Abstract
Direct conversion of somatic cells into neurons holds great promise for regenerative medicine. However, as neuronal conversion is relatively inefficient on human cells compared to mouse cells, it has been unclear what might be key barriers to reprogramming in human cells. We recently elucidated an RNA program mediated by the polypyrimidine tract binding protein PTB to convert mouse embryonic fibroblasts (MEFs) into functional neurons. On human adult fibroblasts (HAFs), however, we unexpectedly find that invoke of the documented PTB-REST-miR-124 loop only generates immature neurons. We now report that the functionality requires sequential inactivation of PTB and the PTB paralog nPTB in HAFs. Inactivation of nPTB triggers another self-enforcing loop essential for neuronal maturation, which comprises nPTB, the transcription factor BRN2, and miR-9. These findings suggest two separate gatekeepers to control neuronal conversion and maturation and consecutively overcoming these gatekeepers enables deterministic reprogramming of HAFs into functional neurons.
INTRODUCTION
Neurogenesis is known to undertake a series of cellular programming from initial morphogenesis to subsequent maturation to form distinct cell subtypes in the nervous system 1,2. These sequential events are likely induced by specific morphogenetic signals in combination with step-wise activation/repression of cell autonomous gene expression programs, as indicated by expression waves of lineage-specific transcription factors (TFs) 3 and their distinct functional requirements 4. Importantly, while most of our current knowledge about neurogenesis in mammals is derived from genetically tractable systems, particularly mice, we know relatively little about the parallel process in humans. Mice and humans are clearly different in brain size and the time required for neural development, which may thus involve distinct regulatory strategies 5, and understanding of such critical differences is vital for utilizing mice as models to study neural development and neurodegenerative diseases in humans 6,7.
Recent breakthroughs in regenerative medicine provide new approaches to study specific human diseases by using patient-derived cells. Through reprogramming 8–10, for example, human fibroblasts can be converted to induced pluripotent stem cells (iPSCs), which can be re-differentiated to specific cell types, such as neurons, for functional studies 11,12. Alternatively, fibroblasts can be directly trans-differentiated into neurons with a set of neuronal-specific transcription factors 13–17. Numerous studies have demonstrated the feasibility of such trans-differential approaches on mouse and human embryonic fibroblasts, but it is a general consensus that human cells are much harder to convert, especially those from aged patients 18. This represents a significant barrier to directly using patient-derived fibroblasts to pursue disease mechanisms.
Most cell differentiation or trans-differentiation protocols developed were inspired by the success in iPSCs, which is to transduce a set of TFs specifically expressed in destination cells to a beginning cell type 19. Such procedure is, however, a largely try-and-error process, although some recent efforts were made to strategize the approach by deducing critical developmentally regulated gene networks 20,21. In principle, any cellular reprogramming would require breakup of the existing homeostatic program followed by establishing a sustainable new program, which likely involves sequential switches in various feedback controls. Notably, some specific small molecular inhibitors are able to replace transcription factors to induce cellular reprogramming 22,23. However, little is known about the mechanisms underlying most induced pluripotency or trans-differentiation procedures.
Interestingly, accompanying switches in the expression of lineage-specific TFs are coordinated changes in the expression of regulatory RNAs and RNA binding proteins, suggesting their key functions in cellular reprogramming 24,25. Indeed, specific miRNAs, such as miR-124, have been shown to contribute to fibroblast conversion to neurons when coupled with certain TFs 26,27. We previously established a regulatory loop in which the transcription repressor REST represses miR-124, which in turn dismantles multiple components of the REST complex, and the RNA binding protein PTB serves both as a substrate for and a key inhibitor to miR-124 targeting. We demonstrated that PTB knockdown is necessary and sufficient to potently activate this loop to turn mouse fibroblasts into functional neurons 28.
Surprisingly, when applying the same protocol to humans, we were able to efficiently convert human fibroblasts to neuronal-like cells with complex morphology, especially when combined with a set of small molecule inhibitors, but all induced neurons were at an immature stage(s), indicating that PTB knockdown is sufficient to activate a full neuronal program in mouse, but not human, cells. We now uncover a second loop consisting of the PTB paralog nPTB, another neuronal specific miR-9, and the transcription activator BRN2 to promote neuronal maturation in human cells. In this loop, BRN2 transcriptionally activates miR-9, which in turn post-transcriptionally diminishes nPTB. These findings highlight the intertwined regulatory loops that are sequentially required for cell fate switch to the neuronal lineage.
RESULTS
Initial barrier to neuronal conversion in human cells
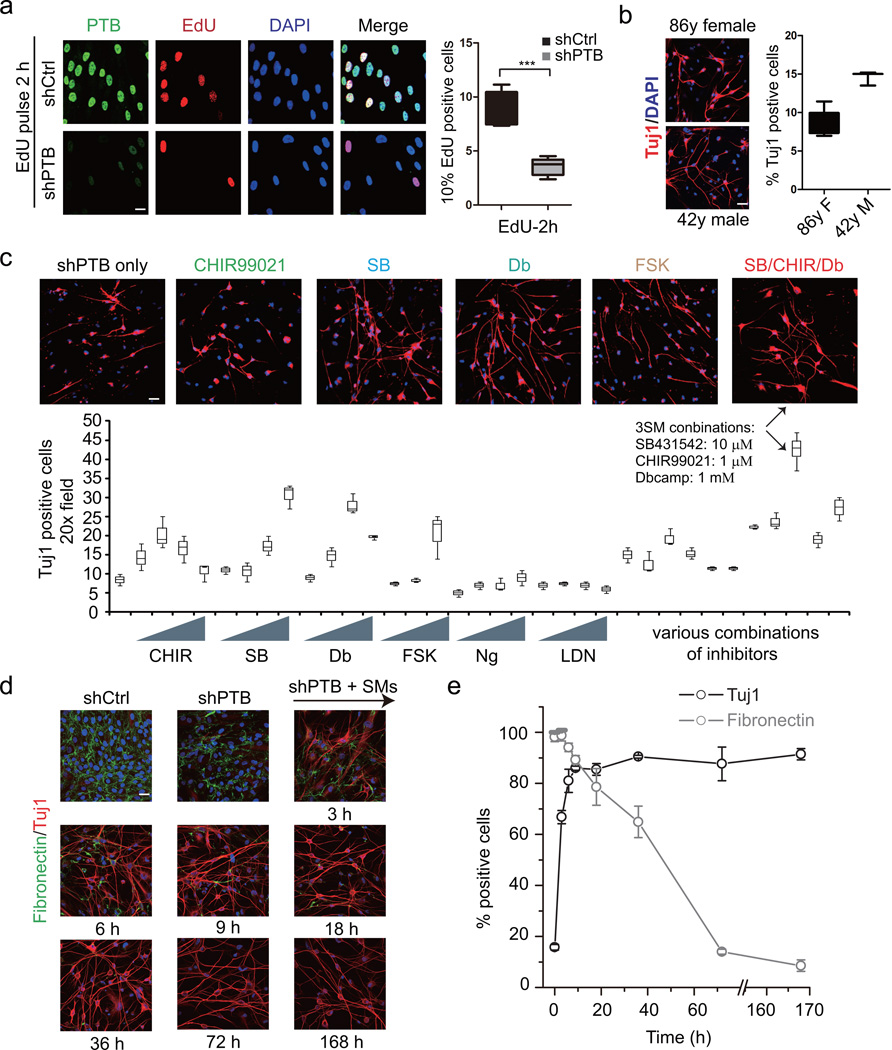
The PTB/miR-124/REST loop is self-sustainable once triggered by initial PTB knockdown. Because the loop is conserved in mammals, we applied the same strategy to two primary human adult fibroblasts (HAFs) derived from a 42-year male and an 86-year female. Both lines showed the expression of fibroblast markers, such as Fibronectin, FSP1 and Vimentin, without detectable contamination of neuronal cells, as indicated by the lack of a series of markers for neurons, neural crest progenitors or neural crest derivatives (Supplementary Fig. 1a). PTB knockdown potently withdrew these cells from cycling, as shown by dramatically reduced EdU signals and increased G2 cells (Fig. 1a and Supplementary Fig. 1b) with 9 to 15% of the seeding cells showing bipolar neuronal morphology and positive staining of the pan-neuronal marker Tuj1 (Fig. 1b). This conversion rate is similar to that on mouse embryonic fibroblasts (MEFs) 28.
Figure 1. Quantitative neuronal induction by shPTB in combination with small molecule inhibitors on HAFs.
(a) EdU pulse labeling in response to PTB depletion. Right panel: Quantification of EdU signals in shPTB and control shRNA-treated cells (n=5 random 20x fields). Two-tailed unpaired Student’s t test was applied to calculate significance. The statistic significance in comparing shCtrl- and shPTB-treated samples is determined: degree of freedom (υ)=8, t = 6.87, P = 0.000127. *** indicates P<0.001.
(b) PTB knockdown in two primary HAF lines and cultured in N3 media for 14 days The efficiency was calculated against total seeding cells in each case (n=6 random 20x fields for 86y female, n=3 random 20x fields for 42y male). All remaining figures were performed on HAFs from the 42y male.
(c) Screen for small molecule inhibitors for enhanced conversion of HAFs to neuronal-like cells in combination with PTB knockdown. Small molecules tested include CHIR (CHIR99021), SB (SB431542), Db (Db-cAMP), FSK (Forskolin), Ng (Noggin), and LDN (LDN-193189). Each compound was titrated in three or four concentrations (see Methods). Tuj1-positive cells were counted from two to four randomly selected fields under 20X magnification. The highest efficiency with CHIR(1 µM)/SB(10 µM)/Db(1 mM) is highlighted by the arrows.
(d) Expression of Tuj1 (red) and the fibroblast marker Fibronectin (green) in HAFs treated with control shRNA or shPTB for 3 days followed by the addition of the three small molecule inhibitors for different periods of time as indicated (n=3 independent biological repeats). Nuclei were stained with DAPI (Blue).
(e) The percentage of cells showing Tuj1 and Fibronectin expression against total seeding cells in different treatment points (n=3 independent biological repeats). All data are presented as mean± SD. All scale bar: 20 µm. See also Supplementary Figure 1.
To increase the conversion efficiency, we tested a set of small molecules (SMs) that have been reported to enhance neuronal differentiation 29–33. While none of these SMs alone was able to turn HAFs into Tuj1-positive cells (Supplementary Fig. 1c), but when individual SMs were combined with PTB knockdown, we detected various levels of enhancement (Fig. 1c). Initially, the induced neuronal-like cells were not healthy looking (Supplementary Fig. 1d). We therefore titrated individual SMs and tested various combinations (Fig. 1c), eventually establishing a 3SM combination (red bar in Fig. 1c) optimal for converting HAFs to Tuj1-positve cells in long-term culture.
We monitored the conversion kinetics: After conditioning the cells by PTB knockdown for 3 days, the cocktail of 3SM was able to boost the number of Tuj1-positive cells as early as 3 hrs, reaching the plateau in ∼1 day, while the level of the fibroblast marker Fibronectin gradually declined to a nearly undetectable level over a period of ∼4 days, resulting in highly efficient conversion (∼90%) of initial HAFs to Tuj1-positive cells (Fig. 1e). In control shRNA-treated cells, while brief 3SM treatment for 3 hrs had no effect (Supplementary Fig. 1e, upper panel), continuous treatment with 3SM alone induced some transient effects on cellular restructuring, as indicated by vaulted nuclei (Supplementary Fig. 1e, upper panels). In contrast, shPTB rapidly induced a persistent switch to neuronal morphology; brief 3SM treatment for 3 hrs clearly enhanced the morphogenetic induction; and the continuous presence of 3SM showed even stronger enhancement effects (Supplementary Fig. 1e, lower panels). These data established that the 3SM alone were insufficient, but their combination with shPTB potently induced cell fate switch. The dramatic reduction in cycling cells coupled with negative Nestin and Sox2 (both neural progenitor makers) staining strongly argue against the possibility of neural stem cell-like intermediates during this conversion process (Fig. 1a and Supplementary Fig. 1f). Interestingly, if we applied the same protocol optimized for HAFs to MEFs, cells died rapidly (data not shown), indicating critical differences in kinetic requirement between mouse and human cells.
Second human-specific barrier to neuronal maturation
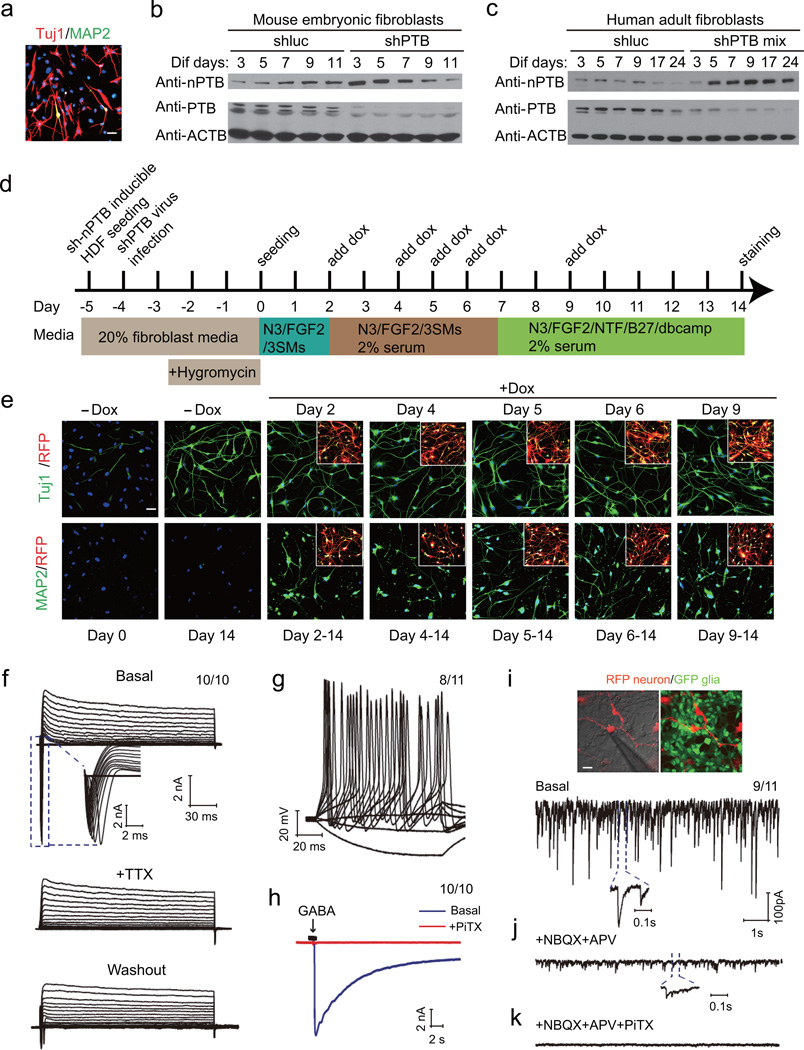
To our surprise, we detected little expression of markers for mature neurons, such as MAP2, NF and NeuN, in shPTB/3SM-treated cells, indicating that the converted neuronal-like cells from HAFs were arrested at a premature stage (Fig. 2a and Supplementary Fig. 2a). As expected, we failed to record any neuronal activity, e.g. Ca2+ influx in response to membrane depolarization or the induction of action potentials upon current injection, even on prolonged culture of the converted cells. This indicates a second barrier to neuronal maturation, which appears to be automatically invoked in MEFs, as we showed previously 28, but not in HAFs.
Figure 2. Generation of functional neurons from HAFs by sequential knockdown of PTB and nPTB.
(a) PTB knockdown-induced neuronal-like cells from HAFs showed extensive Tuj1 (red), but rare MAP2 (green) expression. n=3 independent biological repeats.
(b, c) Time-course analysis of nPTB expression by Western blotting in shPTB-treated MEFs (b) and HAFs (c). PTB and nPTB were examined by Western blotting. β-actin served as loading control, n=2 independent experiments. Uncropped versions of all Western blots are shown in Supplementary Figure 7.
(d) Diagram of the reprogramming strategy by sequential knockdown of PTB and nPTB. HAFs were first infected by an inducible lentivirus containing the expression cassettes for shnPTB and red fluorescence protein (RFP) in combination of an shPTB-expressing lentivirus. After hygromycin selection, cells were treated with the three small molecule inhibitors (SMs). shnPTB expression was induced by the addition of doxycycline (Dox) to the media at different time points. See also Supplementary Figure 2.
(e) Representative images of converted neuronal-like cells from HAFs upon nPTB knockdown at different time points. Top panels: Cells treated with Dox at different days; Tuj1 (green) stained at day 14. Bottom panel: Cells treated with Dox from day 2 to day 9 and MAP2 (green) stained at day 14. RFP signals in the inserts indicate successful shnPTB lentiviral infection. The data were selected from two biological repeats showing similar results.
(f) Representative trace of whole-cell currents under voltage clamp recording mode. Typical voltage-dependent Na+ currents were reversibly blocked by the sodium channel inhibitor TTX (middle panel), which were partially recovered by washing out the drug (bottom panel). 10 out of 10 examined cells showed the recorded activity.
(g) Representative trace of repetitive action potentials induced by current injection. 8 out of 11 examined cells showed the recorded activity.
(h) GABAA receptor-mediated current responses elicited by focal application of GABA (blue), which could be completely blocked with the GABAA receptor specific blocker PiTX. 10 out of 10 examined cells showed the recorded activity.
(i, j, k) Spontaneous postsynaptic currents (PSCs) recorded on RFP-marked neurons co-cultured with GFP-marked glia (i). Insert shows fast-kinetic PSCs. EPSCs were blocked by NBQX+APV (j). Inset highlights GABA-induced IPSCs, blocked with PiTX (k). 9 out of 11 examined cells showed the recorded activity. All scale bar: 20 µm
To search for the missing regulator(s), we performed RNA-seq on converted neuronal-like cells in comparison with beginning HAFs. We noted the induction of a large panel of REST target genes. However, unlike MEFs, we observed a sustained increase in mRNA for the RNA binding protein PTBP2 (nPTB), a PTB paralog in the nervous system, in PTB-depleted HAFs (Supplementary Fig. 2b). As nPTB is known to be induced for several days during initial neuronal induction followed by graduate decline when neuroblasts are differentiated into functional neurons 34,35 or during cortex development 36, we hypothesized that dynamic regulation of nPTB might be a key difference between murine and human cells during trans-differentiation. To test this hypothesis, we monitored nPTB expression after PTB knockdown in both MEFs and HAFs. In contrast to MEFs and consistent with the RNA-seq data, we found that the induced nPTB protein showed persistent expression in neuronal-like cells from HAFs (Fig. 2b,c). This suggests a human-specific barrier to neuronal maturation.
Overcoming both barriers by sequential PTB/nPTB knockdown
To test the 2-barrier hypothesis, we generated an inducible knockdown cell line by infecting HAFs with four validated lentiviral shRNA pools against nPTB (Supplementary Fig. 2c,d). Curiously, we observed cell death if PTB and nPTB were co-depleted in both MEFs and HAFs (data not shown), indicating a critical sequential process required for neuronal differentiation despite the likely timing difference between mouse and human cells. We therefore tested PTB knockdown with shRNA followed by selection with hygromycin for 3 days, and then induced nPTB depletion (indicated by RFP expression from the shnPTB expression unit) at different time points upon doxycycline addition (Fig. 2d). Strikingly, after seeding PTB knockdown cells in differentiation media for 2 days or beyond, nPTB knockdown generated perfectly healthy, MAP2-positive (stained at day 14) cells (Fig. 2e). The resulting neuronal-like cells remained healthy in culture for at least 3 months, the longest time we tested.
We next determined whether such neuronal-like cells by sequential PTB/nPTB knockdowns possess neuronal-specific membrane properties. We detected fast-activating and inactivating inward Na+ currents and outward K+ currents after doxycycline treatment for 4 weeks followed by co-culturing with GFP-marked glial cells for additional 2 weeks (Fig. 2f). The Na+ currents could be blocked by the voltage-gated sodium channel-specific inhibitor tetrodotoxin (TTX), which could be partially relieved after washing out the inhibitor, indicative of an active binding and passive disassociation process (Fig. 2f). We also observed repetitive action potentials upon current injection (Fig. 2g). The cells also expressed functional GABAA-receptors, based on their response to GABA, which could be specifically blocked by picrotoxin (PiTX), a GABAA receptor-specific inhibitor (Fig. 2h). We also detected spontaneous postsynaptic currents of various amplitude and frequencies, which could be sequentially blocked by inhibitors against the excitatory (NBQX+APV) and inhibitory (NBQX+APV+PiTX) receptors (Fig. 2i–k). Collectively, these data demonstrated the functionality of induced neurons from HAFs by sequential PTB/nPTB knockdown in the presence of co-cultured glial cells.
nPTB regulates BRN2 during neuronal maturation
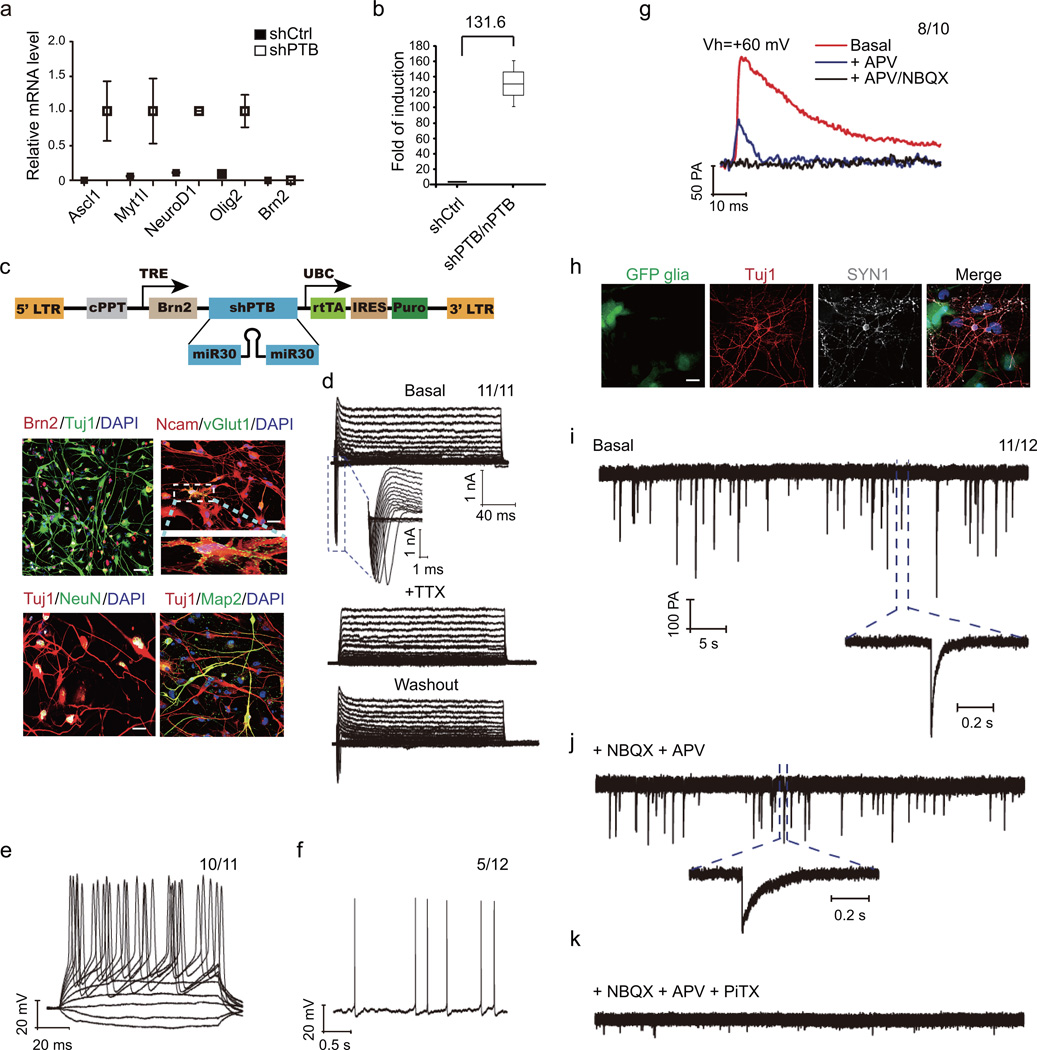
The above data suggest that nPTB knockdown is sufficient to overcome a critical barrier to neuronal maturation, indicating the activation of a new regulatory program. To understand the mechanism, we examined the expression of key TFs previously shown to generate functional neurons on human cells 14, revealing their induction with one exception by PTB knockdown (Fig. 3a), consistent with these genes being target for REST in HAFs (Supplementary Fig. 3a). The exception is BRN2, a cortex-specific transcription factor 37, which remained repressed in PTB knockdown HAFs (Fig. 3a). Importantly, sequential knockdown of PTB and nPTB potently induced BRN2 expression (Fig. 3b).
Figure 3. Neuronal maturation induced by BRN2 in combination with PTB knockdown.
(a, b) mRNA levels of key TFs assessed by RT-qPCR after PTB knockdown (a) and induced BRN2 after sequential knockdown of PTB and nPTB (b). Data were normalized against the internal GAPDH control and presented as mean± SD. Two biological repeat were performed for (a), and three biological repeats were performed for (b). The calculated fold-change is indicated in (b).
(c) Doxycycline-inducible shPTB and BRN2 overexpression units (top). Mature neuronal markers examined (bottom): BRN2 (Red), NCAM (Red), vGlut1 (Green), NeuN (Green) and MAP2 (Green). Nuclei were stained with DAPI (Blue). The data were based from three biological repeats. Scale bar: 20 µm
(d) Representative trace of whole-cell currents in neurons induced by PTB knockdown plus BRN2 overexpression. Insert highlights Na+ currents, reversibly blocked by TTX. 11 out of 11 examined cells showed the recorded activity.
(e, f) Representative trace of repetitive action potentials induced by current injection (e) and spontaneous action potentials (f). 10 out of 11 examined cells showed the recorded activity in (e) and 5 out of 12 examined cells showed the recorded activity (f).
(g) Representative trace of EPSCs. Vh: holding potential (+60mV), revealing both AMPA and NMDA postsynaptic currents (basal line in Red), which could be which could be sequentially blocked with APV (antagonist of NMDA-type glutamate receptors) and NBQX (antagonist of AMPA receptors). 8 out of 10 examined cells showed the recorded activity.
(h) The induced neurons were co-cultured with GFP-marked glia after Dox induction for 50 days. SYN1 (bright, infrared-fluorescence) showed typical punctate staining in Tuj1 (red)-positive cells. The data were based from two biological repeats. Scale bar: 20 µm
(i, j, k) Spontaneous EPSCs and IPSCs recorded on glia co-cultured neurons. Insert shows fast-kinetic AMPA currents (i), blocked by NBQX plus APV (j). The remaining slow response of GABA-induced IPSCs was further blocked with PiTX (k). 11 out of 12 examined cells showed the recorded activity. See also Supplementary Table 1 and Supplementary Figure 3.
We next determined the potential key contribution of activated BRN2 to neuronal maturation. We first tested various combinations of TFs in combined with PTB knockdown, finding that the inclusion of BRN2 was critical for efficient MAP2 expression (Supplementary Fig. 3b) and for invoking Ca2+ influx (Supplementary Fig. 3c). We then engineered an inducible polycistronic unit in a stable HAF line to express both shPTB and BRN2 (Fig. 3c, top panel). Doxycycline treatment induced rapid PTB knockdown and gradual BRN2 expression, and after 2 weeks of such treatment, we detected multiple markers for mature neurons, including MAP2, NCAM, vGLUT1 and NeuN (Fig. 3c, bottom panels).
To demonstrate the functionality of these neurons, we recorded TTX-sensitive sodium currents after 1 week of co-culturing with GFP-marked glial cells (Fig. 3d), current-triggered repetitive action potentials (Fig. 3e), occasionally fired spontaneous action potentials (Fig. 3f), and functional AMPA and NMDA currents characterized by using specific inhibitors (Fig. 3g). After co-culturing with GFP-marked glial cells for 3 to 4 weeks (Fig. 3h), we detected spontaneous postsynaptic activities of variable amplitude and frequencies (Fig. 3i–k). Neurons induced by sequential knockdown of PTB/nPTB or PTB knockdown coupled with BRN2 overexpression were similar based on several key electrophysiological properties (Supplementary Table 1). Collectively, these data demonstrated that PTB knockdown plus BRN2 expression is sufficient to produce functional neurons from HAFs.
BRN2 is essential for NPC differentiation
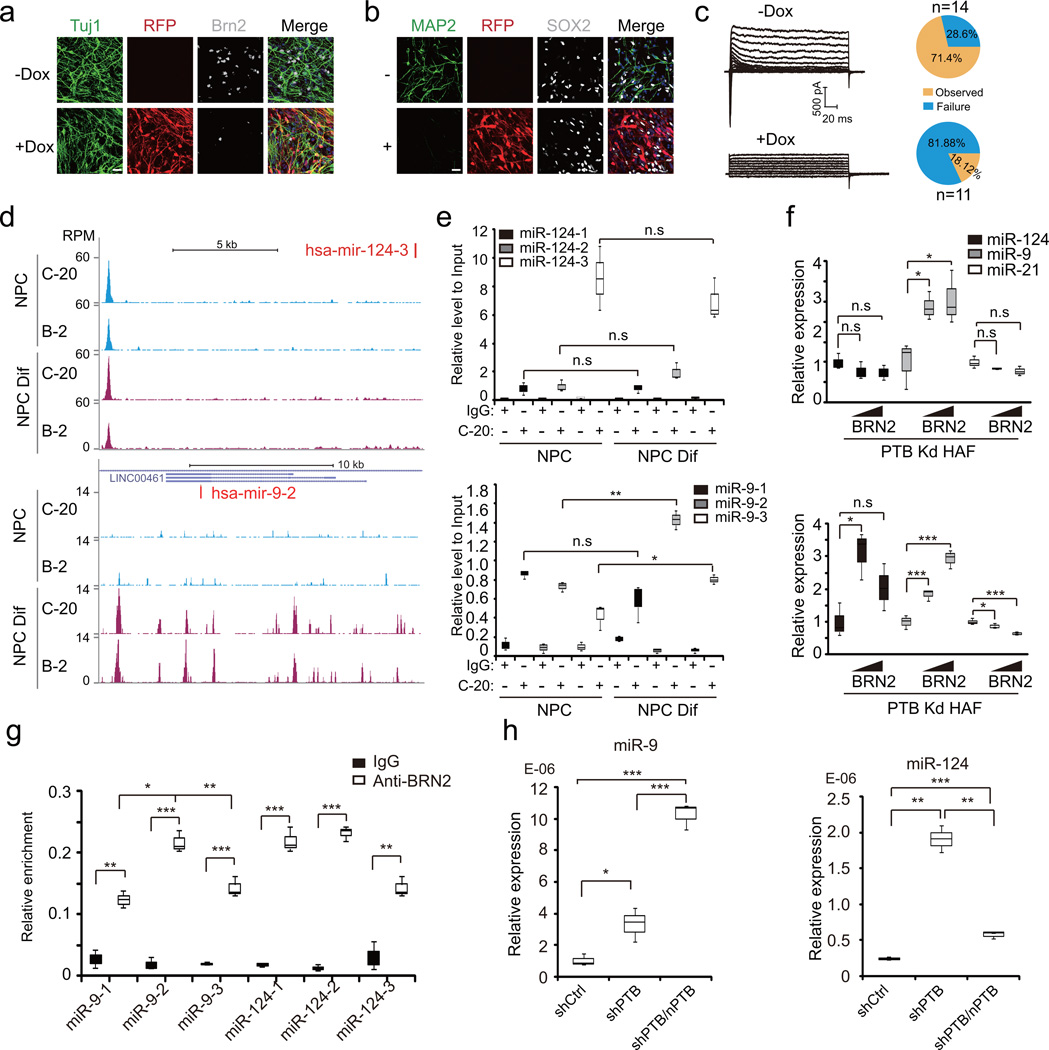
We next wished to establish critical roles of BRN2 in neuronal maturation in a different physiological context, i.e. differentiation of human neural progenitor cells (hNPCs) to cortical neurons upon withdrawing bFGF from culture media 38. These hNPCs showed positive expression of neural stem cell-specific markers, such as Nestin, Sox2, and BRN2 (Supplementary Fig. 4a). Upon BRN2 knockdown, we found that, while early neuronal differentiation events were not affected, as evidenced by normal Tuj1 expression (Fig. 4a), the expression of MAP2, a marker associated with more matured neurons, was severely compromised (Fig. 4b and Supplementary Fig. 4b–c). No inward Na+ currents (Fig. 4c) were hardly detected in BRN2-depleted hNPCs (Supplementary Fig. 4d). These data demonstrated the requirement of BRN2 for hNPC differentiation into functional neurons.
Figure 4. Essential role of BRN2 for neuronal maturation in hNPCs and activation of mature neuron-specific miRNAs.
(a) Immunofluorescence analysis of neurons derived from hNPCs upon BRN2 knockdown. n=3 independent biological repeats. Scale bar: 20 µm
(b) Expression of SOX2 (bright, infrared-fluorescence), but not MAP2 (green), in BRN2-depleted hNPCs. n=3 independent biological repeats. Scale bar: 20 µm
(c) Representative trace of whole-cell currents recorded on neurons derived from hNPCs with or without BRN2 expression. Pie charts: % cells exhibiting the currents before (top) and after (bottom) BRN2 knockdown.
(d) Representative BRN2 ChIP-seq signals on miR-124-3 and miR-9-2 loci before or after hNPC differentiation. Two anti-BRN2 antibodies (Mouse monoclonal IgG B-2 and goat IgG C-20) were used for constructing ChIP-seq libraries. Y-axis: tag density. See also Supplementary Table 2 for the list of identified BRN2 targets.
(e) ChIP-qPCR analysis of BRN2 binding (with C-20 antibody; IgG as negative control) on miR-124 (top) and miR-9 loci (bottom) before and after hNPC differentiation. n=3 independent experiments. Two-tailed unpaired Student’s t-test was applied to calculate significance. For miR-124: box 2/8: υ=4, t=0.0304, P=0.977; box 10/4: υ=4, t=2.365, P= 0.0772: box 12/6: υ=4, t =1.059, P=0.349; For miR-9: box 2/8: υ=4, t=2.339, P=0.0795; box 10/4: υ=3, t=8.583, P=0.00332; box 6/12: υ=4, t=4.577; P=0.0102..
(f) RT-qPCR analysis of miR-124 and miR-9 after induced BRN2 expression for 6 (upper panel) or 12 (lower panel) days in shPTB-treated HAFs. U6 served as internal control. n=3 independent experiments. For 6 days: box 1/2: υ=4, t=1.263, P=0.275; box 1/3: υ=4, t=1.615, P=0.182; box 4/5: υ=4, t=3.529, P=0.0243; box 4/6: υ=4, t=3.059, P=0.0377; box 7/8: υ=4, t=1.802, P=0.146 ; box 7/9: υ=4, t=1.995, P=0.117. For 12 days: box 1/2: υ=4, t=4.069, P=0.0152; box 1/3: υ=4, t=2.051, P=0.110; box 4/5: υ=4, t=5.213, P=0.00646; box 4/6: υ=4. t=9.356, P=0.000727; box 7/8: υ=9, t=3.066, P=0.0134; box 7/9: υ=9, t=8.037; P = 0.0000213.
(g) RRN2 ChIP-qPCR analysis (with C-20 antibody; IgG as negative control) on miR-124 and miR-9 loci in HAF-derived neurons after sequential knockdown of PTB and nPTB. For box 1/2: υ=3, t=6.484, P=0.0074; box 3/4: υ=4, t=16.616, P=0.00076; box 2/4: υ=3, t=5.446, P=0.0122; box 5/6: υ=4, t=12.795, P=0.00022; box 4/6: υ=4, t=5.154, P=0.0067; box 7/8: υ=3, t=13.218, P=0.00094; box 9/10: υ=4, t=30.228, P = 0.000007; box 11/12: υ=4, t=5.684, P = 0.0047. n=3 independent experiments.
(h) RT-qPCR analysis of miR-124 and miR-9 after PTB knockdown or sequential PTB/nPTB knockdown in HAFs. U6 served as internal control. For miR-9: box 1/2: υ=4, t=3.496, P=0.0249; box 1/3: υ=4, t=17.595, P=0.0000613; box 2/3: υ=4, t=8.835, P=0.00091, For miR-124: box 1/2:,υ=3, t=11.886, P=0.00128; box 1/3: υ=4, t= 11.572, P=0.00032; box 2/3: υ=3, t=9.315, P=0.00262. n=3 independent experiments. ns: non-significant; *P<0.05, **P<0.01, ***P<0.001). All data were presented as mean± SD. See also Supplementary Figure 4 and the list of primers used in these analyses in Supplementary Table 3.
BRN2 activates neuronal-specific miRNAs
To understand how BRN2 mediates neuronal maturation, we further took advantage of the hNPC system to perform BRN2 ChIP-seq with two independent antibodies (C-20 and B-2) before and after hNPC differentiation (Supplementary Fig. 4e,f). We detected ~5,000 BRN2 binding peaks in 3,987 genes (Supplementary Table 2). GO term analysis against all expressed genes indicated that BRN2 regulates a large repertoire of genes critical for neurogenesis and maturation (Supplementary Fig. 4g,h).
Among BRN2 targets, we noted that BRN2 directly bound the genes for miR-124-3 on Chr.20 and miR-9-2 on Chr.5 (Fig. 4d). In contrast to the miR-124-3 locus, which showed unaltered BRN2 binding before and after hNPC differentiation, BRN2 on multiple locations in the miR-9-2 locus became detectable only in differentiated hNPCs (Fig. 4d), which we further validated by ChIP-qPCR (Fig. 4e). By monitoring mature miR-124 and miR-9 with miR-21 as a negative control in shPTB-treated HAFs, we again detected the induction of miR-9, but not miR-124, in response to BRN2 overexpression for 6 days, although both miRNAs were induced after prolonged BRN2 expression for 12 days (Fig. 4f). These data suggest that BRN2 binds and regulates miR-124 expression in hNPCs, but such modest level of miR-124 in hNPCs appears insufficient to trigger neuronal differentiation. In contrast, BRN2-regulated miR-9 seems to drive neuronal differentiation, consistent with the burst induction of the miR-9-2 precursor between E12.5 and E13.5 when BRN2 begins to express in the mouse brain 39–41.
We also confirmed BRN2 binding on both miR-9 and miR-124 loci in shPTB/shnPTB-treated HAFs (Fig. 4g). Interestingly, miR-9 showed progressive induction in response to sequential knockdown of PTB and nPTB in HAFs (Fig. 4h, left panel). In contrast, miR-124 was highly induced in response to PTB knockdown, but for an unknown reason, it became somewhat repressed in shPTB/shnPTB-treated HAFs (Fig. 4h, right panel). In any case, these data suggest that miR-124 induction may responsible for neuronal conversion and subsequent miR-9 activation may account for progression of induced neuronal-like cells to functional neurons.
The nPTB-BRN2-miR-9 loop for neuronal maturation
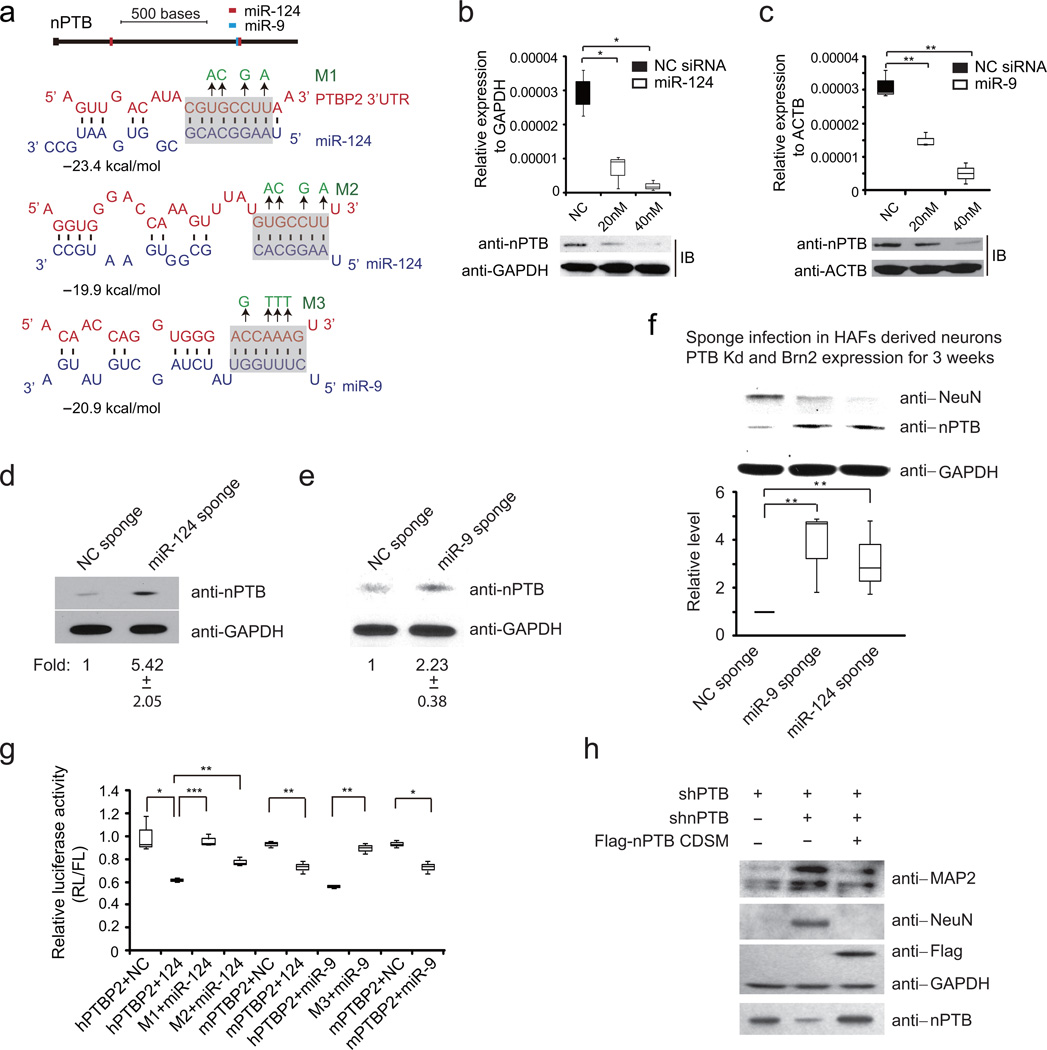
The induction of miR-9 by BRN2 provides an important clue to a new regulatory axis for neuronal maturation, as we noted potential target sites for both miR-9 and miR-124 in the 3’UTR of nPTB with typical base-pairing potentials in prospective seed regions (Fig. 5a). We thus asked whether these two neuronal-specific miRNAs were able to diminish the expression of endogenous nPTB. A previously study showed that miR-124 potently inhibited PTB with relatively minor effects on nPTB 42. In PTB down-regulated HAFs, we found that both miR-124 and miR-9 mimics down regulated nPTB at both mRNA and protein levels (Fig. 5b,c). As expected, shBRN2 prevented nPTB down-regulation while BRN2 overexpression diminished nPTB induction in shPTB-treated HAFs (Supplementary Fig. 5a,b). To further test these miRNAs in post-transcriptional regulation of nPTB, we expressed specific sponges for these miRNAs in hNPCs, finding significantly increased nPTB expression (Fig. 5d,e). The sponges similarly prevented nPTB down-regulation and diminished NeuN expression in HAF-derived neurons (Fig. 5f), consistent with early works that established the essential roles of both miRNAs in maintaining neuron viability of differentiated hNPCs 43,44.
Figure 5. Specific miRNAs repress nPTB during neuronal differentiation.
(a) miR-9/124 target sites in the 3’UTR of nPTB. Illustrated are deduced base pairing potentials, calculated free energies, and specific mutations introduced (green).
(b, c) Changes in nPTB expression examined by RT-qPCR (top) and Western blotting (bottom) in response to transfected miR-124 (b) or miR-9 (c) mimics in HAFs. GAPDH and β−actin served as controls. For miR-124: box 1/2: υ=3, t=3.614, P=0.0364;, box 1/3: υ=3, t=5.328, P=0.012. For miR-9: box 1/2: υ=4, t=6.124, P=0.00360; box 1/3: υ=3, t =6.861, P=0.00634. n=3 independent experiments.
(d, e) The expression levels of nPTB protein (top) and quantified changes (bottom) in response to overexpressed miR-124 (d) or miR-9 (e) sponge in differentiated hNPCs (n=3 independent experiments). GAPDH served as loading control.
(f) nPTB levels in response to the overexpression of the miR-124 or miR-9 sponge in HAF-derived neurons induced by shPTB plus BRN2 expression. GAPDH served as loading control. Quantified changes are shown at bottom (n=3 independent experiments). For miR-9 sponge: υ=4, t=5.503, P=0.0053; for miR-124 sponge: υ=4, t=4.910, P=0.0080.
(g) Luciferase reporter activities in response to co-transfected WT or mutant miRNA mimics in HEK293T cells. hPTBP2 or mPTBP2: Luciferase reporter containing the human or mouse nPTB 3’UTR; NC: Non-specific control mimic. For box 1/2: υ=4, t=4.197, P=0.01373; box 2/3: υ=4, t=10.512, P=0.00046; box 2/4: υ=4, t=6.409, P= 0.00306; box 5/6: υ=4, t=5.827, P=0.00432; box 7/8: υ=3, t=9.095, P=0.00281; box 9/10: υ=4,, t=4.057, P = 0.01539. n=3 independent experiments.
(h) The MAP2 and NeuN expression in HAF-derived neurons upon PTB knockdown or sequential PTB/nPTB knockdown upon the ectopic expression of an shRNA-resistant Flag-tagged nPTB (n=2 independent experiments). All data were presented as mean± SD. All P-values were calculated by two-tailed unpaired Student’s t test (*P<0.05, **P<0.01, ***P<0.001). Uncropped versions of Western blots are shown in Supplementary Figure 7.
We next verified the deduced targeting sites in the 3’UTR of nPTB. We constructed a luciferase reporter containing the nPTB 3’UTR and tested the effects of transfected miRNA mimics with MISSON universal siRNA as a negative control. We found that both miR-124 and miR-9 mimics repressed the reporter activity, and specific mutations in the predicted target sites diminished the miRNA-induced down-regulation of the reporter (Fig. 5g). The data also revealed that, between the two potential miR-124 target sites in the nPTB 3’UTR, the first seemed to be the primary functional site because the mutation at the site largely prevented miR-124-mediated down-regulation (Fig. 5g, compare the 2nd and 3rd bars).
It is important to emphasize that, although forced miR-124 overexpression or miR-124 depletion by sponge were able to affect nPTB expression, it is the BRN2-miR-9-nPTB axis that seems to be critical for neuronal maturation under physiological conditions, because immature neurons induced by shPTB alone clearly expressed a high level of miR-124. To further demonstrate the key contribution of diminished nPTB expression to neuronal maturation, we expressed a shRNA-resistant form of nPTB in shPTB/shnPTB-induced neurons from HAFs, which reduced MAP2 to the basal level, and more importantly, completely prevented the induced expression of NeuN (Fig. 5h). Considered together, the data presented in Fig. 3 through 5 suggest a previously unrecognized loop for driving neuronal maturation in PTB-depleted HAFs (Supplementary Fig. 6). In this loop, nPTB down-regulation directly or indirectly triggers the activation of BRN2; activated BRN2 binds and transcriptionally induces miR-9; and finally, elevated miR-9 post-transcriptionally diminishes nPTB. Therefore, like the PTB-REST-miR-124 loop for neuronal conversion, this nPTB-BRN2-miR-9 loop, once triggered, becomes self-sustainable for neuronal maturation.
DISCUSSION
Physiological relevance of the sequential regulatory loops
Neuronal differentiation is a tightly controlled process as indicated by the sequential expression of transcriptional and post-transcriptional regulators. The two separate yet highly intertwined loops we have elucidated are consistent with the neurogenesis program in vivo. PTB is well known for its role in maintaining neural stem cell pools, as its ablation causes precocious neural differentiation and massive depletion of neural stem cells in the mouse 45. The effect of PTB knockout is largely phenocopied by REST knockout, suggesting their functions in a common pathway during neuronal induction 46. As both PTB and REST are constitutively expressed in non-neuronal cells, it is likely that their inactivation in developing neurons is triggered by induced miR-124 expression 42. Our published work revealed a double negative feedback loop consisting of PTB, miR-124, and REST (Supplementary Fig. 6, left), and once triggered, the loop becomes self-sustainable and dismantled REST is responsible for de-repressing a large number of neuronal-specific genes to drive neurogenesis 28. It is important to note here that REST dismantling induced by miR-124 is fundamentally distinct from REST knockout, as a residual level of REST appears to be required for ensuring the healthy state of differentiated neurons 46.
A key functional consequence of PTB inactivation is de-repression of nPTB due to the inclusion of a PTB-suppressed exon 34,42,47, a process that occurs in the early phase of neural development 35. Our current data show that nPTB is also negatively regulated by miR-124, which may counteract PTB knockdown-induced nPTB expression, thereby maintaining relatively stable nPTB expression during the transition from immature to mature neurons. Importantly, the transit nPTB expression is coincident with the induction of both BRN2 and miR-9 around E13.5; however, it has been unclear how nPTB is switched off during neuronal maturation 39,40. We now establish that these three components form a feed-forward loop (Supplementary Fig. 6, right) and the full activation of this loop underlies gradual nPTB switch-off in further matured neurons 36,48. It remains to be defined in future studies which developmental cue(s) triggers this loop in human brain.
Key difference between mouse and human neurogenic programs
Mouse has been a major model to study neurogenesis and neurodegeneration in humans. However, mouse and human are evolutionally different in many ways. One of the fundamental differences is the neocortex expansion, which is mainly due to increased amplification of neural progenitor cells during embryonic corticogenesis 49. The tightly controlled neuronal conversion and maturation loops might serve as key gatekeepers to ensure programmatic switches in a sequential manner during brain development, as simultaneous knockout of PTB and nPTB produced a lethal phenotype, even on cell lines. Interestingly, while the optimal neuronal conversion protocol developed on mouse cells is insufficient to produce functional neurons from human cells, the optimized protocol developed on human cells causes a lethal phenotype on mouse cells, suggesting that cells from different mammals may have their own built-in timing devices.
Such separate timing devices between mouse and human cells are underscored by the observation that the PTB/nPTB-mediated loops seem to be automatically connected in murine cells, but need to be separately activated in human cells. While we are still early in understanding such species-specific mechanisms, one potential mechanism might be due to distinct regulatory programs for inducing miR-124 and miR-9 between mice and humans. miR-124 is known to be gradually induced during neurogenesis, while the induction of miR-9 expression is largely coincident with neuronal maturation 25. It is therefore conceivable that the initial induction of miR-124 might be responsible for down-regulating both PTB and nPTB in mouse cells, but predominantly for eliminating PTB in human cells. It is likely that miR-124 is more efficient in targeting PTB than nPTB even in mouse cells, as simultaneous inactivation of both PTB and nPTB would severely compromise the viability of differentiating neurons.
Regulated BRN2 expression is an obvious contributor to neuronal maturation, but other neuronal-specific transcription factors, such as NeuroD1, may be also involved 14. One of those mature neuron-specific transcription factors may be responsible for further elevation of miR-124 to join force with BRN2-induced miR-9 in later phases of neurogenesis. Therefore, various combinations of empirically tested TFs might all be able to partially activate both neuronal conversion and maturation programs, but unable to fully activate these programs in all transfected cells. As human cells appear to be stringently controlled by some critical gatekeepers, including the two elucidated in the present study, such opportunistic activation of the neural program may thus account for the overall low conversion efficiency in human cells.
Mechanisms underlying PTB/nPTB-regulated gene expression
PTB and nPTB are well-known splicing regulators 34,36,42,47,48,50. Despite their similar biochemical properties in RNA binding and biological functions in regulated splicing, they clearly have non-redundant functions in development based on knockout studies in mice 36,45,48. Interestingly, our recent study reveals that, besides its traditional role as a splicing regulator, PTB also binds numerous sites in the 3’UTR of many genes to either compete for miRNA targeting or alter RNA secondary structure to promote miRNA targeting, thus modulating miRNA functions in both directions 28. Our early work demonstrated that this new function of PTB is largely responsible for miR-124-mediated dismantling of the REST complex during neuronal conversion.
In the current study, we uncover a critical role of regulated nPTB expression in neuronal maturation. Previous studies have elucidated the function of nPTB in the regulation of neuronal-specific alternative splicing events, which undoubtedly contribute to various phenotypes associated with mature neurons 34,36,48. We now demonstrate a key role of nPTB down-regulation in driving neuronal maturation via the activation of the nPTB-BRN2-miR-9 loop. In this loop, BRN2 likely activates the transcription of miR-9 and induced miR-9 in turn inactivates nPTB via its targeting site in the 3’UTR, but it is currently unclear how down-regulated nPTB leads to BRN2 activation. Future studies will thus address how nPTB down-regulation may activate BRN2 directly via a transcriptional de-repression mechanism or indirectly through induced splicing events of other genes in mature neurons. The established cellular model will empower this and other mechanistic studies in a biologically relevant context.
ONLINE METHODS
Cell culture, virus production and infection
Primary human dermal fibroblasts from a 42 year-old male were purchased from ATCC and expanded in Dulbecco’s Modified Eagle Medium (DMEM)/F12 media (20% FBS, 1X Penicillin/streptomycin) or fibroblast media (DMEM/F12, 5ng/ml of recombinant bFGF, 1µg/ml of hydrocortisone, 50µg/ml of ascorbic acid, 5µg/ml of recombinant human insulin, 2% FBS). Primary human adult fibroblasts from an 86 year-old female were gift of Dr. John Ravits. HEK293T cells were cultured in DMEM containing 10% FBS and 100U of penicillin/streptomycin. Human neural progenitor cells (hNPC) were expanded in neural growth (NG) media containing: DMEM/F12 glutamax, 0.5XN2, 0.5XB27, and 1X Penicillin/streptomycin. Stempro Accutase (Life technologies) was used to detach hNPCs. Plates for passaging NPCs were sequentially treated with 10µg/ml of Poly-L-ornithine hydrobromide overnight and 2.5µg/ml of Laminin for 3 hrs.
Lentiviral shRNAs previously used to target human PTB 28 were shuttled to the pLKO.1-Hygromycin vector (Addgene, #24150). Lentiviral plasmids containing transcription factors ASCL1, BRN2, MYTLl and NeuroD1 were from Addgene (#27150, 27151, 27152, and 30129). Transcription factor NGN2 was constructed by inserting its coding region into the Tet-O-EGFP vector at the EcoR I site. To build inducible shnPTB constructs, the target sequences (GAAGAGAGGATCTGACGAACTA, TAAGAAAGACAGCGCTCTAATA, TTTTAAGAAACCTGGATCCAAA, and CGAGGAAGCAGCTATTACTATG) were inserted into the pTRIPZ-RFP vector between EcoR I and Xho I sites. To simultaneously express shPTB and BRN2 in a doxycycline dependent manner, the target sequence (TCGACATAATCTCTGTATTATA) was inserted into the pTRIPZ-RFP vector first, and RFP was replaced with the BRN2 cDNA between Age I and Cla I sites.
For neuronal reprogramming, human adult fibroblasts were first infected with shPTB lentivirus for 16 hrs. After selection with hygromycin B for 3 days, the cells were switched to N3 media (DMEM/F12 plus 25 µg/ml insulin, 50 µg/ml transferring, 30 nM sodium selenite, 20 nM progesterone, 100 nM putrescrine) for two weeks to induce neuronal morphology. To test the enhancement effects of small molecules, selected cells were primed in N3/basal media (1:1 mix of DMEM/F12 and Neurobasal, 25 µg/ml insulin, 50 µg/ml transferring, 30 nM sodium selenite, 20 nM progesterone, 100 nM putrescrine, 0.4% B27) for two days before the addition of small molecules ChIR99021 (1µM), SB431542 (10µM) and Db-cAMP (1mM) to N3/basal media plus 10ng/ml of FGF2 followed by culturing for additional two days. Cells were then switched to N3/basal media containing FGF2, NTFs (BDNF, GDNF, NT3 and CNTF) and 2% FBS for 8–10 weeks to induce functional neurons. Media were half-changed every the other day. To measure synaptic currents, induced neurons were seeded onto a monolayer of glia cells from P1 GFP transgenic rats for two to three weeks. To differentiate hNPCs into cortex neurons, FGF2 was removed from NG media followed by treatment with 10 µM of ROCK inhibitor Y27632 for 24 hrs and the cells were cultured in NG media for additional 2 to 6 weeks. For infection, viral particles were first concentrated by centrifugation at 20,000rpm for 2 hrs, and then added to NG media with polybrene to a final concentration of 1µg/ml. GFP transgenic rats were group-housed (<4) and under 12/12-h illumination schedule.
Titration of small molecule inhibitors
To increase the conversion efficiency by PTB knockdown, several small molecules beneficial to neuronal differentiation were tested in different concentrations and various combinations. SB431542 is a specific inhibitor of activin receptor-like kinase (ALK5). We tested four concentrations (1 µM, 2 µM, 4 µM, 10 µM) and found the optimal concentration is 10µM, which is the same as the reported working concentration 29. CHIR99021 is the most specific inhibitor to glycogen synthase kinase 3β (GSK-3β). Screening for four different concentrations (0.5 µM, 1 µM, 2 µM, 4 µM), we found the optimal concentration at 1µM, which is lower than the reported working concentration 30,32. Db-cAMP and Forskolin are specific activators of the cAMP dependent protein kinase (PKA). For Db-cAMP, we tested four concentrations (0.1 mM, 0.2 mM, 0.5 mM, 1 mM) and found that the optimal working concentration is 1 mM 51, and for forskolin, we tested three concentrations (5 µM, 10 µM and 20 µM) and identified the optimal working concentration to be 20 µM, which is different from the reported working concentration 31. Noggin is an antagonist of BMP-4, but it can also modulate the activities of other BMPs, including BMP-2, −7,-13,-14 29. We found no effect of this inhibitor in enhancing neuronal programming with all concentrations (50ng/ml, 100ng/ml, 200ng/ml, and 500ng/ml) tested. LDN-193189 is an antagonist of ALK2 and ALK3 32, and we didn’t observe any effect with all concentrations (0.05 mM, 0.1 mM, 0.2 mM, 0.5 mM) tested. Based on the effects of individually tested inhibitors, we made a combination of CHIR99021 (1µM), SB431542 (10µM) and Db-cAMP (1 mM) as the final cocktail to enhance neuronal reprogramming in combination with PTB knockdown.
Dual luciferase assay, Western blot and RT-qPCR
Specific miRNA luciferase reporters were constructed by cloning the 3’UTR of mouse and human nPTB into the psiCheck2 vector between Xho I and Not I sites. 1–5ng of reporters and 30pmol of miRNA mimics were co-transfected and luciferase activity was measured after 24 hrs with a dual-luciferase assay kit (Promega).
For Western blotting, cells were lysed in 1xSDS loading buffer without bromophenol blue, and after quantification, bromophenol blue was added back to a final concentration of 0.1%. 25µg of total protein was resolved on 10% Nupage Bis-Tris gel and probed with following antibodies: Mouse anti-PTBP1 (monoclonal BB7, ATCC, CRL-2501), Rabbit anti-PTBP2 IS2 (gift of Dr. Douglas Black of UCLA)35, PTBP2 monoclonal antibody (M01, clone 2D10-B2, Abnova), Mouse monoclonal to PTBP2 (ab57619, Abcam), Rabbit anti-BRN2 (Cell Signaling Technology, 12137) and anti-GAPDH (14C10, Cell Signaling Technologies), anti-FLAG (sigma, F3165) and Mouse anti-ACTB (A2228, Sigma).
Total RNA was extracted with Trizol (Life Technology), except that 10µg/ml of glycogen was used at the precipitation step to enrich for small RNAs. Total RNA was first treated with DNase I (Promega) and reverse-transcribed with the Superscript III kit (Life Technology). RT-qPCR was performed with gene specific primers and fast start universal SYBR green master mix (Roche) on a step-one plus PCR machine (Applied Biosystems). Statistical significance was determined by Students t-test based on triplicated experiments. All the primers used for plasmid construction and real-time PCR are listed on Supplementary Table 3.
Electrophysiological recording
Whole-cell patch clamp recordings were performed as previous described 28. The only exception was that cultured cells were incubated with oxygenated (95% O2/5% CO2) artificial cerebrospinal fluid (150 mM NaCl, 5 mM KCl, 1 mM CaCl2, 2 mM MgCl2, 10 mM glucose, 10 mM HEPES, pH 7.4) at 37°C for 30min. Recordings under whole-cell voltage clamp mode were performed at a holding potential of −75mV. The pipette resistance is between 5–8 Mohm. Action potentials were elicited by injecting step depolarizing currents under current clamp recording mode. The pipette solution contains 150 mM KCl, 5 mM NaCl, 1 mM MgCl2, 2 mM EGTA, 1 mM Mg-ATP, and 10 mM HEPES (pH 7.2, adjusted with KOH). 1 mM GABA was used to invoke GABA currents. The following concentrations of channel inhibitors were used: TTX: 1 µM; PiTX: 50 µM; NBOX: 20µM; APV: 50 µM APV. All recordings were performed at room temperature (20–22 °C).
Immunocytochemistry
Cells were seeded on coverslip pre-coated with matrigel (BD, 356234) for 1 hr at 37°C, and then fixed in 4% Paraformaldehyde (Electron Microscopy Sciences) for 15min at room temperature followed by permeabilization with 0.1% Triton X-100 in PBS for 15min on ice. After washing twice with PBS, cells were blocked in PBS containing 3% BSA for 1 hr at room temperature. The following primary antibodies were incubated for 1 hr on the cells: Rabbit anti-Tuj1 (Covance, MRB-435P,1:1,000), Mouse anti-Tuj1 (Covance, MMS-435P, 1:1,000), Mouse anti-MAP2 (milipore, MAB3418,1:1000), Mouse anti-NeuN (Milipore, MAB377,1:200), Rabbit anti-Synapsin I (Synaptic System, 106001,1:5000), Rabbit anti-VGLUT1 (Synaptic Systems, 135–303,1:200), Rabbit anti-GABA (Sigma, A2052, 1:1000), Rabbit anti-Brn2/POU3F2 (Cell Signaling Technology, 12137, 1:500), Mouse anti-Fibronectin (DSHB, 13G3B7,1:500), Mouse anti-Vimentin Dyelight 594 (abcam, EPR3776, 1:1000), Mouse anti-P63 (Thermo Fisher, MA1–21871, 1:300), Goat anti-MITF (Santa Cruz, sc-10999, 1:100) , Rabbit anti-K5 (Thermo Fisher, RM2106S0, 1:300), Rabbit anti-NGF receptor P75 (Milipore, AB 1554, 1:100), Goat anti-Sox2 (Santa Cruz, sc-17319, 1:200), Mouse anti-Pax3 (DSHB, PAX3, 1:250), Mouse anti-Pax7 (DSHB, PAX7-c, 1:250), Mouse anti-NKX2.2 (DSHB, 74.5A5, 1:100), Mouse anti-NCAM (DSHB, 5A5-c, 1:100), Mouse anti-Olig1 (Neuromab, 75–180, 1:100), Mouse anti-GFAP (Neuromab, 75–240, 1:100), Rabbit anti-Tbr1(Milipore, AB10554, 1:300), Moue anti-GFP (Life Technologies, A-11120, 1:400), Rabbit anti-SOX10 (Abgent, AP5854c, 1:50), Rabbit anti-BRN3A (Bioss USA, bs-3669R, 1:50). Secondary antibodies and DAPI were applied to the cells for another 1hr and 20min respectively. After washing six times with PBS, Fluoromount-G mounting media was applied onto the glass slides, which were examined under Olympus FluoView FV1000.
Identification of BRN2 targets by ChIP-seq
Human NPCs were cross-linked with 1% formaldehyde and quenched by adding 1/20th volume of 2.5M glycine to a final concentration of 0.125M. After washing three times with ice-cold PBS, cells were scraped off from the plate and collected by centrifugation at 2500rpm. Cell pellets were suspended in 1 ml of Cyto lysis buffer, mixed briefly, and incubated on ice for 10–15min with occasional inversion every 2min. Cells were pelleted for 5min at 3500rpm, 4°C, supernatant discarded, and the nuclear pellets were resuspended in 500µl of nuclear lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-Cl, pH8.1, 1Xprotease inhibitor cocktail) and incubated on ice for 10min. The pellets were sonicated seven times at the 10 sec interval at the maximum setting (Branson, Sonifer cell disruptor 185). The lystate was cleared by centrifugation at 14000rpm for 10min in 4°C and the supernatant was collected in 15ml tube. The supernatant was diluted 1:10 with the dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-Cl, pH8.1, 1Xprotease inhibitor cocktail) to yield the soluble chromatin fraction, 100µg-200µg of which was used for each ChIP reaction. Lystate was pre-cleared by adding 20µl of Protein A/G beads followed by rotation for 1 hr at 4°C. 5µg of anti-Brn2 (Cell Signaling Technology) were applied to the supernatant and incubated overnight at 4°C. 50 µl of Protein A&G magnetic beads were blocked with (1xPBS+5mg/ml BSA-fraction V) and incubated at 4°C for another 4 hrs. Magnetic beads were harvested on a magnetic stand and beads wished sequentially at 4°C, twice in 0.7 ml of TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH8.1, 150 mM NaCl), twice in TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH8.1, 500 mM NaCl), twice in Buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, pH8.0, 10 mM Tris-HCl, pH8.1), and finally, twice with 1ml of TE buffer at 4°C. Eluted beads were twice washed with 150 µl of elution buffer (TE buffer, 1%SDS) by votexing at 70°C, 1000rpm for 10min. The eluents were heated at 65°C overnight to reverse formaldehyde-induced crosslinks. For input sample, 150–300 µl of soluble chromatin was also heated at 65°C overnight. DNA fragments were purified with QIAquick Spin PCR purification Kit (Qiagen, CA).
ChIP-seq libraries were constructed by using the TruSeq ChIP Sample Prep Kit (Illumina). To include spike-in control, 700ng of soluble chromatin from Drosophila was added to 200µg of the soluble chromatin from hNPCs before and after differentiation at immunoprecipitation step followed by the addition of 5µg of anti-BRN2 and 1µg of Drosophila-specific H2AV antibodies. After deep sequencing, total sequencing tags from Drosophila were used for normalization. ChIP-seq reads were aligned to the human genome with Bowtie 2 (ref. 52) using default parameters with local alignment model. Duplicated reads and low-mapping-quality reads were removed with Samtools 53. Binding peaks were identified by MACS 54 with default parameters. The identified Brn2 targets are listed in Supplementary Table 2.
Statistical analysis
Statistical differences were calculated by two-tailed Student t-test with SigmaPlot 12 software and the degree of freedom (υ) was usually defined by the number of experiments in group 1 plus the number of experiments in group 2 minus two (n1+n2-2). . The variance is similar between the groups that are being statistically compared. Data distribution was assumed to be normal but this was not formally tested. All data collection and analyses were performed by experimenters blind to the treatment conditions. The image fields were randomly selected under 20X magnification. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those generally employed in the field. A Supplementary Methods Checklist is available.
Supplementary Material
Acknowledgments
We thank Zhen-Ning Zhang from the Alysson R. Muotri lab for sharing hNPCs and culture protocols and Kang Zhang for sharing various antibodies and GFP transgenic rats. We are grateful to Steven Dowdy for help on FACS analysis, to John Ravits for human adult fibroblasts, to Yang Yu from the Gregory Hannon lab for the pTRIPZ lentiviral plasmid. This work was supported by the National Natural Science Foundation of China (91440101) to Y.C.X, by key programs of Chinese Academy of Sciences to X.D.F and Y.C.X (KJZD-EW-L12) and by NIH grants (GM049369 and HG004659) to X.D.F.
Footnotes
AUTHOR CONTRIBUTIONS
Y.C.X and X.D.F conceived the project. Y.C.X designed and performed most biochemical experiments. H.Q was responsible for all electrophysiology analysis and NPC differentiation with input of A.K and Z.P.P. B.Z performed bioinformatics analysis, Y.Z and X.H.H contributed to RNA-seq analysis. J.H performed EdU labeling, ChIP-qPCR, and FACS analysis. Y.C.X and X.D.F. wrote the manuscript. High-throughput data have been deposited in Gene Expression Omnibus under accession number of GSE74814.
COMPETING FINANCIAL INTERESTS
Authors declare no competing financial interests.
REFERENCES
- 1.Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- 2.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 3.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 4.Bang AG, Goulding MD. Regulation of vertebrate neural cell fate by transcription factors. Current opinion in neurobiology. 1996;6:25–32. doi: 10.1016/s0959-4388(96)80005-5. [DOI] [PubMed] [Google Scholar]
- 5.Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015 doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 6.Herculano-Houzel S. The human brain in numbers: a linearly scaled-up primate brain. Frontiers in human neuroscience. 2009;3:31. doi: 10.3389/neuro.09.031.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lepski G, et al. Delayed functional maturation of human neuronal progenitor cells in vitro. Molecular and cellular neurosciences. 2011;47:36–44. doi: 10.1016/j.mcn.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 11.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 12.Wernig M, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci U S A. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caiazzo M, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 14.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfisterer U, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Son EY, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broccoli V, Caiazzo M, Dell’Anno MT. Setting a highway for converting skin into neurons. Journal of molecular cell biology. 2011;3:322–323. doi: 10.1093/jmcb/mjr029. [DOI] [PubMed] [Google Scholar]
- 19.Amamoto R, Arlotta P. Development-inspired reprogramming of the mammalian central nervous system. Science. 2014;343:1239882. doi: 10.1126/science.1239882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cahan P, et al. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris SA, et al. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158:889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou P, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011;21:196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cimadamore F, Amador-Arjona A, Chen C, Huang CT, Terskikh AV. SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc Natl Acad Sci U S A. 2013;110:E3017–E3026. doi: 10.1073/pnas.1220176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;11:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- 26.Ambasudhan R, et al. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A. 2011;108:8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu ML, et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ladewig J, et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods. 2012;9:575–578. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- 33.Prasad KN, Hsie AW. Morphologic differentiation of mouse neuroblastoma cells induced in vitro by dibutyryl adenosine 3’:5’-cyclic monophosphate. Nature: New biology. 1971;233:141–142. doi: 10.1038/newbio233141a0. [DOI] [PubMed] [Google Scholar]
- 34.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng S, et al. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–388. doi: 10.1038/nn.3026. S381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, et al. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife. 2014;3:e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagino-Yamagishi K, et al. Predominant expression of Brn-2 in the postmitotic neurons of the developing mouse neocortex. Brain Res. 1997;752:261–268. doi: 10.1016/s0006-8993(96)01472-2. [DOI] [PubMed] [Google Scholar]
- 38.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cerebral cortex. 2013;23:2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. Rna. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akerblom M, et al. MicroRNA-124 is a subventricular zone neuronal fate determinant. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giusti SA, et al. MicroRNA-9 controls dendritic development by targeting REST. eLife. 2014;3 doi: 10.7554/eLife.02755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibasaki T, et al. PTB deficiency causes the loss of adherens junctions in the dorsal telencephalon and leads to lethal hydrocephalus. Cerebral cortex. 2013;23:1824–1835. doi: 10.1093/cercor/bhs161. [DOI] [PubMed] [Google Scholar]
- 46.Gao Z, et al. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Licatalosi DD, et al. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez S, et al. A cAMP-activated pathway, including PKA and PI3K, regulates neuronal differentiation. Neurochemistry international. 2004;44:231–242. doi: 10.1016/s0197-0186(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 52.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.