Abstract
Targeting the clinically unvalidated reverse transcriptase (RT) associated ribonuclease H (RNase H) for human immunodeficiency virus (HIV) drug discovery generally entails chemotypes capable of chelating two divalent metal ions in the RNase H active site. The hydroxypyridone carboxylic acid scaffold has been implicated in inhibiting homologous HIV integrase (IN) and influenza endonuclease via metal chelation. We report herein the design, synthesis and biological evaluations of a novel variant of the hydroxypyridone carboxylic acid scaffold featuring a crucial N-1 benzyl or biarylmethyl moiety. Biochemical studies show that most analogues consistently inhibited HIV RT-associated RNase H in the low micromolar range in the absence of significant inhibition of RT polymerase or IN. One compound showed reasonable cell-based antiviral activity (EC50 = 10 µM). Docking and crystallographic studies corroborate favorable binding to the active site of HIV RNase H, providing a basis for the design of more potent analogues.
Graphical Abstract
1. Introduction
Current HIV antivirals target three of the four virally encoded enzymes (RT polymerase, IN and protease), as well as viral entry proteins and cellular co-receptors.1 These drugs form the basis for current combination drug therapy of HIV infection (HAART)2 by virtue of their distinct mechanisms of action. However, since current HIV therapeutics involve only a few of possible targets, the efficacy of HAART is plagued by the inevitable emergence of HIV mutants resistant to current drugs. Thus, the development of antivirals targeting underexplored and unvalidated processes required for HIV replication may be key to combating current drug-resistant viral strains. One such target is the RT associated RNase H activity.3 RT encodes two distinct enzymatic functions4: the polymerase activity that carries out both RNA and DNA dependent DNA polymerization; and the RNase H function that selectively degrades RNA from the RNA/DNA heteroduplex intermediate. The well-known drug families of nucleoside RT inhibitors (NRTIs)5 and non-nucleoside RT inhibitors (NNRTIs)6 all target the DNA polymerase function of RT; the RNase H function remains a clinically unvalidated target as none of its inhibitors has advanced to development pipeline.7 Nevertheless, the crucial role of RNase H in HIV replication has been well recognized as research showed that active site mutations associated with reduced RNase H biochemical activity conferred attenuated HIV replication in cell culture.8 This suggests that potent and selective RNase H inhibition should lead to effective antiviral activity.
The main medicinal chemistry challenge of targeting HIV RNase H is the the lack of appropriate chemical scaffolds supporting specific RNase H inhibition and potent antiviral activity. Reported RNase H inhibitors9 generally target the active site by using a chelating triad (Figure 1a; in magenta) for competitive binding to divalent metals, e.g. hydroxyisoquinolinedione (HID, 1),10 β-thujaplicinol (2),11 and dihydroxycoumarin (3).12 More potent and selective RNase H inhibition was achieved with structurally more elaborate chemotypes, such as diketoacid 4,13 pyrimidinol carboxylic acid 5,14 hydroxynaphthyridine 615 and pyridopyrimidone 7,16 all featuring a hydrophobic aromatic moiety seemingly important for antiviral activity. Compounds 6–7 are among the very few RNase H inhibitors with reported antiviral activity. Exploiting a chelating core for competitive metal binding is a general medicinal chemistry approach for inhibiting a wide array of enzymes homologous to HIV RNase H.17 For example, the hydroxypyridone carboxylate core (blue, Figure 1b) for current study has been implicated in influenza endonuclease inhibitor 818 as well as the FDA-approved HIV IN strand transfer inhibitor (INSTI) dolutegravir (DTG)19 9 (Figure 1b). Based on the pharmacophore model of RNase H inhibitors 4—7, chemotype 10 was designed for selectively inhibiting RNase H. We report herein the chemical synthesis, biochemical and antiviral evaluations, molecular modeling, and co-crystal structure of 10.
Figure 1.
Design of active site RNase H inhibitors. (a) Major chemotypes reported as HIV RNase H active site inhibitors. All chemotypes contain a chelating triad (magenta); scaffolds 4–7 also feature an aryl or biaryl moiety (cyan) connected through a methylene or amino linker; (b) hydroxypyridone carboxylate (blue) is implicated in influenza endonuclease inhibitor 8 and HIV INSTI 9. Active site RNase H inhibitor chemotype 10 based on the same chelting core (blue) is designed. An N-1 aryl or biaryl methyl group is included to mimick scaffolds 5–7.
2. Results and Discussion
Chemistry
The pyridone carboxylate chelating core was constructed using a modification of a previously reported approach.18 The synthesis began with a benzyl etherification of the commercially available ethyl chloroacetoacetate (12). The resulting ether intermediate (13) was subjected to a one-carbon homologation to yield enamine 14. The subsequent Claisen condensation followed by an immediate cyclization produced the pyrone intermediate 15. The conversion of the pyrone ring into pyridone (16) was effected by heating with an aromatic amine (R = aryl), which upon saponification afforded pyridone carboxylic acid 17. At this stage, biaryl N-1 substituents were constructed via a Suzuki coupling reaction with a para-bromopenyl substrate (17, R = 4-bromobenzyl). The final step of the synthesis involved a debenzylation to yield the designed analogues (10a–z).
Additional carboxylate derivatives were prepared to provide preliminary structure-activity-relationship (SAR) data (Scheme 2). Direct debenzylation of pyridone carboxylate 16d provided an ester analogue (18), and a carboxamide analogue (20) was prepared from 16d via amination and debenzylation. A structurally more elaborate N-benzyl substituted carboxamide (22) was synthesized via a three step sequence involving saponification, amide coupling and debenzylation.
Scheme 2a.
Synthesis of hydroxylpyridinone carboxylate analogues.
a Reagents and conditions: a) 7N NH3/ MeOH, r.t, 48 h, 70%; b) TFA, 80 °C, 20 min, MW, 65—78% c) 2N NaOH, EtOH, 90 °C, 4 h, 87% d) benzyl amine, HATU, DIPEA, DMF, r.t, 12 h, 72%.
Biology
All compounds were evaluated biochemically for inhibition in HIV RT RNase H and polymerase assays. Selected compounds were also tested in an HIV IN strand transfer assay. Antiviral screening was conducted using a HIV-1 single replication cycle assay.
RNase H inhibition
Inhibition of RT-associated RNase H inhibitory activity was evaluated in a biochemical assay20 using three different oligonucleotide duplexes as substrates to probe three distinct modes of RNase H cleavages: HTS-1 for internal cleavages which likely represent the primary mode of RNase H cleavage; HTS-2 for the DNA 3’ end-directed cleavages allowing specific RNA cuts 17—18 nucleotides downstream from the polymerase active site; and HTS-3 for the RNA 5’ end-directed cleavages of recessed RNA templates. Assay results are summarized in Table 1. Most of the hydroxypyridone carboxylate analogues inhibited the internal cleavage (HTS-1) by RNase H in the low- to sub-micromolar range (IC50 = 0.65–18 µM), with some exceptions (10a–c, 10n–p). Secondly, analogues bearing a two-ring (10r–t) or a biaryl (10r–z) substituent at N-1 position demonstrated significantly more potent inhibition (IC50 = 0.65–7.7 µM) than those with a single-ring substituent (10b–p, IC50 = 12–>25 µM), with 10q being the sole exception. N-1 unsubstituted (10a) and phenyl substituted (10b) analogues were both inactive. These observations strongly corroborate the pharmacophore model that an aryl or biaryl ring separated from the chelating core with a one-atom linker is required for RNase H inhibition and that the biaryl confers better inhibition than the aryl (Fig. 1a, 5–7). Third the two submicromolar inhibitors, 10x and 10y, both have H-bond acceptors and / or donors at the terminus of the biaryl substituent, suggesting interaction of the biaryl with the nucleic acid substrate.
Table 1.
Biochemical assay results of N substituted pyridone-carboxylic acid analogues (10) against HIV RT
 | ||||||
|---|---|---|---|---|---|---|
| Compd | R | RNH IC50 a (µM) | RNH Fragmente IC50 (µM) |
RT Pol IC50a (µM) |
||
| HTS-1b | HTS-2c | HTS-3d | ||||
| 10a | H | >25 | >25 | >25 | 22 ± 2.3 | >25 |
| 10b | >25 | >25 | >25 | >25 | >25 | |
| 10c | >25 | >25 | >25 | 11 ± 1.5 | >25 | |
| 10d | 17 | >25 | >25 | 7.8 ± 0.6 | >25 | |
| 10e | 16 ± 4.5 | >25 | >25 | 5.3 ± 0.5 | >25 | |
| 10f |  |
17 ± 5.2 | >25 | >25 | 9.8 ± 0.7 | >25 |
| 10g |  |
13 ± 2.1 | >25 | >25 | 6.8 ± 0.9 | >25 |
| 10h | 17 | >25 | >25 | 7.8 ± 0.8 | >25 | |
| 10i |  |
14 | >25 | >25 | 4.7 ± 0.5 | >25 |
| 10j |  |
12 | >25 | >25 | 4.2 ± 0.4 | >25 |
| 10k |  |
18 ± 2.8 | >25 | >25 | 9.8 ± 0.5 | >25 |
| 10l | >25 | >25 | >25 | >25 | >25 | |
| 10m |  |
18 ± 4.5 | >25 | >25 | 11 ± 1.9 | >25 |
| 10n | >25 | >25 | >25 | >25 | >25 | |
| 10o |  |
>25 | >25 | >25 | 25 ± 2.0 | >25 |
| 10p | >25 | >25 | >25 | 14 ± 1.1 | >25 | |
| 10q | 2.5 ± 0.2 | 15 ± 0.5 |
20 ± 2.2 |
4.8 ± 0.2 | >25 | |
| 10r |  |
2.7 ± 0.6 | 4.3 ± 0.4 |
5.4 ± 1.1 |
1.9 ± 0.1 | 8.5 ± 1.5 |
| 10s |  |
7.0 ± 0.7 | >25 | 13 ± 2.7 |
3.7 ± 0.6 | 19 ± 5.5 |
| 10t |  |
1.5 ± 0.2 | 5.1 ± 0.5 |
4.9 ± 2.2 |
8.2 ± 0.1 | 1.8 ± 0.1 |
| 10u |  |
7.7 ± 2.4 | 25 ± 1.5 |
>25 | 5.1 ± 0.3 | >25 |
| 10v | 2.5 ± 0.4 | 2.8 ± 0.6 |
2.9 ± 0.3 |
1.8 ± 0.3 | 2.2 ± 0.1 | |
| 10w | 3.2 ± 0.3 | 4.4 ± 0.6 |
3.9 ± 0.4 |
4.4 ± 0.5 | 3.2 ± 0.8 | |
| 10x |  |
0.90 ± 0.05 | 1.2 ± 0.1 |
0.90 ± 0.2 |
1.0 ± 0.01 | 1.1 ± 0.1 |
| 10y |  |
0.65 ± 0.05 | 1.5 ± 0.2 |
1.0 ± 0.2 |
1.3 ± 0.05 | 1.2± 0.1 |
| 10z | 3.6 ± 0.2 | 10 ± 0.6 |
15 ± 6.2 |
2.6 ± 0.6 | >25 | |
| 18 | -- | >25 | >25 | >25 | >25 | >25 |
| 20 | -- | 12 ± 1.8 | >25 | >25 | 6.8 ± 0.7 | >25 |
| 22 | -- | 7.5 ± 0.4 | 4.5 ± 0.8 |
4.9 ± 1.0 |
3.2 ± 0.2 | 2.4 ± 0.1 |
IC50: concentration of a compound producing 50% inhibition, expressed as mean ± standard deviation from at least three independent experiments.
Substrate that measures internal cleavage.
Substrate that measures DNA 3’ end directed cleavage.
Substrate that measures RNA 5’ end directed cleavage.
Reconstituted HIV RNase H domain.
The broadest range of inhibition across all compounds was noted with the HTS-1 substrate, which is not surprising since this substrate was designed to enable highly sensitive detection of inhibitors.21 No inhibition was observed using the more rigorous substrates HTS-2 or HTS-3 for analogues with a single ring N-1 substituent (10b–p). For analogues with a two-ring N-1 substituent, the same substrate bias was observed with compounds 10r–u and 10z, whereas others (10v–y) showed nearly equal inhibition with all three substrates.
In addition, the biochemical assessment of RNase H inhibition was also carried out with a catalytically active RNase H domain fragment (Table 1, RNase H Fragment). Nearly all analogues inhibited the RNase H fragment in low micromolar range. The data with this fragment confirmed that biaryl substitution confers better inhibition than aryl.
Our SAR analysis was expanded by exploring the importance of the carboxylate functional group by synthesizing analogues of 10d: the ester 18, the carboxamide 20 and the 2,4-diflurobenzyl carboxamide 22. The biochemical assay results showed that esterification abrogated inhibitory activity (compound 18), whereas the inhibitory activity was preserved with carboxamide modification (20 and 22). Importantly, the 2,4-difluorobenzyl carboxamide 22 provided significantly better inhibition of RT polymerase and RNase H activities compared to the parent carboxylic acid 10d. This same modification may also provide integrase inhibition, leading to multi-target efficacy in a single compound.
Polymerase inhibition
All compounds were evaluated for inhibition of RT RNA-dependent DNA polymerase activity. A sharp SAR trend was noted in which the N-1 unsubstituted analogue 10a and all one-ring substituted analogues (10b–q) showed no inhibitory activity (at concentrations up to 25 µM) whereas most two-ring substituted analogues (10r–t and 10v–y) inhibited RT pol with potencies nearly equal to RNase H inhibition. Although the mechanism of this polymerase inhibition remains unclear, we speculate that these inhibitors may block the polymerase active site metal-binding triad to prevent facile catalysis of nucleotide incorporation.
Order-of-addition RNase H inhibition assays
To determine whether lead analogues retain inhibitory activity when added after the RNA/DNA substrate we performed order-of-addition assays to examine the effect of various pre-incubation conditions on RT-associated RNase H inhibition. We tested six of the most potent inhibitors of RT-associated RNase H using β-thujaplicinol for comparison under conditions similar to those run for the in vitro RNase H inhibition assays in which the RNA/DNA substrate is in excess of the RT (Table 1). Generally, the inhibitors are most potent against RNase H activity when they are added to RT before the RNA/DNA substrate (Table 2). When the RT/substrate complex is formed before addition of compound, the inhibitor potency is decreased in all cases, including the previously published β-thujaplicinol.22 Under these conditions, analogue 10x is the most potent of the inhibitors against cleavage of the pre-formed RT/substrate complex (>50% inhibition), suggesting it can compete with the substrate for binding to RT.
Table 2.
Order-of-addition RNase H inhibition assay results
| % Inhibition | ||||
|---|---|---|---|---|
| 1. Pre-Incubation (10 min at 37 °C) |
RT + Inhibitor | RT + MgCl2 + Inhibitor |
RT + Inhibitor + HTS-1 |
RT + HTS-1 |
| 2. Reaction Intiation (10 min at 37 °C) |
HTS-1 + MgCl2 |
HTS-1 | MgCl2 | Inhibitor + MgCl2 |
| 10qa | 69% ± 5%b | 52% ± 17% | 27% ± 4% | 21% ± 13% |
| 10r | 58% ± 0% | 51% ± 7% | 35% ± 3% | 28% ± 5% |
| 10t | 75% ± 5% | 84% ± 1% | 38% ± 2% | 18% ± 13% |
| 10v | 68% ± 9% | 36% ± 12% | 37% ± 7% | 21% ± 19% |
| 10x | 95% ± 1% | 68% ± 10% | 66% ± 8% | 50% ± 7% |
| 10y | 97% ± 1% | 79% ± 9% | 68% ± 6% | 14% ± 3% |
| β-thujaplicinol | 100% ± 1% | 99% ± 1% | 79% ± 8% | 12% ± 4% |
Final reaction conditions: 100 nM HIV RT, 250 nM HTS-1, 6 mM MgCl2, 30 µM inhibitor.
Values are reported as mean ± standard deviation from two independent experiments.
When these assays were carried out using RT in excess of the RNA/DNA substrate, which more closely resembles actual conditions in the virion, generally the analogues more efficiently maintain effective inhibition against the pre-formed RT/substrate complex. Under conditions of excess RT compared to RNA/DNA substrate, 10y is the most potent inhibitor of the RT/substrate complex, as it blocks 80% of the RT-asociated RNase H activity (Table 3).
Table 3.
Order-of-addition RNase H inhibition assay results in the presence of excess RT
| % Inhibition | ||
|---|---|---|
| 1. Pre-Incubation (10 min at 37 °C) |
RT + Inhibitor | RT + HTS-1 |
| 2. Reaction Intiation (2 min at 37 °C) |
HTS-1 + MgCl2 |
Inhibitor + MgCl2 |
| 10qa | 70% ± 9%b | 58% ± 3% |
| 10r | 82% ± 1% | 61% ± 8% |
| 10t | 42% ± 1% | 15% ± 5% |
| 10v | 78% ± 2% | 59% ± 6% |
| 10x | 91% ± 0% | 74% ± 4% |
| 10y | 92% ± 1% | 80% ± 1% |
| β-thujaplicinol | 100% ± 0% | 98% ± 4% |
Final reaction conditions: 300 nM HIV RT, 50 nM HTS-1, 0.5 mM EDTA, 6 mM MgCl2, 30 µM inhibitor.
Values are reported as mean ± standard deviation from two independent experiments.
HIV integrase strand transfer inhibition
HIV IN and RNase H share similar active site configurations, thus inhibitors of these enzymatic functions entail overlapping pharmacophore features. Selective inhibition of RNase H over INST is therefore particularly important in RNase H inhibitor discovery. To assess the selectivity profile of the new pyridone chemotype, all analogues were tested for INST activity (Table 4).
Table 4.
Comparison of biochemical inhibition against RT RNase H, RT pol and INST
| Compd | RT RNase H IC50a(µM) | RT Pol IC50 (µM) | IN IC50 (µM) |
|---|---|---|---|
| 10a | >25 | >25 | >100 |
| 10b | >25 | >25 | 37 |
| 10c | >25 | >25 | 35 |
| 10d | 17 | >25 | 35 |
| 10e | 16 ± 4.5 | >25 | 34 |
| 10f | 17 ± 5.2 | >25 | >100 |
| 10g | 13 ± 2.1 | >25 | >100 |
| 10h | 17 | >25 | 67 |
| 10i | 14 | >25 | 36 |
| 10j | 12 | >25 | 25 |
| 10k | 18 ± 2.8 | >25 | 44.5 ± 18.1 |
| 10l | >25 | >25 | >100 |
| 10m | 18 ± 4.5 | >25 | >100 |
| 10n | >25 | >25 | >100 |
| 10o | >25 | >25 | >100 |
| 10p | >25 | >25 | >100 |
| 10q | 2.5 ± 0.2 | >25 | >100 |
| 10r | 2.7 ± 0.6 | 8.5 ± 1.5 | 23 |
| 10s | 7.0 ± 0.7 | 19 ± 5.5 | 2.26 ± 0.87 |
| 10t | 1.5 ± 0.2 | 1.8 ± 0.1 | 14 |
| 10u | 7.7 ± 2.4 | >25 | 17 |
| 10v | 2.5 ± 0.4 | 2.2 ± 0.1 | 22 |
| 10w | 3.2 ± 0.3 | 3.2 ± 0.8 | 7.79 ± 1.93 |
| 10x | 0.90 ± 0.05 | 1.1 ± 0.1 | 16.0 ± 6.5 |
| 10y | 0.65 ± 0.05 | 1.2± 0.1 | >100 |
| 10z | 3.6 ± 0.2 | >25 | 35.0 ± 10.0 |
| 18 | >25 | >25 | >100 |
| 20 | 12 ± 1.8 | >25 | 32 |
| 22 | 7.5 ± 0.4 | 2.4 ± 0.1 | >100 |
Data with HTS-1 as substrate
While most of the compounds showed some INST inhibitory activity, this inhibitory activity was much weaker (14 µM – > 100 µM) than that provided against RT RNase H, suggesting that the hydroxypyridone carboxylate core represents an interesting chemotype for future development of specific inhibitors of HIV RNase H.
Antiviral assay
All compounds were screened in a previously reported single replication cycle antiviral assay23 to gauge their ability in inhibiting HIV replication in cell culture. Although most compounds did not inhibit HIV-1 replication in this assay, analogue 10r inhibited HIV-1 replication in a dose-dependent manner (Figure 2, EC50 = 10 µM) without cytotoxicity at concentrations up to 25 µM (CC50 > 25 µM). This is an important and exciting observation as very few RNase H inhibitors to date have been reported to possess antiviral activity in cell-based assays.
Figure 2.
Dose response antiviral effect of 10r against HIV-1. Experiments were done in triplicate with each data point. The antiviral EC50 is calculated to be 10 µM.
Molecular modeling
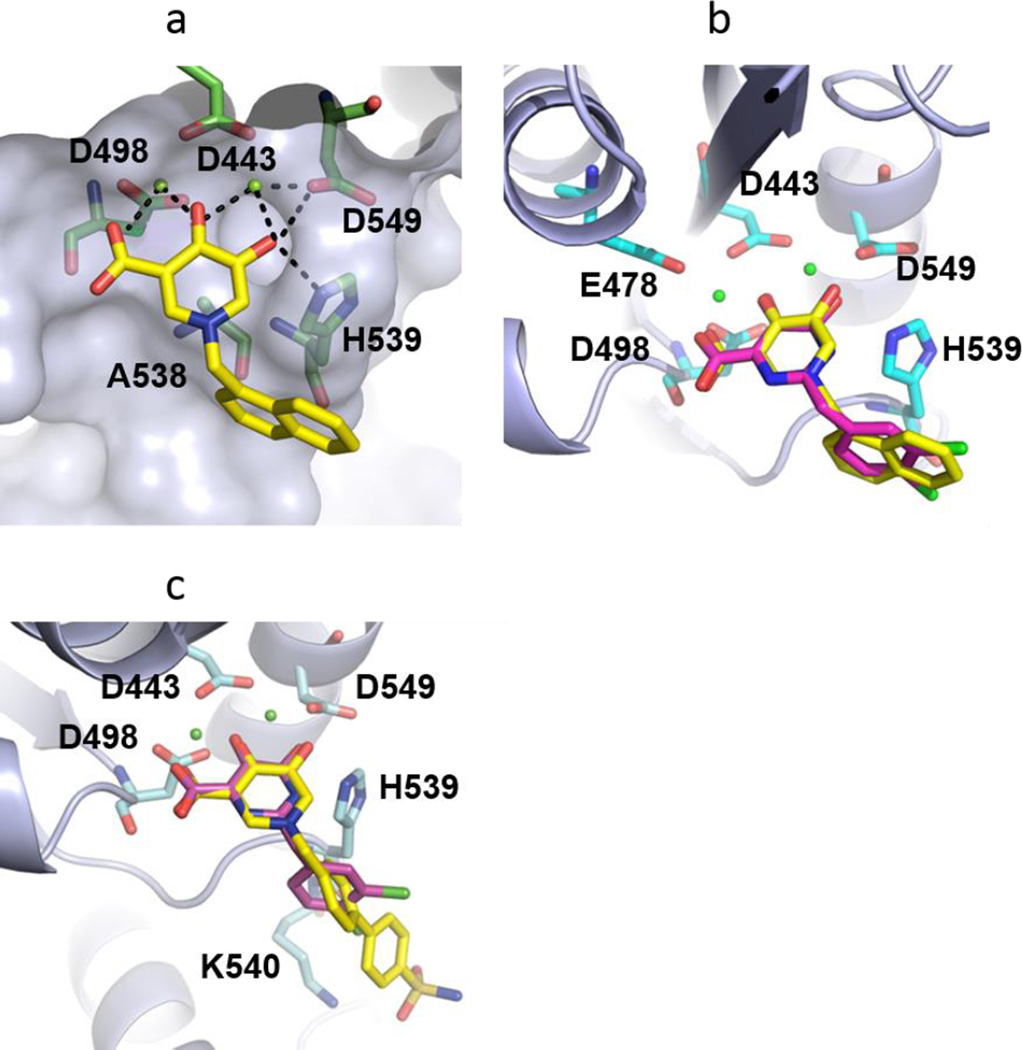
Docking analysis was performed with Glide XP(v.6.4)24–25 using two metal sites as a constraint. The predicted binding mode of compound 10r in the active site of RNase H suggests an interaction between the 4-oxo-1,4-dihydropyridine carboxylic acid core (chelating triad) and the two active site metal cations (coordinated to essential active site residues D443, E478, D498 and D549). The hydroxyl group at 5-position of 10r can form H-bonding interactions with H539 which would stabilize inhibitor positioning near the active site metal-chelating residues D443, E478, D498 and D549, all of which are crucial for RT-RNase H activity. The flexible methylene linker at N1 position of the pyridone core induces a conformational flexibility in the inhibitor that allows Π-stacking interaction between the naphthalene ring of the ligand and RNase H residues H539 and A538 and L560. This interaction may limit the conformational flexibility of the loop with H539 and thus impact on efficiency of RNase H catalytic activity. Notably, a similar binding mode was observed for RT complexes with dsDNA or RNA-DNA hybrids as reported by Harrison et.al26 and Sarafianos et.al.27 The predicted binding mode of 10r is shown in Figure 3a within the active site of RNase H. Compound 10y was predicted to bind in an identical fashion to that of 10r. Compounds 10r and 10y were observed to overlay with the pyrimidinol carboxylic acid core (5) and mimic the interactions within the RNaseH to a greater extent as depicted in Figure3b and 3c.
Figure 3.
Molecular modeling of analogues 10r and 10y at the RNase H active site and comparison to crystal structure of the HIV RNase H domain in complex with pyrimidinol carboxylic acid. a) Predicted binding mode of 10r within active site of RNase H (PDB code: 3HYF). Active site residues are highlighted in green sticks with metal ions as spheres. Some chelating and H-bond interactions are depicted as black dotted lines. b) Overlay of 10r (yellow) on the crystal structure of RNase H with pyrimidinol carboxylic acid (magenta). c) Overlay of 10y (yellow) on the crystal structure of RNase H with pyrimidinol carboxylic acid (magenta).
Crystal structure of HIV RT in complex with 10y
In order to further understand the molecular details of HIV RT-associated RNase H inhibition by hydroxypyridone carboxylate analogues, we solved the crystal structure of HIV RT in complex with the most potent RNase H inhibitor from the in vitro assays, 10y, at 2.9 Å resolution. The asymmetric unit comprises two RT molecules, and therefore two distinct RNase H active sites. Analogue 10y is only observed at one RT active site in the asymmetric unit, most likely due to partial occlusion of one RNase H active site by the fingers subdomain of the second RT molecule. Unlike other RT/RNase H active site-directed inhibitor complex structures,28–29 the RT/10y complex was crystallized without a non-nucleoside RT inhibitor (NNRTI), leaving the positions of the two p66 Thumb domains in a closed conformation, similar to other reported unliganded RT structures.30–34
In the occupied RNase H active site, two Mg2+ ions are bound by conserved active site residues D443, E478, D498, and D549. Analogue 10y chelates both Mg2+ ions through the carboxylate, carbonyl, and hydroxyl groups of the pyrimidone (Figure 4), in a manner similar to that of a previously reported RT/pyrimidinol carboxylic acid inhibitor.29 10y interacts directly with RT through interactions between the hydroxyl group of the pyridine and H539, and the sulfonamide group of the N-1 substituted biaryl moiety with K540 (Figure 4). These additional interactions with the RT enzyme likely provide increased stability to the RT/10y complex and may potentially explain the potent RNase H inhibition observed for 10y in the in vitro assays (Table 1).
Figure 4.
X-ray crystal structure of HIV RT in complex with analogue 10y. Cross-eyed stereo view of 10y (cyan) bound at the RNase H active site of HIV RT. The RNase H domain of RT is shown in orange, the p51 in light gray. Conserved active site residues are shown as sticks, and Mg2+ ions are shown as magenta spheres. Chelating and H-bond interactions are indicated by dashed lines. Cartoon was prepared by PyMOL35 and crystallographic coordinates have been submitted to the Protein Data Bank (PDB ID: 5J1E).
Compared to other analgoues, 10y was found to be the most potent inhibitor of RT-associated RNase H inihibiton. Structural insights suggest that the length provided by the N-1 substituted biaryl moiety could be important for RNase H inhibition, since many of the shorter phenyl-substituted analogues (10b–q) were less potent. It also appears that charge may also contribute to potent inhibition, as other biaryl-substituted compounds without charged groups (such as 10w) were less effective inhibitors of RNase H activity. Furthermore, para substitution of the biaryl moiety relative to the pyridone ring seems to position the biaryl group in a favorable position to have potential interactions with RT, which may not be achievable with ortho-substituted analogues such as 10u and may explain this analogue’s decreased potency.
Conclusions
New hydroxypyridone carboxylate analogues featuring an N-1 benzyl or biarylmethyl moiety were designed and synthesized as potential inhibitors of HIV RT associated RNase H. In vitro biochemical assays showed that analogues with a two-ring substituent at N-1 are significantly more potent than those with a one-ring substituent against all three modes of RNase H cuts as well as the RT polymerase function. While some analogues also inhibited strand transfer activity of HIV IN, this inhibition was substantially less than that for RT RNase H inhibition, suggesting that the pyridone chemotype may represent an interesting scaffold for development of RNase H-specific inhibitors. Importantly, compound 10r exhibited significant inhibitory activity in a cell-based antiviral assay with an EC50 of 10 µM. Molecular docking of 10r and the crystal structure of RT/10y corroborate for hydroxypyridone carboxylate analogues a mechanism of active site binding for RNase H inhibition. The mechanism of the observed polymerase inhibition remains unclear. These results indicate that the hydroxypyridone carboxylate chemotype previously implicated in the inhibition of INST and influenza endonuclease can be valuable in the discovery of HIV antivirals targeting the RT-associated RNase H.
Experimental
Chemistry: General Procedures
All commercial chemicals were used as supplied unless otherwise indicated. Flash chromatography was performed on a Teledyne Combiflash RF-200 with RediSep columns (silica) and indicated mobile phase. All moisture sensitive reactions were performed under an inert atmosphere of ultra-pure argon with oven-dried glassware. 1H and 13C NMR spectra were recorded on a Varian 600 MHz spectrometer. Mass data were acquired on an Agilent TOF II TOS/MS spectrometer capable of ESI and APCI ion sources. Analysis of sample purity was performed on a Varian Prepstar SD-1 HPLC system with a Phenomenex Gemini, 5 micron C18 column (250mm × 4.6 mm). HPLC conditions: solvent A = H2O, solvent B = MeCN; flow rate = 1.0 mL/min; compounds were eluted with a gradient of 5% MeCN/H2O for 0–5 min and to 95% MeCN/ H2O from 5–30 min followed by 100% MeCN from 35–40 min. Purity was determined by total absorbance at 254 nm. All tested compounds have a purity ≥ 98%.
Ethyl-4-(benzyloxy)-3-oxobutanoate (13)
To a suspension of 60% NaH (66.83 mmol, 2.2 eq) in THF (20 mL) at 0 °C was added benzyl alcohol (30.37 mmol, 1.05 eq) and the resulting mixture was stirred at r.t for 2h. To this mixture was then added ethyl-4-chloroacetoacetate (30.37 mmol, 1.0 eq) dropwise over 30 min and the resulting clear yellow solution was stirred at r.t overnight before it was cooled to 5 °C, acidified to pH = 4 using 5% HCl and extracted with the EtOAc (2 × 25 mL). The combined organics were washed with brine, dried over Na2SO4 and concentrated in vacuo to yield the desired product (13) as orange oil. The crude material was used for next step without further purification. Yield: 95% (6.8 g). 1H NMR (600 MHz, CDCl3) δ 7.40–7.28 (m, 5H), 4.59 (s, 2H), 4.18 (q, J = 7.1 Hz, 2H), 4.14 (s, 2H), 3.54 (s, 2H), 1.25 (t, J = 7.1 Hz, 3H).
4-Benzyloxy-2-(1-dimethylaminomethylidene)-3-oxo-butyric acid ethyl ester (14)
To a solution of 13 (28.36 mmol, 1.0 eq) in toluene (30 mL) was added DMF DMA (42.54 mmol, 1.5 eq) and the resulting mixture was stirred at r.t for 12 h. The reaction mixture was quenched by adding water and was acidified (pH = 4) using 5% HCl. The aqueous solution was extracted using EtOAc (2 × 20 mL). The combined organics were washed with NaHCO3, and brine, dried over Na2SO4 and concentrated in vacuo. The crude mixture was purified using CombiFlash with eluent starting from hexane/EtOAc (4:1) to hexane/EtOAc (1:1) to yield the desired product (14) as yellow oil. Yield: 30% (2.45 g); 1H NMR (600 MHz, DMSO-d6) δ 7.67 (s, 1H), 7.38–7.30 (m, 4H), 7.28 (t, J = 6.8 Hz, 1H), 4.45 (s, 2H), 4.30 (s, 2H), 4.06 (q, J = 7.1 Hz, 2H), 3.24 (brs, 3H), 2.73 (brs, 3H), 1.17 (t, J = 7.1 Hz, 3H).
General method for the synthesis of pyrone: Ethyl-5-(benzyloxy)-4-oxo-4H-pyran-3-carboxylate (15)
To a suspension of potassium tert-butoxide (1M in THF; 15.79 mmol, 2.0 eq) in THF (14 mL) was added ethyl formate (33.8 mmol, 4.3 eq) and the mixture was stirred at r.t for 30 min before compound 13 (7.89 mmol, 1.0 eq) in THF (14 mL) was added dropwise over 10 min. The resulting mixture was stirred at r.t for 8 h and quenched by adding 2N HCl (12 mL). After extraction with the EtOAc (2×20 mL), the combined organics were washed with NaHCO3 (2 × 10 mL), water (30 mL), brine (30 mL), dried over Na2SO4 and concentrated in vacuo to leave brownish yellow oil. This crude product was triturated by adding hexanes to yield the desired product (15) as brown solid. Yield: 75% (1.62 g); 1H NMR (600 MHz, DMSO-d6) δ 8.72 (d, J = 0.8 Hz, 1H), 8.30 (d, J = 0.8 Hz, 1H), 7.49 –7.33 (m, 5H), 4.95 (s, 2H), 4.22 (q, J = 7.1 Hz, 2H), 1.25 (t, J = 7.1, 3H).
General method for the synthesis of 4-oxo-dihydropyridine esters (16a–16s)
To a solution of 15 (1.0 eq) in EtOH (5 mL) was added amine (2.0 eq) and the resulting mixture was heated at 100 °C for 40 min under microwave conditions. After the solvent was removed in vacuo the residue was purified using CombiFlash to produce the crude product as colorless oil which was triturated with hexanes/EtOAc to yield the titled compound as colorless solid (16a–16s).
Ethyl-1-benzhydryl-5-(benzyloxy)-4-oxo-1,4-dihydropyridine-3-carboxylate (16a)
1H NMR (600 MHz, DMSO-d6) δ 8.18 (d, J = 1.8 Hz, 1H), 7.48 – 7.40 (m, 6H), 7.36 – 7.31 (m, 3H), 7.29–7.26 (m, 3H), 7.11 (d, J = 7.0 Hz, 4H), 6.92 (s, 1H), 4.93 (s, 2H), 4.14 (q, J = 7.1 Hz, 2H), 1.19 (t, J = 7.1 Hz, 3H).
General procedure for saponification: (17a–17s)
To a solution of 16 (1.0 eq) in EtOH (5 mL) was added 2N NaOH (10 mL). The resulting mixture was heated at 90 °C for 6 h and was then cooled to r.t and concentrated in vacuo. The resulting solution was acidified (pH = 3) using 2N HCl. The precipitate was collected via filtration, washed with water, and then dried to obtain the titled compound as colorless solid (17).
1-Benzhydryl-5-(benzyloxy)-4-oxo-1, 4-dihydropyridine-3-carboxylic acid (17a)
1H NMR (600 MHz, CD3OD) δ 8.41 (s, 1H), 7.48 (s,1H), 7.42–7.39 (m, 6H), 7.27–7.26 (m, 5H), 7.06 (d, J = 3.9 Hz, 4H), 6.92 (s, 1H), 5.05 (s, 2H).
General procedure for Suzuki coupling (17w–17z)
The mixture of 17l (1.0 eq), boronic acid (1.6 eq), Pd (PPh3)4 (0.065 eq), K2CO3 (3.6 eq) in DME/H2O (4:1) was irradiated at 150 °C for 30 min under microwave conditions. The black residue formed was filtered through celite and the solvent was concentrated in vacuo. The resulting aqueous solution was acidified (pH = 3) using 2N HCl. A white precipitate was obtained via filtration, washed with water and dried under vacuum to furnish the desired compound as colorless solid (17w–17z).
1-((4'-(Aminomethyl)-[1,1'-biphenyl]-4-yl)methyl)-5-(benzyloxy)-4-oxo-1,4-dihydropyridine-3-carboxylic acid (17w)
1H NMR (600 MHz, DMSO-d6): δ 8.79 (s, 1H), 8.18 (s, 1H), 7.67 (d, J = 7.8 Hz, 2H), 7.75 – 7.60 (m, 4H), 7.52 – 7.35 (m, 4H), 7.35 – 7.17 (m, 5H), 5.41 (s, 2H), 5.11 (s, 2H), 4.08 (s, 2H).
5-(Benzyloxy)-1-(4-fluorobenzyl)-4-oxo-1,4-dihydropyridine-3-carboxamide (19)
To a solution of 16d (90 mg) in methanol was added NH3 in MeOH (7N, 4 mL) and the resulting solution was stirred at r.t for 48 h. A white precipitate was collected via filtration, washed with methanol, and then dried under high vacuum to yield the carboxamide as colorless solid (19). Yield: 70%. 1H NMR (600 MHz, DMSO-d6): δ 9.47 (s, 1H), 8.52 (s, 3H), 7.81 (s, 1H), 7.44 (s, 1H), 7.35 (m, 7H), 7.24 – 7.15 (m, 2H), 5.22 (s, 2H), 5.00 (s, 2H).
General procedure for amide coupling: 5-(benzyloxy)-N-(2, 4-difluorobenzyl)-1-(4-fluorobenzyl)-4-oxo-1, 4-dihydropyridine-3-carboxamide (21)
To a mixture of 17d (1.0 eq) in DMF (5 mL) was added HATU (1.2 eq), DIPEA (1.2 eq), amine (1.2 eq) and the resulting reaction mixture was stirred at r.t for 12 h. The reaction was quenched with water and extracted with the EtOAc (2 × 20 mL). The combined organics were washed with 1N HCl (2 × 10 mL), NaHCO3 (2 × 10 mL), water (30 mL), brine (30 mL) and dried over Na2SO4. The solvent was removed in vacuo to produce the crude product as yellow solid which was purified using CombiFlash with 0–100% EtOAc as an eluent to furnish the titled compound as colorless solid (21). Yield: 72%. 1H NMR (600 MHz, DMSO-d6) δ 10.61 (t, J = 5.8 Hz, 1H), 8.57 (d, J = 1.7 Hz, 1H), 7.87 (d, J = 1.7 Hz, 1H), 7.37 (m, 7H,), 7.22 (m, 4H), 7.05 (dd, J = 8.4, 6.4 Hz, 1H), 5.26 (s, 2H), 5.02 (s, 2H), 4.51 (d, J = 5.8 Hz, 2H).
General procedure for O-debenzylation: (10a–10z, 18, 20, 22)
A mixture of 17 (1.0 eq) in TFA (2 mL) was irradiated at 80 °C for 20 min under microwave conditions. The solvent was removed in vacuo to produce the crude product as pale yellow solid which was triturated in methanol and ether to produce the titled compound as colorless solid (10a–10z, 18, 20, 22).
5-Hydroxy-4-oxo-1, 4-dihydropyridine-3-carboxylic acid (10a)
1H NMR (600 MHz, DMSO-d6): δ 12.74 (s, 1H), 9.84 (s, 1H), 8.31 (s, 1H), 7.69 (s, 1H). 13C NMR (150 MHz, DMSO-d6) δ 171.8, 166.6, 148.1, 137.0, 122.4, 113.3. HRMS-ESI (−) m/z calculated for C6H4NO4 154.0146 [M-H]−, found: 154.0148.
5-Hydroxy-4-oxo-1-phenyl-1,4-dihydropyridine-3-carboxylic acid (10b)
1H NMR (600 MHz, DMSO-d6): 10.22 (s, 1H), 8.46 (d, J = 2.3 Hz, 1H), 8.05 (d, J = 2.3 Hz, 1H), 7.74–7.68 (m, 2H), 7.63–7.52 (m, 3H). 13C NMR (150 MHz, DMSO-d6) δ 171.8, 166.0, 148.8, 142.9, 139.6, 130.3, 129.6, 125.8, 123.9, 113.8. HRMS-ESI (−) m/z calculated for C12H8NO4 230.0459 [M-H]−, found: 230.0468.
1-Benzyl-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10c)
1H NMR (600 MHz, DMSO-d6): δ 10.06 (s, 1H), 8.67 (d, J = 2.0 Hz, 1H), 7.83 (d, J = 2.0 Hz, 1H), 7.51 – 7.31 (m, 5H), 5.35 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 170.8, 165.0, 148.3, 139.9, 135.4, 128.8, 128.4, 127.9, 125.5, 113.0, 59.8. HRMS-ESI (−) m/z calculated for C13H10NO4 244.0615 [M-H]−, found: 244.0627.
1-(4-Fluorobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10d)
1H NMR (600 MHz, DMSO-d6): 10.06 (s, 1H), 8.68 (d, J = 2.0 Hz, 1H), 7.84 (d, J = 2.0 Hz, 1H), 7.51 (dd, J = 8.5, 5.5 Hz, 2H), 7.45 (t, J = 8.8 Hz, 2H), 5.33 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.3, 166.3, 148.9, 140.6, 132.2, 130.7, 116.5, 115.8, 115.6, 113.5, 59.5. HRMS-ESI (−) m/z calculated for C13H9FNO4 262.0521 [M-H]−, found: 262.0531.
1-(1-(4-Fluorophenyl) ethyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10e)
1H NMR (600 MHz, DMSO-d6): δ 10.05 (s, 1H), 8.64 (d, J = 2.2 Hz, 1H), 7.83 (d, J = 2.2 Hz, 1H), 7.61 – 7.51 (m, 2H), 7.35 – 7.08 (m, 2H), 5.78 (q, J = 7.1 Hz, 1H), 1.86 (d, J = 7.1 Hz, 3H). 13C NMR (150 MHz, DMSO-d6) δ 171.1, 166.1, 163.0, 148.9, 138.9, 135.3, 129.5, 123.6, 116.1, 113.3, 64.9, 19.3. HRMS-ESI (−) m/z calculated for C14H11FNO4 276.0678 [M-H]−, found: 276.0682.
1-(4-Fluoro-3-methylbenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10f)
1H NMR (600 MHz, DMSO-d6): δ 10.06 (s, 1H), 8.66 (d, J = 2.1 Hz, 1H), 7.82 (d, J = 2.1 Hz, 1H), 7.39 (dd, J = 7.2, 1.6 Hz, 1H), 7.34 (m, 1H), 7.21 (m, 1H), 5.29 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.1, 166.2, 161.8, 160.2, 148.7, 140.2, 131.9, 131.6, 128.2, 127.9, 125.2, 113.3, 59.5, 14.2. HRMS-ESI (−) m/z calculated for C14H11FNO4 276.0678 [M-H]−, found: 276.0687.
1-(2,4-Difluorobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10g)
1H NMR (600 MHz, DMSO-d6): δ 10.11 (s, 1H), 8.60 (d, J = 1.7 Hz, 1H), 7.76 (d, J = 2.0 Hz, 1H), 7.57 (dd, J = 15.2, 8.6 Hz, 1H), 7.41 – 7.31 (m, 1H), 7.17 (td, J = 8.5, 2.2 Hz, 1H), 5.42 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.2, 166.1, 161.5, 160.1, 148.7, 140.6, 140.4, 132.3, 125.6, 119.2, 113.3, 112.4, 104.6, 54.2. HRMS-ESI (−) m/z calculated for C13H8F2NO4 280.0427 [M-H]−, found: 280.0425.
1-(4-Chlorobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10h)
1H NMR (600 MHz, DMSO-d6): 10.08 (s, 1H), 8.69 (d, J = 1.8 Hz, 1H), 7.83 (d, J = 1.8 Hz, 1H), 7.48 (d, J = 8.5 Hz, 2H), 7.45 (d, J = 8.5 Hz, 2H), 5.34 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.5, 166.5, 149.1, 140.6, 135.1, 133.8, 130.5, 129.4, 126.0, 113.7, 59.6. HRMS-ESI (−) m/z calculated for C13H9ClNO4 278.0226 [M-H]−, found: 278.0222.
1-(3,4-Dichlorobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10i)
1H NMR (600 MHz, dmso): 10.08 (s, 1H), 8.72 (s, 1H), 7.87 (s, 1H), 7.81 (d, J = 8.5 Hz, 2H), 7.68 (s, 1H), 7.43 (d, J = 8.5 Hz, 2H), 5.33 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.6, 166.5, 149.1, 140.9, 140.7, 137.0, 131.9, 131.6, 131.0, 128.9, 126.0, 113.8, 59.1. HRMS-ESI (−) m/z calculated for C13H8Cl2NO4 311.9836 [M-H]−, found: 311.9843.
1-(2,4-Dichlorobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10j)
1H NMR (600 MHz, DMSO-d6): 10.12 (s, 1H), 8.58 (d, J = 2.1 Hz, 1H), 7.74 (d, J = 2.2 Hz, 1H), 7.50 (dd, J = 8.3, 2.2 Hz, 1H), 7.30 (d, J = 8.3 Hz, 1H), 5.47 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.7, 166.4, 149.0, 141.2, 134.7, 134.1, 132.7, 131.8, 129.8, 128.6, 126.2, 113.7, 57.8. HRMS-ESI (−) m/z calculated for C13H8Cl2NO4 311.9836 [M-H]−, found: 311.9844.
1-(3-Chloro-4-fluorobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10k)
1H NMR (600 MHz, DMSO-d6): δ 10.07 (s, 1H), 8.72 (s, 1H), 7.87 (s, 1H), 7.80 (d, J = 6.5 Hz, 1H), 7.49–7.45 (m, 2H), 5.31 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 170.9, 165.9, 158.0, 148.5, 140.1, 133.3, 130.7, 129.2, 125.3, 119.8, 117.4, 113.2, 58.5. HRMS-ESI (−) m/z calculated for C13H8ClFNO4 296.0131 [M-H]−, found: 296.0134.
1-(4-Bromobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10l)
1H NMR (600 MHz, DMSO-d6): δ 10.09 (s, 1H), 8.68 (d, J = 1.5 Hz, 1H), 7.83 (d, J = 1.5 Hz, 1H), 7.61 (d, J = 8.3 Hz, 2H), 7.38 (d, J = 8.3 Hz, 2H), 5.33 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 170.9, 165.9, 148.5, 140.9, 131.8, 130.2, 125.5, 121.9, 113.1, 59.1. HRMS-ESI (−) m/z calculated for C13H9BrNO4 321.9720 [M-H]−, found: 321.9727.
5-Hydroxy-4-oxo-1-(3-(trifluoromethyl)benzyl)-1,4-dihydropyridine-3-carboxylic acid (10m)
1H NMR (600 MHz, DMSO-d6): δ 10.09 (s, 1H), 8.75 (d, J = 2.1 Hz, 1H), 7.90–7.89 (m, 2H), 7.75 (d, J = 7.7 Hz, 1H), 7.72 (d, J = 7.8 Hz, 1H), 7.65 (t, J = 7.8 Hz, 1H), 5.43 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.0, 165.9, 148.5, 140.2, 136.8, 132.2, 130.0, 129.5, 125.4, 125.0, 124.9, 113.2, 59.1. HRMS-ESI (−) m/z calculated for C14H9F3NO4 312.0489 [M-H]−, found: 312.0484.
5-Hydroxy-4-oxo-1-(pyridin-4-ylmethyl)-1,4-dihydropyridine-3-carboxylic acid (10n)
1H NMR (600 MHz, CD3OD): 8.74 (d, J = 4.9 Hz, 2H), 8.60 (d, J = 2.0 Hz, 1H), 7.69 (d, J = 2.0 Hz, 1H), 7.64 (d, J = 4.9 Hz, 2H), 5.57 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ HRMS-ESI (−) m/z calculated for C12H9N2O4 245.0568 [M-H]−, found: 245.0570.
5-Hydroxy-1-(4-(methylsulfonyl)benzyl)-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10o)
1H NMR (600 MHz, DMSO-d6): δ 10.11 (s, 1H), 8.73 (d, J = 2.1 Hz, 1H), 7.96 (d, J = 8.3 Hz, 2H), 7.86 (d, J = 2.1 Hz, 1H), 7.64 (d, J = 8.3 Hz, 2H), 5.48 (s, 2H), 3.21 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 171.6, 166.5, 141.8, 141.3, 140.9, 129.2, 128.1, 126.1, 113.8, 87.2, 59.9, 44.4. HRMS-ESI (−) m/z calculated for C14H12NO6S 322.0391 [M-H]−, found: 322.0382.
1-(4-Carboxybenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10p)
1H NMR (600 MHz, DMSO-d6): δ 13.04 (s, 1H), 10.09 (s, 1H), 8.69 (d, J = 2.0 Hz, 1H), 7.96 (d, J = 8.2 Hz, 2H), 7.83 (d, J = 2.1 Hz, 1H), 7.48 (d, J = 8.2 Hz, 2H), 5.44 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.2, 167.0, 166.2, 148.8, 140.5, 131.0, 130.1, 128.4, 128.0, 125.9, 113.4, 59.7. HRMS-ESI (−) m/z calculated for C14H10NO6 288.0514 [M-H]−, found: 288.0515.
5-Hydroxy-1-(4-methoxybenzyl)-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10q)
1H NMR (600 MHz, DMSO-d6): δ 10.04 (s, 1H), 8.64 (d, J = 2.1 Hz, 1H), 7.82 (d, J = 2.1 Hz, 1H), 7.40 (d, J = 8.7 Hz, 2H), 6.96 (d, J = 8.7 Hz, 2H), 5.26 (s, 2H), 3.75 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 171.0, 166.3, 159.7, 148.6, 139.7, 130.0, 127.6, 125.7, 114.5, 113.3, 59.8, 55.4. HRMS-ESI (−) m/z calculated for C14H12NO5 274.0721 [M-H]−, found: 274.0722.
5-Hydroxy-1-(naphthalen-1-ylmethyl)-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10r)
1H NMR (600 MHz, DMSO-d6): δ 10.09 (s, 1H), 8.64 (d, J = 1.8 Hz, 1H), 8.07 (d, J = 8.3 Hz, 1H), 8.02 (d, J = 8.3 Hz, 1H), 8.00 (d, J = 8.4 Hz, 1H), 7.82 (d, J = 1.8 Hz, 1H), 7.64 (t, J = 7.3 Hz, 1H), 7.60 (t, J = 7.3 Hz, 1H), 7.56 (t, J = 7.6 Hz, 1H), 7.39 (d, J = 7.6 Hz, 1H), 5.90 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.3, 166.4, 148.8, 140.4, 133.7, 131.2, 130.7, 129.7, 129.1, 127.4, 127.3, 126.7, 126.1, 126.0, 123.2, 113.5, 58.1. HRMS-ESI (−) m/z calculated for C17H12NO4 294.0772 [M-H]−, found: 294.0776.
5-Hydroxy-1-(1-(naphthalen-1-yl)ethyl)-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10s)
1H NMR (600 MHz, DMSO-d6): δ 10.06 (s, 1H), 8.58 (d, J = 1.8 Hz, 1H), 8.05 (d, J = 8.2 Hz, 1H), 8.01 (dd, J = 8.0, 4.0 Hz, 2H), 7.86 (d, J = 7.1 Hz, 1H), 7.82 (d, J = 1.8 Hz, 1H), 7.66 (dd, J = 16.0, 8.3 Hz, 1H), 7.58 (dt, J = 14.7, 6.9 Hz, 2H), 6.59 (q, J = 6.8 Hz, 1H), 1.98 (d, J = 6.8 Hz, 3H). 13C NMR (150 MHz, DMSO-d6) δ 171.2, 166.4, 149.0, 138.4, 133.9, 133.0, 130.7, 130.4, 129.4), 127.6, 126.6, 126.1, 125.8, 124.3, 122.8, 113.6, 62.1, 20.7. HRMS-ESI (−) m/z calculated for C18H14NO4 308.0928 [M-H], found: 308.0936.
5-Hydroxy-4-oxo-1-((1-phenylcyclopentyl)methyl)-1,4-dihydropyridine-3-carboxylic acid (10t)
1H NMR (600 MHz, DMSO-d6): δ 9.82 (s, 2H), 7.70 (s, 2H), 7.29 (t, J = 7.3 Hz, 5H), 7.21 (dd, J = 15.2, 7.4 Hz, 7H), 7.09 (s, 2H), 4.32 (s, 5H), 1.94 (d, J = 5.1 Hz, 8H), 1.81 (s, 5H), 1.58 (s, 4H). HRMS-ESI (−) m/z calculated for C18H18NO4 312.1241 [M-H]−, found: 312.1253.
1-([1,1’-Biphenyl]-2-ylmethyl)-5-hydroxy-4-oxo-1, 4-dihydropyridine-3-carboxylic acid (10u)
1H NMR (600 MHz, DMSO-d6): δ 9.94 (s, 1H), 8.04 (s, 1H), 7.50 – 7.44 (m, 2H), 7.44 – 7.39 (m, 2H), 7.38 (d, J = 6.9 Hz, 1H), 7.28 (dd, J = 14.2, 7.0 Hz, 5H), 5.36 (s, 2H). HRMS-ESI (−) m/z calculated for C19H14NO4 320.0928 [M-H]−, found: 320.0936.
1-([1,1’-Biphenyl]-4-ylmethyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10v)
1H NMR (600 MHz, DMSO-d6): δ 8.71 (s, 1H), 7.88 (s, 1H), 7.70 (d, J = 7.7 Hz, 2H), 7.66 (d, J = 7.3 Hz, 2H), 7.51 (d, J = 7.6 Hz, 2H), 7.46 (t, J = 7.3 Hz, 2H), 7.38 (d, J = 7.2 Hz, 1H), 5.40 (s, 2H). HRMS-ESI (−) m/z calculated for C19H14NO4 320.0928 [M-H]−, found: 320.0936.
1-((4'-(Aminomethyl)-[1,1'-biphenyl]-4-yl)methyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylic acid (10w)
1H NMR (600 MHz, DMSO-d6): δ 10.10 (s, 1H), 8.71 (d, J = 2.0 Hz, 1H), 8.19 (s, 3H), 7.90 (d, J = 2.0 Hz, 1H), 7.74 (dd, J = 8.2, 4.1 Hz, 4H), 7.54 (dd, J = 8.2, 5.3 Hz, 4H), 5.40 (s, 2H), 4.08 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.6, 166.7, 149.2, 140.7, 140.3, 140.2, 135.7, 134.0, 130.1, 129.5, 127.9, 127.5, 126.4, 113.9, 60.3. HRMS-ESI (−) m/z calculated for C20H17N2O4 349.1194 [M-H]−, found: 349.1196.
5-Hydroxy-4-oxo-1-(4-(quinolin-3-yl) benzyl)-1,4-dihydropyridine-3-carboxylic acid (10x)
1H NMR (600 MHz, DMSO-d6): δ 9.32 (d, J = 2.1 Hz, 1H), 8.80 (s, 1H), 8.74 (d, J = 2.0 Hz, 1H), 8.10 (t, J = 7.9 Hz, 2H), 7.96 (d, J = 8.2 Hz, 2H), 7.91 (d, J = 2.0 Hz, 1H), 7.84 (t, J = 7.1 Hz, 1H), 7.71 (t, J = 7.4 Hz, 1H), 7.63 (d, J = 8.2 Hz, 2H), 5.44 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 170.8, 166.0, 148.1, 148.0, 144.2, 139.9, 136.5, 135.7, 135.2, 132.2, 130.7, 128.9, 128.5, 127.7, 127.6, 126.5, 125.6, 113.1, 59.5. HRMS-ESI (−) m/z calculated for C22H15N2O4 371.1037 [M-H], found: 371.1039.
5-Hydroxy-4-oxo-1-((4'-sulfamoyl-[1,1'-biphenyl]-4-yl)methyl)-1,4-dihydropyridine-3-carboxylic acid (10y)
1H NMR (600 MHz, DMSO-d6): δ 10.08 (s, 1H), 8.73 (d, J = 2.0 Hz, 1H), 7.89 (d, J = 2.0 Hz, 1H), 7.88 (d, J = 8.6 Hz, 2H), 7.86 (d, J = 8.6 Hz, 2H), 7.78 (d, J = 8.2 Hz, 2H), 7.55 (d, J = 8.2 Hz, 2H), 7.39 (s, 2H), 5.41 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 170.8, 165.9, 148.2, 142.9, 142.4, 139.9, 138.7, 135.5, 128.7, 127.0, 126.0, 125.5, 113.0, 59.4. HRMS-ESI (−) m/z calculated for C19H15N2O6S 399.0656 [M-H], found: 399.0667.
5-Hydroxy-4-oxo-1-(4-(pyridin-3-yl) benzyl)-1,4-dihydropyridine-3-carboxylic acid (10z)
1H NMR (600 MHz, DMSO-d6): δ 9.01 (s, 1H), 8.73 (d, J = 1.9 Hz, 1H), 8.68 (d, J = 4.9 Hz, 1H), 8.33 (d, J = 7.9 Hz, 1H), 7.90 (d, J = 2.0 Hz, 1H), 7.82 (d, J = 8.2 Hz, 2H), 7.70 (dd, J = 7.8, 5.1 Hz, 1H), 7.59 (d, J = 8.1 Hz, 2H), 5.42 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 171.1, 166.2, 148.7, 145.3, 144.4, 140.2, 138.3, 136.4, 136.3, 136.0, 129.1, 127.9, 125.8, 125.4, 113.4, 59.7. HRMS-ESI (−) m/z calculated for C18H13N2O4 321.0881 [M-H], found: 321.0887.
Ethyl-1-(4-fluorobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxylate (18)
1H NMR (600 MHz, CD3OD): 8.50 (d, J = 1.2 Hz, 1H), 7.58 (d, J = 1.2 Hz, 1H), 7.41 (dd, J = 8.2, 5.4 Hz, 2H), 7.16 (t, J = 8.2 Hz, 2H), 5.25 (s, 2H), 4.34 (q, J = 7.1 Hz, 2H), 1.36 (t, J = 7.1 Hz, 3H). 13C NMR (150 MHz, DMSO-d6) δ 161.2, 156.1, 154.1, 142.5, 133.5, 123.4, 122.1, 113.2, 107.9, 105.7, 52.7, 52.1, 9.3. HRMS-ESI (−) m/z calculated for C15H13FNO4 290.0834 [M-H], found: 290.0837.
1-(4-Fluorobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxamide (20)
1H NMR (600 MHz, DMSO-d6): δ 9.44 (d, J = 4.2 Hz, 1H), 9.13 (s, 1H), 8.49 (d, J = 2.1 Hz, 1H), 7.65 (d, J = 2.1 Hz, 1H), 7.53 – 7.39 (m, 3H), 7.23 (t, J = 8.8 Hz, 2H), 5.23 (s, 2H). 13C NMR (150 MHz, DMSO-d6) δ 169.7, 165.6, 163.0, 161.4, 149.5, 139.9, 132.6, 130.5, 122.1, 116.0, 58.8. HRMS-ESI (−) m/z calculated for C13H10FN2O3 261.0681 [M-H], found: 261.0681.
N-(2,4-difluorobenzyl)-1-(4-fluorobenzyl)-5-hydroxy-4-oxo-1,4-dihydropyridine-3-carboxamide (22)
1H NMR (600 MHz, DMSO-d6): δ 10.60 (t, J = 5.7 Hz, 1H), 9.20 (s, 1H), 8.51 (d, J = 1.8 Hz, 1H), 7.67 (d, J = 1.8 Hz, 1H), 7.46 (dd, J = 8.2, 5.8 Hz, 2H), 7.41 (dd, J = 15.5, 8.4 Hz, 1H), 7.23 (dd, J = 14.5, 5.8 Hz, 3H), 7.08 – 7.03 (m, 1H), 5.25 (s, 2H), 4.53 (d, J = 5.7 Hz, 2H). HRMS-ESI (−) m/z calculated for C20H14N2O3 387.0962 [M-H], found: 387.0967.
Biology
Reagents
Biologicals
Recombinant HIV-1 reverse transcriptase (RT) was expressed and purified as previously described.36 The catalytically active RNase H domain fragment of HIV-1 RT was expressed from plasmid pCSR231 (Dr. Daria Hazuda, Merck, West Point, PA) and purified as previously described.37 P4R5 HIV infection indicator cells were obtained from the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH (p4R5.MAGI from Dr. Nathaniel Landau). These cells express CD4, CXCR4 and CCR5 as well as a β-galactosidase reporter gene under the control of an HIV LTR promoter.
Chemicals
DNA and RNA oligonucleotides for the preparation of RNA/DNA duplexes for assay of RNase H activity were purchased from Trilink (San Diego, CA).
RNase H assay
RNase H activity was measured essentially as previously described.21 Three different RNA/DNA duplex substrates were used, each assessing a different mode of RNAse H cleavage. HTS-1 (RNA 5’-gaucugagccugggagcu -3’-fluorescein annealed to DNA 3’-CTAGACTCGGACCCTCGA -5’-Dabcyl) is a high sensitivity duplex that assesses non-specific internal cleavage. HTS-2 (RNA 5’–cugguuagaccagaucugagccugggagcu–3’-fluorescein annealed to DNA 3’-GGTCTAGACTCGGACCCTCGA–5’-Dabcyl) provides a duplex with a recessed DNA 3’-terminus and measures 3’-DNA directed or polymerase directed RNase H cleavage. HTS-3 (RNA 5’–accagaucugagccugggagcu–3-fluorescein annealed to DNA 3’-GACCAATCTGGTCTAGACTCGGACCCTCGA–5’-Dabcyl) measures 5’-RNA-directed RNase H cleavage.
RT polymerase assay
HIV RT polymerase activity was determined in the presence and the absence of inhibitor using 10 µM [3H]-TTP and 40nM poly(rA)-oligo(dT)16 (both obtained from Perkin Elmer, Waltham, MA) in 50 mM Tris-HCl, pH 7.4 (37°C) containing 60 mM KCl and 5 mM MgCl2. Reactions were initiated by the addition of 10 nM WT or mutant RT and carried out for 20 min at 37°C. Reactions were quenched by 200 µl ice cold 10% TCA containing 20 mM sodium pyrophosphate and filtered using a 1.2 µm glass fiber filter 96-well plates (Millipore, Billerica, MA) followed by sequentially wash with 10% TCA and ethanol. The extent of radionucleotide incorporation was determined by liquid scintillation spectrometry.
Order-of-addition RNase H inhibition assays
Order-of-addition RNase H inhibition assays were carried out as previously described with some minor modifications.21, 38 Each reaction contained 100 nM HIV-1 RT, 6 mM MgCl2, 250 nM RNA/DNA (HTS-1) substrate, 30 µM inhibitor, and 1% DMSO. HIV-1 RT was pre-incubated with inhibitor, MgCl2, and/or RNA/DNA (HTS-1) substrate in the following conditions at 37 °C for 10 min prior to starting the reaction: RT + inhibitor, RT + inhibitor + MgCl2, RT + inhibitor + HTS-1, RT + HTS-1, and RT + HTS-1 + MgCl2 + DMSO (no inhibitor control). The reactions were initiated by the addition of the missing components, and were run at 37 °C for 10 min. A control with only DMSO carrier was used to adjust the fluorsecnece signal of the samples that contained inhibitor. Values are reported as mean ± standard deviation from two independent experiments.
To mimic conditions similar to those in the virion, order-of-addition reactions containing 300 nM HIV-1 RT, 6 mM MgCl2, 50 nM RNA/DNA (HTS-1) substrate, 0.5 mM EDTA, 30 µM inhibitor, and 1% DMSO were also performed. HIV-1 RT was pre-incubated with inhibitor, MgCl2, and/or RNA/DNA (HTS-1) substrate in the following conditions at 37 °C for 10 min prior to starting the reaction: RT + inhibitor, RT + HTS-1, and RT + HTS-1 + MgCl2 + DMSO (no inhibitor control). The reactions were initiated by the addition of the missing components, and were run at 37 °C for 2 min. A control with only DMSO carrier was used to adjust the fluorsecnece signal of the samples that contained inhibitor. Values are reported as mean ± standard deviation from two independent experiments.
HIV IN assay
HIV integrase was expressed and purified as previously reported.39 Inhibition assays were performed using a modified protocol of our reported method.39 Briefly, 2.1 µL of compound suspended in DMSO was placed in duplicate into a Black 96 well non-binding plate (corning 3991). Compounds were plated in duplicate to a final concentration of 0.13 — 100 µM. To each well of the plate 186.9 µL of reaction mixture without DNA substrate was added (10 mM HEPES pH 7.5, 10 % glycerol w/v, 10 mM MnCl2, 1 mM DTT, 1 µM integrase). The enzyme was incubated with inhibitor for 10 min at 25 °C after which the reaction was initiated by the addition of 21 µL of 500 nM oligo (5’ biotin ATGTGGAAAATCTCTAGCA annealed with ACTGCTAGAGATTTTCCACAT 3’ Cy5). Reactions were incubated at 37 °C for 30 min and then quenched by the addition of 5.2 µL 500 mM EDTA. Each reaction was moved (200 µL) to a MultiScreen HTS PCR plate (Millipore MSSLBPC10) containing 20 µL streptavidin agarose beads (Life Technologies S951) and incubated with shaking for 30 min. A vacuum manifold was used to remove the reaction mixture and the beads were similarly washed 3 times with wash buffer (.05% SDS, 1 mM EDTA in PBS). The plates were further washed 3 times with 200 µL 50 mM NaOH to denature DNA not covalently linked to the biotin modification. For each denaturation step the plate was incubated with shaking at 25 °C for 5 min and the NaOH was removed by centrifugation at 1000 g for 1 min. The reaction products were eluted from the beads by the addition of 150 µL formamide. The plate was incubated at 25 °C for 10 min and read directly at 635/675 in a SpectraMax i3 plate reader (Molecular Devices).
Antiviral assays
Antiviral assays were carried out using P4R5 indicator cells essentially as previously described.23 P4R5 cells were cultured in 96-well microplates (5×103 cells per well and maintained in DMEM/10% FBS supplemented with puromycin (0.5 µg/ml). Cells were incubated in the presence or the absence of drug for 16h then exposed to HIV followed by an additional incubation period of 48h. The extent of infection was assessed using a fluorescence-based β-galactosidase detection assay, as previously described.40
Modeling and docking analysis
Molecular modeling was performed using the Schrodinger small molecule drug discovery suite 2014-3. The crystal structure of HIV-1 RNase H p15 with engineered E.coli loop co-crystallized with pyrimidinol carboxylic acid (5) was extracted from the protein data bank (PDB code: 3HYF31) as reported by Lansdon et.al.14 The above structure was subjected to analysis and found that the native ligand, pyrimidinol carboxylic acid was bound to the active site of RNase H. This model was subjected to protein preparation wizard32–33 (Schrödinger Inc) in which missing hydrogens atoms were added, zero-order bonds to metals were created followed by the generation of metal binding states. The structure of protein was minimized using OPLS 2005 force field34 to optimize hydrogen bonding network and converge heavy atoms to the RMSD of 0.3 Å. The processed model indicates that the interaction between the pyrimidinol carboxylic acid (5) and RNase H is mediated by two metal cations (Mn2+) coordinated by the active site residues D443, E478, D498 and D549.
The receptor grid generation tool in Maestro (Schrodinger Inc) was used to define an active site around the native ligand (5) to cover all the residues within 12 Å from it with both the metal cofactors (Mn2+) as a constraint to identify the chelating triad during docking. Compounds 10r and 10y were drawn using Maestro and subjected to Lig Prep35 to generate conformers, possible protonation at pH of 7±3 and metal binding states which serves as an input for docking process. All the dockings were performed using Glide XP22–23 (Glide, version 6.4) mode with both the Mn2+ metal cofactors as a constraint. The van der Waals radii of non-polar atoms for each of the ligands were scaled by a factor of 0.8. The predicted favorable binding mode of compounds 10r and 10y features the critical interaction between the chelating triad and to the divalent metal cofactors. All the ligands within the active site of RNase H were further refined post docking by minimizing under implicit solvent to account for the local protein flexibility.
HIV-1 RT crystallization and data collection
HIV-1 RT containing C280S mutations in the p66 and p51 subunits was expressed and purified as previously described.38, 41 Co-crystallization of HIV-1 RT with 10y was acheived by mixing a solution of RT at a final concentration of 10 mg/ml, 50 mM MgCl2, 5 mM tris(2-carboxyethyl)phosphine (TCEP) HCl, 0.5% β-octylglucoside, and 1 mM 10y in a 1:1 ratio with a solution of 15% PEG 3500, 0.1 M sodium potassium phosphate, 5% ethylene glycol, and 0.1 M MES pH 6.1. Large, blocky crystals grew by hanging drop vapor diffusion at 18 °C in 2–3 days.
HIV-1 RT/10y co-crystals were additionally soaked with 1.5 mM 10y, 5 mM TCEP HCl, 0.6% β-octylglucoside, 8% ethylene glycol, and 50 mM MgCl2 for 20 minutes before brief cryoprotection in a solution containing 23% ethylene glycol and 4% trimethylamine N-oxide, followed by fast cryo-cooling in liquid nitrogen. Four data sets collected at Beamline 4.2.2 of the Advanced Light Source at Lawrence Berkeley National Laboratory were processed, scaled, and merged to 2.9 Å resolution using XDS.42 The HIV-1 RT/10y crystals were of space group P1, and had unit cell dimensions of a = 69.1 Å, b = 89.3 Å, c = 108.2 Å and α = 105.5°, β = 92.7°, γ = 110.8°. Two RT molecules were present in the asymmetric unit, and the Matthews coefficient was 2.5 Å3/Da, corresponding to a solvent content of 51.6%.43 Crystal data and statisitcs are listed in SI Table 1.
Phase determination and structure refinement
The structure was determined by molecular replacement using Phaser.44 A crystal structure of unliganded HIV-1 RT (Protein Data Bank code 1DLO) was used as an initial search model for a related higher resolution structure (2.28 Å) of an unpublished P1 RT/RNase H inhibitor complex with two molecules in the asymmetric unit. Rigid-body, simulated annealing (to remove bias), atomic displacement parameter (ADP), real-space and restrained refinement were perfomed on this initial model using Phenix.45 Several cycles of model building and refinement were carried out using Coot46 and Phenix or Refmac,47 respectively, for the related higher resolution RT/RNase H inhibior complex, and this final model was used as the search model for molecular replacement of the RT/10y complex using the above protocols. The final RT/10y structure was validated using MolProbity.48 Final refinement statistics are listed in SI Table 1.
Supplementary Material
Scheme 1a.
Synthesis of hydroxylpyridinone carboxylic acids 10a–z.
a Reagents and conditions: a) NaH, THF, r.t, 16 h, 95% ; b) DMF DMA, toluene, r.t, 30%; c) Ethyl formate, KOtBu, THF, r.t, 75%; d) R-NH2, EtOH, 100 °C, 30–40 min, MW, 40–85%; e) 2N NaOH, EtOH, 90 °C, 4–6 h, 65–95%; f) ArB(OH)2, K2CO3, Pd(PPh3)4, DME/H2O (4:1), 150 °C, 30–40 min, MW, 60–80%; g) TFA, 80 °C, 20 min, MW, 40–87%.
Acknowledgments
This research was supported in part by the National Institutes of Health (AI100890 to SGS, MAP and ZW), and by the Research Development and Seed Grant Program of the Center for Drug Design, University of Minnesota. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
ABBREVIATIONS USED
- RT
reverse transcriptase
- RNase H or RNH
ribonuclease H
- HIV
human immunodeficiency virus
- IN
integrase
- NRTIs
nucleoside RT inhibitors
- NNRTIs
nonnucleoside RT inhibitors
- INST
integrase strand transfer
- SAR
structure-activity-relationship
- MW
Microwave
Footnotes
ASSOCIATED CONTENT
Supporting Information Available. Chemical synthesis, general procedures, spectral data, including 1H NMR of intermediates (16a–16s; 17a–17s, 17w–17z), and crystallographic data and refinement statistics are available in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
PDB ID. The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (PDB ID: 5J1E). Authors will release the atomic coordinates and experimental data upon article publication.
Reference
- 1. http://www.unaids.org/en/dataanalysis/datatools/aidsinfo/ [Google Scholar]
- 2.Yeni P. Update on HAART in HIV. J. Hepatol. 2006;44:S100–S103. doi: 10.1016/j.jhep.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 3.Tramontano E, Di Santo R. HIV-1 RT-associated RNase H function inhibitors: recent advances in drug development. Curr. Med. Chem. 2010;17:2837–2853. doi: 10.2174/092986710792065045. [DOI] [PubMed] [Google Scholar]
- 4.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cihlar T, Ray AS. Nucleoside and nucleotide HIV reverse transcriptase inhibitors: 25 years after zidovudine. Antiviral Res. 2010;85:39–58. doi: 10.1016/j.antiviral.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 6.de Bethune MP. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989–2009) Antiviral Res. 2010;85:75–90. doi: 10.1016/j.antiviral.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 7.HIV drugs in development. [accessed 2016]; http://www.thebody.com/index/treat/newdrugs.html. [Google Scholar]
- 8.Parniak MA. Unpublishd data. [Google Scholar]
- 9.Cao L, Song W, De Clercq E, Zhan P, Liu X. Recent progress in the research of small molecule HIV-1 RNase H inhibitors. Curr. Med. Chem. 2014;21:1956–1967. doi: 10.2174/0929867321666140120121158. [DOI] [PubMed] [Google Scholar]
- 10.Klumpp K, Hang JQ, Rajendran S, Yang YL, Derosier A. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. In: P WK, Overton H, Parkes KEB, Cammack N, Martin JA, editors. Nucleic Acids Res. Vol. 31. 2003. pp. 6852–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himmel DM, Maegley KA, Pauly TA, Bauman JD, Das K, Dharia C, Clark AD, Jr, Ryan K, Hickey MJ, Love RA, Hughes SH, Bergqvist S, Arnold E. Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site. Structure. 2009;17:1625–1635. doi: 10.1016/j.str.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Himmel DM, Myshakina NS, Ilina T, Van Ry A, Ho WC, Parniak MA, Arnold E. Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNase H domain of HIV-1 reverse transcriptase and structure-activity analysis of inhibitor analogs. J. Mol. Biol. 2014;426:2617–2631. doi: 10.1016/j.jmb.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw-Reid CA, Munshi V, Graham P, Wolfe A, Witmer M, Danzeisen R, Olsen DB, Carroll SS, Embrey M, Wai JS, Miller MD, Cole JL, Hazuda DJ. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem. 2003;278:2777–2780. doi: 10.1074/jbc.C200621200. [DOI] [PubMed] [Google Scholar]
- 14.Kirschberg TA, Balakrishnan M, Squires NH, Barnes T, Brendza KM, Chen X, Eisenberg EJ, Jin W, Kutty N, Leavitt S, Liclican A, Liu Q, Liu X, Mak J, Perry JK, Wang M, Watkins WJ, Lansdon EB. RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information. J. Med. Chem. 2009;52:5781–5784. doi: 10.1021/jm900597q. [DOI] [PubMed] [Google Scholar]
- 15.Williams PD, Staas DD, Venkatraman S, Loughran HM, Ruzek RD, Booth TM, Lyle TA, Wai JS, Vacca JP, Feuston BP, Ecto LT, Flynn JA, DiStefano DJ, Hazuda DJ, Bahnck CM, Himmelberger AL, Dornadula G, Hrin RC, Stillmock KA, Witmer MV, Miller MD, Grobler JA. Potent and selective HIV-1 ribonuclease H inhibitors based on a 1-hydroxy-1,8-naphthyridin-2(1H)-one scaffold. Bioorg. Med. Chem. Lett. 2010;20:6754–6757. doi: 10.1016/j.bmcl.2010.08.135. [DOI] [PubMed] [Google Scholar]
- 16.Beilhartz GL, Ngure M, Johns BA, Deanda F, Gerondelis P, Gotte M. Inhibition of the ribonuclease H activity of HIV-1 reverse transcriptase by GSK5750 correlates with slow enzyme-inhibitor dissociation. J. Biol. Chem. 2014;289:16270–16277. doi: 10.1074/jbc.M114.569707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujishita T, Mikamiyama M, Kawai M, Akiyama T. Substituted 3-hydroxy-4-pyridone derivative. 2010 WO2010110231. [Google Scholar]
- 19.Johns BA, Kawasuji T, Weatherhead JG, Taishi T, Temelkoff DP, Yoshida H, Akiyama T, Taoda Y, Murai H, Kiyama R, Fuji M, Tanimoto N, Jeffrey J, Foster SA, Yoshinaga T, Seki T, Kobayashi M, Sato A, Johnson MN, Garvey EP, Fujiwara T. Carbamoyl pyridone HIV-1 integrase inhibitors 3. A diastereomeric approach to chiral nonracemic tricyclic ring systems and the discovery of dolutegravir (S/GSK1349572) and (S/GSK1265744) J. Med. Chem. 2013;56:5901–5916. doi: 10.1021/jm400645w. [DOI] [PubMed] [Google Scholar]
- 20.Vernekar SKV, Liu Z, Nagy E, Miller L, Kirby KA, Wilson DJ, Kankanala J, Sarafianos SG, Parniak MA, Wang ZQ. Design, synthesis, biochemical, and antiviral evaluations of C6 benzyl and C6 biarylmethyl substituted 2-hydroxylisoquinoline-1,3-diones: dual inhibition against HIV reverse transcriptase-associated RNase H and polymerase with antiviral activities. J. Med. Chem. 2015;58:651–664. doi: 10.1021/jm501132s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parniak MA, Min KL, Budihas SR, Le Grice SF, Beutler JA. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal. Biochem. 2003;322:33–39. doi: 10.1016/j.ab.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Beilhartz GL, Wendeler M, Baichoo N, Rausch J, Le Grice S, Gotte M. HIV-1 reverse transcriptase can simultaneously engage its DNA/RNA substrate at both DNA polymerase and RNase H active sites: implications for RNase H inhibition. J. Mol. Biol. 2009;388:462–474. doi: 10.1016/j.jmb.2009.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirivolu VR, Vernekar SKV, Ilina T, Myshakina NS, Parniak MA, Wang ZQ. Clicking 3 '-azidothymidine into novel potent inhibitors of human immunodeficiency virus. J. Med. Chem. 2013;56:8765–8780. doi: 10.1021/jm401232v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Small-molecule drug discovery suite 2014-3: Glide, version 6.4. New York: Schrödinger LLC; 2014. [Google Scholar]
- 25.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 26.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 27.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su H-P, Yan Y, Prasad GS, Smith RF, Daniels CL, Abeywickrema PD, Reid JC, Loughran HM, Kornienko M, Sharma S, Grobler JA, Xu B, Sardana V, Allison TJ, Williams PD, Darke PL, Hazuda DJ, Munshi S. Structural Basis for the Inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors. J. Virol. 2010;84:7625–7633. doi: 10.1128/JVI.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lansdon EB, Liu Q, Leavitt SA, Balakrishnan M, Perry JK, Lancaster-Moyer C, Kutty N, Liu X, Squires NH, Watkins WJ, Kirschberg TA. Structural and binding analysis of pyrimidinol carboxylic acid and N-hydroxy quinazolinedione HIV-1 RNase H inhibitors. Antimicrob. Agents Chemother. 2011;55:2905–2915. doi: 10.1128/AAC.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC. The structure of unliganded reverse-transcriptase from the human-immunodeficiency-virus Type-1. Proc. Natl. Acad. Sci. USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsiou Y, Ding J, Das K, Clark AD, Hughes SH, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 angstrom resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 32.Sarafianos SG, Das K, Clark AD, Jr, Ding J, Boyer PL, Hughes SH, Arnold E. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA. 1999;96:10027–10032. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsiou Y, Ding JP, Das K, Clark AD, Boyer PL, Lewi P, Janssen PAJ, Kleim JP, Rosner M, Hughes SH, Arnold E. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J. Mol. Biol. 2001;309:437–445. doi: 10.1006/jmbi.2001.4648. [DOI] [PubMed] [Google Scholar]
- 34.Tu XY, Das K, Han QW, Bauman JD, Clark AD, Hou XR, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, Hughes SH, Sarafianos SG, Arnold E. Structural basis of HIV-1 resistance to AZT by excision. Nat. Struct. Mol. Biol. 2010;17:1202–1209. doi: 10.1038/nsmb.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.PyMOL: [accessed 2016]; www.pymol.org. [Google Scholar]
- 36.Fletcher RS, Holleschak G, Nagy E, Arion D, Borkow G, Gu Z, Wainberg MA, Parniak MA. Single-step purification of recombinant wild-type and mutant HIV-1 reverse transcriptase. Protein Expres. Purif. 1996;7:27–32. doi: 10.1006/prep.1996.0004. [DOI] [PubMed] [Google Scholar]
- 37.Gong Q, Menon L, Ilina T, Miller LG, Ahn J, Parniak MA, Ishima R. Interaction of HIV-1 reverse transcriptase ribonuclease H with an acylhydrazone inhibitor. Chem. Biol. Drug Des. 2011;77:39–47. doi: 10.1111/j.1747-0285.2010.01052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirby KA, Marchand B, Ong YT, Ndongwe TP, Hachiya A, Michailidis E, Leslie MD, Sietsema DV, Fetterly TL, Dorst CA, Singh K, Wang Z, Parniak MA, Sarafianos SG. Structural and inhibition studies of the RNase H function of xenotropic murine leukemia virus-related virus reverse transcriptase. Antimicrob. Agents Chemother. 2012;56:2048–2061. doi: 10.1128/AAC.06000-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Bennett EM, Wilson DJ, Salomon C, Vince R. Rationally designed dual inhibitors of HIV reverse transcriptase and integrase. J. Med. Chem. 2007;50:3416–3419. doi: 10.1021/jm070512p. [DOI] [PubMed] [Google Scholar]
- 40.Abram ME, Parniak MA. Virion instability of human immunodeficiency virus type 1 reverse transcriptase (RT) mutated in the protease cleavage site between RT p51 and the RT RNase H domain. J. Virol. 2005;79:11952–11961. doi: 10.1128/JVI.79.18.11952-11961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauman JD, Das K, Ho WC, Baweja M, Himmel DM, Clark AD, Oren DA, Boyer PL, Hughes SH, Shatkin AJ, Arnold E. Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design. Nucleic Acids Res. 2008;36:5083–5092. doi: 10.1093/nar/gkn464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthews BW. Determination of Protein Molecular-weight, hydration, and packing from crystal density. Method Enzymol. 1985;114:176–187. doi: 10.1016/0076-6879(85)14018-8. [DOI] [PubMed] [Google Scholar]
- 44.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.