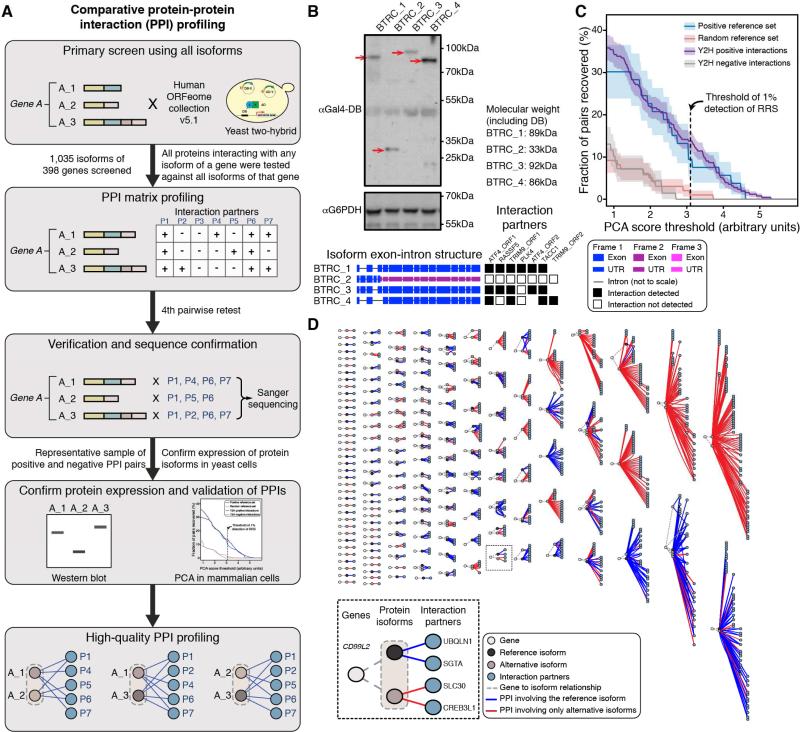
Figure 2. Comprehensive Binary PPI Mapping for Protein Isoforms.
(A) Comparative binary PPI profiling pipeline.
(B) Western blot analysis showing comparable expression of four DB-BTRC alternative protein isoforms using an anti-Gal4-DB antibody (see red arrows). Interaction profiles are shown at bottom right with black and white boxes representing positive and negative interactions, respectively.
(C) Validation of protein isoform interaction dataset. Shown is the fraction of pairs recovered by an orthogonal protein complementation assay (PCA) relative to increasing assay stringency. Shading indicates the standard error of the fraction.
(D) Protein isoform interactome subnetworks. Each subnetwork displays relationships between genes, isoforms, and interaction partners with interactions mediated by reference protein isoforms shown in blue, and those mediated by novel alternatively spliced protein isoforms shown in red. See also Figure S2 and Table S2.
First we performed yeast two-hybrid (Y2H) screens in which all protein isoforms, fused to the Gal4 DNA binding domain (DB), were tested against proteins encoded by the hORFeome v5.1 collection of ~15,000 ORF clones fused to the Gal4 activation domain (AD) (Dreze et al., 2010; Rolland et al., 2014; Rual et al., 2005). Following first-pass screening, each protein isoform was pairwise tested for interaction with the candidate partners identified not only for itself but also for all first-pass partners of all other protein isoforms encoded by the same gene, thus minimizing biases due to incomplete sampling sensitivity (Venkatesan et al., 2009). To generate a final dataset of verified Y2H pairs, pairs showing a positive result in at least two out of the three pairwise tests were subjected to a fourth pairwise retest, and PCR products amplified from the final positive pairs were sequenced to confirm the identity of clones encoding each interacting protein (Figure 2A; Table S2B). Western blots were performed for all protein isoforms of a subset of randomly picked genes, demonstrating comparable heterologous protein expression of all isoforms of the same gene tested by Y2H (Figures 2B and S2A-H). Finally, to validate the overall quality of the PPI dataset of human protein pairs identified by Y2H, we selected a representative sample of the isoform-partner interacting and non-interacting pairs and subjected them to orthogonal validation in human HEK293T cells using a protein complementation assay (PCA) (Dreze et al., 2010; Rolland et al., 2014) (Figure 2C; Table S2C). The isoform-partner positive pairs were recovered at a rate similar to that seen for pairs from a well-described positive reference set (PRS) (Venkatesan et al., 2009), while isoform-partner negative pairs validated at a rate similar to that seen for pairs from a random reference set (RRS) (Figure 2C; Tables S2C and S2D).
In total, we obtained high-quality PPI profiles for 366 protein isoforms encoded by 161 genes (Figure 2D; Table S2B). While 118 isoforms returned no binary PPIs, 248 isoforms had one or more interactions for a total of 1,043 binary PPIs with 381 proteins. Less than one third of these PPIs (323/1043) involve reference isoforms (Figure S2I). When compared to a network mapped with a single isoform per gene, including PPIs detected by novel isoforms led to a 3.2-fold increase in the number of interactions (Figure S2I). This strongly suggests that sequence differences between alternative isoforms underlie substantial functional differences.
Isoform-Specific Regions Associated with Isoform-Specific PPIs
To identify isoform-specific regions (ISRs) that might mediate isoform-specific interactions, we searched for contiguous sequence regions of at least 40 amino acids, slightly shorter than the average human exon length, that are present in only one, a subset, or all isoforms of the genes tested here. This method allowed identification of any isoform-specific sequence region, enabling us to go beyond the analysis of simple exon inclusion or exclusion events to detect more complex splicing patterns.
We examined the patterns of correspondence between ISRs and isoform-specific interactions for all groups of isoforms, including cases of two isoforms per gene (n = 495) and three isoforms per gene (n = 266) (see online Supplemental Material), and distinguished four interaction classes according to their effects on PPIs: promoting, inhibiting, promoting or inhibiting, and complex (Figures 3A and 3B; Table S3A). “Promoting” occurs when the partner interacts exclusively with isoforms that contain a given ISR. “Inhibiting” occurs when the partner interacts with only those isoforms lacking a given ISR. “Promoting or inhibiting” occurs when the partner's interaction is positively correlated with both the presence of an ISR and the absence of a different ISR. Finally, “complex” represents cases where there is no perfectly associated single ISR and may represent scenarios where an interaction is regulated by exon-exon junctions or by combinations of alternatively spliced regions. The many cases of “complex” associations (n = 133, 27% of the set of two or more isoforms) suggest that PPIs may be modulated by the combined actions of multiple ISRs. Hence, studies on full-length protein isoforms coupled with unbiased screens for all possible biophysical isoform-specific interactions are necessary to fully understand how differences in protein sequences affect interactions and functions.
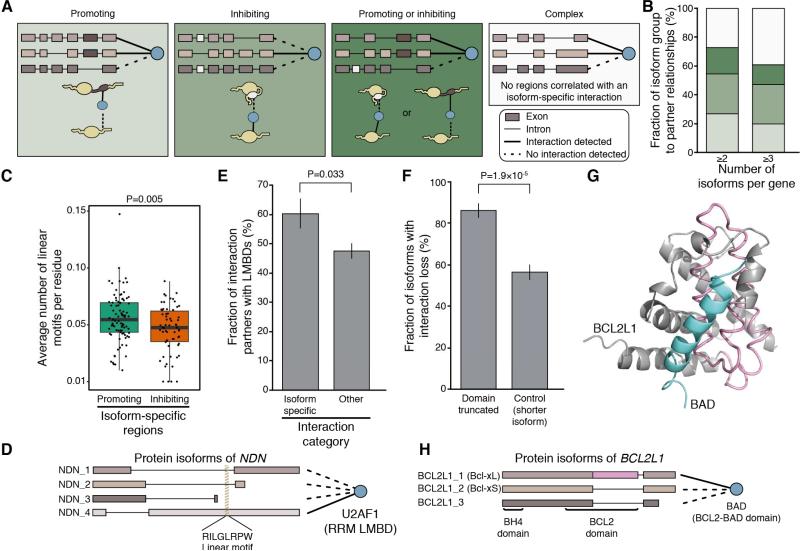
Figure 3. Contiguous Sequence Regions Associated with Isoform-Specific PPIs.
(A) Four categories of isoform specific regions (ISRs) according to their effects on interactions (promoting, inhibiting, promoting or inhibiting, or complex).
(B) Fraction of interaction partners classified in each of the four categories for all genes encoding at least two (left) or three (right) isoforms.
(C) Box plot showing the average number of linear motifs per residue in promoting and inhibiting ISRs. P values from two-sided Wilcoxon rank test.
(D) Schematic diagram illustrating interaction modulation potentially explained by differential splicing of linear motifs within exons.
(E) Histogram showing the fraction of interaction partners that contain linear motif binding domains (LMBDs) and exhibit isoform-specific interactions associated with promoting regions or not. P values from two-sided Fisher's exact test; error bars represent the standard error of the fraction, estimated using bootstrapping with 100 resamplings.
(F) Histogram showing the fraction of isoforms with interaction loss where a predicted interaction domain was disrupted by alternative splicing. P values from two-sided Fisher's exact test; error bars represent the standard error of the fraction, estimated using bootstrapping with 100 resamplings.
(G) Three dimensional structure of BCL2-xL (grey; PDB code 1g5j) in complex with BAD (blue). The interaction interface is disrupted in the BCL2-xS isoform with the 3′-end of the first exon spliced out (pink). See Figure S3 for more structure examples.
(H) Schematic diagram illustrating the interaction modulation of alternatively spliced isoforms of BCL2L1 potentially explained by differential splicing of BCL2 domain. See also Figure S3 and Table S3.
Isoform-Specific PPIs Mediated by Linear Motifs
Linear motifs are short contiguous stretches of amino acids that interact with linear motif binding domains (LMBDs) (Dinkel et al., 2012; Neduva and Russell, 2006). Therefore, ISRs that contain linear motifs and are excluded or included by alternative splicing may modulate PPIs. Because linear motifs are short, many non-functional motifs can occur throughout the proteome by chance; hence, they are typically difficult to identify. Despite this challenge, a high density of linear motif matches can indicate the presence of functional linear motifs. We scanned ISRs for linear motifs from the Eukaryotic Linear Motif (ELM) database, excluding extremely short or frequent motifs. Using our isoform PPI dataset, we found that the density of linear motifs, i.e., the number of motifs per number of residues, was greater in interaction-promoting ISRs than in interaction-inhibiting ISRs (two-sided Wilcoxon rank sum test, P = 0.005, Figures 3C and S3A; Table S3B), suggesting that some isoform-specific interactions are mediated by the presence of linear motifs.
LMBDs interact with linear motifs with remarkable selectivity; therefore, interactions should tend to occur exclusively between partners containing an LMBD and the subset of isoforms that contain its cognate linear motif, or vice-versa. For example, of the four protein isoforms from the NDN gene, only one interacts with U2AF1, which contains the RRM LMBD, and this is the only isoform to contain the sequence ‘RILGLRPW’ which matches the RRM-interacting ELM motif x[I/L/V/M]LGxxPx (Rideau et al., 2006) (Figure 3D). Globally, we found that isoform-specific interaction partners associated with interaction promoting regions are more likely to contain an LMBD than are other interaction partners (two-sided Fisher's exact test, P = 0.033, Figures 3E and S3B; Table S3C).
Splicing-Mediated Disruption of Interaction Domains
Binary PPIs are frequently governed by interactions between globular domains, and many domain-domain interactions (DDIs) have been predicted based on three-dimensional structures of protein complexes or other computational approaches (Finn et al., 2005; Mosca et al., 2014). The alternative inclusion or exclusion of domains participating in DDIs could modulate PPIs. To investigate the link between splicing-mediated domain disruptions and loss of interactions involving such domains, we searched our dataset for cases where the interacting partner contains a domain predicted to interact with a domain in one or more isoforms of the bait protein. We then considered each pair of isoforms of the gene where the partner protein interacts with only one of the two isoforms. From these isoform pairs, we derived two sets: (1) cases in which one isoform lacks at least 50 amino acids (chosen based on the average size of domains (Jones et al., 1998)) of the predicted interaction domain relative to the other isoform, and (2), cases where one isoform is shorter than the other by 50 or more residues, regardless of domain content. In 87% of cases (52/60) with the ≥50 residue domain deletion/truncation, the loss or truncation is associated with the concomitant loss of the interaction (Figures 3F and S3C; Table S3D). By comparison, one isoform simply being shorter than the other by ≥50 residues, irrespective of domain content, is associated with the loss of interaction in only 57% of cases (100/176; two-sided Fisher's exact test, P = 1.9 × 10−5). This suggests that some interaction differences between isoforms of the same gene may be explained by alternative splicing of protein domains associated with DDIs. For example, partial truncation of the BCL2 domain in a BCL2L1 protein isoform results in the loss of an interaction with the protein BAD (Figures 3G and 3H). The relevant ISR that interacts with the protein partner BAD is present in the longer isoform (Bcl-xL) but missing in the shorter isoform (Bcl-xS) (Figures 3G and 3H). In this well-studied example, the inclusion of this ISR makes Bcl-xL pro-survival, and exclusion of it makes Bcl-xS pro-apoptotic (Schwerk and Schulze-Osthoff, 2005), demonstrating the importance of alternative splicing in regulating gene function. Finally, we mapped 55 unique interactions between proteins of two genes (without considering different isoforms) onto three-dimensional structures to define the interaction interface. Using a local pairwise alignment between the structure sequence and the corresponding isoform, we mapped isoform sequences onto the structures for a total of 125 interactions involving 55 unique reference isoforms. The vast majority of isoforms that are able to interact retain the interface, while only half of the interactions are maintained when interface residues are lost (Figure S3D). See Figure S3E for more examples of the structural basis of alternative-splicing-mediated interaction modulations.
These results provide unbiased evidence at a large scale that gene function(s) can be mediated through alternative splicing by alternative inclusion and/or exclusion of regions that contain interacting linear motifs or interaction domains.
Widespread Expansion of Protein Interaction Capabilities
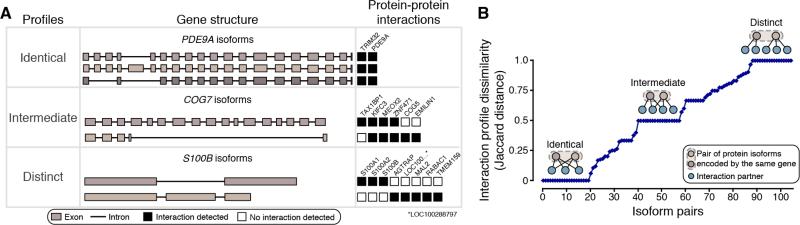
To investigate the extent to which any two isoforms encoded by the same gene mediate interactions with different partners, we calculated the dissimilarity of their interaction profiles (Jaccard distance) by comparing all possible pairs of isoforms and calculating the fraction of total interacting partners that are specific to an isoform. We restricted our analysis to pairs of isoforms where both exhibit at least one interaction and where the interactions were verified as either positive or negative for each of the two isoforms (n = 105, Table S4). Only 21% of isoform pairs exhibit identical interaction profiles, i.e., a Jaccard distance of 0. For example, all protein isoforms encoded by the PDE9A gene exhibit physical interaction with the exact same protein partners, a ‘homodimeric’ interaction with PDE9A (the form corresponding to the reference ORF) and TRIM32 (Figure 4A). Strikingly, 16% of pairs exhibit completely distinct PPI profiles, yielding the maximal Jaccard distance of 1. For example, one isoform encoded by the S100B gene interacts with three partners while the other isoform interacts with a distinct set of five other partners. For the majority (63%) of isoform pairs, the situation is intermediate with some specific interactions, referred to below as “isoform-specific interactions”, and others that are shared between isoform pair members. For example, the two isoforms encoded by the COG7 gene share three interaction partners and, in addition, exhibit interactions with one and two specific partners, each. Collectively, comparative interactome profiles differ by 50% or more for about half of the tested isoform pairs (Figures 4B and S4). This striking result suggests a widespread expansion of protein interaction capabilities by alternative splicing.
Figure 4. Comparison of Interaction Profiles for Alternative Isoforms.
(A) Representative examples of alternative isoforms displaying identical, intermediate, or distinct interaction profiles.
(B) Distribution of interaction profile differences between all possible pairs of alternative isoforms as measured by Jaccard distance. A Jaccard distance of 0 means that both isoforms share all interaction partners, whereas a distance of 1 means the isoforms have no shared partners. Isoforms for which no interactions were detected were omitted from the graph. See also Figure S4 and Table S4.
Interactome Network Analysis of Isoform-Specific Interaction Partners
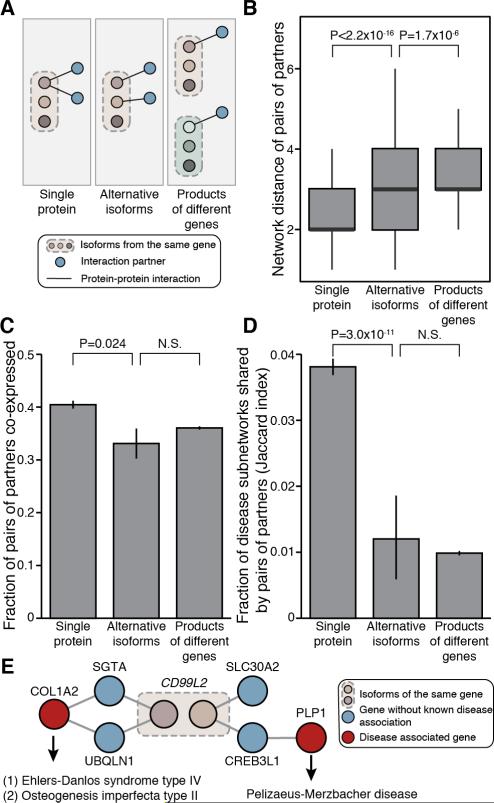
To better understand the functional divergence between alternative isoforms, we analyzed their protein partners in the context of global interactome network maps (Figure 5A). It is well documented that the interaction partners of a single protein and those of proteins encoded by separate genes have strikingly different properties in the context of interactome networks. For example, the partners of a single protein tend to be “closer” to each other than those of proteins encoded by separate genes, as measured by the minimal number of links between them (Vidal et al., 2011). We reasoned that the global functional diversity mediated by alternative splicing could be approximated by comparing the partners of alternative isoforms encoded by the same gene to those of single proteins and of proteins encoded by separate genes.
Figure 5. Functional Differences between Isoforms Revealed by Properties of Isoform Interaction Partners.
(A) Schematic showing two different partners (blue nodes) interacting with either a single protein (left panel), alternative isoforms encoded by a common gene (middle panel), or the protein products of different genes (right panel).
(B) Average network distance of pairs of partners interacting with a single protein, alternative isoforms, or the protein products of different genes. Error bars represent standard errors of the mean.
(C) Fraction of pairs of partners interacting with a single protein, alternative isoforms, or the protein products of different genes, and showing positively correlated mRNA levels across 16 human tissues (Illumina Human Body Map 2.0). Error bars represent standard errors of the mean.
(D) Mean Jaccard index of disease subnetwork co-occurrence of pairs of partners interacting with a single protein, alternative isoforms, or the protein products of different genes. Error bars represent standard errors of the mean.
(E) Example of alternative isoforms interacting exclusively with proteins from different disease subnetworks. Pink nodes represent two protein isoforms encoded by the CD99L2 gene. Blue nodes represent the respective isoform interaction partners. Red nodes represent two different proteins encoded by genes associated with distinct diseases. See also Figure S5.
First, we used a recent systematic, unbiased binary PPI dataset referred to as HI-II-14 (Rolland et al., 2014) to examine the network properties of interacting partners. In this context, the difference was striking between partners that interact with a single protein and those that interact with proteins encoded by separate genes (Figure 5B). Partners that interact with alternative isoforms (n = 256) tend to be further apart than partners that interact with any single protein (n = 4,655; two-sided Wilcoxon rank sum test, P < 2.2 × 10−16, Figures 5B and S5A) and only marginally closer to each other than partners that interact with proteins encoded by separate genes (n = 45,560) (two-sided Wilcoxon rank sum test, P = 1.7 × 10−6, Figures 5B and S5A).
Next, we examined co-expression relationships between interaction partners using the Illumina Body Map 2.0 dataset across 16 human tissues to quantify mRNA expression levels, followed by calculation of the Pearson correlation coefficient between all genes. As expected, the difference between pairs of partners interacting with a single protein and partners interacting with proteins encoded by separate genes was highly significant (two-sided Fisher's exact test, P = 7.7 × 10−9). We found that pairs of partners that interact with alternative isoforms (n = 248) are significantly less likely to be co-expressed than those that interact with a single protein (n = 4,694; two-sided Fisher's exact test, P = 0.024, Figures 5C and S5B). Furthermore, no significant difference was observed in the fraction of co-expressed pairs between partners interacting with alternative isoforms and partners interacting with proteins encoded by separate genes (n = 69,220).
Finally, we examined the extent to which pairs of interaction partners belong to common disease subnetworks, as defined by the set of disease-associated genes from GeneCards (Safran et al., 2010) and their first-degree neighbors in the human interactome (Rolland et al., 2014). We measured the similarity (Jaccard index) of the disease-association profiles between any two partner proteins. We found that partners interacting with alternative isoforms (n = 125) were less likely to be associated with the same diseases or interact with proteins associated with the same diseases than partners interacting with any given protein (n = 3,873; two-sided Wilcoxon rank sum test, P = 3.0 × 10−11, Figures 5D and S5C). Importantly, there was no significant difference in disease association between interaction partners of alternative isoforms and those of proteins encoded by separate genes (n = 28,081; two-sided Wilcoxon rank sum test, P = 0.47, Figures 5D and S5C). For example, one protein isoform encoded by CD99L2 was connected to the COL1A2 disease subnetwork, which is associated with connective tissue disorders such as Ehlers-Danlos syndrome (Raff et al., 2000) and osteogenesis imperfecta (Pollitt et al., 2006). The other isoform from CD99L2 was connected to the PLP1 disease subnetwork associated with Pelizaeus-Merzbacher disease (Inoue, 2005) (Figure 5E).
The observed isoform-specific differences demonstrate that interaction differences between isoforms are not random, but rather reflect distinct functions of individual isoforms. Furthermore, knowledge of isoform specificity can provide useful information about the interaction partners themselves, with important consequences for applications such as inferring new disease-gene associations or identifying potential drug targets.
Patterns of Alternative Splicing-Mediated Interaction Profile Differences
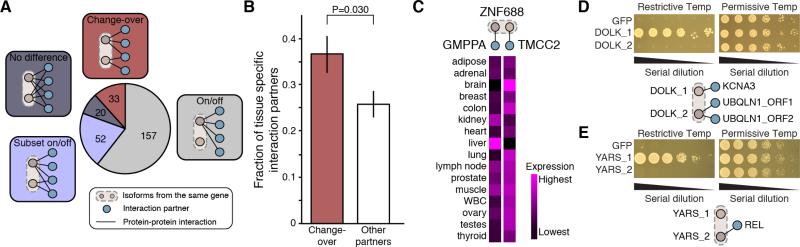
To examine the consequences of alternative splicing-mediated differences in the interaction profiles of alternative isoform pairs, we first performed a pairwise comparison of isoforms and classified isoform pairs into four groups (Figure 6A; Table S4): i) no difference, where the pair of isoforms shared the same set of interaction partners; ii) on/off, where one of the two protein isoforms possessed no interactions; iii) subset on/off, where one protein isoform interacts with a subset of interaction partners of the other isoform but had no unique interaction partners; and iv) change-over, where each protein isoform possessed one or more unique interactions (with or without any shared interactions; the set of isoform pairs with a Jaccard distance of 1 (Table S4) exhibited the highest degree of change-over). For protein isoforms that are on/off or subset on/off, alternative splicing can regulate protein function simply by inhibiting or promoting some or all PPIs through alternative inclusion of exons (Buljan et al., 2012; Ellis et al., 2012). In contrast, a change-over pattern, with one or more unique interaction partners for each isoform, suggests that each isoform may have a distinct function, more similar to the relationship expected between protein products from different genes. By interacting with different partners, change-over isoforms can potentially be involved in different network modules or cellular processes or be associated with different diseases, as shown in Figures 5D and 5E. Interestingly, the ISRs from the “change-over” isoform pairs had the greatest predicted disorder content while the ISRs from the “no difference” isoform pairs had the lowest (Figures S6A). This finding is consistent with previous observations that intrinsically disordered regions tend to be involved in protein-protein interactions (Buljan et al., 2012; Ellis et al., 2012; Haynes and Iakoucheva, 2006) and are frequently alternatively spliced (Romero et al., 2006). While protein partners of different isoforms tended to be expressed in different tissues as compared to partners of the same isoform (Figure 5C), we also observed that partners responsible for the change-over classification of a pair of isoforms (n = 148) were expressed in an even more highly tissue-specific manner than other partners (n = 241) (range of expression levels across 16 tissues, two-sided Fisher's exact test, P = 0.030; Figure 6B and S6B). Such differences in tissue-localized expression of interaction partners were observed despite similar average expression levels overall (two-sided Wilcoxon rank sum test, P = 0.99). Figure 6C shows an example of two “change-over” protein partners with vastly different expression profiles across 16 tissues. These results indicate that change-over isoform interactions may play an important role in tissue specialization and that change-over interaction differences may allow different isoforms of a gene to adopt distinct functions in distinct tissues.
Figure 6. Protein Isoforms with Change-Over Interaction Profiles Exhibit Different Tissue-Specificities.
(A) Distribution of four types of PPI differences exhibited by protein isoform pairs: change-over, each protein isoform has at least one exclusive interaction partner; on/off, one protein isoform lacks all interactions relative to another protein isoform from the same gene; subset on/off, one protein isoform lacks a subset of interactions; no difference, no differences observed in interaction partners for protein isoform pairs.
(B) Comparison of the fraction of tissue-specific interaction partners, as estimated from the range of normalized log2 RNA-Seq read counts from 16 human tissues (Illumina Human Body Map 2.0) for change-over interaction partners and other partners. P value from Fisher's exact test; error bars represent the standard error of the fraction, estimated using bootstrapping with 100 resamplings.
(C) Example of a change-over isoform pair from the ZNF688 gene where each isoform interacts with a different protein whose mRNA is detected in very distinct sets of tissues.
(D, E) Yeast complementation assays. Pictures on top show the growth status of yeast thermosensitive mutants transformed with different isoforms of the DOLK (panel D) or YARS (panel E) genes. GFP is used as negative control. Diagrams at the bottom show interactions and complementation mediated by the two isoforms. See also Figure S6 and Table S4.
To further investigate functional differences between isoforms exhibiting different PPI profiles, we exploited a cross-species complementation assay to measure the ability of each isoform to rescue phenotypic defects of a loss-of-function mutation in a cognate yeast gene. We found eight cases of described human-to-yeast complementation relationships (Kachroo et al., 2015) among 138 genes with at least two isoforms showing different interaction profiles, altogether corresponding to 19 distinct isoforms. Yeast-based functional complementation assays were performed for these 19 isoforms. Isoforms of two genes, DOLK and YARS, showed differential abilities to rescue the corresponding yeast temperature sensitive mutants, strongly suggesting the appearance of a genuine functional divergence between these isoforms during evolution (Figures 6D and 6E).
CONCLUDING REMARKS
Transcriptomic analyses have highlighted the tremendous potential proteome diversity generated by alternative splicing (Barbosa-Morais et al., 2012; Pan et al., 2008; Wang et al., 2008). However, the functional divergence between alternatively spliced protein isoforms remained unclear on a proteomic scale. Although systematic functional studies of protein isoforms have been described for selected groups of genes (Corominas et al., 2014), most recorded functional annotations and protein interactions are at gene-level resolution.
Systematic cloning of native splice isoforms and proteome-scale mapping of isoform interactions has enabled us to capture a wide range of interaction profile differences between protein isoforms, providing deeper insight into the global influence of alternative splicing on the interactome. We have established that PPI network expansion is a major consequence of alternative splicing, and that different isoforms from the same gene can give rise to different local features within interactome networks. We found differences in interaction profiles for a majority of isoform pairs (Figure 4B), suggesting widespread functional differences between isoforms encoded by the same gene. Our analyses of the functional properties of isoform interaction partners further demonstrate a continuum of functional divergence between isoforms, up to the extreme degree where two different isoforms encoded by the same gene appear to functionally behave like two different proteins (Figure 5). This in turn strongly suggests that the “functional alloform” model of alternative isoforms should not be excluded and in fact might more accurately reflect the reality of the whole human proteome than the “functional isoform” model (Figure 1A).
Global functional divergence between isoforms may explain how organisms like humans, with vast splicing diversity, can generate greater network complexity and thus potentially greater phenotypic complexity from only about 20,000 protein-coding genes. This functional divergence also suggests that each protein isoform needs to be studied individually to understand its unique roles, including contributions to disease pathogenesis or potential as a drug target. The mapping of isoform-specific protein interactions can also reveal valuable information about isoforms of the same gene and their interaction partners. Significant functional divergence between isoform pairs as shown in Figure 5E may not be unusual. We found that a sizeable fraction of isoform pairs interact with distinct groups of proteins (Figures 4B and 6A), exhibiting an interaction profile pattern we have termed “change-over”. Each isoform in these “change-over” isoform pairs possesses unique interaction partners that show localized expression in specific tissues (Figures 6B and 6C) and tend to be members of distinct disease modules (Figure 5E). These findings suggest that the change-over pattern of splicing-mediated PPI networks is a key driver of functional divergence between isoforms and may contribute to functional specialization of tissues.
We were able to identify alternatively spliced regions containing potential interaction determinants that “promote” or “inhibit” interactions (Figures 3A and 3B). Many “interaction-promoting” regions contain linear motifs, and isoform-specific interaction partners contain LMBDs (Figures 3C-3E), consistent with previous findings that tissue-specific exons often contain linear motifs (Buljan et al., 2012; Ellis et al., 2012; Merkin et al., 2012). Similarly, interaction-promoting regions tend to contain predicted interaction domains based on known or predicted domain-domain interactions (Figures 3F-3H). The fact that linear motifs and interaction-associated domains tend to be found in “interaction promoting” regions offers a mechanistic explanation for the interaction differences between isoforms.
Alternative splicing is a major mechanism in the production of diverse protein isoforms with different primary sequence. Beyond the primary sequence, each protein isoform can be further processed through post-translational modifications (PTMs), producing many more distinct polypeptides or “proteoforms” (Smith and Kelleher, 2013). In the present study, we measured each protein isoform's PPIs in a heterologous expression system (Y2H), and thus could have missed interactions modulated by a protein's PTMs, subcellular location, stability, and other factors unique to the protein's endogenous environment. Although it is beyond the scope of the present study, PTMs, such as phosphorylation, can lead to differences in protein-protein interactions or other functional properties. For example, deep transcriptome sequencing across different tissues and different species reveals that tissue-specific exons are enriched in phosphorylation sites (Merkin et al., 2012), suggesting that alternative splicing may be involved in both the regulation of protein interactions as well as the modulation of phosphorylation potential. Therefore, compiling a comprehensive catalog of different proteoforms and subsequently studying their distinct functions will be necessary for full understanding of normal cellular biology as well as disease pathogenesis at the systems level.
In summary, our results support a central role for alternative splicing in network organization, function, and cross-tissue dynamics, demonstrating the importance of an isoform-resolved global view of interactome networks. They also support a paradigm in which most genes encode multiple distinct protein isoforms, each of which potentially yields multiple proteoforms, and where each proteoform possesses a potentially unique set of functions. Collectively, this process would generate a vast diversity of “functional alloforms”, contributing to vastly different physiological and developmental outcomes, disease pathologies, and potentials for therapeutic development.
EXPERIMENTAL PROCEDURES
See Supplemental Experimental Procedures for additional details. Schematic diagrams of isoform exon-intron structures, ORF sequences, and isoform interaction profiles are available at http://isoform.dfci.harvard.edu/.
ORF Cloning
ORF cloning and sequencing was carried out as described (Salehi-Ashtiani et al., 2008).
RNA Abundance
The RNA-Seq Expectation Maximization program (RSEM, v1.1.21) was used to estimate transcriptional abundances separately for each tissue (Li and Dewey, 2011).
Binary Interaction Mapping and Validation
Y2H screening was performed as described (Dreze et al., 2010; Rolland et al., 2014; Rual et al., 2005). All isoforms of the same gene were pairwise tested against all possible interaction partners of any isoform for the same gene. PPI validation by a protein complementation assay was performed as described (Rolland et al., 2014).
Isoform Features
An ISR is defined as the longest contiguous region shared by a subset of isoforms. Regions mapping to all isoforms of a gene are considered constitutive regions. We calculated whether isoform-specific interactions were more likely to be associated with a potential promoting or inhibiting region than expected by chance.
Linear motifs and LMBDs
For each interaction partner in our dataset, we determined the linear motif density in the longest ISR associated with that partner (Dinkel et al., 2012). To quantify the enrichment of LMBDs in isoform partners exhibiting isoform-specific interactions, Pfam-A domains (Finn et al., 2014) were mapped to all interaction partners using Hmmer 3.0 (e-value = 10−2) (Finn et al., 2011) and each partner was classified as either containing an LMBD, as annotated in the ELM (Dinkel et al., 2012) or Dilimot (Neduva and Russell, 2006) databases, or not. Interaction partners were then assigned either as exhibiting an isoform-specific interaction associated with a promoting ISR, or not.
Domain-Domain Interactions
Pfam-A domains (Finn et al., 2014) were mapped to all isoforms and interaction partners using Hmmer 3.0 (e-value = 10−5) (Finn et al., 2011). We identified isoform-partner pairs encoding a predicted DDI from iPfam (Finn et al., 2005), 3Did (Mosca et al., 2014), or Domine (Yellaboina et al., 2011).
Structural Analysis of Isoform-Specific Interactions
Interactome3D (Mosca et al., 2013) was queried for PPI pairs. The interaction interface is defined as those residues that had a heavy atom at a distance < 6 Å to the binding partner. Local pairwise alignment between the structure sequence and the corresponding isoform identified interface residues.
Interactome Network Analysis of Isoform Interaction Partners
The mean shortest path distance in HI-II-14 (Rolland et al., 2014) between any two proteins that interact with the same single protein, interact with alternative isoforms, or interact with proteins encoded by separate genes was calculated. Path lengths involving the tested protein isoform were excluded. P values were calculated using the t-test.
Reads from the Illumina Body Map 2.0 16-tissue RNA-Seq dataset (Illumina BodyMap 2.0) were mapped to all hORFeome clone sequences and the log2 read count was calculated for each gene for each tissue. Pearson correlation coefficients were calculated on all pairs of interaction partners after filtering out genes with a maximal expression below 1/32nd of the upper-quartile gene expression. The fraction of pairs co-expressed (i.e., having a positive Pearson correlation coefficient greater than 0.15) was calculated for each of the three groups of pairwise proteins described above. P values were derived using Fisher's exact test.
Disease subnetworks were created by mapping the set of disease associated genes from GeneCards (Safran et al., 2010) onto HI-II-14 (Rolland et al., 2014) and retrieving the disease genes and their first degree PPI neighbors. The mean of the Jaccard index of disease subnetwork co-occurrence for all protein pairs within each class was then calculated. P values were calculated using Wilcoxon rank sum test.
Tissue-Specificity of Isoform Interaction Partners
We measured the range of normalized log2 expression levels in the Illumina Body Map 2.0 16-tissue RNA-Seq dataset (Illumina BodyMap 2.0) and considered genes with a range greater than 7 as tissue-specific.
Yeast-Based Functional Complementation Assays
Selected ORFs were expressed from low-copy expression vectors in temperature sensitive (ts) yeast strains. The complementation status was determined by comparing the growth of yeast ts strains at restrictive and permissive temperatures.
Supplementary Material
Highlights.
Alternative splicing can produce isoforms with vastly different interaction profiles.
These differences can be as great as those between proteins encoded by different genes.
Isoform-specific partners exhibit discrete expression and functional characteristics.
ACKNOWLEDGMENTS
This work was supported by NHGRI CEGS grant P50HG004233 (M.V., F.P.R.); NHGRI grant U01HG001715 (M.V., D.E.H, F.P.R.); the Ellison Foundation (M.V.), NCI grant R33CA132073 (M.V.); the Krembil Foundation (Canada) (F.P.R.); a Canada Excellence Research Chair Award (F.P.R.); an Ontario Research Fund-Research Excellence Award (F.P.R.); E. K. Shriver NICHD grant R01HD065288 (L.M.I., K.S.-A.); NIMH grants R01MH091350 (L.M.I., T.H.), R01MH105524 (L.M.I), and R21MH104766 (L.M.I.); NSF grant CCF-1219007, NSERC grant RGPIN-2014-03892 (Canada), Canada Foundation for Innovation grant JELF-33732 and Canada Research Chairs Program (Y.X.); NIH training grant T32CA009361 (G.M.S.); a NSERC fellowship (Canada) (J.C.-H.); NIGMS grant R01GM105431 (X.J.Z.); and a Swedish Research Council International Postdoc Grant (S.S.). M.V. is a FRS-FNRS Chercheur Qualifié Honoraire (Belgium).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.V. conceived the project. X.Y., A.R., S.S., F.Y., K.S., B.E.B., R.R.M., A.M., Q.Z., S.A.W., S.T., L.G., N.S., S.Y., M.D.R., D.B., G.T., and M.C. performed experiments. J.C.-H., S.K., G.M.S., T.H., M.D.-F., and P.A. performed computational analysis with contributions from X.Y., Y.A.S., S.J.P., X.J.Z., B.C., F.P.R., and Y.X.. X.Y., T.H., K.S.-A., B.A., C.B., M.A.C., P.A., F.P.R., D.E.H., L.M.I., Y.X., and M.V. designed and/or advised research. X.Y., J.C.-H., S.K., G.M.S., B.C., A.A.C., M.A.C., P.A., F.P.R., D.E.H., L.M.I., Y.X., and M.V. wrote the paper.
ACCESSION NUMBERS
The GenBank accession numbers for the data reported in this paper are KU177872-KU178906.
SUPPLEMENTAL INFORMATION
Supplemental information includes extended experimental procedures, six figures, and four tables.
REFERENCES
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell. 2012;46:871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas R, Yang X, Lin GN, Kang S, Shen Y, Ghamsari L, Broly M, Rodriguez M, Tam S, Trigg SA, et al. Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat. Commun. 2014;5:3650. doi: 10.1038/ncomms4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, et al. ELM–the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40:D242–251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O'Hanlon D, Kim PM, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol. Cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Marshall M, Bateman A. iPfam: visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34:305–312. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K. PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics. 2005;6:1–16. doi: 10.1007/s10048-004-0207-y. [DOI] [PubMed] [Google Scholar]
- Jones S, Stewart M, Michie A, Swindells MB, Orengo C, Thornton JM. Domain assignment for protein structures using a consensus approach: characterization and analysis. Protein Sci. 1998;7:233–242. doi: 10.1002/pro.5560070202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo AH, Laurent JM, Yellman CM, Meyer AG, Wilke CO, Marcotte EM. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348:921–925. doi: 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. Function of alternative splicing. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca R, Ceol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat. Methods. 2013;10:47–53. doi: 10.1038/nmeth.2289. [DOI] [PubMed] [Google Scholar]
- Mosca R, Ceol A, Stein A, Olivella R, Aloy P. 3did: a catalog of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2014;42:D374–379. doi: 10.1093/nar/gkt887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neduva V, Russell RB. DILIMOT: discovery of linear motifs in proteins. Nucleic Acids Res. 2006;34:W350–355. doi: 10.1093/nar/gkl159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pollitt R, McMahon R, Nunn J, Bamford R, Afifi A, Bishop N, Dalton A. Mutation analysis of COL1A1 and COL1A2 in patients diagnosed with osteogenesis imperfecta type I-IV. Hum. Mutat. 2006;27:716. doi: 10.1002/humu.9430. [DOI] [PubMed] [Google Scholar]
- Raff ML, Craigen WJ, Smith LT, Keene DR, Byers PH. Partial COL1A2 gene duplication produces features of osteogenesis imperfecta and Ehlers-Danlos syndrome type VII. Hum. Genet. 2000;106:19–28. doi: 10.1007/s004390051004. [DOI] [PubMed] [Google Scholar]
- Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Huttelmaier S, Singer RH, Matthews S, Curry S, Smith CW. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat. Struct. Mol. Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J-F, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010. 2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Yang X, Derti A, Tian W, Hao T, Lin C, Makowski K, Shen L, Murray RR, Szeto D, et al. Isoform discovery by targeted cloning, 'deep-well' pooling and parallel sequencing. Nat. Methods. 2008;5:597–600. doi: 10.1038/nmeth.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 2013;31:1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Kelleher NL. Proteoform: a single term describing protein complexity. Nat. Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple G, Gerhard DS, Rasooly R, Feingold EA, Good PJ, Robinson C, Mandich A, Derge JG, Lewis J, Shoaf D, et al. The completion of the Mammalian Gene Collection (MGC). Genome Res. 2009;19:2324–2333. doi: 10.1101/gr.095976.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Jahanbani F, Blauwkamp T, Moshrefi A, Jaeger E, Chen F, Harel I, Bustamante CD, Rasmussen M, Snyder MP. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat. Biotechnol. 2015;33:736–742. doi: 10.1038/nbt.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K, Rual J-F, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat. Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabási A-L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellaboina S, Tasneem A, Zaykin DV, Raghavachari B, Jothi R. DOMINE: a comprehensive collection of known and predicted domain-domain interactions. Nucleic Acids Res. 2011;39:D730–735. doi: 10.1093/nar/gkq1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.