Abstract
Objective
The metabolically healthy obesity (MHO) phenotype is an important obesity subtype in which obesity is not accompanied by any metabolic comorbidity. However, the underlying molecular mechanisms remain elusive. In this study, a shotgun proteomics approach to identify circulating biomolecules and pathways associated with MHO was used.
Methods
The subjects were 20 African‐American women: 10 MHO cases and 10 metabolically abnormal individuals with obesity (MAO) controls. Serum proteins were detected and quantified using label‐free proteomics. Differential expression of proteins between the two groups was analyzed, and the list of differentially expressed proteins was analyzed to determine enriched biological pathways.
Results
Twenty proteins were differentially expressed between MHO and controls. These proteins included: hemoglobin subunits (HBA1, P = 6.00 × 10−18), haptoglobin‐related protein (HPR, P = 1.2 × 10−15), apolipoproteins (APOB‐100, P = 1.50 × 10−40; APOA4, P = 1.1 × 10−14), retinol‐binding protein 4 (RBP4, P = 7.1 × 10−08), and CRP (P = 2.0 × 10−04). MHO was associated with lower levels of proinflammatory and higher levels of anti‐inflammatory biomarkers when compared with MAO. Pathway analysis showed enrichment of lipids and inflammatory pathways, including LXR/RXR and FXR/RXR activation, and acute phase response signaling.
Conclusions
These findings suggested that protection from dysregulated inflammatory and lipid processes were primary molecular hallmarks of MHO. The candidate biomarkers (AHSG, RBP4, and APOA4) identified in this study are potential prognostic markers for MHO.
Introduction
Obesity has reached epidemic proportion globally 1, 2. Annually, 300,000 excess deaths are linked to obesity and its complications, making obesity the second leading cause of premature death 3. Obesity is a major contributor to the global burden of disabilities and chronic diseases including high blood pressure (BP), type 2 diabetes (T2D), and heart diseases 1, 2. While obesity triggers a range of complications, the most common complication is T2D, which is usually preceded by a state of insulin resistance (IR) before T2D is actually diagnosed 4. Paradoxically, it has been documented that not all individuals develop IR or T2D or other cardiometabolic co‐morbidities as they gain weight, a phenomenon referred to as metabolically healthy obesity (MHO). This paradox has led to the claim that “not all obese humans are created equal” 5. MHO subjects were first described about 30 years ago 6 with recent extensive description by Karelis et al. and many other investigators 6, 7, 8, 9, 10. Using the presence of metabolic abnormalities as a classifying variable, individuals with obesity can be grouped into: MHO (also known as the insulin sensitive individuals with obesity) and the metabolically abnormal subjects with obesity (MAO), or the insulin resistant individuals with obesity. Reported prevalence of MHO varies between 10% and 40% of all subjects with obesity and with prevalence varying by many factors including age, ethnic background, and level of physical activity 11, 12. Interestingly, interventions such as weight loss in MHO had no apparent benefit or may even be detrimental as shown by a study conducted by Shin et al. and others 13, 14.
There is evidence that the MHO phenotype is not stable and that many such individuals eventually develop metabolic abnormalities 15, 16, 17. While many MHO may convert to MAO over time, there is a substantial proportion of MHO who remain protected from obesity‐related abnormalities 18. The MHO phenotype seems to span the entire life‐span and has been observed in individuals in their ninth decade of life, as demonstrated in the National Health and Nutrition Examination Survey (NHANES) study in which MHO was 22.1% among individuals with obesity 80 years and older 19. These observations suggest that the phenotype provides an opportunity to investigate the factors that distinguish MHO from MAO and thereby gain insight into biomarkers and molecular mechanisms that are involved in the development of cardiometabolic abnormalities. Most studies of the MHO phenotype have been epidemiological studies 7, 8, 18, 20, 21, 22, and only a few have focused on the molecular basis of MHO.
In the present study, we use a shotgun proteomics approach to evaluate differential protein expression between MHO and MAO individuals. By identifying differentially expressed proteins (DEPs) in serum samples of MHO compared with MAO, we aim to gain biological insights into the underlying biological mechanisms that mediate the MHO phenotype.
Methods
Subject selection and study design
The individuals included in this study were selected from a well‐phenotyped cohort of African Americans (AA) recruited in the Washington, DC, area to study the genetic epidemiology of complex diseases in populations of African descent, the Howard University Family Study (HUFS). Briefly, the main objective of the HUFS was to enroll and examine a randomly obtained sample of African‐American families along with a set of unrelated individuals for study of the genetic and environmental bases of common complex traits including hypertension, obesity, diabetes, and associated phenotypes. In order to maximize the utility of this cohort for the study of multiple common traits, families and individuals were not selected based on any phenotype. All participants were recruited after an overnight fast of at least 8 hours prior to the blood draw and all collected samples were stored in a −80°C freezer pending measurement of biochemical parameters 22. This study was approved by Howard University's institutional review board (IRB) and informed consent was obtained from each participant.
Case definition of MHO
Since this study is exploratory, we used very stringent definitions to select individuals included as MHO or MAO. A combination of three definitions of MHO was applied as follows:
Definition 1: No hypertension (BP ≤ 130/85, no BP medication), no diabetes (glucose ≤ 126 mg/dL), HDL‐C ≥ 40 mg/dL for men and ≥ 50 mg/dL for women, all conditions have to be met. This is the basic MHO definition.
Definition 2: No hypertension (BP ≤ 130/85, no BP medication), no diabetes (glucose ≤ 100 mg/dL), HOMA ≤ 5.1, TG/HDL ≤ 1.65 for men and ≤ 1.32 for women, all conditions have to be met. This is the modified Wildman et al. definition 19.
Definition 3: All definition 2 criteria + hsCRP ≤ 0.3 mg/dL, all conditions have to be met. This third definition takes into account inflammatory status. The C‐reactive protein (CRP) cutoff used was based on Karelis et al. recommendations 9.
The subjects included had to meet all three definitions, that is, these individuals (cases) would be identified as MHO by all three definitions (Figure 1, Supporting Information Table S1).
Figure 1.
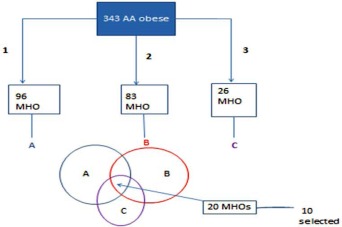
Flowchart for MHO case selection. 1, definition 1: No hypertension (BP ≤ 130/85, no BP medication), no diabetes (glucose ≤ 126 mg/dL), HDL‐C ≥ 40 mg/dL for men and ≥50 mg/dL for women, all conditions have to be met. 2, definition 2: No hypertension (BP ≤ 130/85, no BP medication), no diabetes (glucose ≤ 100 mg/dL), HOMA ≤ 5.1, TG/HDL ≤ 1.65 for men and TG/HDL ≤ 1.32 for women, all conditions have to be met. 3, definition 3: definition 2 + hsCRP ≤ 0.3 mg/dL. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Definition of controls (MAO)
MAOs were selected if they were individuals with obesity who had four or all five of the following abnormalities: hypertension, no diabetes but glucose level higher than 100 mg/dL, HOMA >5.1, TG/HDL >1.65 for men and >1.32 for women, and hsCRP >0.3 mg/dL).
About 10 MHO cases and 10 MAO controls meeting these criteria were studied.
Serum proteomics
Serum shotgun proteomics were done at the National Cancer Institute, Proteomics Core Lab. Serum samples were depleted of 14 highly abundant proteins using Agilent's Multiple Affinity Removal System (MARS) spin cartridges which are 0.45 mL immunoaffinity cartridges packed with antibody‐modified resin (Agilent Technologies, Santa Clara, CA) (list of the 14 proteins depleted, Supporting Information Table S2). The enriched pool of low abundant proteins were then collected and combined from several flow‐through fractions. Quality control measures were used to assess depletion efficiency by removing an aliquot of both the low and high abundant pools and subsequently resolving the proteins on a 4%‐12% SDS‐PAGE gel. Proteins were visualized by staining with Coomassie blue.
A BCA assay was used to determine the protein concentration of the low abundant protein pools. The samples were desalted and concentrated by buffer exchange using 5,000 Da molecular weight cutoff (MWCO) spin concentrators followed by protein digestion using sequencing grade modified trypsin at a 1:20 trypsin‐to‐protein ratio. Resultant peptides were then subjected to solid phase extraction followed by fractionation using strong cation exchange (SCX) chromatography.
Each SCX peptide fraction was analyzed by microcapillary reversed‐phase liquid chromatography tandem mass spectrometry (µRPLC‐MS/MS) using an Agilent 1100 capillary LC system (Agilent Technologies, Santa Clara, CA) coupled online to a linear ion trap mass spectrometer (LTQ, Thermo Electron, Waltham, MA). Separations were performed using 75 μm i.d. × 360 μm o.d. × 10 cm long fused silica capillary ESI emitter columns (Polymicro Technologies, Phoenix, AZ) slurry‐packed in‐house with 3 μm, 300 Å pore size C‐18 silica (Vydac, Hysperia, CA). The mass spectrometer was operated in data‐dependent MS/MS mode with each full MS scan being followed by seven MS/MS scans where the seven most intense peptide molecular ions in the MS scan were sequentially and dynamically selected for collision‐induced dissociation (CID). Dynamic exclusion was employed to minimize redundant acquisition of tandem mass spectra.
Mass spectra were searched against a Uniprot human proteomic database using SEQUEST. A tryptic enzyme restriction with a maximum of two internal missed cleavage sites was used and methionine residues were considered modified in the database search. For a fully tryptic peptide to be considered legitimately identified, it had to achieve stringent charge state and proteolytic cleavage‐dependent cross correlation (Xcorr) scores of 1.9 for [M + H]1+, 2.2 for [M + 2H]2+, 3.1 for [M + 3H]3+, and 4.5 for [M + 4H]4+, and a minimum delta correlation (ΔCn) of 0.08. SEQUEST results were further filtered and analyzed using software developed in‐house. The output is peptide count (PC) data in which columns represent peptide counts (peptide abundance) in each experimental sample and rows represent distinct proteins detected in the samples.
Data quality control and data analysis
A total of 16,553 proteins were identified, at least once, among the 20 subjects. The proteomics data were analyzed using DanteR/InfernoRDN R‐based software packages designed to analyze quantitative proteomics data 23. Only proteins detected in both groups (n = 7,836 proteins) were retained for analysis. The proteomics data were reported as PC which follows a Poisson distribution. To determine DEPs among the two groups, analysis of variance (ANOVA) was carried out using Poisson regression. P‐values were corrected for multiple testing using Benjamin and Hochberg (FDR) and the threshold for significance was P < 0.05. The list of DEPs was submitted to Ingenuity pathway analysis (IPA) to determine biological functions and pathways that were enriched (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). The IPA biological functions analysis calculates the probability of a set of DEPs being associated with known biological functions/canonical pathways by chance alone. The significance of the association between the DEPs and the pathways was measured in two ways: (a) A ratio of the number of molecules from the DEP data set that map to the pathway divided by the total number of molecules that map to the canonical pathways, (b) P‐values obtained from Fisher's exact tests used to assess the probability that observed associations between the DEPs and the canonical pathway were due to chance alone; P‐values <0.05 were considered significant.
Anthropometric and clinical parameters were compared between the two groups by Student's t‐test using SPSS package. Data are presented as mean± standard deviation or median and interquartile range when appropriate.
Results
Characteristics of study groups
The anthropometric and clinical characteristics of the study subjects are summarized in Table 1. The cases (MHO) and the controls (MAO) individuals were not statistically different from each other in key parameters including age, BMI, PFM, and hip circumference. As expected, the six parameters (BP, glucose, HOMA‐IR, HDL‐C, TG, and hsCRP) used in the selection of the two groups are statistically different between the two groups. Notably, MHO had healthier lipid profiles with considerably lower TG and higher HDL‐C levels compared with MAO. Inflammation, measured by adiponectin and hsCRP, was minimal in MHO compared with MAO. Notably, adiponectin, an adipokine decreased in obesity, is paradoxically higher in this group of MHO, about 1.8 times higher than in the MAO group (8,074.0 ± 3,977.4 μg/mL vs. 4,486.8 ± 2,535.9 μg/mL).
Table 1.
Characteristics of study subjects (MAO vs. MHO)
| Variable | MAO | MHO | P a |
|---|---|---|---|
| Age (years) | 43.3 ± 7.8 | 41.3 ± 10.1 | 0.6 |
| BMI (kg/m2) | 44.7 ± 13.0 | 36.5 ± 5.7 | 0.08 |
| Waist (cm) | 120.8 ± 21.1 | 102.5 ± 16.0 | 0.04 |
| Hip (cm) | 134.3 ± 20.4 | 125.1 ± 15.0 | 0.3 |
| WHR | 0.90 ± 0.05 | 0.82 ± 0.06 | 0.004 |
| PFM | 49.1 ± 5.4 | 47.1 ± 5.8 | 0.4 |
| SBPS (mmHg) | 145.8 ± 25.1 | 118.2 ± 6.3 | 0.007 |
| DBPS (mmHg) | 86.4 ± 13.6 | 73.8 ± 4.8 | 0.01 |
| Glucose (mg/dL) | 103.0 ± 16.2 | 83.8 ± 4.5 | 0.004 |
| Insulin (μU/mL) | 67.1 ± 86.2 | 8.9 ± 3.9 | 0.04 |
| HOMA‐IR | 17.3 ± 23.6 | 1.8 ± 0.9 | 0.05 |
| TG (mg/dL) | 233.5 ± 186.6 | 83.8 ± 34.9 | 0.03 |
| LDL‐C (mg/dL) | 103.4 ± 35.3 | 130.5 ± 51.3 | 0.2 |
| HDL‐C (mg/dL) | 34.1 ± 8.6 | 62.2 ± 8.7 | <0.001 |
| Adiponectin (μg/mL) | 4,486.8 ± 2,535.9 | 8,074.0 ± 3,977.4 | 0.03 |
| hsCRP (mg/dL) | 2.2 ± 1.5 | 0.2 ± 0.08 | 0.02 |
Values in this table represent mean ± standard deviation.
Student's t‐test compared means between the two groups. P‐value <0.05 in bold.
Identification of DEPs among MHO and MAO
At total of 56 proteins were differentially expressed (33 upregulated and 23 downregulated) at an unadjusted P < 0.05 as shown on the volcano plot (Supporting Information Figure S1 and Supporting Information Table S3). After adjusting for multiple testing by the Benjamini and Hochberg method and using a P‐value threshold of <0.05, 20 proteins were differentially expressed between MHO and MAO including 8 downregulated and 12 upregulated in MHO compared with MAO (Table 2, Supporting Information Figure S2). Among the most upregulated in MHO were APOB‐100 (P = 1.5 × 10−40, FC = 1.2) and alpha‐2‐HS‐glycoprotein (ASHG, P = 3.00 × 10−29, FC = 1.7). Hemoglobin subunit alpha (HBA, P = 6.0 × 10−18, FC= 0.44) and haptoglobin‐related protein (HPR, P = 1.2 × 10−15, FC = 0.6) were downregulated in MHO compared with MAO. CRP was also confirmed to be downregulated in MHO by the proteomic data (P = 2.0 × 10−04). Supporting Information Figure S3 is a principal components plot of the DEPs that shows separation of MHO and MAO subjects by PC1, PC2, and PC3, indicating the discriminant ability of these 20 proteins.
Table 2.
Differentially expressed proteins among MHO and MAO African‐American women determined by label‐free serum proteomics
| Accession # | Protein name | Gene symbol | Log effect size (log2FC) | P MHO unadjusted | P MHO BH | Average PC in MHO | Average PC in MAO |
|---|---|---|---|---|---|---|---|
| 8 under‐expressed proteins in MHO compared with MAO | |||||||
| P69905 | Hemoglobin subunit alpha | HBA1 | −1.17 | 3.1 × 10−21 | 6.00 × 10−18 | 8.5 | 27.5 |
| P00739 | Haptoglobin‐related protein | HPR | −0.78 | 7.6 × 10−19 | 1.2 × 10−15 | 19.2 | 42.1 |
| P68871 | Hemoglobin subunit beta | HBB | −0.80 | 4.6 × 10−15 | 4.5 × 10−12 | 13.8 | 30.8 |
| P00751 | Complement factor B | CFB | −0.21 | 1.4 × 10−10 | 1.1 × 10−07 | 172.7 | 212.6 |
| Q14624 | Inter‐alpha‐trypsin inhibitor heavy chain H4 | ITIH4 | −0.16 | 3.0 × 10−09 | 1.70 × 10−06 | 242 | 285.1 |
| P02741 | C‐reactive protein | CRP | −2.13 | 4.00 × 10−07 | 2.0 × 10−04 | 1.2 | 10.1 |
| P27169 | Serum paraoxonase1 | PON1 | −1.16 | 1.5 × 10−06 | 7.0 × 10−04 | 2.44 | 7.8 |
| P0C0L4 | Complement C4‐A | C4A | −0.081 | 2.8 × 10−06 | 1.0 × 10−03 | 638.4 | 692.5 |
| 12 over‐expressed proteins in MHO compared with MAO | |||||||
| P04114 | Apolipoprotein B‐100 | APOB | 0.28 | 1.9 × 10−44 | 1.5 × 10−40 | 563.1 | 423.7 |
| P02765 | Alpha‐2‐HS‐glycoprotein | AHSG | 0.75 | 7.7 × 10−33 | 3.00 × 10−29 | 79.1 | 37.4 |
| P01008 | Antithrombin‐III | SERPINC1 | 0.41 | 3.50 × 10−26 | 9.2 × 10−23 | 163.8 | 108.2 |
| P06727 | Apolipoprotein A‐IV | APOA4 | 0.40 | 8.5 × 10−18 | 1.10 × 10−14 | 113.8 | 76.1 |
| P05155 | Plasma protease C1 inhibitor | SERPING1 | 0.71 | 4.6 × 10−15 | 4.5 × 10−12 | 37 | 18.2 |
| P02753 | Retinol‐binding protein 4 | RBP4 | 0.46 | 8.1 × 10−11 | 7.1 × 10−08 | 51.5 | 32.5 |
| P19823 | Inter‐alpha‐trypsin inhibitor heavy chain H2 | ITIH2 | 0.19 | 2.3 × 10−10 | 1.6 × 10−07 | 249.5 | 206.6 |
| P06396 | Gelsolin | GSN | 0.25 | 3.4 × 10−10 | 2.25 × 10−07 | 135.1 | 104.3 |
| P04196 | Histidine‐rich glycoprotein | HRG | 0.30 | 8.9 × 10−10 | 5.3 × 10−07 | 99.4 | 73.8 |
| P19827 | Inter‐alpha‐trypsin inhibitor heavy chain H1 | ITIH1 | 0.15 | 7.5 × 10−07 | 0.0004 | 228.9 | 196.6 |
| P02774 | Vitamin D‐binding protein | GC | 0.17 | 1.7 × 10−06 | 0.0007 | 165.8 | 139.3 |
| P10643 | Complement component C7 | C7 | 0.36 | 1.8 × 10−05 | 0.007 | 34.9 | 24.4 |
Cutoff P‐value < 0.05.
Enriched biological pathways in the most differentially expressed proteins in MHO compared with MAO
We used IPA to identify enriched biological pathways in the 20 DEPs. The most significant enriched canonical pathways include LXR/RXR activation (P = 1.4 × 10−15), FXR/RXR activation (P = 1.9 × 10−15), acute phase response signaling (2.9 × 10−14), and complement system (P = 4.6 × 10−08) (Figure 2, Table 3). Interestingly, a number of the enriched pathways are interconnected through DEPs that are common to more than one canonical pathway. For example, acute phase response signaling has three and four DEPs in common with complement system (C4A/C4B, CFB, SERPING1) and LXR/RXR activation (AHSG, C4A/C4B, ITIH4, RBP4), respectively (Figure 3, Table 3). Figure 4 depicts DEPs mapped to the most significant enriched canonical pathways.
Figure 2.
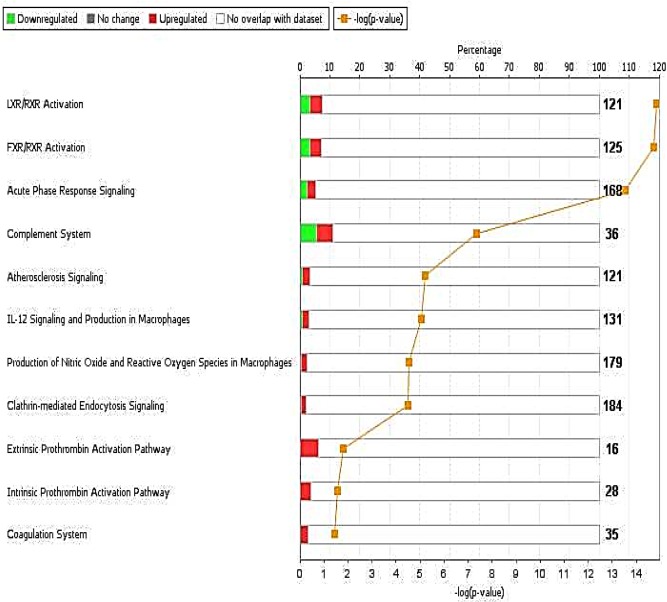
Canonical pathways significantly enriched in the 20 DEPs. Bars represent total number of molecules in a pathway; upper Y‐axis represents the ratio between the number of molecules in our data set that are associated with a pathway and the total number of molecules in that pathway; lower Y‐axis represents –log of the P‐value associated with each pathway. X‐axis lists the top pathways associated with the DEPs. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table 3.
DEPs associated with the most significantly enriched canonical pathways
| Name of canonical pathways | Associated DEPs | Overlap ratioa | P‐valueb |
|---|---|---|---|
| LXR/RXR activation |
AHSG, APOA4, APOB, C4A/C4B GC, HPR, ITIH4, PON1, RBP4 |
9/121 (7.4%) | 1.4 × 10−15 |
| FXR/RXR activation |
AHSG, APOA4, APOB, C4A/C4B, GC, HPR ITIH4, PON1, RBP4 |
9/125 (7.2%) | 1.9 × 10−15 |
| Acute phase response signaling |
AHSG, C4A/C4B, CFB, CRP, HRG, ITIH2, ITIH4, RBP4, SERPING1 |
9/168 (5.4%) | 2.9 × 10−14 |
| Complement system | C4A/C4B, CFB, SERPING1 | 4/36 (11.1%) | 4.6 × 10−08 |
| Atherosclerosis signaling | APOA4, APOB, PON1, RBP4 | 4/121 (3.3%) | 6.3 × 10−06 |
Numerator of ratio represents the number of DEPs in our data set that map to the canonical pathway; the denominator represents the total number of molecules/proteins that map to the pathway.
P‐value obtained by Fisher's exact test.
Figure 3.
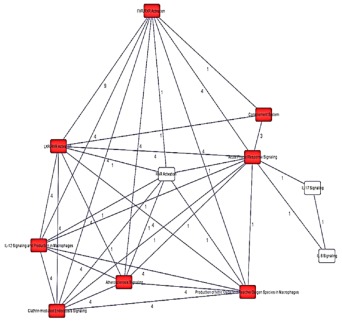
Overlap between the top canonical pathways enriched in the 20 DEPs. Black lines: interconnectivity between canonical pathways; red boxes: significantly enriched canonical pathways in the study. Numbers on black lines represent the number of proteins common to two interconnected pathways. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 4.
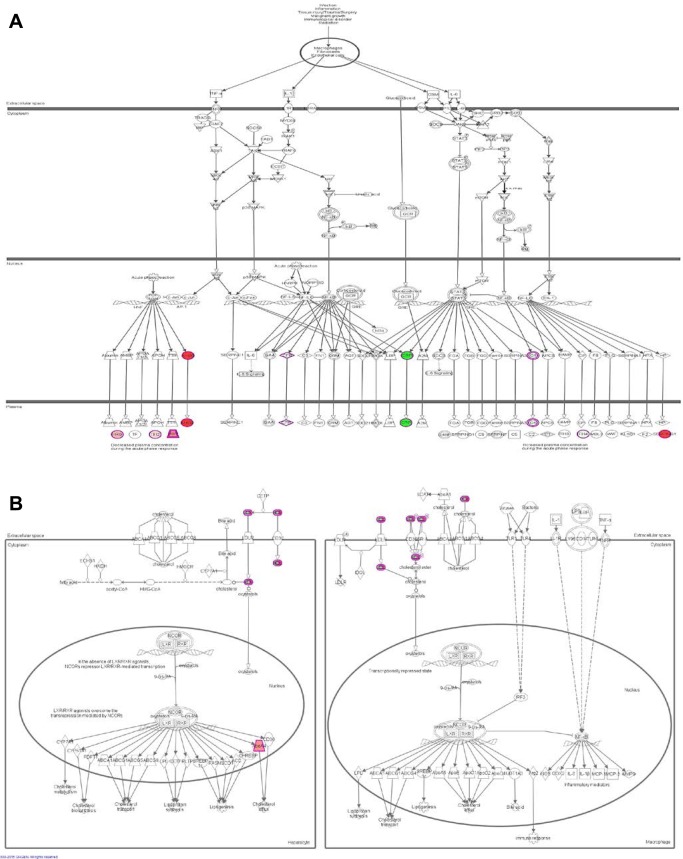
Two of the most significantly enriched canonical pathways in MHO. (A) Acute phase reactant signaling in MHO overlapped with DEPs. (B) LXR/RXR activation in MHO overlapped with DEPs. Red: proteins upregulated in our data set; the intensity of the color indicates the degree of upregulation. Green: proteins downregulated in our data set; the intensity of the color indicates the degree of downregulation. White: proteins not specific to our data set but incorporated as part of the network. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Discussion
Despite considerable interest in this subset of MHO subjects, the underlying molecular mechanisms remain largely unknown. Understanding the pathophysiology of MHO is important not only for this subset of individuals but also for obesity as a whole. Here, we describe the results of the first study to use high‐throughput omics technology, namely shotgun label‐free quantitative proteomics, to profile the proteome and obtain a comprehensive snapshot of differences in protein expression between MHO and MAO African‐American women.
The DEPs between MHO and MAO underscored important inflammation‐related proteins including AHSG, CRP, histidine‐rich glycoprotein (HRG), retinol‐binding protein 4 (RBP4), complement factor‐4A (C4A), and inter‐alpha‐trypsin inhibitor heavy chain H4 (ITIH4). These molecules belong to the family of acute phase reactant proteins (APP) and are primarily produced by the liver in response to injury and infection 24, but some (e.g., RBP4) are also produced by adipocytes 25. It is well documented that inflammation is a key component of obesity and has been associated with the progression and development of obesity‐related complications 26. In this study, the APPs differentially expressed can be categorized into two groups, the negative APPs (AHSG, HRG, and RBP4) all of which were over‐expressed in MHO, and the positive APPs (HRP, CRP, C4A, and ITIH4) which in contrast to the negative APPs were downregulated in MHO. These changes suggest a favorable inflammatory profile associated with MHO.
Some of these molecules have also been involved in adverse metabolic events such as impairment of insulin signaling, fatty liver diseases, and suppression of adiponectin production 25, 27, 28, 29, 30, 31. For example, high levels of AHSG, a glycoprotein produced by hepatocytes, have been linked to metabolic syndrome in certain populations 28, 30. However, the association seems to vary with gender as one report showed an association between high AHSG and metabolic syndrome in Japanese men but not in women 32. While our study showed that AHSG is upregulated in MHO, we did not see any of the other reported changes associated with elevated AHSG. For example, the MHO in this study had high adiponectin levels compared with MAO (8,074.0 ± 3,977.4 μg/mL vs. 4,486.8 ± 2,535.9 μg/mL) and remained insulin sensitive based on their HOMA‐IR values (1.8 vs. 17.3). These results suggest at least two hypotheses—(a) AHSG is regulating its effectors differently in this MHO cohort or (b) a threshold in AHSG concentration needs to be reached to see its effects on metabolic markers such adiponectin and insulin. In the latter scenario, AHSG may be a potential biomarker on its own or in combination with other proteins (e.g., RBP4, APOA4, APOB) to identify MHO that will convert into MAO.
RBP4, an adipokine with differential regulation in animals and humans and involved in the transport of vitamin A to peripheral tissues, has been associated with dysfunction of insulin signaling especially in animal models. However, in humans the positive association between RBP4 levels and IR has not been consistent 25, 29, 33. RBP4 could also be a promising marker to follow the changes in MHO especially now that the MHO phenotype has been shown to be a transient state for many affected individuals 12.
The complement system pathway, part of the humoral immune system and involved in inflammatory processes, was over‐represented among the DEPs associated with MHO. As expected, all DEPs (C4A/C4B, CFB, SERPING1) associated with the complement system were also associated with the acute phase reactant pathway showing a commonality between the over‐represented pathways. All but SERPING1 were downregulated, thus agreeing with the lower inflammation profile seen in MHO. In fact, SERPING1 is an inhibitory regulator of the complement cascade and may play a role in inflammation suppression. SERPING1 has been shown to be downregulated in patients with T2D compared with healthy controls 34.
Nine of the DEPs (AHSG, APOA4, APOB, C4A/C4B, GC, HPR, ITIH4, PON1, and RBP4) were associated with lipid metabolism. Four of these DEPs are involved in both inflammatory and lipid processes (AHSG, C4A/C4B, ITIH4, and RBP4), underscoring the pleiotropic function of some of the DEPs. Previous studies have found an association between AHSG and RBP4 and serum lipids (mainly triglycerides [TG] and HDL‐C) 27, 35. APOB‐100, APOA4, and PON1 are all members of the HDL/LDL family. APOA4 is secreted by enterocytes along with TG and chylomicrons. For example, APOA4 has anti‐oxidant and anti‐inflammatory properties and is proposed to be protective against cardiovascular diseases. It also plays a role in glucose homeostasis and satiety (reduces food intake) 36. Interestingly in T2D and obesity, increased levels of APOA4 have been found to be mainly related to hypertriglyceridemia 37. In this study, APOA4 is upregulated without hypertriglyceridemia; while the mechanisms involved need further investigation, the absence of hypertriglyceridemia seems to be protective in MHO.
A major strength of the study is the stringent and conservative approaches used to define MHO individuals (cases). Three distinct definitions of MHO were used and only individuals who were classified as MHO by all three definitions were selected and studied. Though this approach decreases the pool of MHO, it is more effective in reducing misclassification. In contrast to a recently published metabolomics study in MHO in which the definition of MAO used may have resulted in heterogeneous metabolic phenotypes 38, this study included only MAO with all four or five metabolic abnormalities. Secondly, the differential proteomic method combined with bioinformatics analysis (IPA) demonstrated changes in serum protein profiles in MHO individuals that were not previously reported.
While very promising, the insights provided by this study into the molecular basis of MHO should be interpreted within the following context—(a) by design, this study is a discovery investigation and as such the findings need to be confirmed in larger and diversified populations; (b) the cross‐sectional design provides only a snapshot of the proteome and causality should not be inferred; and (c) important confounding factors that may affect the serum proteome including level of physical activity and diet were not analyzed in this study. While earlier studies did not find any difference in dietary profile and physical activity between MHO and MAO 11, 13, 18, 26, a recent study with larger sample sizes and the use of healthy eating index (HEI) showed that MHO have a better dietary compliance to the US guidelines than MAO including intake of more whole grains, fruits, and beans. However, the authors acknowledged that the effect seen may be due to reverse causality 39. Although diet and exercise are likely to affect biological functions in MHO, studies available to date have not specifically assessed inflammation and other biological functions in a systematic manner. Future omics studies, including whole genome microarray, miRNA profiling, and whole exome sequencing, investigating the determinants of the MHO phenotype will benefit from having data on potential lifestyle confounding variables such as physical activity and diets.
Conclusion
Our study is the first to use proteomics to investigate the mechanisms that are involved in the MHO phenotype. Our findings confirm previous reports that inflammation is a key hallmark of metabolic health in individuals with obesity but also identified a network of APPs, more than what was previously reported, to be involved in the inflammatory processes associated with MHO. Some of these proteins, mainly RBP4, AHSG, APOA4, PON1, and APOB‐100, could be good candidates to follow up in longitudinal studies to determine their ability to predict which MHO individuals convert to MAO. We also found that differential regulation of lipid metabolism is also important to the MHO phenotype.
Supporting information
Supporting Information
Acknowledgments
Raw proteomics study data set available at http://crggh.nih.gov/resources.cfm under HUFS.
Funding agencies: Support for this study is provided by National Institutes of Health (NIH) grant No. 3T37TW00041‐03S2 from the Office of Research on Minority Health, the National Human Genome Research Institute (NHGRI), the National Center for Research Resources (NCRR), and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant DK‐54001. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of the NIH. This research was supported in part by the Intramural Research Program of the Center for Research on Genomics and Global Health. The Center for Research on Genomics and Global Health is supported by the NHGRI, the NIDDK, the Center for Information Technology, and the Office of the Director at the NIH (Z01HG200362).
Disclosure: The authors declare no conflict of interest.
Author contributions: AD—conceived the study, was responsible for study coordination, performed the statistical and proteomics analyses, drafted the manuscript, and interpreted the data. AA—participated in the design of the study, analyzed the proteomics data, and edited and approved the manuscript. JZ—cleaned and managed the data. MZ, DP—coordinated and carried out the shotgun proteomics in the proteomics core lab. CR—participated in the conception and design of the study, guided the research, and approved the final manuscript. All authors read and approved the final manuscript.
References
- 1. Bastien M, Poirier P, Lemieux I, Despres JP. Overview of epidemiology and contribution of obesity to cardiovascular disease. Prog Cardiovasc Dis 2014;56:369–381. [DOI] [PubMed] [Google Scholar]
- 2. Shamseddeen H, Getty JZ, Hamdallah IN, Ali MR. Epidemiology and economic impact of obesity and type 2 diabetes. Surg Clin N Am 2011;91:1163–1172. [DOI] [PubMed] [Google Scholar]
- 3. Allison DB, Fontaine KR, Manson JE, Stevens J, VanItallie TB. Annual deaths attributable to obesity in the United States. JAMA 1999;282:1530–1538. [DOI] [PubMed] [Google Scholar]
- 4. Vague J, Vague P, Tramoni M, Vialettes B, Mercier P. Obesity and diabetes. Acta Diabetol Lat 1980;17:87–99. [DOI] [PubMed] [Google Scholar]
- 5. Reaven G. All obese individuals are not created equal: insulin resistance is the major determinant of cardiovascular disease in overweight/obese individuals. Diabetes Vasc Dis Res 2005;2:105–112. [DOI] [PubMed] [Google Scholar]
- 6. Sims EA. Are there persons who are obese, but metabolically healthy? Metabolism 2001;50:1499–1504. [DOI] [PubMed] [Google Scholar]
- 7. Roberson LL, Aneni EC, Maziak W, et al. Beyond BMI: the “Metabolically healthy obese” phenotype & its association with clinical/subclinical cardiovascular disease and all‐cause mortality ‐ a systematic review. BMC Public Health 2014;14:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boonchaya‐anant P, Apovian CM. Metabolically healthy obesity‐does it exist? Curr Atheroscler Rep 2014;16:441 [DOI] [PubMed] [Google Scholar]
- 9. Karelis AD, Brochu M, Rabasa‐Lhoret R, Garrel D, Poehlman ET. Clinical markers for the identification of metabolically healthy but obese individuals. Diabetes Obes Metab 2004;6:456–457. [DOI] [PubMed] [Google Scholar]
- 10. Karelis AD, Faraj M, Bastard JP, et al. The metabolically healthy but obese individual presents a favorable inflammation profile. J Clin Endocrinol Metab 2005;90:4145–4150. [DOI] [PubMed] [Google Scholar]
- 11. Phillips CM. Metabolically healthy obesity: definitions, determinants and clinical implications. Rev Endocr Metab Disord 2013;14:219–227. [DOI] [PubMed] [Google Scholar]
- 12. Bluher M. Are metabolically healthy obese individuals really healthy? Eur J Endocrinol 2014;171:R209–R219. [DOI] [PubMed] [Google Scholar]
- 13. Karelis AD, Messier V, Brochu M, Rabasa‐Lhoret R. Metabolically healthy but obese women: effect of an energy‐restricted diet. Diabetologia 2008;51:1752–1754. [DOI] [PubMed] [Google Scholar]
- 14. Shin MJ, Hyun YJ, Kim OY, et al. Weight loss effect on inflammation and LDL oxidation in metabolically healthy but obese (MHO) individuals: low inflammation and LDL oxidation in MHO women. Int J Obes 2006;30:1529–1534. [DOI] [PubMed] [Google Scholar]
- 15. Achilike I, Hazuda HP, Fowler SP, Aung K, Lorenzo C. Predicting the development of the metabolically healthy obese phenotype. Int J Obes 2015;39:228–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soriguer F, Gutierrez‐Repiso C, Rubio‐Martin E, et al. Metabolically healthy but obese, a matter of time? Findings from the prospective Pizarra study. J Clin Endocrinol Metab 2013;98:2318–2325. [DOI] [PubMed] [Google Scholar]
- 17. Kramer CK, Zinman B, Retnakaran R. Are metabolically healthy overweight and obesity benign conditions?: a systematic review and meta‐analysis. Ann Intern Med 2013;159:758–769. [DOI] [PubMed] [Google Scholar]
- 18. Plourde G, Karelis AD. Current issues in the identification and treatment of metabolically healthy but obese individuals. Nutr Metab Cardiovasc Dis 2014;24:455–459. [DOI] [PubMed] [Google Scholar]
- 19. Wildman RP, Muntner P, Reynolds K, et al. The obese without cardiometabolic risk factor clustering and the normal weight with cardiometabolic risk factor clustering: prevalence and correlates of 2 phenotypes among the US population (NHANES 1999‐2004). Arch Intern Med 2008;168:1617–1624. [DOI] [PubMed] [Google Scholar]
- 20. Fabbrini E, Yoshino J, Yoshino M, et al. Metabolically normal obese people are protected from adverse effects following weight gain. J Clin Investig 2015;125:787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van der AD, Nooyens AC, van Duijnhoven FJ, Verschuren MM, Boer JM. All‐cause mortality risk of metabolically healthy abdominal obese individuals: the EPIC‐MORGEN study. Obesity 2014;22:557–564. [DOI] [PubMed] [Google Scholar]
- 22. Doumatey AP, Bentley AR, Zhou J, et al. Paradoxical hyperadiponectinemia is associated with the metabolically healthy obese (MHO) phenotype in African Americans. J Endocrinol Metab 2012;2:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taverner T, Karpievitch YV, Polpitiya AD, et al. DanteR: an extensible R‐based tool for quantitative analysis of ‐omics data. Bioinformatics 2012;28:2404–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jain S, Gautam V, Naseem S. Acute‐phase proteins: as diagnostic tool. J Pharm Bioall Sci 2011;3:118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park SE, Park CY, Sweeney G. Biomarkers of insulin sensitivity and insulin resistance: past, present and future. Crit Rev Clin Lab Sci 2015;4:1–11. [DOI] [PubMed] [Google Scholar]
- 26. Navarro E, Funtikova AN, Fito M, Schroder H. Can metabolically healthy obesity be explained by diet, genetics, and inflammation? Mol Nutr Food Res 2015;59:75–93. [DOI] [PubMed] [Google Scholar]
- 27. Dabrowska AM, Tarach JS, Wojtysiak‐Duma B, Duma D. Fetuin‐A (AHSG) and its usefulness in clinical practice ‐ review of the literature. Biomedical papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia; 2015. [DOI] [PubMed]
- 28. Stefan N, Hennige AM, Staiger H, et al. Alpha2‐Heremans‐Schmid glycoprotein/fetuin‐A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care 2006;29:853–857. [DOI] [PubMed] [Google Scholar]
- 29. Janke J, Engeli S, Boschmann M, et al. Retinol‐binding protein 4 in human obesity. Diabetes 2006;55:2805–2810. [DOI] [PubMed] [Google Scholar]
- 30. Ix JH, Shlipak MG, Brandenburg VM, et al. Association between human fetuin‐A and the metabolic syndrome: data from the Heart and Soul Study. Circulation 2006;113:1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hennige AM, Staiger H, Wicke C, et al. Fetuin‐A induces cytokine expression and suppresses adiponectin production. PLoS One 2008;3:e1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Obuchi A, Adachi H, Enomoto M, et al. High plasma fetuin‐A levels are associated with metabolic syndrome among males but not females in a Japanese general population. Diabetes Res Clin Pract 2014;106:128–135. [DOI] [PubMed] [Google Scholar]
- 33. Kotnik P, Fischer‐Posovszky P, Wabitsch M. RBP4: a controversial adipokine. Eur J Endocrinol 2011;165:703–711. [DOI] [PubMed] [Google Scholar]
- 34. Zhang Q, Fillmore TL, Schepmoes AA, et al. Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. J Exp Med 2013;210:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Broch M, Gomez JM, Auguet MT, et al. Association of retinol‐binding protein‐4 (RBP4) with lipid parameters in obese women. Obes Surg 2010;20:1258–1264. [DOI] [PubMed] [Google Scholar]
- 36. Wang F, Kohan AB, Lo CM, et al. Apolipoprotein A‐IV: a protein intimately involved in metabolism. J Lipid Res 2015;56:1403–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verges B. Apolipoprotein A‐IV in diabetes mellitus. Diabetes Metab 1995;21:99–105. [PubMed] [Google Scholar]
- 38. Chen HH, Tseng YJ, Wang SY, et al. The metabolome profiling and pathway analysis in metabolic healthy and abnormal obesity. Int J Obes 2015;39:1241–1248. [DOI] [PubMed] [Google Scholar]
- 39. Camhi SM, Whitney Evans E, Hayman LL, Lichtenstein AH, Must A. Healthy eating index and metabolically healthy obesity in U.S. adolescents and adults. Prev Med 2015;77:23–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


