Summary
Antibiotic-resistant bacterial infection is a serious threat to public health. Peptidoglycan biosynthesis is a well-established target for antibiotic development. MraY (phospho-MurNAc-pentapeptide translocase) catalyzes the first and an essential membrane step of peptidoglycan biosynthesis. It is considered a very promising target for the development of new antibiotics, as many naturally occuring nucleoside inhibitors with antibacterial activity target this enzyme1-4. However, antibiotics targeting MraY have not been developed for clinical use mainly due to a lack of structural insight into inhibition of this enzyme. Here we present the crystal structure of MraY from Aquifex aeolicus (MraYAA) in complex with its naturally occurring inhibitor, muraymycin D2 (MD2). Upon binding MD2, MraYAA undergoes remarkably large conformational rearrangements near the active site, which lead to the formation of a nucleoside-binding pocket and a peptide-binding site. MD2 binds the nucleoside-binding pocket like a two-pronged plug inserting into a socket. Additional interactions it makes in the adjacent peptide-binding site anchor MD2 to and enhance its affinity for MraYAA. Surprisingly, MD2 does not interact with three acidic residues or the Mg2+ cofactor required for catalysis, suggesting that MD2 binds to MraYAA in a manner that overlaps with, but is distinct from its natural substrate, UDP-MurNAc-pentapeptide. We have deciphered the chemical logic of MD2 binding to MraYAA, including how it avoids the need for pyrophosphate and sugar moieties, which are essential features for substrate binding. The conformational plasticity of MraY could be the reason that it is the target of many structurally distinct inhibitors. These findings can inform the design of new inhibitors targeting MraY as well as its paralogs, WecA and TarO.
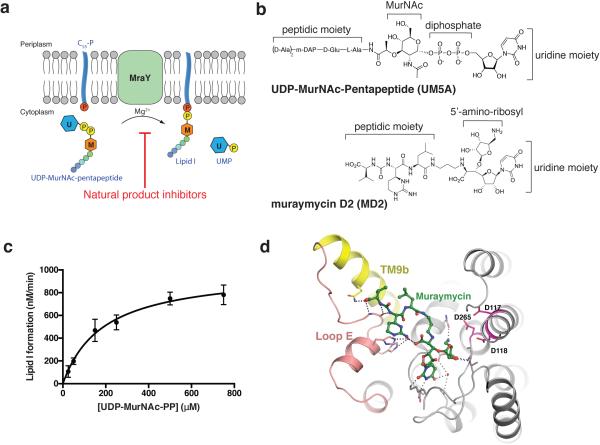
MraY is a member of the polyprenylphosphate N-acetyl hexosamine 1-phosphate transferase (PNPT) superfamily. The PNPT superfamily includes bacterial and eukaryotic integral membrane enzyme families such as MraY, WecA, TarO, WbcO, WbpL, RgpG, and GPT, which are involved in cell envelope polymer synthesis and protein N-linked glycosylation 1. WecA and TarO are also targets for antibiotic development. MraY catalyzes the transfer of phospho-MurNAc-pentapeptide from UDP-MurNAc-pentapeptide (UM5A) to the lipid carrier undecaprenyl phosphate (C55-P), yielding undecaprenyl-pyrophosphoryl-MurNAc-pentapeptide, known as Lipid I (Extended Data Fig. 1a). This step is essential, rate limiting, and Mg2+-dependent 1. It is blocked by five classes of natural nucleoside antibiotics (e.g. muraymycin and tunicamycin), and bacteriolytic protein E from bacteriophage phiX174, with various modes of inhibition (Extended Data Fig. 1a) 5-8. MraY-targeted natural products have gained attention because of their in vivo efficacy against pathogenic bacteria including M. tuberculosis, methicillin-resistant S. aureus (MRSA), and vancomycin-resistant Enterococcus (VRE) 6,9-12. Despite their promise, no antibacterial natural products that target MraY have been developed for clinical use, in part due to a lack of structural information on MraY catalysis and inhibition. We carried out structural studies of MraY in complex with a naturally occurring inhibitor of MraY, muraymycin, which shows antibacterial effects against MRSA, VRE, and P. aeuruginosa, and S. aureus 11,13-18. We used muraymycin D2 (MD2) for our structural, enzymatic, and biophysical studies (Extended Data Fig. 1b) 17. The muraymycins are known to be competitive inhibitors for the natural substrate UM5A 14. Unlike UM5A, the muraymycins do not have pyrophosphate and sugar moieties and have a 5-amino ribosyl group (Extended Data Fig. 1b). Using a radiochemical transfer assay 19, we determined the KM for UM5A with purified MraYAA to be ~190 μM (Extended Data Fig 1c), which is within the range of measured cellular UM5A concentrations 20. MraYAA activity is significantly reduced upon addition of 0.3 μM MD2 (Fig. 1a). Using isothermal titration calorimetry (ITC), we measured the KD of MD2 for MraYAA to be ~20 nM (Fig. 1b). We generated crystals of MraYAA in the presence of MD2, which diffracted to 2.95 Å. Phasing was obtained by molecular replacement (MR) using the apoMraYAA structure (PDB ID: 4J72) with all the cytoplasmic loops and TM9b removed as a search model. The structure was refined to good statistics (R/Rfree = 0.247/0.261) (Extended Data Table 1).
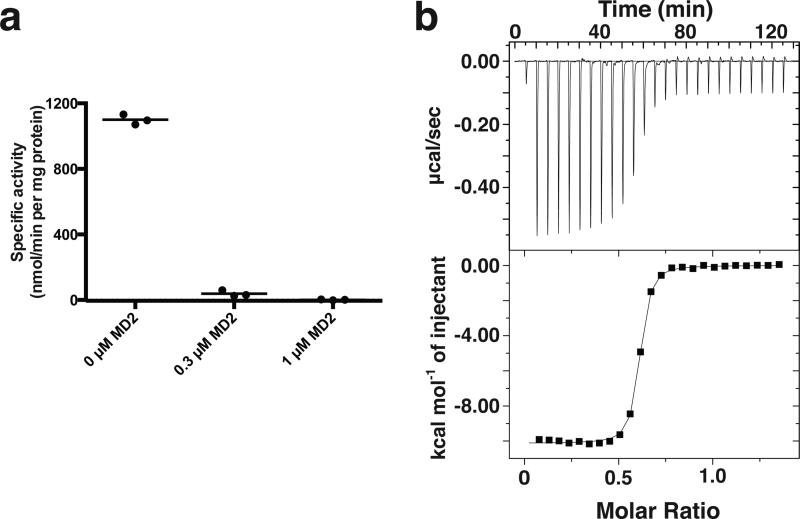
Figure 1. The natural product muraymycin D2 (MD2) binds to and inhibits MraYAA.
a, Specific activity measurements of MraYAA WT in the presence and absence of MD2 using radiolabeled substrate, [14C]UM5A. The radiolabeled product, [14C]Lipid I, was quantified using a liquid scintillation counting method (dpm). Three technical replicates are shown with the mean value indicated by a line. b, Representative ITC raw data and binding isotherm for MD2 interacting with MraYAA WT in the presence of 10 mM MgCl2; KD = 17.2 nM, ΔH° = −10.1 kcal/mol. This ITC experiment was performed in triplicate (technical replicates) and mean thermodynamic parameters are shown in Extended Data Table 2.
MraYAA in complex with MD2 crystallizes as a dimer, as does apoMraYAA 21. Each protomer contains ten transmembrane helices (TM1-TM10) and five cytoplasmic loops (loops A-E) (Fig. 2a). TM9 breaks into two helical fragments (TM9a and TM9b) and TM9b bends outward toward the membrane (Fig. 2b). We previously outlined the active site as a cleft formed by the inner-leaflet membrane regions of TMs 3, 4, 5, 8, and 9b and cytoplasmic loops B, C, D, and E. Many absolutely conserved polar/charged amino acid residues are localized in this cleft, including three catalytically critical acidic residues: Asp117, Asp118, and Asp265 (Fig. 2a and 2c), which are conserved in the PNPT superfamily 1,19,21-23. Asp265 interacts with Mg2+ in the apoMraYAA structure and Asp117 has been proposed to bind to the phophate moiety of C55-P 21,22. Situated in the active site cleft, the nucleoside portion of MD2 is inserted between loop C and loop D and the peptide portion interacts with TM9b and loop E (Fig. 2c). High sequence conservation is observed around the MD2-binding region in the active site (Fig. 2d). Interestingly, MD2 does not interact with any of the three catalytically critical acidic residues (Fig. 2c and Extended Data Fig. 1d). MraY undergoes remarkable conformational rearrangements near the active site upon binding MD2 (Fig. 3). The amphipathic TM9b rotates away from the active site while loop E rearranges, packs against the hydrophilic part of TM9b (Extended Data Fig. 2 and Supplementary video), and a helical segment of the conserved HHH motif (PXHHHXEXXG) extends 1,21. This TM9b-loop E rearrangement widens and reshapes the active site, allowing the peptidic moiety of MD2 to bind to the side of TM9b and loop E (Extended Data Fig 3). The C-terminal portion of TM5 and loop C unwinds (cyan, Fig 3, a-b) and loop D rearranges (magenta, Fig 3, a-b), creating a pocket where the 5-aminoribosyl moiety and the uracil base of uridine interact (Fig. 3d and Extended Data Fig. 4a-c). The concerted motions of TM5 and loops C and D lead to the rearrangement of loop A and part of TM1 (green, Fig. 3b), although these regions do not appear to interact with MD2 directly. It is noteworthy that the amino acid residues interacting with the uracil base move large distances (5-17 Å) while the residues interacting with the 5-aminoribosyl moiety move shorter distances (Extended Data Fig. 4a-c and Supplementary video). The active-site structural rearrangement leads to substantial changes in its electrostatic potential, including enlargement of the acidic milieu around the nucleoside-binding pocket, which may play a role in MD2 binding (Fig. 3, c-d).
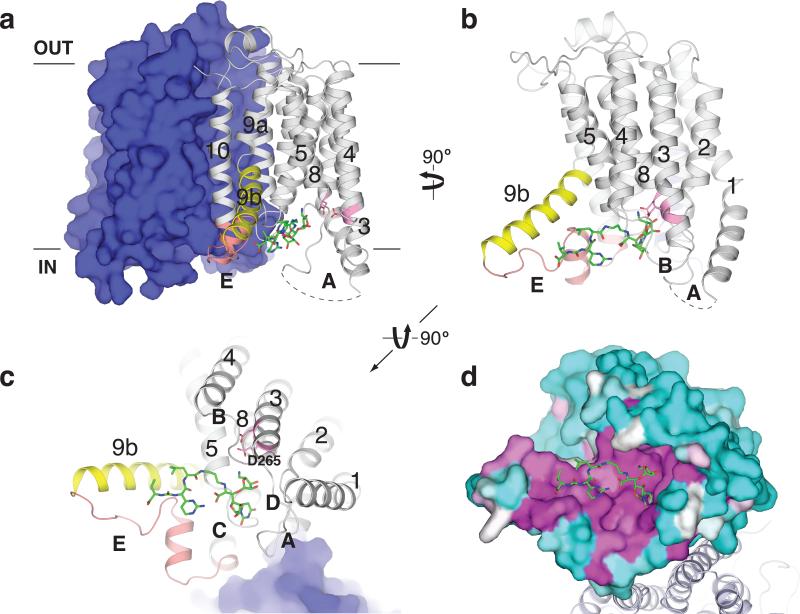
Figure 2. MD2 binds to a conserved site in MraYAA.
a, The MD2-bound MraYAA dimer viewed from the membrane. One protomer is shown as surface representation and the other as a cartoon. MD2 (green sticks) resides in the pocket formed by TMs 3-5 and 8-9b and cytoplasmic loops B-E. Conserved catalytic aspartic acid residues at the active site are shown in pink. b, View from the membrane rotated 90° about a vertical axis relative to (a). One protomer is shown for clarity. c, Cytosolic view of the MraYAA-MD2 complex, rotated 90° about a horizontal axis relative to (b). Part of MD2 is near the putative substrate recognition site formed by TM9b (yellow) and loop E (orange). d, Conservation mapping of MraYAA from high (magenta) to low (cyan) sequence identity, based on the alignment of 28 MraY homologs 21.
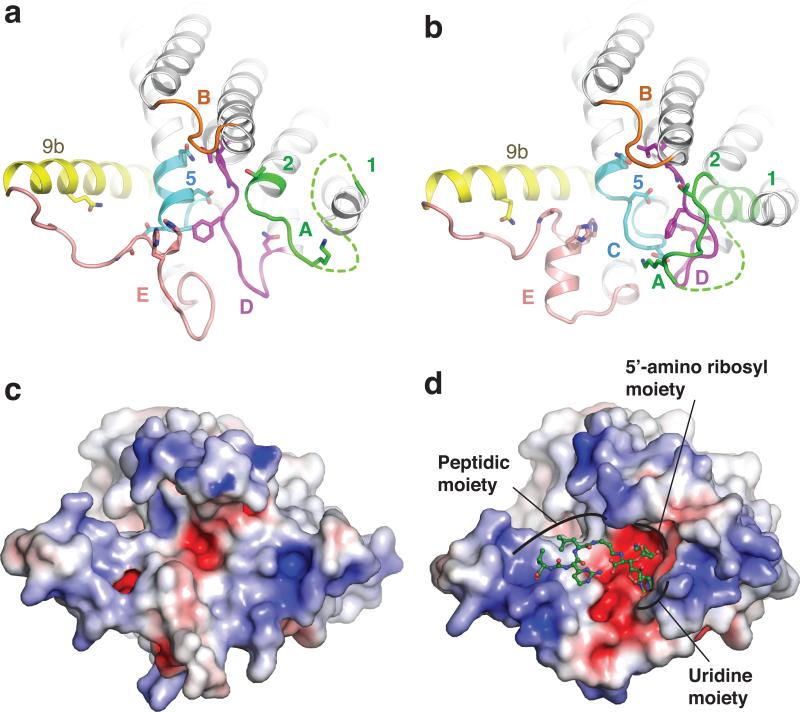
Figure 3. Conformational rearrangement of MraYAA upon MD2 binding.
a, apoMraYAA (PDB ID: 4J72) viewed from the cytoplasm, as in Fig. 2c. Residues involved in interactions with MD2 are shown as sticks. b, MD2-bound MraYAA with MD2 omitted. Part of TM1 (light green) is transparent due to its absence in the apoMraYAA structure. c, Electrostatic surface representation of apoMraYAA, viewed from the cytoplasm as in a-b. d, Electrostatic surface representation of MraYAA in complex with MD2. MD2 is green and shown in ball-and-stick representation.
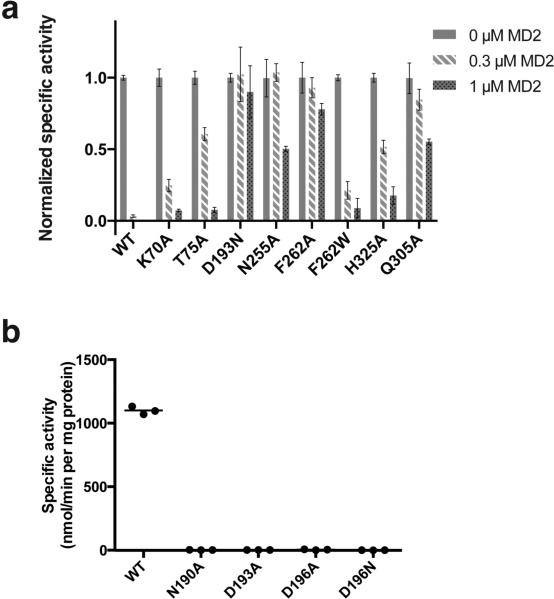
Developing nucleotide-sugar mimicking inhibitors for glycosyltransferases has been challenging largely due to the difficulty of developing pyrophosphate mimics capable of cellular entry with high affinity for the target enzyme 24-27. MraY is a phosphoglycosyltransferase that shares a common nucleotide sugar substrate with glycosyltransferases. Nucleoside antibiotics targeting MraY have garnered interest because they can enter the cell with high affinity for MraY 24,25,28. MD2 does not contain a pyrophosphate or sugar moiety and has a 5-amino ribosyl group, which was thought to mimic the pyrophosphate in the natural substrate UM5A (Extended Data Fig. 1b) 23,29. To understand the principle of MD2 inhibition of MraYAA, we performed mutagenesis and measured the mutational effects on MD2 binding using ITC. Amino acid residues involved in binding MD2 were grouped based on the substructures with which they interact: 1) uracil (Lys70, Gly194, Asp196, Asn255, and Phe262), 2) 5-amino ribose (Thr75, Asn190, Asp193, and Gly264), and 3) peptidic sidechain (Gln305, Ala321, and His325). We performed site-directed mutagenesis on the residues that form side-chain interactions with MD2 (Lys70, Thr75, Asn190, Asp193, Asp196, Asn255, Phe262, Gln305, His325) (Fig. 4). The MraYAA mutants that showed significant enzyme activity (Extended Data Fig. 5) were used for ITC experiments with MD2 (Extended Data Fig. 6 and Extended Data Table 2).
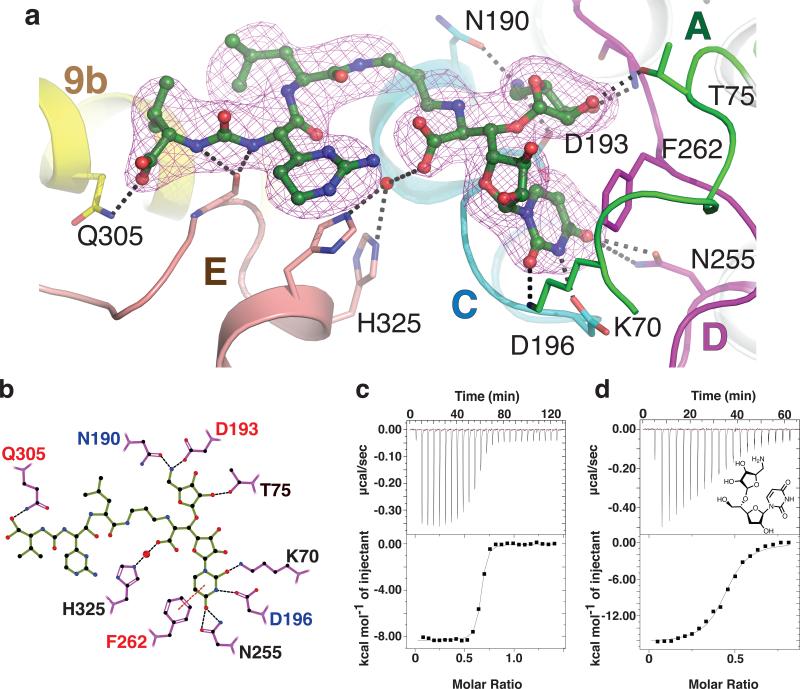
Figure 4. Dissection of the interactions between MD2 and MraYAA elucidates the chemical logic of MraYAA inhibition.
a, Composite simulated annealing 2Fo–Fc omit electron density of MD2 in the crystal structure of MD2-bound MraYAA at 1.7σ. The TMs are colored as in Fig. 3, a-b. The residues forming side chain interactions with MD2 are labeled. b, A two-dimensional representation of the interactions between MD2 and MraYAA. Hydrogen bonds (3.2 Å cutoff) are indicated with black dashed lines and π–π contacts are indicated with red dashes. Mutation of residues with red colored labels resulted in a larger than five-fold increase in the KD of MD2 and those with blue residue labels are nearly inactive. c, Representative ITC raw data and binding isotherm for MD2 titrated into MraYAA in the absence of added Mg2+; KD = 14.8 nM, ΔH° = −8.3 kcal/mol. A similar KD is observed for MD2 titrated into MraYAA with added Mg2+. d, Representative ITC raw data and binding isotherm for 5-aminoribosyl-3-deoxy uridine titrated into MraYAA WT; KD = 283 nM, ΔH° = −16.4 kcal/mol. Each ITC experiment was performed in triplicate (technical replicates) and mean thermodynamic parameters are shown in Extended Data Table 2.
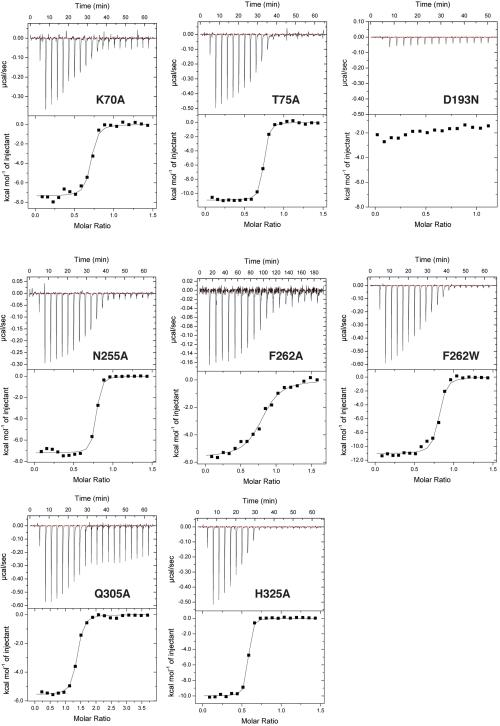
The affinity of MD2 for MraYAA was most perturbed with D193N and F262A, mutations that disrupt interactions with the 5-aminoribose and uracil moieties of MD2, respectively (Fig. 4, Extended Data Table 2, and Extended Data Fig. 6). Phe262 interacts with the uracil base via a π-π interaction (Fig. 4 and Extended Data Fig. 4d). When Phe262 is mutated to another aromatic amino acid, such as tryptophan, there is a smaller effect on KD relative to the alanine mutation, indicating the importance of this π-π interaction. Residue Asp193 makes sidechain interactions with the 5-amino ribose moiety of MD2 (Extended Data Fig. 4e). Because the D193A mutant is nearly inactive (Extended Data Fig. 5b), we used functionally competent D193N for ITC with MD2 (Extended Data Fig. 5a). However, the heat associated with binding was too low to measure, suggesting the D193N mutation greatly reduces the affinity of MD2 for MraYAA (Extended Data Fig. 6). This observation is consistent with previous studies indicating the antibacterial activity of MraY inhibitors with a 5-aminoribose is dependent on the amino group of that moiety 29,30. The Q305A mutant exhibits a larger than five-fold increase in KD (Fig. 4 and Extended Data Table 2), indicating that the interactions formed by the peptidic moiety of MD2 contribute to the binding affinity. Asp193, Phe262, and Gln305 are absolutely conserved in MraY orthologs 21. The results from the equilibrium binding experiments are consistent with the enzymatic inhibition experiments because the F262A mutation results in partial inhibition and the D193N mutant is not inhibited in the presence of 1 μM MD2 (Extended Data Fig. 5a).
We infer that MD2 and the natural substrate, UM5A, utilize different strategies for binding MraY. First, the three catalytically critical acidic residues, including the Mg2+-binding Asp265, do not participate in direct interactions with MD2 (Extended Data Fig. 1d). Second, the D193N mutant remains functionally active, although it disrupts an interaction MraY makes with the 5-aminoribosyl group and affects the binding affinity of MD2 dramatically (Extended Data Table 2 and Extended Data Fig. 6). This suggests the 5-aminoribosyl group does not function as a pyrophosphate mimic and instead forms interactions that are not present in or important for UM5A binding. If MD2 lacks a pyrophosphate mimic, it is unlikely that Mg2+ plays an important role in MD2 binding. To test this idea, we performed ITC in the absence of Mg2+ and found that MD2 does not require Mg2+ for MraY binding (Fig. 4c). It is possible that the amino group of the 5-aminoribose mimics Mg2+ and Asp193 interacts with Mg2+, as previously suggested 14. However, in such a case, we would expect D193N to be inactive. Furthermore, the amino group of the 5-aminoribose is not near the Asp265, which coordinates Mg2+ in the apoMraY structure 21.
In summary, the 5-aminoribosyl and uracil moieties of MD2 bind to the nucleoside-binding pocket like a two-pronged electrical plug inserts into a socket, and these interactions are the most critical for binding. The peptidic moiety also contributes to the binding energy by anchoring MD2 to MraY, likely contributing to its specificity for MraY. With the 5-aminoribose moiety of MD2 positioned as a second prong alongside uridine in the nucleoside binding site, MD2 binds to MraY with increased affinity, making the pyrophosphate and sugar moieties unnecessary for binding. To confirm the importance of the 5-aminoribosyl and uracil moieties of MD2, we synthesized 5-aminoribosyl-3-deoxy uridine 29 and found it retains substantial binding affinity for MraYAA (Fig 4d). Our structural and biochemical studies demonstrate the principles of MD2 inhibition of MraY and explain why MD2 does not require pyrophosphate and sugar moieties for binding, unlike the natural substrate, UM5A. This illustrates an example of nature circumventing a long-standing problem in chemical biology: developing a nucleotide-sugar-like inhibitor for glycosyltransferases 24-27. Finally, the large conformational arrangement observed in MraY implies conformational plasticity, which could be the reason why MraY accommodates so many structurally different nucleoside inhibitors, as well as protein E, with distinct modes of action 7.
Methods
Crystallization
Wild-type MraYAA and mutants were expressed and purified as described 21. All MraYAA bacterial expression plasmids were 10x histidine (His10)-maltose binding protein (MBP) fusion constructs expressed in C41 (DE3) cells. The cells were lysed by microfluidizer and the protein was extracted from the crude lysate using 40 mM dodecyl-maltoside. The lysates were centrifuged to remove the insoluble fraction and the supernatant was applied to a Co2+-affinity column for purification. The His10-MBP tag was cleaved overnight by PreScission Protease and MraYAA was isolated by gel filtration using a Superdex 200 10/300 GL column in the presence of 5 mM decyl-maltoside, 150mM NaCl, 20mM Tris-HCl pH 8.0, and 2mM DTT. All purification steps were performed at 4 °C. After gel filtration, the protein was concentrated to ~10 mg/ml (~250 μM) and MD2 was added to a final concentration of 400-500 μM before crystallization. Crystals were grown using sitting-drop vapor diffusion in the presence of 50 mM MgCl2, 40% PEG400, and 100 mM sodium cacodylate pH 5.6. Crystals were harvested after 10–14 days and flash frozen in liquid nitrogen.
Data Collection and Structure Determination
X-ray data were collected at beamlines 22-ID-D and 24-ID-C at the Advanced Photon Source in Argonne National Laboratory at a wavelength of 1.0 Å and processed using iMosflm. The data were ellipsoidally truncated at 3.0 Å on the c axis and anisotropically scaled using the UCLA anisotropy diffraction server (http://services.mbi.ucla.edu/anisoscale/). Phases of the MD2 complex structures were solved by molecular replacement using PHASER 31 with a partial apoMraYAA structure (PDB ID: 4J72, TM9b and cytoplasmic loops removed) as the search model. The omit electron density peaks corresponding to MD2 were prominent from the beginning of the refinement. After the protein model was built, a simulated annealing omit map was generated and MD2 was modeled. PHENIX 32 and Coot 33 were used to refine the structure. The final model has good geometry with 98.8%/1.2%/0.0% Ramachandran favored/allowed/outliers.
Enzymatic Assays
All enzymatic assays monitored the MraYAA-mediated transfer of [14C]phospho-MurNAc-pentapeptide from [14C]UDP-MurNAc-pentapeptide(DAP) ([14C]UM5A) to undecaprenyl phosphate (C55-P), forming [14C]Lipid I. A radiochemical transfer assay 19 was used, which was optimized for the MraYAA-catalyzed reaction as follows. Reaction mixtures (20 μL each) containing 100 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM MgCl2, 20 mM CHAPS, and 250 μM C55-P were incubated at 45 °C for 5-6 min in the presence of 0 μM, 0.3 μM, or 1.0 μM MD2. For the specific activity assays, 150 μM [14C]UM5A (3.6 nCi/assay) was used. For MraYAA WT KM and kcat determination, [14C]UM5A concentration varied from 25 μM to 750 μM (0.6 – 17.9 nCi/assay). The substrate [14C]UM5A (specific radioactivity: 1.19 × 10−3 Ci/mmol) was purchased from the BaCWAN facility at the University of Warwick 34. MraYAA WT or mutant MraYAA was added to the reaction mixture to a final concentration that enabled product detection within the enzymatic linear range: 50 nM (WT), 500 nM (K70A), 400 nM (T75A), 350 nM (D193N), 250 nM (N255A), 200 nM (F262A), 50 nM (F262W), 400 nM (Q305A), and 500 nM (H325A). The mutant MraYAA enzymes N190A, D193A, and D196A were added to the reaction mixture at a final concentration of 500 nM. All WT and mutant MraYAA enzymes were purified as previously described 21. Each reaction was initiated with the addition of enzyme and it was quenched with 20 μL of 6 M pyridinium acetate, pH 3.0. The radiolabeled product, [14C]Lipid I, was isolated from the hydrophilic substrate, [14C]UM5A, with butanol extraction (200 μL). After vortexing (30 s) and centrifugation at 3000 ×g (5 min), the upper butanol phase was removed, added to 5 mL scintillation fluid (Fisher Chemical), and analyzed using a liquid scintillation counting method (dpm) for 14C detection (Packard 2500 TR Liquid Scintillation Analyzer). Control reactions lacking enzyme and inhibitor were incubated, extracted, and analyzed following same protocol described above and were used for background subtraction. Each reaction rate was calculated by converting the dpm measured (with background subtraction) to moles of [14C]-Lipid I formed using the specific radioactivity and dividing by the reaction time. All experiments were performed in triplicate (technical replicates).
Chemical Synthesis
Muraymycin D2 (MD2) was synthesized as published 17. The MD2 analog, 5-aminoribosyl 3-deoxyuridine, was synthesized as published 35, with a minor modification as follows. In the coupling of the hexose with the ribose, we replaced 5-azido-5-deoxy-D-ribofuranosyl chloride 2, 3-diacetate in the original procedure with a synthetically equivalent 5-azido-5-deoxy-2,3-O-(1-methylethylidene)-D-ribofuranosyl fluoride since the ribofuranosyl fluoride was previously prepared in our laboratory 36. Except for this minor modification, our synthesis of the MD2 analog was identical to the original synthesis by Dini et al 35.
Isothermal Titration Calorimetry
MraYAA WT and mutants (K70A, T75A, D193N, N255A, F262A, F262W, Q305A, and H325A) were purified as previously described 21 in a buffer containing 150mM NaCl, 20mM Tris-HCl pH 8.0, 4 mM DM, 2mM DTT and 10 mM MgCl2. This same buffer was used to dilute the ligand, MD2. One triplicate set of titrations with MraYAA WT and MD2 did not include MgCl2. For MraYAA WT, 145-240 μM MD2 or 118-130 μM 5-aminoribosyl-3-deoxy uridine was titrated into 6.6-35 μM enzyme. For MraYAA mutants K70A, T75A, D193N, N255A, F262W, and H325A, 210 μM MD2 was titrated into 30 μM enzyme. For MraYAA Q305A, 315-430 μM MD2 was titrated into 25-27 μM enzyme. For MraYAA F262A, 80-110 μM MD2 was titrated into 7-10.5 μM enzyme. All titrations were performed in triplicate (technical replicates) at 37°C using either a MicroCal iTC200 or VP-ITC system (GE Healthcare). The total heat exchanged during each injection was fit to a single-site binding isotherm with KD and ΔH° as independent parameters. Data were analyzed and figures were generated using Origin software (OriginLab Corp).
Extended Data
Extended Figure 1.
MraY catalyzes the formation of Lipid I and binds MD2. a, Scheme of the reaction catalyzed by MraY. The U-labeled blue hexagon represents uridine and the M-labeled orange hexagon represents MurNAc. The phosphates associated with the lipid carrier C55-P are shown as red circles, and the phosphates from the substrate, UDP-MurNAc-pentapeptide (UM5A), are shown as yellow circles. b, Chemical structures of the substrate, UM5A (top) and the inhibitor MD2 (bottom). c, Michaelis-Menten kinetic characterization of MraYAA translocase activity. The reaction monitored is the MraYAA-catalyzed transfer of [14C]phospho-MurNAc-pentapeptide from [14C]UM5A to C55-P, forming [14C]Lipid I. The enzymatic parameters measured are as follows: KM = 190 ± 60 μM, kcat = 20 ± 2 min−1, kcat/KM = 0.11 ± 0.3 μM−1 min−1. Data are shown as means of three technical replicates ± s.e.m. d, MD2 (green) in complex with MraYAA. The distances between MD2 and the three catalytic acidic residues D117, D118 and D265 (magenta) are all greater than 4.5 Å.
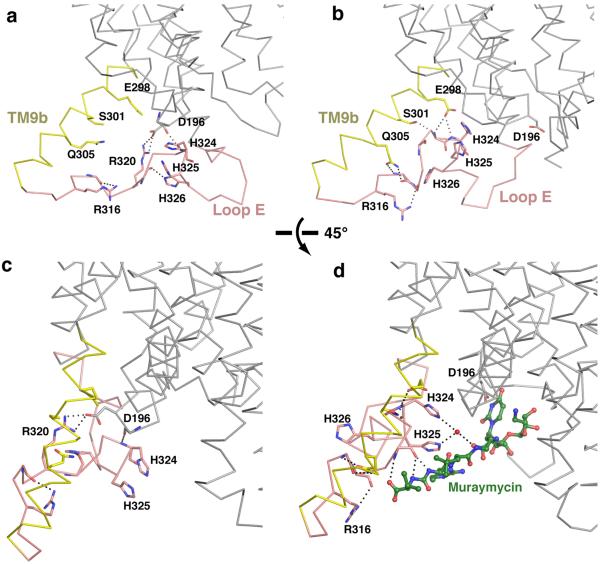
Extended Figure 2.
Conformational changes of the TM9b and loop E region of MraYAA upon MD2 binding. a, Zoomed-in view of TM9b (yellow) and loop E (pink) of apoMraYAA, viewed from within the membrane. b, TM9b and loop E of MD2-bound MraYAA viewed from within the membrane. MD2 is omitted to illustrate the conformational change of TM9b and loop E associated with MD2 binding. Conserved amino acid residues and their interactions are shown in stick representation as dotted lines, respectively. c, 45-degree rotated view of (a) about a horizontal axis. d, 45-degree rotated view of (b) about a horizontal axis, including the model of MD2 (green). The rotation of TM9b and rearrangement of loop E, including the HHH motif, allows for MD2 binding, especially its peptidic moiety.
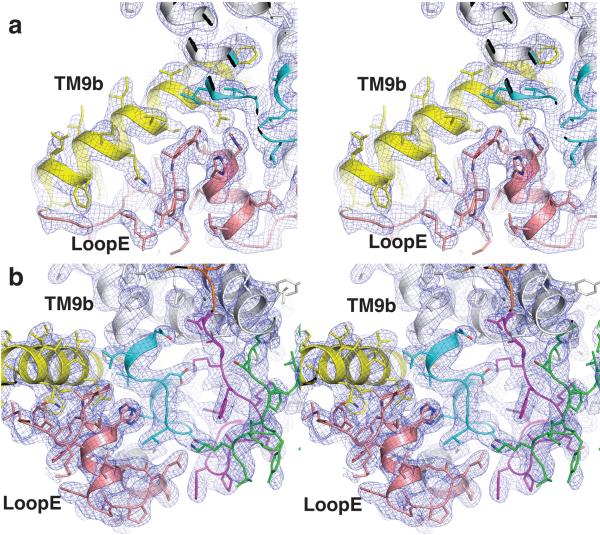
Extended Figure 3.
Quality of electron density map surrounding MD2. a, Stereo view of 2Fo–Fc electron density map at 1σ for TM9b and loop E. b. Stereo view of 2Fo–Fc electron density map at 1σ for the MD2 binding pocket. The electron density peaks corresponding to MD2 are carved for clarity and all TMs are colored as in Fig. 3.
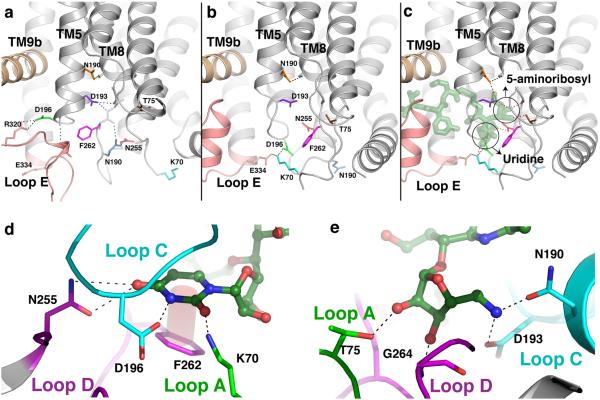
Extended Figure 4.
Conformational changes in MraYAA that create binding pockets for the uridine and 5-amino ribosyl groups of MD2. a, A close-up view of apoMraYAA with key residues that participate in conformational changes upon MD2 binding shown as sticks in various colors. b, A close-up view of the nucleoside-binding pocket in the MraYAA-MD2 complex with MD2 omitted. Key residues are colored as in (a). c, A close-up view of the interactions MD2 (green) makes with the nucleoside-binding pocket of MraYAA. Interactions between MraYAA and MD2 are shown as dotted lines. It is noteworthy that residues interacting with the uridine moiety of MD2 move large distances (5-17 Å for residues K70, D196, N255, and F262), while the residues binding the 5-aminoribosyl group of MD2 (T75, N190, and D193) do not make large sidechain movements upon MD2 binding. The uridine and 5-amino ribosyl groups of MD2 are circled. d, Interactions between the uracil base of MD2 (green) and the nucleoside-binding pocket of MraYAA. The uracil base forms H-bonds with side chains of N255, D196 and K70 and forms a π - π interaction with F262. d, The 5-amino ribosyl group of MD2 forms H-bond interactions with side chains of T75, N190, and D193, and the backbone amide of G264.
Extended Figure 5.
Specific activity of MraYAA WT and mutants in the presence and absence of MD2. a, Normalized specific activity of MraYAA WT and enzymatically active mutants with and without MD2 treatment. MraYAA WT or mutant MraYAA was added to the reaction mixture to a final concentration that enabled product detection within the enzymatic linear range: 50 nM (WT), 500 nM (K70A), 400 nM (T75A), 350 nM (D193N), 250 nM (N255A), 200 nM (F262A), 50 nM (F262W), 400 nM (Q305A), and 500 nM (H325A). Each reaction was carried out in the presence of either 0 μM, 0.3 μM, or 1 μM MD2. Data are shown for three technical replicates ± s.e.m. Specific activity measurements for each mutant were normalized relative to that without added MD2. b, Specific activity of MraYAA WT and enzymatically inactive mutants. MraYAA N190A, D193A, D196A, and D196N were each added to a final concentration of 500 nM, while MraYAA WT was present at 50 nM. All enzymatic reactions were conducted with a radiochemical assay monitoring the transfer of [14C]phospho-MurNAc-pentapeptide from [14C]UM5A to C55-P, forming [14C]Lipid I. The radiolabeled product, [14C]Lipid I, was quantified using a liquid scintillation counting method (dpm). Specific activity was calculated by determining moles of [14C]-Lipid I formed, divided by the reaction time and the quantity of enzyme added. Three technical replicates are shown with the mean value indicated by a line.
Extended Figure 6.
Representative ITC raw data and binding isotherms for MD2 interacting with mutant MraYAA. All titrations were performed in triplicate (technical replicates); see source data for all titrations. Representative data is shown. For MraYAA mutants K70A, T75A, D193N, N255A, F262W, and H325A, 210 μM MD2 was titrated into 30 μM enzyme. For MraYAA Q305A, 315-430 μM MD2 was titrated into 25-27 μM enzyme. For MraYAA F262A, 80-110 μM MD2 was titrated into 7-10.5 μM enzyme. Mean thermodynamic parameters for triplicate titrations are shown in the Extended Data Table 2. Mean KD values for each triplicate are as follows: 63.9 ± 4.7 nM for K70A; 27.4 ± 1.5 nM for T75A; KD not determined for D193N; 29.7 ± 0.8 nM for N255A; 228 ± 4 nM for F262A; 68.4 ± 0.9 nM for F262W; 117 ± 10 nM for Q305A; 24.4 ± 0.4 nM for H325A.
Extended Data Table 1.
Data collection and refinement statistics
| MraYAA-Muraymycin D2 | |
|---|---|
| Data collection | |
| Space group | C 2 2 21 |
| Cell dimensions | |
| a, b, c (Å) | 94.48, 102.05, 135.8 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 2.95 (3.06 – 2.95)* |
| Rmerge (%) | 27.7 (>100) |
| Rpim (%) | 11.6 (64.5) |
| I/σ/ | 4.6 (1.2) |
| CC1/2 (%) | 87.4 (50.2) |
| Completeness (%) | 100 (100) |
| Redundancy | 7.0 (6.3) |
| Refinement | |
| Resolution (Å) | 2.95 (3.18 – 2.95)† |
| No. reflections | 14053 |
| Completeness (%) | 99.40 (93.90) |
| Rwork / Rfree(%) | 24.7/26.1 |
| No. atoms | |
| Protein | 2576 |
| Ligand/ion | 64 |
| Water | 9 |
| B-factors | |
| Protein | 63.50 |
| Ligand/ion | 60.50 |
| Water | 50.00 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.14 |
| Molprobity | |
| Overall | 1.75 |
| Ramachandran (%) | |
| Favored | 99 1 |
| Allow | 0.9 |
| Outlier | 0 |
Values in parentheses are for highest-resolution shell.
Anisotropic truncation at 3.0 Å on c axis by the UCLA Diffraction Anisotropy Server http://services.mbi.ucla.edu/anisoscale/
Extended Data Table 2.
Equilibrium dissociation constants (KDs) and binding parameters demonstrating the effect of mutation in MraYAA on MD2 binding.
| KD (nM) | N (sites) | ΔH (kcal/mol) | ΔS (cal/mol/deg) | |
|---|---|---|---|---|
| WT +Mg2+ | 20.4 ± 1.9 | 0.62 ± 0.02 | −15.0 ± 3.5 | −11.5 ± 9.8 |
| WT −Mg2+ | 15.1 ± 0.2 | 0.65 ± 0.05 | −9.1 ± 0.6 | 6.5 ± 2.0 |
| K70A | 63.9 ± 4.7 | 0.72 ± 0.05 | −7.2 ± 0.2 | 9.8 ± 0.7 |
| T75A | 27.4 ± 1.5 | 0.71 ± 0.01 | −11.7 ± 0.6 | −3.3 ± 1.7 |
| D193N | - | - | - | - |
| N255A | 29.7 ± 0.8 | 0.68 ± 0.06 | −7.6 ± 0.3 | 10.1 ± 1.1 |
| F262A | 228 ± 4 | 0.77 ± 0.07 | −4.9 ± 0.8 | 14.7 ± 2.5 |
| F262W | 68.4 ± 0.9 | 0.81 ± 0.03 | −10.4 ± 0.7 | −0.7 ± 2.2 |
| Q305A | 117 ± 10 | 0.73 ± 0.08 | −9.8 ± 0.3 | 0.1 ± 1.0 |
| H325A | 24.4 ± 0.4 | 0.56 ± 0.07 | −11.3 ± 0.7 | −1.7 ± 2.2 |
| WT + 5-aminoribosyl-3-deoxy uridine | 283 ± 3 | 0.44 ± 0.02 | −15.0 ± 1.5 | −19.9 ± 4.8 |
Data are shown as means of three technical replicates ± s.e.m.
Supplementary Material
Acknowledgements
Data for this study were collected at beamlines NE-CAT 24-ID-C and SER-CAT 22-ID-D and at the Advanced Photon Source. We thank K. Yokoyama for advice and guidance throughout the project and Z. Johnson for manuscript reading. Initial X-ray screening of crystals was performed at the Duke macromolecular crystallography facility. This work was supported by NIH R01 GM100984 (S.-Y.L.) and Duke startup funds (S.-Y.L.). This work was also supported by the JSPS Grant-in-Aid for Scientific Research on Innovative Areas “Chemical Biology of Natural Products” (S.I., Grant Number 24102502) and Scientific Research (B) (S.I., Grant Number 25293026).
Footnotes
Author Contributions B.C. solved the structures and performed some of ITC experiments and E.M. carried out the enzymatic assays and performed most of ITC experiments, both under the guidance of S.-Y.L. T.T. carried out chemical synthesis of MD2 under the guidance of S.I. and A.M. M.K. synthesized 5-aminoribosyl-3-deoxyuridine under the guidance of J.H. S.-Y.L., E.M., and B.C. wrote the paper. All authors discussed the results and commented on the manuscript.
Atomic coordinates and structure factors for the reported crystal structure are deposited in the Protein Data Bank under accession code 5CKR. The authors declare no competing financial interests.
A supplementary video is included in a separate file.
References
- 1.Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. doi:10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 2.Bugg TD, Lloyd AJ, Roper DI. Phospho-MurNAc-pentapeptide translocase (MraY) as a target for antibacterial agents and antibacterial proteins. Infect Disord Drug Targets. 2006;6:85–106. doi: 10.2174/187152606784112128. [DOI] [PubMed] [Google Scholar]
- 3.Lecercle D, et al. Bacterial transferase MraY inhibitors: synthesis and biological evaluation. Bioorganic & medicinal chemistry. 2010;18:4560–4569. doi: 10.1016/j.bmc.2010.04.023. doi:10.1016/j.bmc.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro AB, Jahic H, Gao N, Hajec L, Rivin O. A high-throughput, homogeneous, fluorescence resonance energy transfer-based assay for phospho-N- acetylmuramoyl-pentapeptide translocase (MraY). J Biomol Screen. 2012;17:662–672. doi: 10.1177/1087057112436885. doi:10.1177/1087057112436885. [DOI] [PubMed] [Google Scholar]
- 5.Walsh CT, Zhang W. Chemical logic and enzymatic machinery for biological assembly of peptidyl nucleoside antibiotics. ACS chemical biology. 2011;6:1000–1007. doi: 10.1021/cb200284p. doi:10.1021/cb200284p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winn M, Goss RJ, Kimura K, Bugg TD. Antimicrobial nucleoside antibiotics targeting cell wall assembly: recent advances in structure-function studies and nucleoside biosynthesis. Nat Prod Rep. 2010;27:279–304. doi: 10.1039/b816215h. doi:10.1039/b816215h. [DOI] [PubMed] [Google Scholar]
- 7.Brandish PE, et al. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob Agents Chemother. 1996;40:1640–1644. doi: 10.1128/aac.40.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernhardt TG, Struck DK, Young R. The lysis protein E of phi X174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. The Journal of biological chemistry. 2001;276:6093–6097. doi: 10.1074/jbc.M007638200. doi:10.1074/jbc.M007638200. [DOI] [PubMed] [Google Scholar]
- 9.Bogatcheva E, et al. Chemical modification of capuramycins to enhance antibacterial activity. J Antimicrob Chemother. 2011;66:578–587. doi: 10.1093/jac/dkq495. doi:10.1093/jac/dkq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koga T, et al. Activity of capuramycin analogues against Mycobacterium tuberculosis, Mycobacterium avium and Mycobacterium intracellulare in vitro and in vivo. J Antimicrob Chemother. 2004;54:755–760. doi: 10.1093/jac/dkh417. doi:10.1093/jac/dkh417. [DOI] [PubMed] [Google Scholar]
- 11.McDonald LA, et al. Structures of the muraymycins, novel peptidoglycan biosynthesis inhibitors. J Am Chem Soc. 2002;124:10260–10261. doi: 10.1021/ja017748h. [DOI] [PubMed] [Google Scholar]
- 12.Nikonenko BV, et al. Activity of SQ641, a capuramycin analog, in a murine model of tuberculosis. Antimicrob Agents Chemother. 2009;53:3138–3139. doi: 10.1128/AAC.00366-09. doi:10.1128/AAC.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeoka Y, et al. Expansion of Antibacterial Spectrum of Muraymycins toward Pseudomonas aeruginosa. ACS medicinal chemistry letters. 2014;5:556–560. doi: 10.1021/ml5000096. doi:10.1021/ml5000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanino T, et al. Mechanistic analysis of muraymycin analogues: a guide to the design of MraY inhibitors. Journal of medicinal chemistry. 2011;54:8421–8439. doi: 10.1021/jm200906r. doi:10.1021/jm200906r. [DOI] [PubMed] [Google Scholar]
- 15.Tanino T, et al. Synthesis and Biological Evaluation of Muraymycin Analogues Active against Anti-Drug-Resistant Bacteria. ACS medicinal chemistry letters. 2010;1:258–262. doi: 10.1021/ml100057z. doi:10.1021/ml100057z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanino T, Ichikawa S, Matsuda A. Synthesis of L-epi-capreomycidine derivatives via C-H amination. Organic letters. 2011;13:4028–4031. doi: 10.1021/ol201527k. doi:10.1021/ol201527k. [DOI] [PubMed] [Google Scholar]
- 17.Tanino T, Ichikawa S, Shiro M, Matsuda A. Total synthesis of (−)-muraymycin D2 and its epimer. The Journal of organic chemistry. 2010;75:1366–1377. doi: 10.1021/jo9027193. doi:10.1021/jo9027193. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita A, et al. Muraymycins, novel peptidoglycan biosynthesis inhibitors: synthesis and SAR of their analogues. Bioorganic & medicinal chemistry letters. 2003;13:3345–3350. doi: 10.1016/s0960-894x(03)00671-1. [DOI] [PubMed] [Google Scholar]
- 19.Lloyd AJ, Brandish PE, Gilbey AM, Bugg TD. Phospho-N-acetyl-muramyl pentapeptide translocase from Escherichia coli: catalytic role of conserved aspartic acid residues. Journal of bacteriology. 2004;186:1747–1757. doi: 10.1128/JB.186.6.1747-1757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mengin-Lecreulx D, Flouret B, van Heijenoort J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. Journal of bacteriology. 1982;151:1109–1117. doi: 10.1128/jb.151.3.1109-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung BC, et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–1016. doi: 10.1126/science.1236501. doi:10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Dabbagh B, et al. Active site mapping of MraY, a member of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily, catalyzing the first membrane step of peptidoglycan biosynthesis. Biochemistry. 2008;47:8919–8928. doi: 10.1021/bi8006274. doi:10.1021/bi8006274. [DOI] [PubMed] [Google Scholar]
- 23.Price NP, Momany FA. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology. 2005;15:29R–42R. doi: 10.1093/glycob/cwi065. doi:10.1093/glycob/cwi065. [DOI] [PubMed] [Google Scholar]
- 24.Izumi M, Yuasa H, Hashimoto H. Bisubstrate analogues as glycosyltransferase inhibitors. Current topics in medicinal chemistry. 2009;9:87–105. doi: 10.2174/156802609787354351. [DOI] [PubMed] [Google Scholar]
- 25.Wang R, et al. A search for pyrophosphate mimics for the development of substrates and inhibitors of glycosyltransferases. Bioorganic & medicinal chemistry. 1997;5:661–672. doi: 10.1016/s0968-0896(97)00005-9. [DOI] [PubMed] [Google Scholar]
- 26.Gloster TM, Vocadlo DJ. Developing inhibitors of glycan processing enzymes as tools for enabling glycobiology. Nature chemical biology. 2012;8:683–694. doi: 10.1038/nchembio.1029. doi:10.1038/nchembio.1029. [DOI] [PubMed] [Google Scholar]
- 27.Rillahan CD, Brown SJ, Register AC, Rosen H, Paulson JC. High- throughput screening for inhibitors of sialyl- and fucosyltransferases. Angewandte Chemie. 2011;50:12534–12537. doi: 10.1002/anie.201105065. doi:10.1002/anie.201105065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodolis MT, et al. Mechanism of action of the uridyl peptide antibiotics: an unexpected link to a protein-protein interaction site in translocase MraY. Chemical communications. 2014;50:13023–13025. doi: 10.1039/c4cc06516f. doi:10.1039/c4cc06516f. [DOI] [PubMed] [Google Scholar]
- 29.Dini C, et al. Synthesis of the nucleoside moiety of liposidomycins: elucidation of the pharmacophore of this family of MraY inhibitors. Bioorganic & medicinal chemistry letters. 2000;10:1839–1843. doi: 10.1016/s0960-894x(00)00349-8. [DOI] [PubMed] [Google Scholar]
- 30.Ii K, Ichikawa S, Al-Dabbagh B, Bouhss A, Matsuda A. Function-Oriented Synthesis of Simplified Caprazamycins: Discovery of Oxazolidine-Containing Uridine Derivatives as Antibacterial Agents against Drug-Resistant Bacteria. Journal of medicinal chemistry. 2010;53:3793–3813. doi: 10.1021/jm100243n. doi:Doi 10.1021/Jm100243n. [DOI] [PubMed] [Google Scholar]
References for methods
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. doi:10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. doi:S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Clarke TB, et al. Mutational analysis of the substrate specificity of Escherichia coli penicillin binding protein 4. Biochemistry. 2009;48:2675–2683. doi: 10.1021/bi801993x. [DOI] [PubMed] [Google Scholar]
- 35.Dini C, et al. Synthesis of sub-micromolar inhibitors of MraY by exploring the region originally occupied by the diazepanone ring in the liposidomycin structure. Bioorganic & medicinal chemistry letters. 2002;12:1209–1213. doi: 10.1016/s0960-894x(02)00109-9. [DOI] [PubMed] [Google Scholar]
- 36.Hirano S, Ichikawa S, Matsuda A. Total synthesis of caprazol, a core structure of the caprazamycin antituberculosis antibiotics. Angewandte Chemie. 2005;44:1854–1856. doi: 10.1002/anie.200462439. doi:10.1002/anie.200462439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.