Abstract
Herein, we report studies leading to the discovery of the neoseptins and a comprehensive examination of the structure–activity relationships of this new class of small molecule mouse Toll-like receptor 4 (mTLR4) agonists. The compound class, which emerged from screening a α-helix mimetic library, stimulate the immune response, act by a well-defined mechanism (mouse TLR4 agonist), are easy to produce and structurally manipulate, exhibit exquisite structure–activity relationships, are non-toxic, and elicit improved and qualitatively different responses than lipopolysaccharide (LPS) even though they share the same receptor.
Graphical Abstract
Introduction
The human immune system is divided into innate and adaptive components. The innate immune system represents the first line of defense in mammals, responding to pathogens and abnormal cells through activation of multiple cell types including dendritic cells, macrophages, neutrophils, and natural killer cells. The adaptive immune system entails antigen-specific T cell and B cell responses mediated by antigen-presenting dendritic cells, and serves to protect the host in the longer term by neutralizing the pathogens and abnormal cells through the action of T cell receptors and antibodies.1,2 The Toll-like receptors (TLRs),3–6 the most widely recognized subset of the pathogen-associated or damage-associated molecular pattern (PAMPs and DAMPs) receptors, act as part of the initial innate immune response system, recognizing molecular signatures of infectious pathogens or abnormal cells.3,5 Their activation not only initiates an intracellular signaling cascade that leads to the activation of transcription factors that trigger the production of cytokines and chemokines,3,5 but it also induces the adaptive immune response.6 In fact, the action of vaccines is due in part to the activation of the TLR system.7 TLR agonists are immune system enhancers and could be useful in the treatment of not only infectious diseases but also cancer, representing a complementary small molecule approach in the emerging area of immunotherapy.8–11 They would also be attractive as new vaccine adjuvants for both infectious diseases and oncology that act by well-defined mechanisms.8–11 However, a relatively small number of small molecule classes have been found to behave as TLR agonists.8–11 Notable examples include the TLR7 agonists imiquimod,12 isotoribine,13 and 8-oxo-9-benzyladenine14 as well as the TLR7/8 agonist resiquimod15 that today still serve as the inspiration for nearly all such work on small molecule TLR agonists.16,17 Recent additional work has described TLR2/TLR1 agonists.18,19 Relevant to the studies detailed herein and among the TLRs, TLR4 selectively responds to LPS (lipopolysaccharide or endotoxin, Figure 1)5 extracted from aggregated LPS by LPS binding protein (LBP),20 delivered by cluster of differentiation 14 (CD14)21 to the deep hydrophobic pocket of myeloid differentiation protein 2 (MD-2)22 in complex with TLR4 that leads to receptor dimerization and adoption of the conformation that characterizes the activated complex (LPS–MD2/TLR4)2.23
Figure 1.
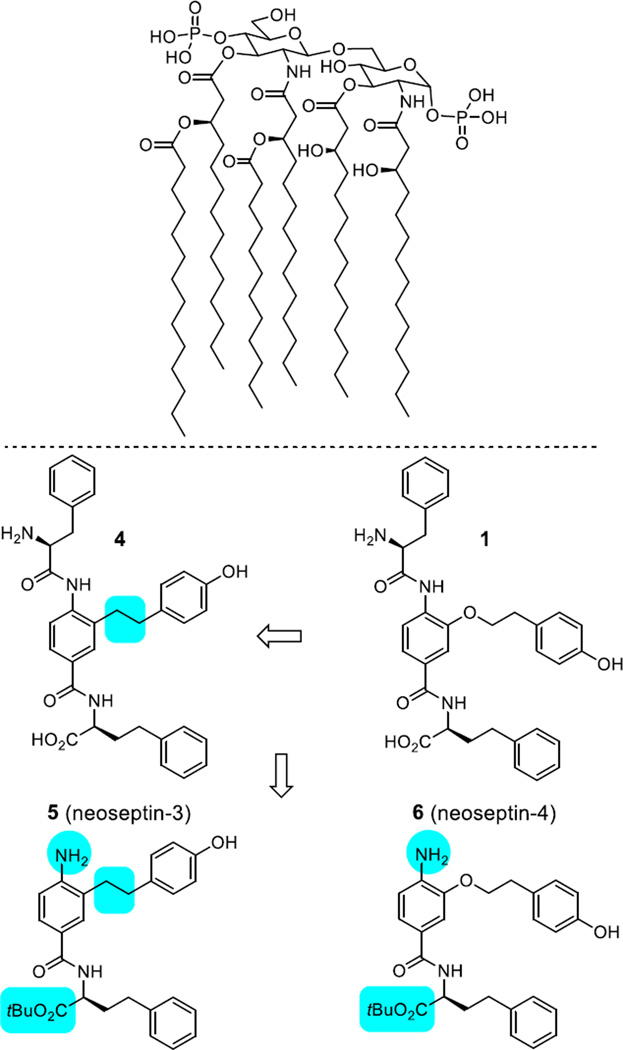
Top: Structure of Lipid A, a component of lipopolysaccharide (LPS, endotoxin), which is responsible for the toxicity of Gram-negative bacteria. It is the innermost of three regions of LPS. While its toxic effects are damaging, the sensing of Lipid A by the human immune system (TLR4/MD-2) is critical for the onset of immune responses to Gram-negative bacteria, and the fight against the infection. Bottom: Key steps in the initial elaboration of a screening lead, compound 1, to the compounds 5 and 6 examined herein.
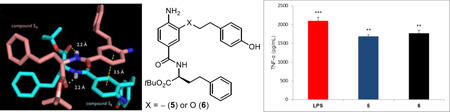
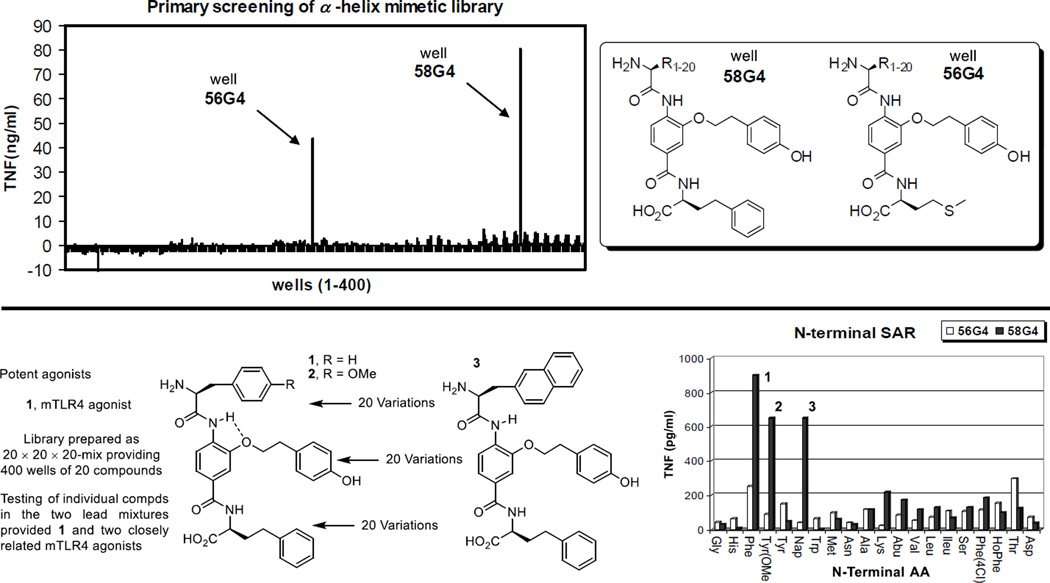
Complementary to a genetic approach in whole organisms that induce random nucleic acid mutations in search of phenotypic changes derived from altered genes impacting the immune system, which was used to discover that LPS acts through activation of a Toll-like receptor (TLR4),5 we explored the use of an alternative chemical genetics approach.24 The approach entailed screening libraries of compounds in cell-based functional assays that serve as a read out of stimulated signaling of an immune response. In a screening campaign conducted with nearly 100,000 compounds25 using a functional assay that measures the stimulated release of TNF-α from treated mouse macrophages, we identified a new class of activators of the immune response.26 The libraries screened in our efforts represent a unique compound collection populated by nontraditional compounds25 designed to target protein–protein27,28 or protein–DNA interactions29 as well as the major enzyme classes.30 The library screening for mouse macrophage TNF-α production resulted in the discovery of a single class of active compounds found in an 8,000-membered α-helix mimetic library, prepared as 400 mixtures of 20 compounds.27 It is worth noting that the functional activity measured in the screening assay is both a rare activation event (activator/agonist vs inhibitor/antagonist) and was conducted with an assay that is extraordinarily sensitive, such that even weak and rare activation for TNF-α release proved detectable and clear (Figure 2). The two active pools from the α-helix mimetic library observed in the screen (56G4 and 58G4) were retested as their individual compounds providing three closely related active compounds 1–3 (Figure 2). From the initial screening results, it was also determined that even small deviations from their structure resulted in no detection of activity, providing a first level structure–activity relationship (SAR) directly from screen. Representative of this SAR data available from the initial screen, activity was observed only with a central tyrosine side chain substituent and all other closely related substituents in the library,27 including phenyl, 4-chlorophenyl, naphthyl, 4-methoxyphenyl, and indolyl, were inactive. Similarly, the two active mixtures contained either a homophenylalanine (HoPhe) or methionine (Met) side chain at the carboxylic acid terminus and all other 18 residues in the library27 were inactive, including phenylalanine, 4-chlorophenylalanine, 4-methoxyphenylalanine, naphthylalanine, tyrosine, tryptophan, leucine, valine, isoleucine, alanine, glycine, asparagine, lysine, serine, threonine, aspartate, histidine, and aminobutyric acid (Abu). Equipped with this information, approximately 220 compounds in a library exploring the C-terminus of 1 and 225 compounds varying the N-terminus of 1 were prepared. These libraries provided analogues that approached the activity of 1, and others that exhibited an altered profile of TLR4 dependence. However, the removal of the entire N-terminus subunit along with capping the C-terminus carboxylic acid as its t-butyl ester, providing 6, resulted in both a significant structure simplification as well as a further enhancement in efficacy. As these studies progressed, it was also established that most initial changes to the central residue of 1 resulted in a loss in activity except the removal of the linking oxygen atom in 1 to give 4, which provided an enhancement in mouse TLR4 (mTLR4) agonist activity. These initial optimizations not only led to simplifying structural modifications, but they also provided two efficacious mTLR4 agonists 5 and 6 (neoseptin-326 and neoseptin-426), nearly matching the maximal agonist efficacy of LPS (Figure 1).26
Figure 2.
Top: mTLR4 agonists emerged from screening an 8000-membered α-helix mimetic library designed to represent the i, i + 4, and i + 7 residues of all α-helices. The library, consisting of 400 wells of 20 compounds each and prepared as a 20 × 20 × 20-mix, was tested at 50 µM for stimulated release of TNF-α against mouse macrophages. Bottom: mTLR4 agonists identified in the library. The individual compounds found in the two active wells (56G4 and 58G4) were prepared and tested (50 µM) for stimulated release of TNF-α against mouse macrophages providing the lead compounds 1–3.
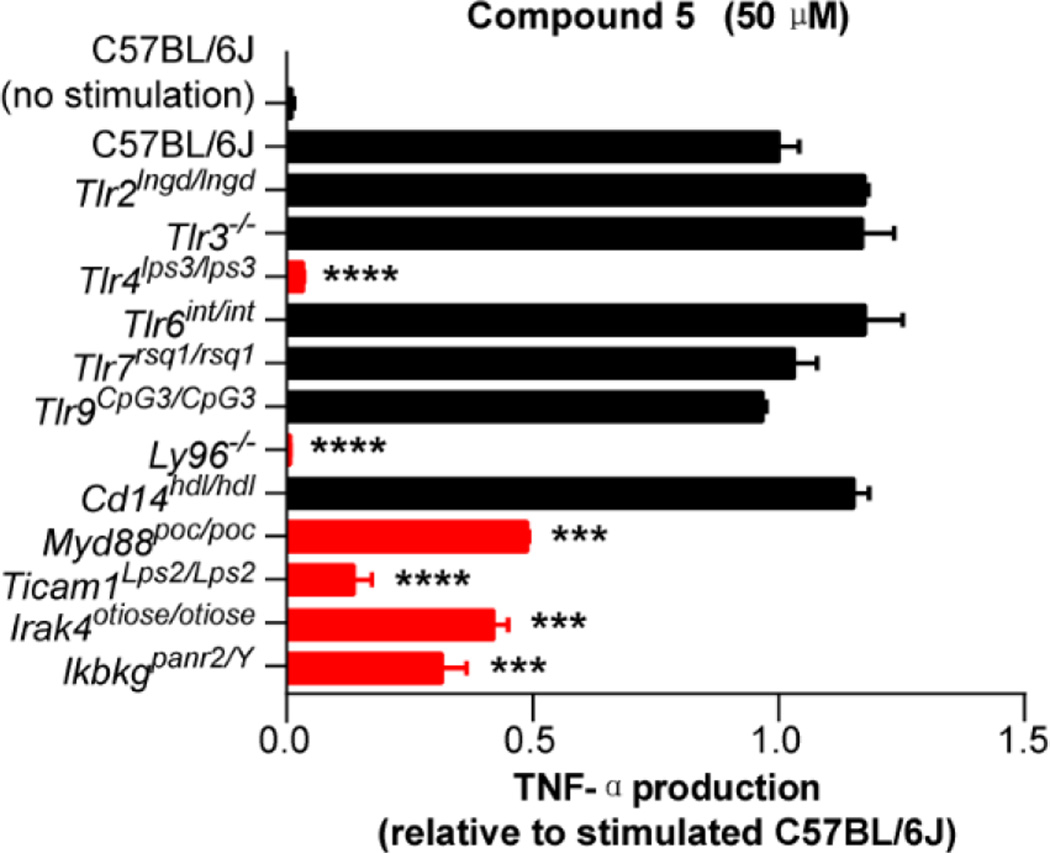
Compounds 5 and 6 exhibited well-defined dose-dependent activation and stimulated release of TNF-α (IC50 = 18.5 and 20.7 µM, respectively) in the functional assay and both were selective for mTLR4 activation among all mTLRs and other pattern recognition receptors. Although the target identification for leads that emerge from an undirected functional assay such as the one used in this work is usually challenging, especially if the lead compounds bear no structural resemblance to known effector molecules, it proved straightforward for 5 and 6. By design, it was recognized that the follow up assay of lead compounds against macrophages available from mice bearing germline genetic defects or knockouts of the genes encoding each of the TLRs, other pattern recognition receptors, or downstream signaling molecules could be used to establish whether the activity was derived from known or presently unrecognized targets.31 By employing this approach, the TNF-α production induced by 5 was established to be solely dependent on both mTLR4 and mMD-2, and independent of other mTLRs (Figure 3) and other pattern recognition receptors (not shown). Moreover, 5 proved nearly identical to LPS in its dependency on not only mTLR4 and mMD-2, but also the downstream signaling molecules MyD88, TRIF, IRAK4, and IKBKG (Figure 3). Unlike LPS,32 the activity of 5 was independent of CD14, which serves to deliver LPS to mMD-2 bound to mTLR4. Like LPS, 5 has been additionally shown to activate the NF-κB, P38 MAPK, JNK, and ERK signaling pathways (not shown).26 Compound 5 more effectively stimulates a type I interferon response than LPS or MPLA, resulting in release of IFN-β but not IFN-α (5 > 6), activates IL-6 production, but fails to induce pro-IL-1β expression and subsequent capase-1 mediated IL-1β release thought to contribute to the higher toxicity of LPS relative to MPLA.34 This IFN-β production was also shown to be dependent on mTLR4, mMD-2 and TRIF, whereas LPS but not 5 additionally required CD14.26 These features represent potential advantages to the use of 5 as a stimulator of the immune system or as a vaccine adjuvant. Like LPS and monophosphoryl Lipid A (MPLA), the binding of 5 with mMD-2 was first inferred by inhibition of its mTLR4 agonist activity (stimulated TNF-α release) by eritoran, a competitive antagonist of the binding of LPS/MPLA with mMD-2,33 supported by a NMR detected interaction, and unambiguously established in a X-ray co-crystal structure of 5 bound to mMD-2 in complex with the mTLR4 ectodomain.26 Like 5 and 6,26 the analogues disclosed herein are inactive against human TLR4, but the results of their examination define structural regions, structural insights, and approaches (e.g., dimerization) available for modifications to address this species dependent activity.
Figure 3.
Assay for TNF-α release upon treatment with 5 (50 µM) using macrophages from mice containing disabling germline mutations or knockouts of genes encoding mTLRs and downstream signaling proteins. All results are representative of two independent experiments. Error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001; **** = P ≤ 0.0001.
When examined in vivo in mice as detailed herein, 5 was found to be an effective adjuvant and, unlike LPS, is nontoxic.26 Unlike LPS or its active core Lipid A on which substantial work has been conducted,35 5 and 6 are easy to prepare and structurally manipulate, are intrinsically much more stable, and exhibit exquisite structure–activity relationships as detailed herein. Compound 5 also was found to elicit an improved and qualitatively different response than LPS even though they share the same receptor, including an altered profile of downstream cytokine release. With a few notable exceptions, some of which were disclosed as our own studies were underway, 5 and 6 are among only a handful of small molecule agonists capable of TLR4 activation beyond those related to LPS and Lipid A.36–38 Herein we report the synthesis and the results of a systematic study of the structure–activity relationships of 5 and 6, and the ability of 5 to stimulate the immune response in vivo in mice as established by its ability to serve as an efficacious adjuvant.
Results and Discussion
Compound Assessment
Mouse macrophages (1 × 105 cells) were stimulated with 5, 6 or the candidate analogues (dissolved in DMSO at the concentrations indicated, <0.5% DMSO in assay) and ultra-pure LPS (dissolved in H2O, Enzo Life Sciences) for 4 h. Mouse TNF-α in the supernatants was measured by ELISA kits according to the manufacturer’s instructions (eBioscience and PBL Assay Science). All data reported in Figure 4–Figure 12 is the mean of triplicates and SD is within +10% of mean and all samples were assessed alongside LPS, 5, and DMSO controls. Mouse cells were from wild type C57BL/6J mice.
Compound Synthesis and Results of Their Assessment
Synthesis of 5 and 6
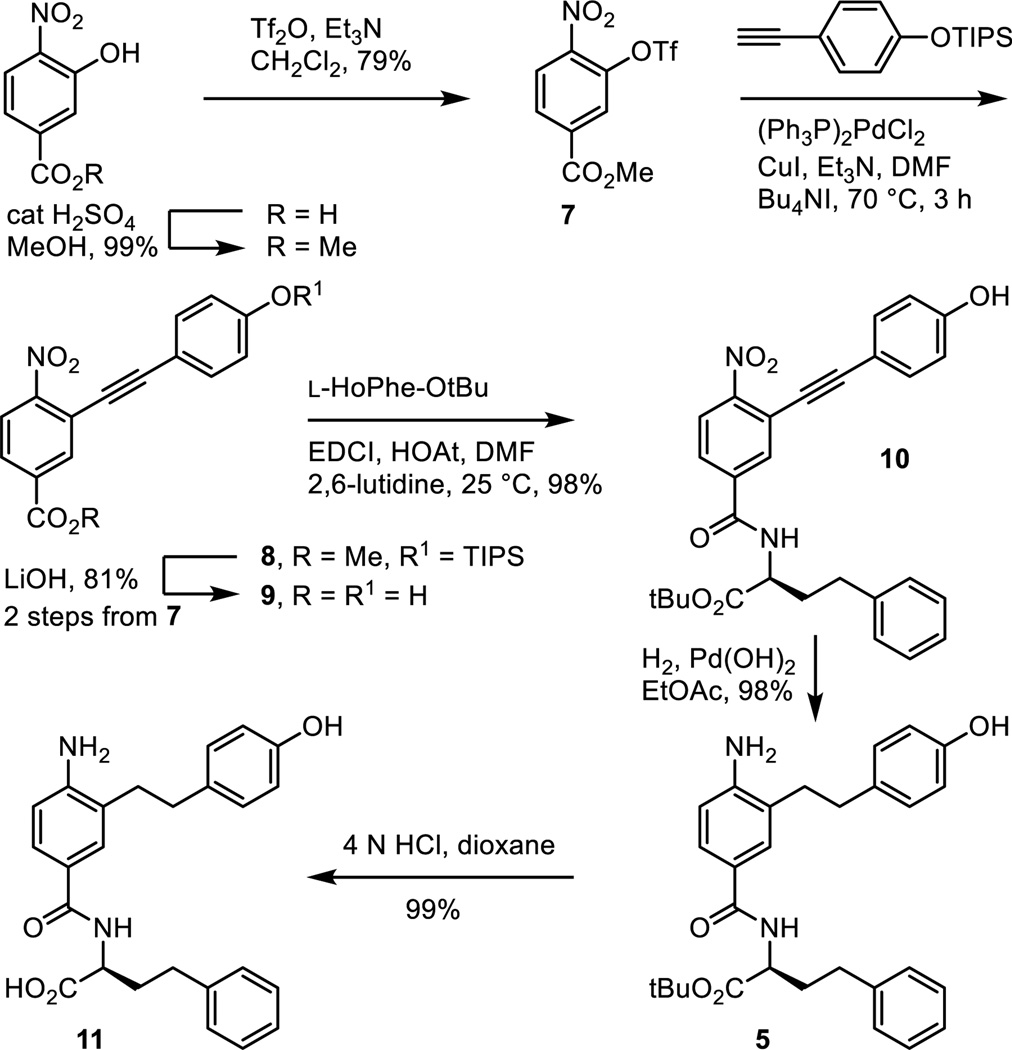
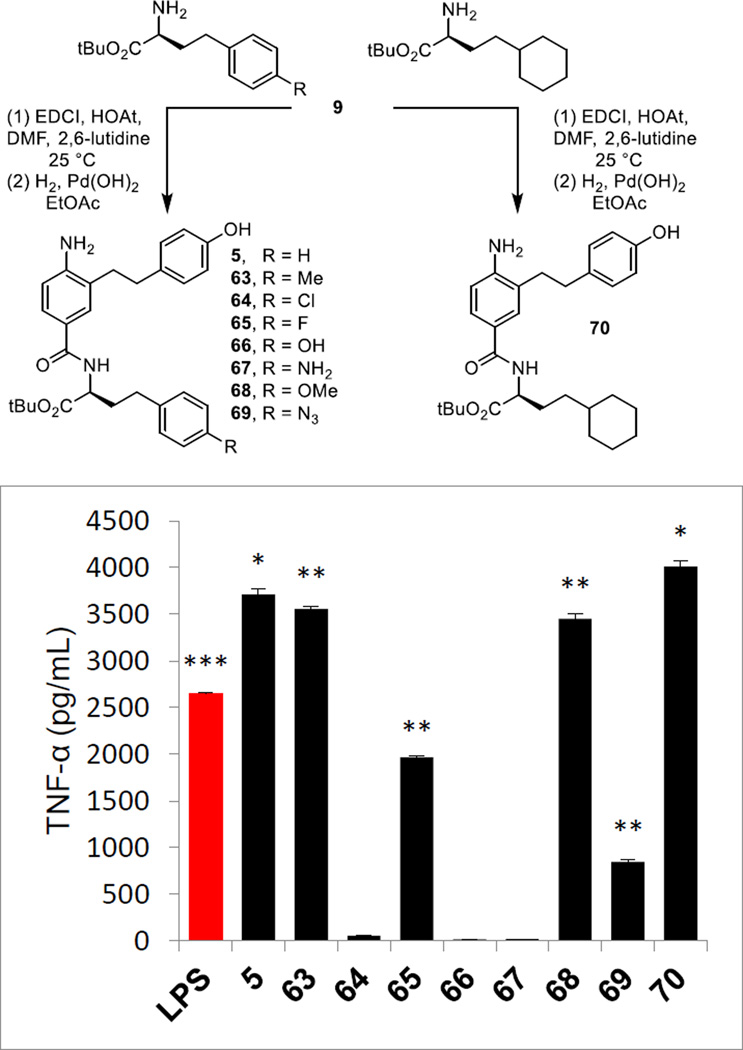
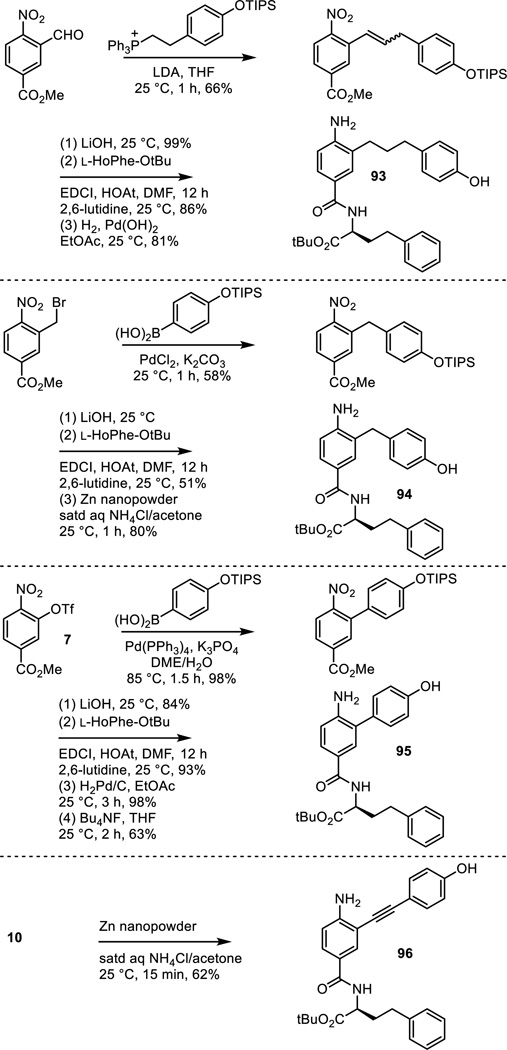
Fischer esterification of commercially available 3-hydroxy-4-nitrobenzoic acid provided methyl 3-hydroxy-4-nitrobenzoate (cat. H2SO4, MeOH, reflux, 18 h, 99%; also commercially available) and was followed by formation of the aryl triflate 7 (Tf2O, Et3N, CH2Cl2, 0 °C, 18 h, 79%) (Scheme 1). Sonogashira cross-coupling of 7 with [(4-triisopropylsilyloxy)phenyl]acetylene, silyl ether deprotection and concurrent hydrolysis of the methyl ester 8 (LiOH, THF/MeOH/H2O 4:1:1, 25 °C, 16 h, 81% for 2 steps), and coupling of the resulting carboxylic acid 9 with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 18 h, 98%) provided 10. Alkyne hydrogenation and concurrent nitro group reduction with Pearlman’s catalyst (H2, Pd(OH)2/C, EtOAc, 25 °C, 16 h, 98%) afforded 5.
Scheme 1.
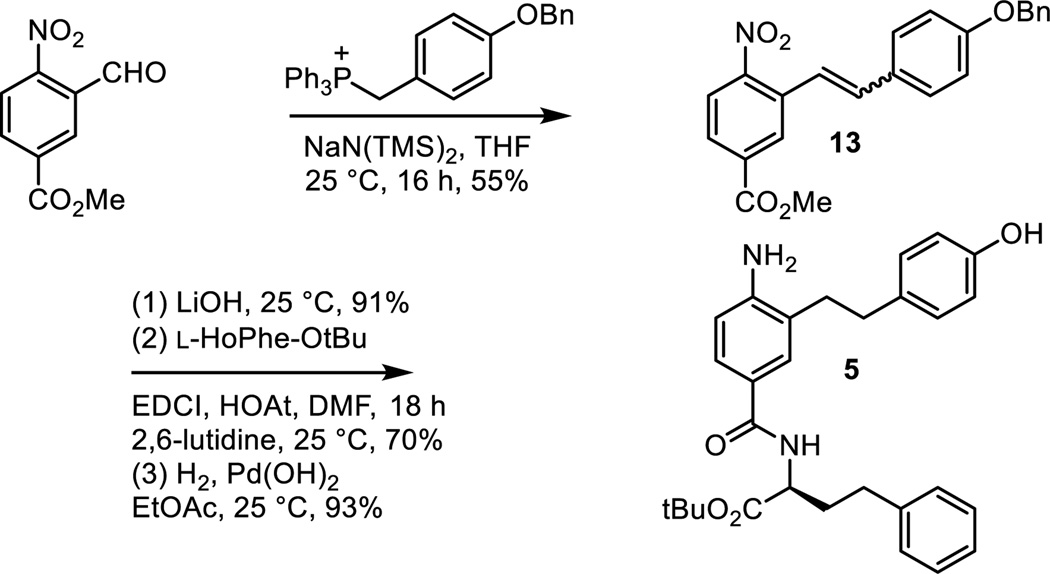
Subsequent to the completion of much of the work detailed herein, an alternative synthesis of 5 was developed (Scheme 2). Wittig reaction of (4-benzyoxyphenylmethylene)triphenylphosphorane with methyl 3-formyl-4-nitrobenzoate (NaN(SiMe3)2, THF, 25 °C, 16 h, 55%) provided 13. Methyl ester hydrolysis (LiOH, THF/MeOH/H2O 3:1:1, 25 °C, 3 h, 91%), coupling of the resulting carboxylic acid with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 18 h, 70%), and single-step nitro group reduction, alkene hydrogenation, and benzyl ether hydrogenolysis (H2, Pd(OH)2, EtOAc, 25 °C, 12 h, 93%) provided 5.
Scheme 2.
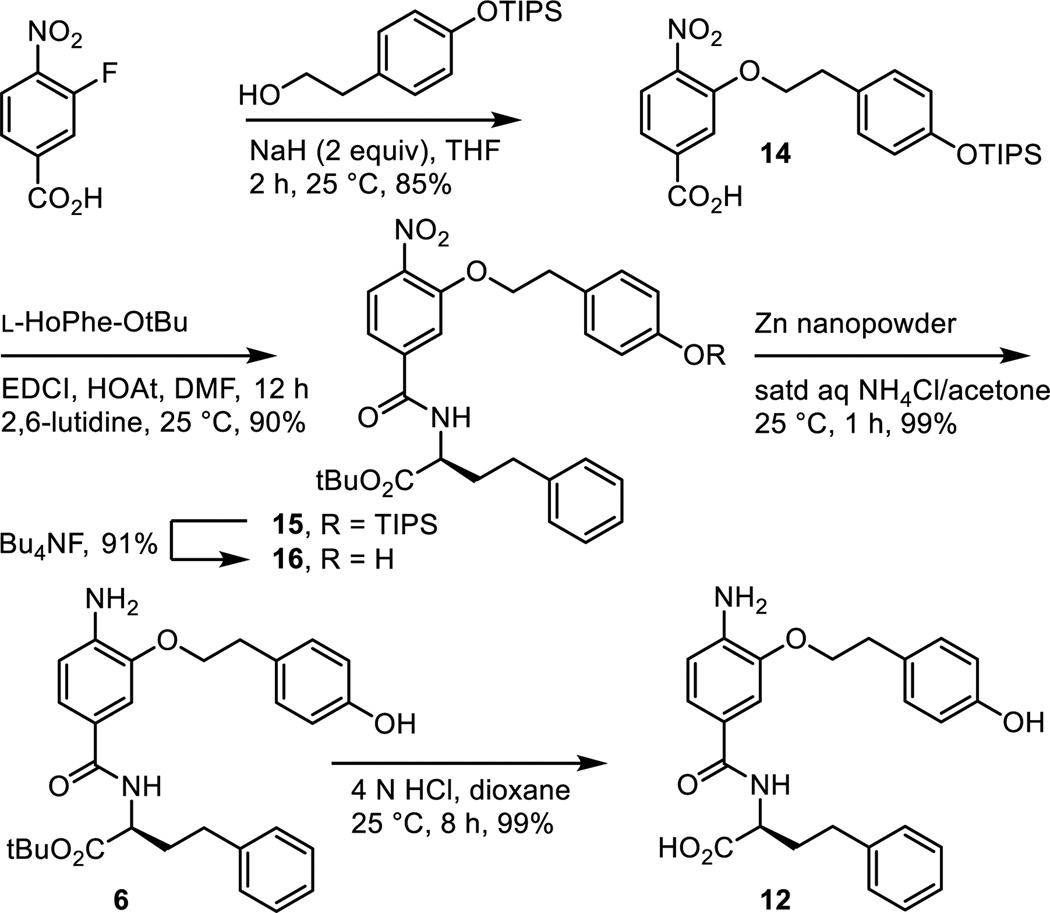
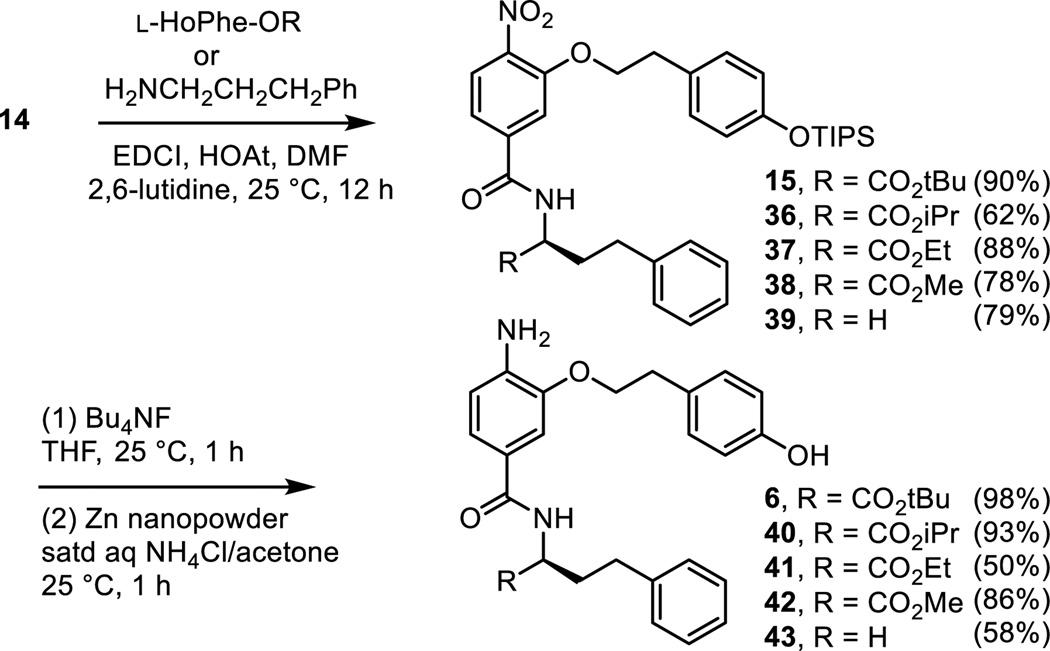
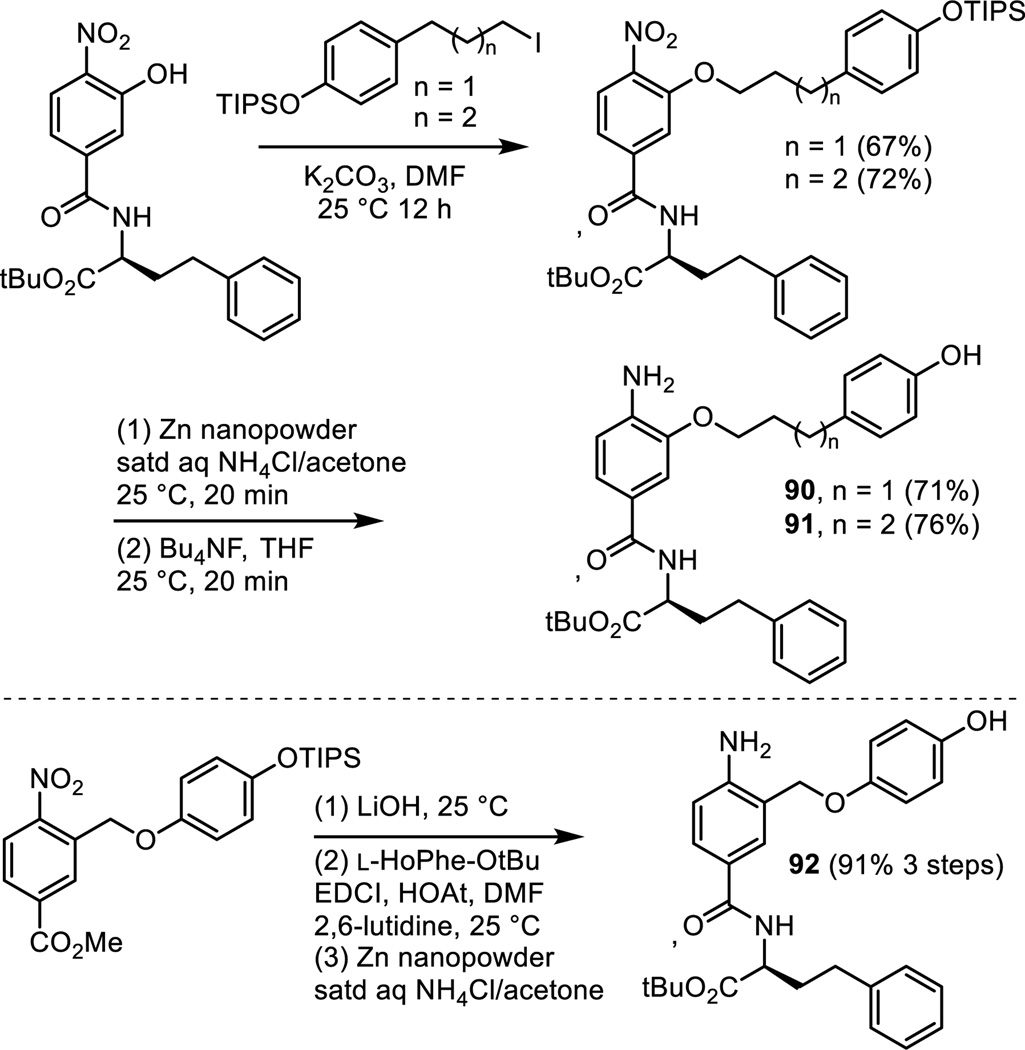
As detailed in our work that provided the α-helix mimetic library,27 aromatic nucleophilic substitution of commercially available 3-fluoro-4-nitrobenzoic acid with the sodium alkoxide derived from 2-[4-(triisopropylsilyloxy)phenyl]ethanol (NaH, THF, 25 °C, 2 h, 85%) provided 14 in a reaction conducted at room temperature and on the free carboxylic acid (Scheme 3). Coupling of carboxylic acid 14 with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12 h, 90%) afforded 15. Silyl ether deprotection (Bu4NF, THF, 25 °C, 1 h, 91%) and subsequent nitro group reduction of 16 (Zn nanopowder, saturated aqueous NH4Cl/acetone, 25 °C, 1 h, 99%) provided 6.
Scheme 3.
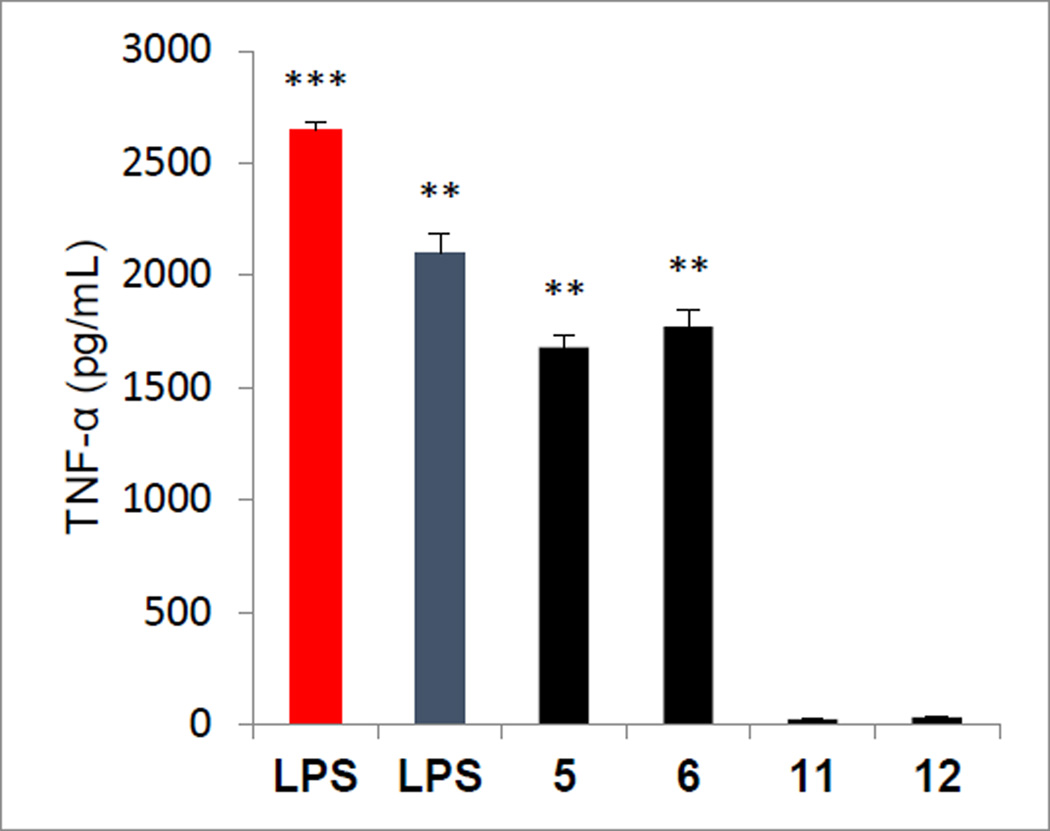
Both 5 and 6, but not their corresponding carboxylic acids 11 and 12, exhibited well-defined, dose-dependent stimulated TNF-α release from treated mouse macrophages (Figure 4). As summarized above,31 the TNF-α production induced by 5 and 6 was established to be nearly identical to LPS in its dependency in mice cell lines on TLR4, MD-2, MyD88, TRIF, TRAM, MAL, IRAK4, and IKBKG (Figure 3), and in their independency of all other TLRs. Not surprisingly and unlike LPS, their activity proved independent of CD14, which serves to deliver LPS to the cell surface receptor complex. Finally, as detailed herein, 5 proved to be a robust, in vivo active adjuvant in a mouse model of vaccination.
Figure 4.
Activity (TNF-α release) of 5 and 6 versus the corresponding carboxylic acids 11 and 12, assayed at 50 µM against mouse macrophages alongside LPS (5 ng/mL in red, and 1 ng/mL in blue). All data is the mean of triplicate assay versus vehicle control, error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001.
Scan of the C-terminus residue
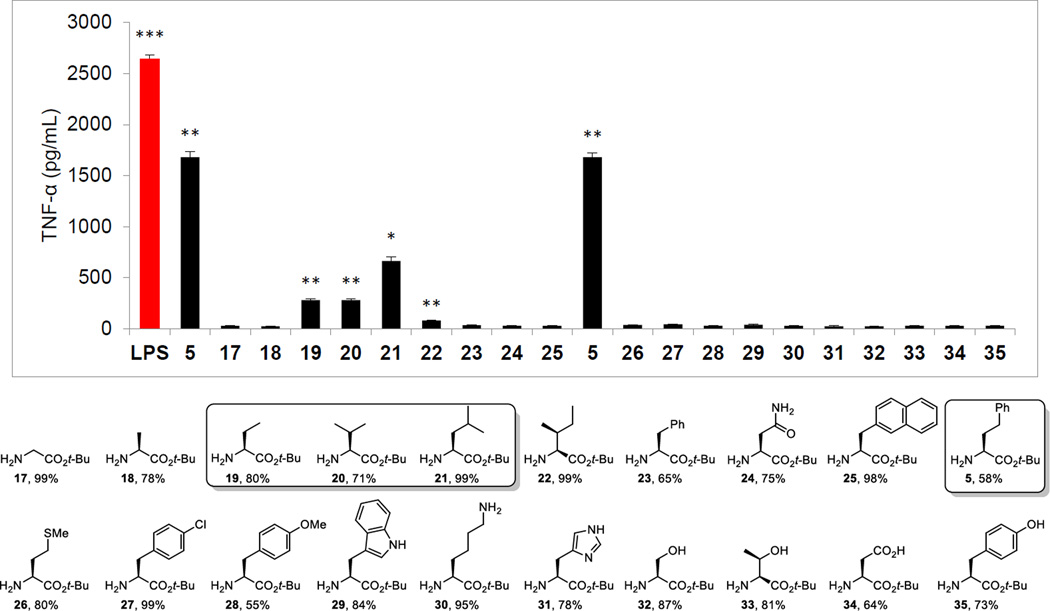
Although not the first of the modifications to be conducted on 5 or 6, we reestablished the unique activity of the L-HoPhe-OtBu C-terminus residue when coupled to the core amine terminus found in 5, examining all C-terminus residues that were present in the original α-helix mimetic library27 (Figure 5). Thus, coupling of 9 with the L-α-amino acid t-butyl esters (H2N-Xxx-OtBu, EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12–24 h), bearing hydrogenolysis labile side chain protecting groups as needed (H-Lys(Z)-OtBu and H-Asp(OBn)-OtBu), followed by concurrent alkyne and nitro group reduction (H2, Pd(OH)2/C, EtOAc, 25 °C, 16 h) provided 17–35. Included in this group was L-HoPhe-OtBu, reconfirming its activity when tested alongside the companion analogues. Most notable in the set of analogues was the inactivity of the compounds containing other aryl residues (e.g., L-Phe-OtBu, 23), highlighting that the activity of 5 is not observed if the linker to the aryl ring is shortened by even a single carbon regardless of the extended size of such aryl residues (e.g., NapAla-OtBu, 25). Interestingly, 19–21, which progressively contain more elements of this linking chain but lack the attached phenyl group, exhibited weak and progressively stronger agonist activity, whereas Met-OtBu (26) was inactive.
Figure 5.
C-terminus scan, compounds prepared from 9 (overall % yields) and assayed (50 µM) against mouse macrophages alongside LPS (5 ng/mL) for stimulated release of TNF-α. All data is the mean of triplicate assay versus vehicle control, error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001.
Analogues defining the importance of the C-terminus t-butyl ester
Within the structure of 6, five systematic variations on the t-butyl ester were examined including the free carboxylic acid 12. These were prepared by a synthetic route analogous to that used for the preparation of 6 itself, coupling intermediate 14 with the corresponding i-Pr, Et, or Me ester of L-HoPhe or 3-phenyl-1-propylamine followed by subsequent tri-isopropylsilyl (TIPS) ether deprotection and nitro group reduction with these latter two reactions carried out in an exchangeable order for the ethyl ester 41 (Scheme 4). Like the observations made with 5 below, replacement of the t-butyl ester with smaller ester groups led to progressive losses in activity with only the i-Pr ester 40 exhibiting weak agonist activity (Figure 6). Both the removal of the t-butyl ester altogether with 43 or its cleavage to provide the carboxylic acid 12 led to a complete loss in activity.
Scheme 4.
Figure 6.
Preparation of t-butyl ester analogues of 5 and the assessment of the ester analogues of 5 (above) and 6 (Scheme 4) assayed (50 µM) against mouse macrophages alongside LPS (1 ng/mL) for stimulated release of TNF-α. All data is the mean of triplicate assay versus vehicle control, error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001.
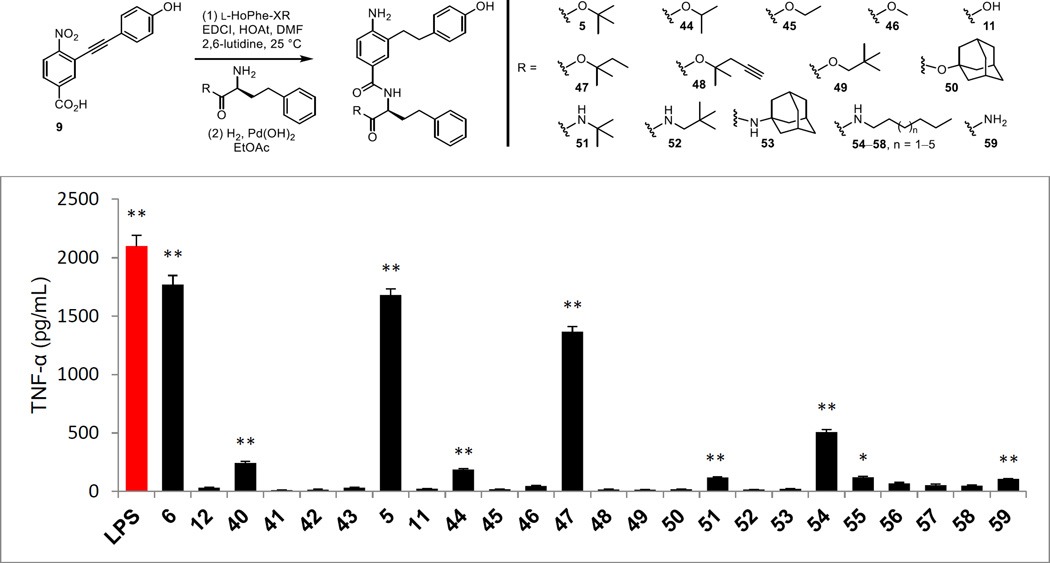
A more extensive series of C-terminus analogues of 5 was prepared and was conducted by coupling intermediate 9 with a series of alternative ester or amide derivatives of L-HoPhe-XR (X = O or NH) followed by concurrent alkyne and nitro group reduction by the protocol developed for 5 itself (Figure 6). Like the observations made with 6, replacement of the t-butyl ester with smaller esters led to progressive and complete loss in agonist activity with only the i-Pr ester 44 maintaining weak activity (Figure 6). Similarly, its hydrolysis to the carboxylic acid 11 also resulted in a complete loss in activity. Only the closely related ester 47 approached the activity of 5, whereas the related but slightly larger ester 48 bearing an alkyne was inactive. Even extending the t-butyl group by a single carbon with ester 49 resulted in a complete loss in agonist activity as did its replacement with the larger adamantyl ester 50. Replacement of the t-butyl ester with an amide was not productive. Only a trace of activity was observed with the corresponding t-butyl amide 51, representing a single heavy atom change in 5 (amide NH for ester O), and perhaps a trace of activity was observed with the primary carboxamide 59. Without knowledge at the time of the nature of the binding of 5 to MD-2 and inspired by the lipid chains found in LPS, we also examined a series of long chain alkyl amides 54–58. Although a hint of activity was seen with a selected member in the group (54), the very modest level of activity observed and the lack of a SAR trend of increasing activity with the longer alkyl amides 55–58 did not inspire continued work on this series.
Synthesis of analogues probing the HoPhe absolute stereochemistry and linker length
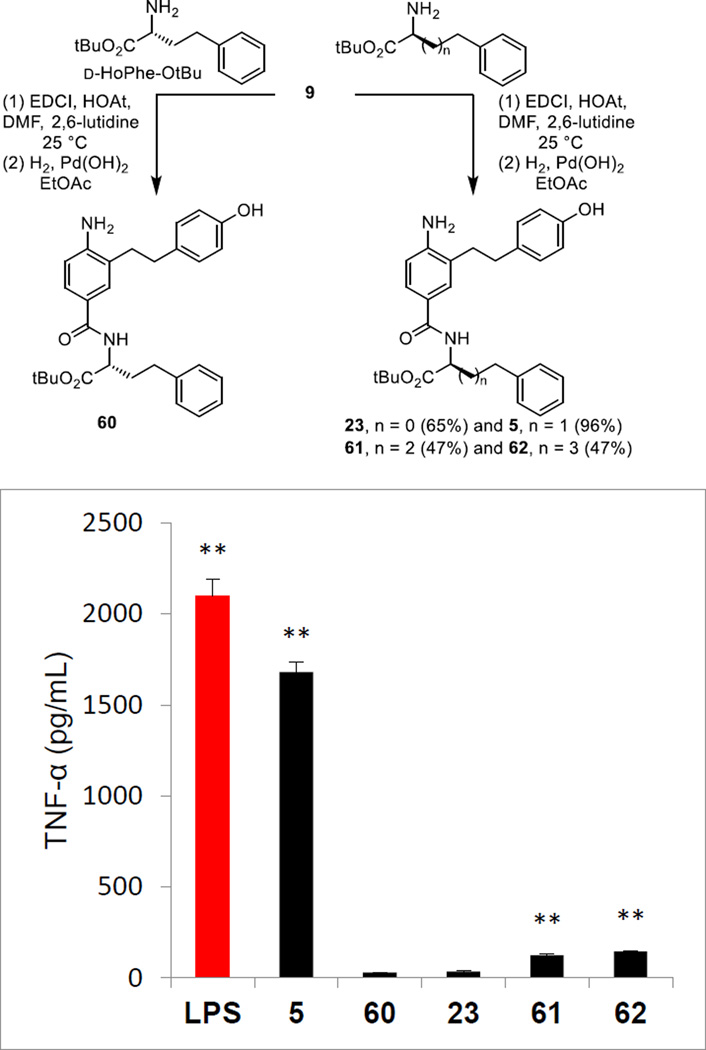
Replacement of the C-terminus L-HoPhe-OtBu with its enantiomer D-HoPhe-OtBu as well as the extension of the linker chain by one or two additional carbons was conducted by coupling 9 with the corresponding amines following the protocols developed for 5 (Figure 7). The D-enantiomer 60 proved inactive confirming that the interaction of 5 with the mTLR4/mMD-2 complex is diastereospecific (Figure 7). Remarkably and like the reduction in the length of the linker chain to the phenyl group by one carbon with 23 that was inactive, its extension by either one or two carbons with 61 and 62 also led to essentially complete loss in agonist activity.
Figure 7.
Preparation of the enantiomer (60) of 5 and analogues altering the HoPhe chain length and their assessment assayed (50 µM) against mouse macrophages alongside LPS (1 ng/mL) for stimulated release of TNF-α. All data is the mean of triplicate assay versus vehicle control, error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001.
Synthesis of analogues containing L-HoPhe aryl substituents and aryl ring replacements
A small series of p-substituted L-HoPhe-OtBu derivatives were examined as well as a single analogue in which the phenyl ring was replaced by a saturated cyclohexyl ring. These analogues were prepared by coupling 9 with the corresponding amines followed by alkyne and nitro group reduction (Figure 8). Although the number of compounds in the series was not large, the results of their examination were especially revealing. Incorporation of polar p-substituents (R) were not tolerated, resulting in a complete loss in agonist activity with 66 and 67 (R = OH or NH2). Even benign but electron dense halide substituents led to a substantial reduction (65, R = F) or loss (64, R = Cl) in activity. However, nonpolar substituents represented by the analogue 63 (R = Me) exhibited the robust activity of 5, surpassing its efficacy and indicating that there are likely substantial nonpolar modifications to this region of the molecules that are possible. Consistent with such expectations, the methyl ether derivative 68 (R = OMe) proved equally active with 5, recovering the full agonist activity lost with the corresponding phenol 66 (R = OH). These latter results indicate that it is the nature of the substituent (hydrophobic vs hydrophilic) and not its presence (steric) that influences activity, likely the result of binding deep in the hydrophobic pocket of mMD-2. Especially revealing was the potent agonist activity of 70, surpassing the activity of 5. Not only does this indicate that even the unsaturation of the phenyl ring is not necessary and that hydrophobic saturated replacements are well tolerated, but also that this is a region of the molecule that can be substantially modified in continued optimization efforts. Dose-response studies established that 63, 68, and 70 exhibit IC50’s (21.6, 22.2, and 29.5 µM respectively) comparable to those found for 5 and 6.
Figure 8.
Preparation of analogues of 5 bearing HoPhe phenyl substituents or replacements and their assessment assayed (50 µM) against mouse macrophages alongside LPS (5 ng/mL) for stimulated release of TNF-α. All data is the mean of triplicate assay versus vehicle control, error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001.
Synthesis of analogues probing the amide
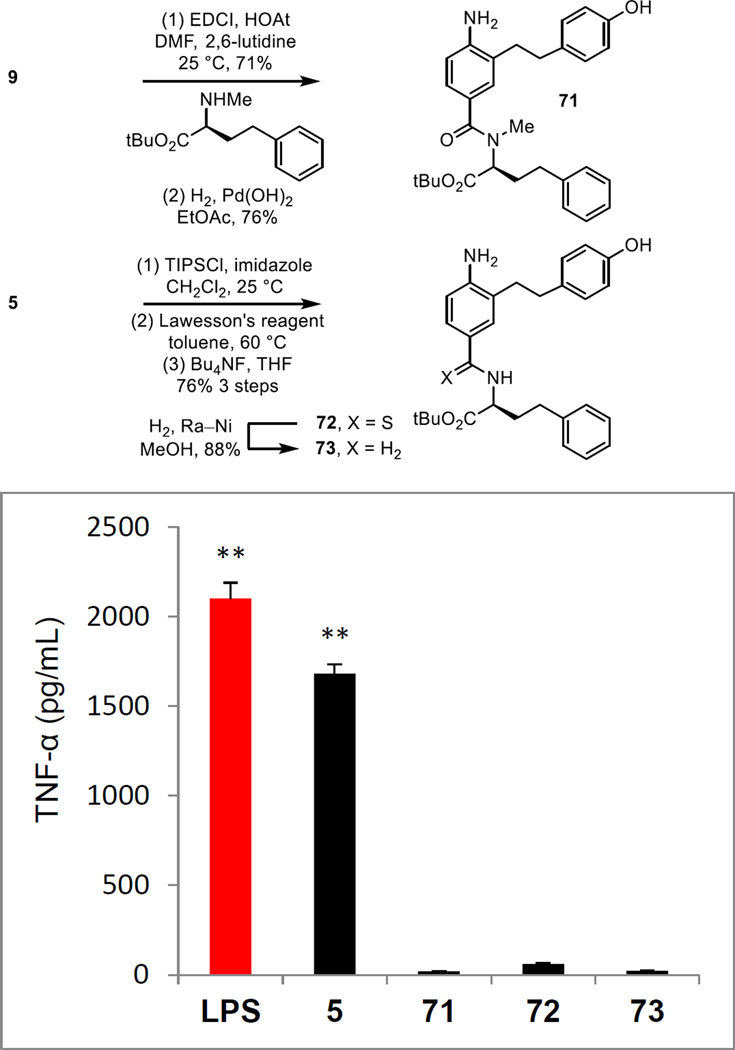
The importance of the linking amide was established with a small set of key analogues. The N-methyl amide 71 of 5 was prepared by coupling L-NMe-HoPhe-OtBu with 9 followed by nitro group and alkyne reduction (Figure 9). In addition, the corresponding thioamide 72 was prepared from 5 itself by TIPS ether protection of the phenol, Lawesson’s reagent conversion to the thioamide (toluene, 60 °C, 12 h) and subsequent silyl ether deprotection (Bu4NF, THF, 25 °C, 6 h, 76% for 3 steps). Reductive removal of the thiocarbonyl (H2, Raney Ni, MeOH, 18 h) provided the amine 72, lacking the amide carbonyl. Each modification of the linking amide resulted in a complete loss in activity (Figure 9). Not only does this highlight the essential contribution of the secondary amide to the expression of the properties of 5, but it also underscores how remarkable the behavior of 5 is where even the conservative single atom replacement (thioamide S for amide O) in the amide completely disrupted the observed activity.
Figure 9.
Preparation of analogues of 5 bearing changes to the amide and their assessment assayed (50 µM) against mouse macrophages alongside LPS (1 ng/mL) for stimulated release of TNF-α. All data is the mean of triplicate assay versus vehicle control, error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001.
Synthesis of analogues probing the importance of the presence, position, and substitution of the aniline amine
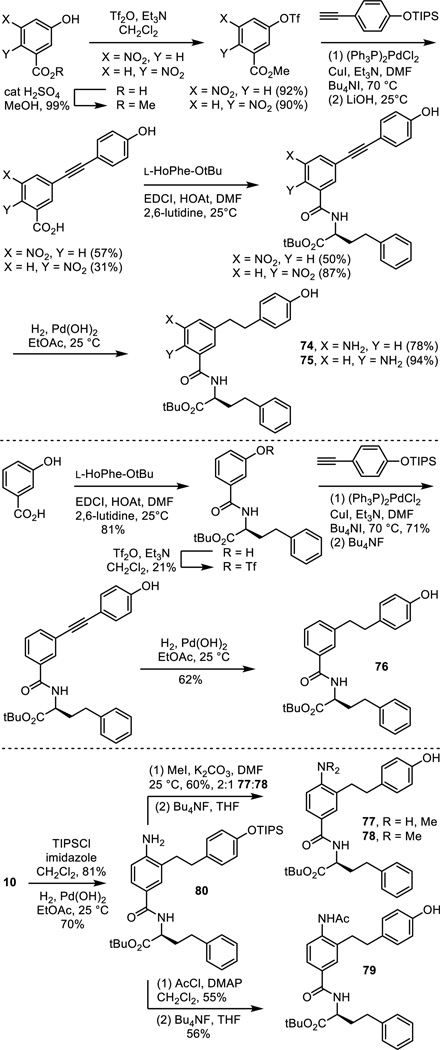
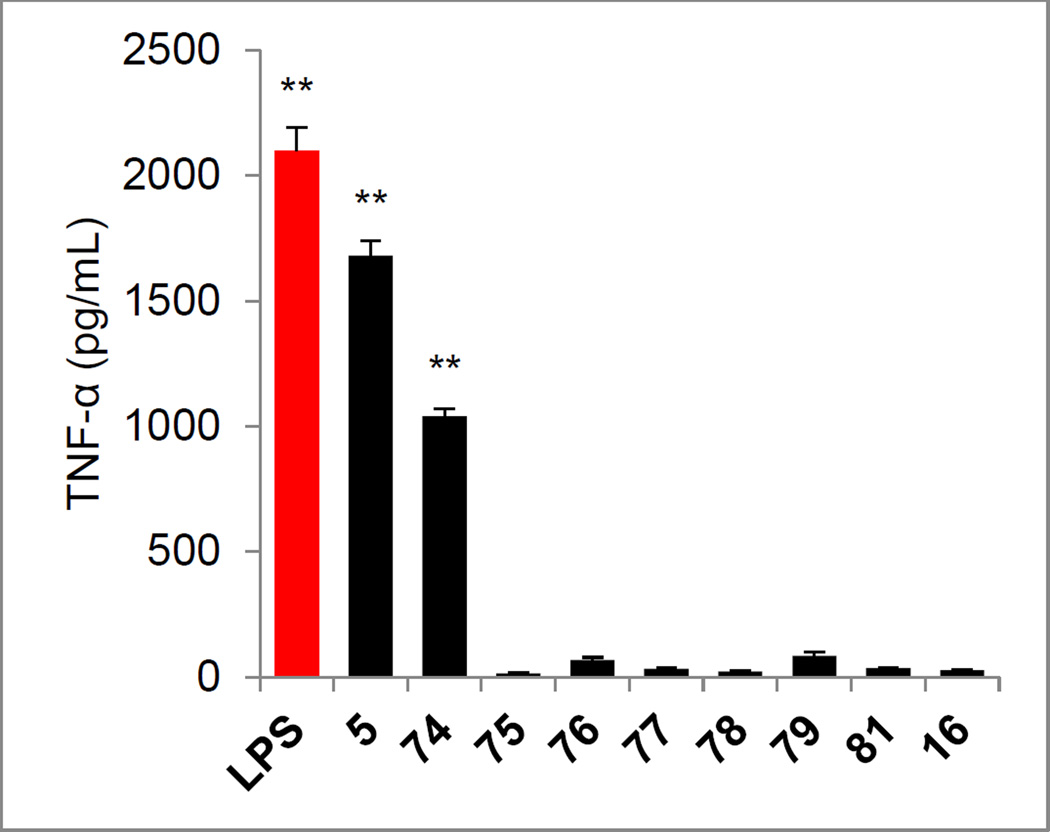
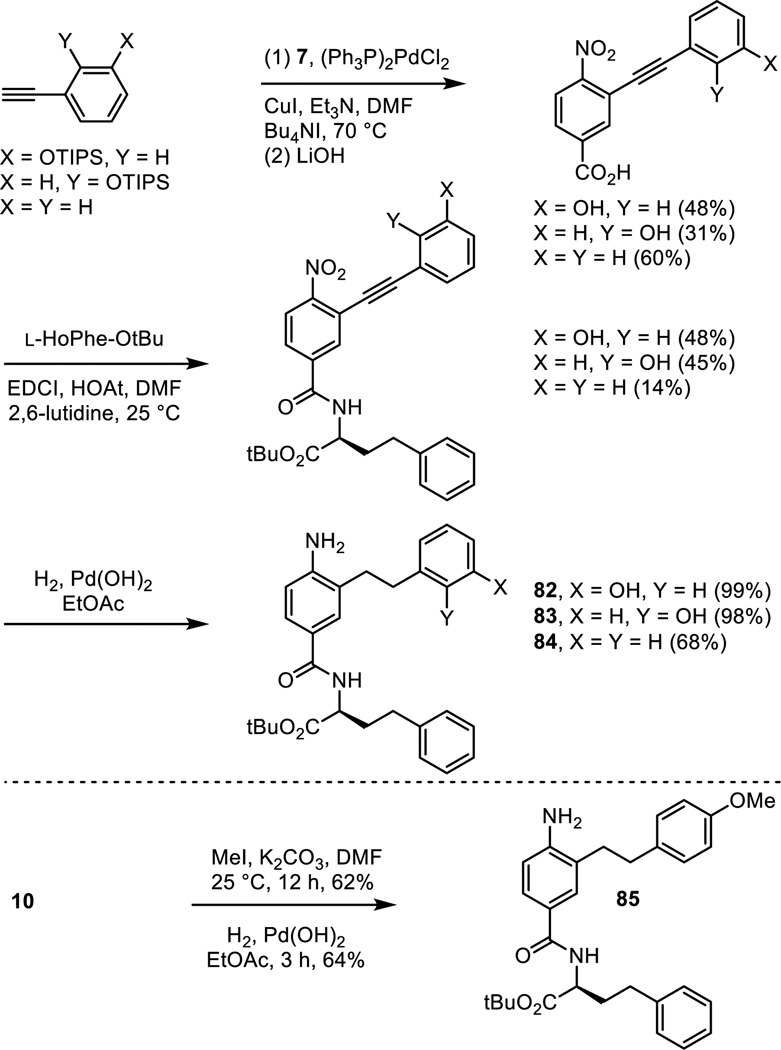
A similarly key series of analogues of 5 and 6 were prepared to establish the importance of the presence and position of the aniline amine and the impact of its substitution. Those prepared that are analogues of 5 include the aniline positional isomers 74 and 75, requiring their synthesis from the commercially available 5-nitro- or 6-nitro-3-hydroxybenzoic acids by a synthetic route modeled on that used for 5 itself (Scheme 5). Similarly, the analogue 76 of 5 lacking the aniline amine was prepared from commercially available 3-hydroxybenzoic acid. Finally, the monomethyl and dimethyl anilines 77 and 78 were prepared by nonselective methylation of 80, available in 2 steps from 10, followed by silyl ether deprotection. The acetamide 79 was prepared from the same intermediate by acylation with acetyl chloride followed by silyl ether deprotection.
Scheme 5.
The nitro precursor 16 (Scheme 3) to compound 6 was examined as an additional probe of the importance of the presence of the aniline and the analogue 81 of 6 lacking the aniline amine was prepared by phenol alkylation with 2-(4-triisopropylsilyloxyphenyl)ethyl bromide (K2CO3, refluxing acetone) followed by silyl ether deprotection (Bu4NF, THF 25 °C) (Scheme 6).
Scheme 6.
Removal of the aniline amine (76 and 81), its mono- or di-methylation (77 and 78), its simple acetylation (79), its replacement with a nitro group (16), and its movement to the 6-position on the central ring (75) resulted in a complete loss in agonist activity (Figure 10). However and interestingly, relocating the aniline amine in 5 to the adjacent 5-position on the central aromatic ring provided an analogue 74 that retained much of the activity of 5. Finally, the corresponding carboxylic acids derived from both series (74–79 or 16 and 81) were also prepared by acid-catalyzed t-butyl ester deprotection (4 N HCl, dioxane, 25 °C) and all were found to be inactive (not shown).
Figure 10.
Evaluation of analogues of 5 (Scheme 5) and 6 (Scheme 6) used to probe the importance of the presence, position, and substitution of the aniline amine assayed (50 µM) against mouse macrophages alongside LPS (1 ng/mL) for stimulated release of TNF-α. All data is the mean of triplicate assay versus vehicle control, error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001.
Synthesis of analogues probing the importance of the presence and position of the phenol hydroxyl group
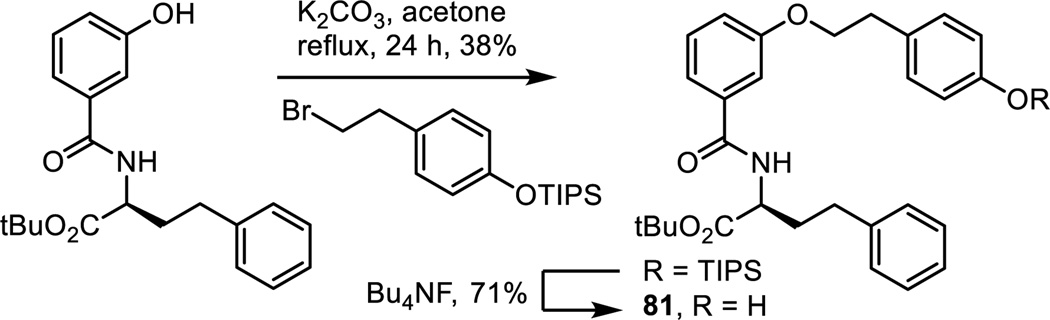
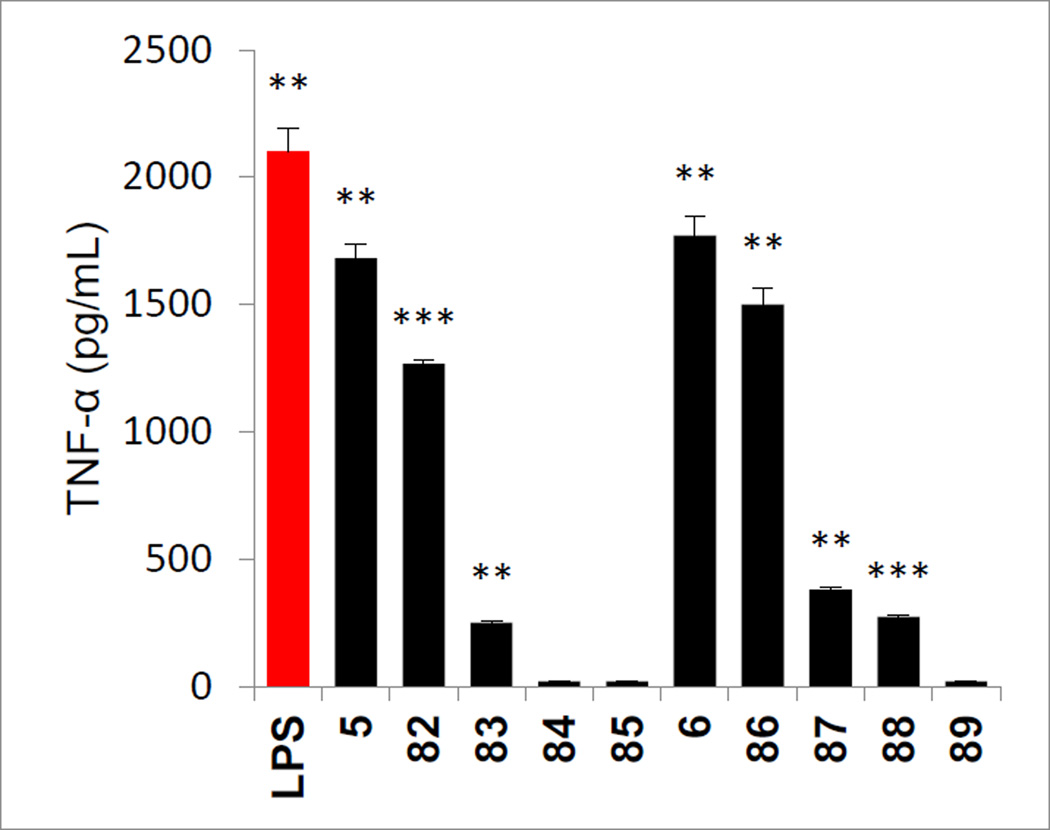
Positional isomers of the phenol substitution and the removal of the phenol hydroxyl group altogether were examined with the analogues 82–84 of 5 prepared as detailed in Scheme 7. Sonogashira cross-coupling of the appropriately substituted alkynes with triflate 7, LiOH promoted hydrolysis of the methyl ester and simultaneous phenol silyl ether deprotection, and coupling of the resulting carboxylic acids with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12–24 h) followed by alkyne and nitro group reduction (H2, Pd(OH)2/C, EtOAc, 25 °C, 3 h) provided 82–84. The corresponding methyl ether 85 of the phenol found in 5 was prepared by O-methylation of intermediate 10 (MeI, K2CO3, DMF, 25 °C, 12 h) followed by alkyne and nitro group reduction.
Scheme 7.
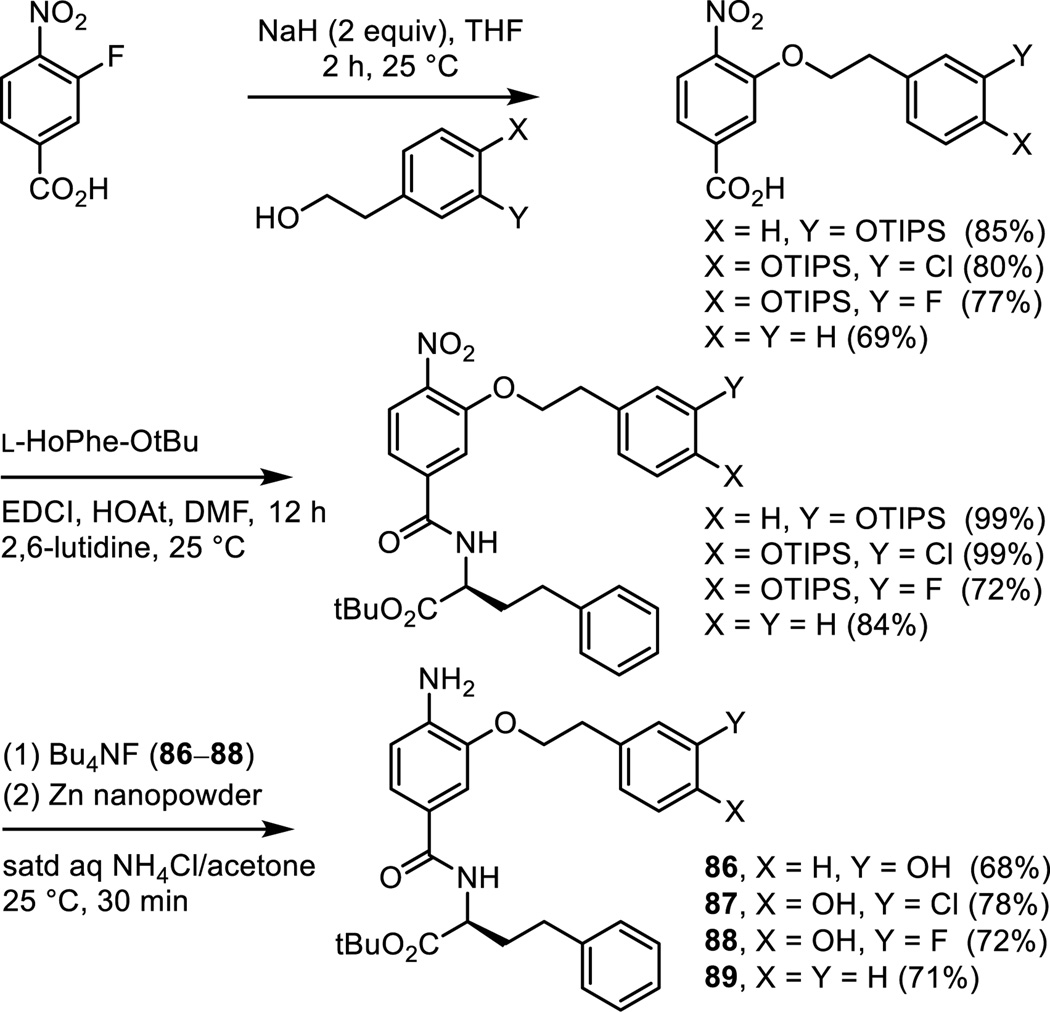
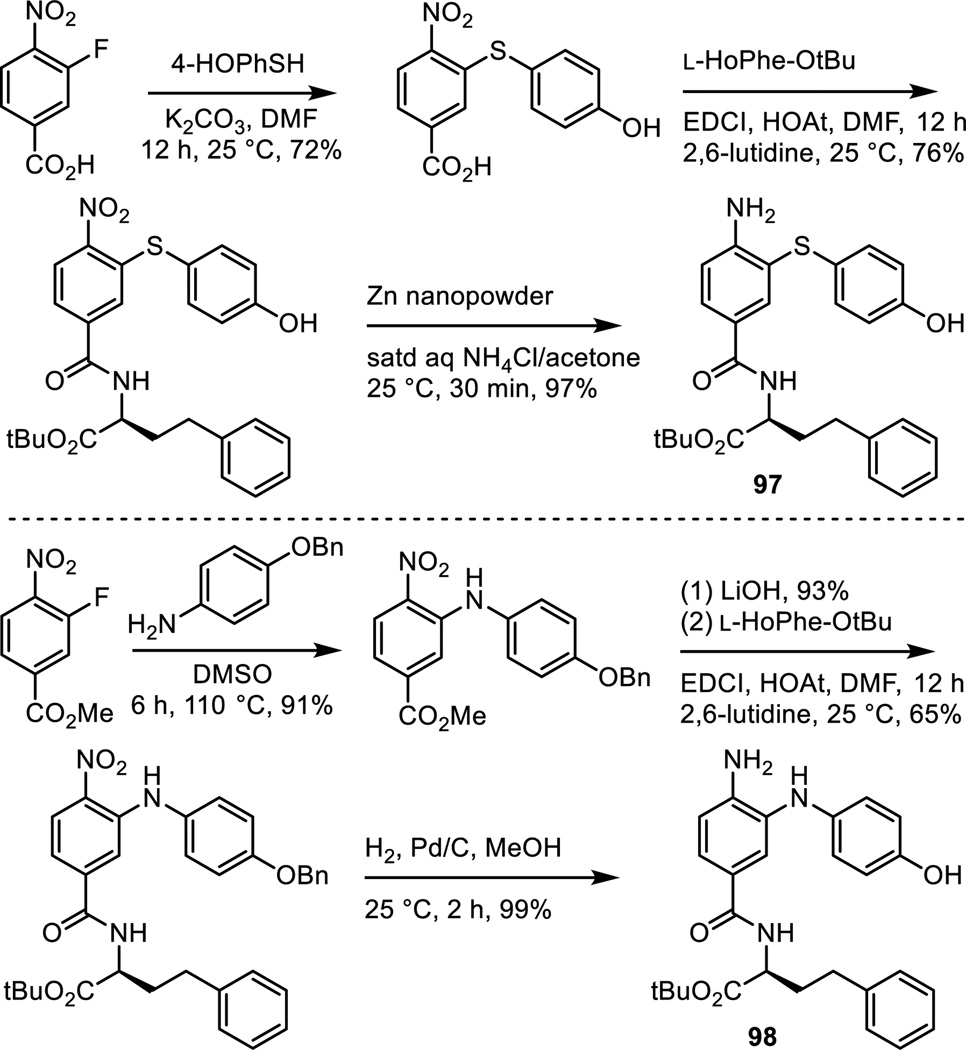
Analogues of 6 were prepared in which the phenol was moved from the p-position to the m-position and in which it was removed altogether. Additionally, two analogues that contain a benign substituent (Cl and F) adjacent to the p-phenol hydroxyl group were also prepared. Thus, aromatic nucleophilic substitution of 3-fluoro-4-nitrobenzoic acid conducted at room temperature and on the free carboxylic acid with a series of four sodium alkoxides provided the product carboxylic acids (Scheme 8). Their subsequent coupling with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12 h), silyl ether deprotection (Bu4NF, THF, 25 °C, 2 h if needed) and subsequent nitro group reduction (Zn nanopowder, saturated aqueous NH4Cl/acetone, 25 °C, 30 min) provided 86–89.
Scheme 8.
Removal of the phenol hydroxyl group (84 and 89), its conversion to a methyl ether (85), and its movement to the o-position (83) led to complete loss (84, 85, and 89) or near complete loss (83) in agonist activity, indicating its importance to the expression of the properties of 5 and 6 (Figure 11). Even the addition of rather benign substituents adjacent to the p-phenol (87 and 88, F and Cl) led to the sharp reduction in the agonist activity of 6, suggesting that there may be constraints on further substituting the aryl ring. Only the corresponding m-phenol positional isomers 82 and 86 retained the activity of 5 and 6, exhibiting slightly reduced agonist activity and suggesting that the connecting flexible linking chain allows adjustments in the aryl positioning to accommodate the relocated m-phenol. The carboxylic acids derived from both series (82–85 and 86–89) were also prepared by acid-catalyzed t-butyl ester deprotection (4 N HCl, dioxane, 25 °C) and all were found to be inactive (not shown).
Figure 11.
Evaluation of analogues of 5 (Scheme 7) and 6 (Scheme 8) used to probe the importance of the presence, position, and substitution of the phenol hydroxyl group assayed (50 µM) against mouse macrophages alongside LPS (1 ng/mL) for stimulated release of TNF-α. All data is the mean of triplicate assay versus vehicle control, error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001.
Synthesis of analogues probing the central residue linker to the phenol
Among the most carefully explored regions of 5 and 6 was the linker extending off the central residue to the phenol. This is the site that distinguishes 5 and 6, differing by the insertion of an oxygen atom in the linker for 6. Thus, both a two-atom and three-atom linker proved effective in early studies. A series of additional linkers containing an oxygen atom were prepared as shown in Scheme 9. Two extend the linker found in 6 by one or two additional carbons (90 and 91) and were prepared by O-alkylation of a common intermediate with the respective iodides followed by nitro group reduction (Zn nanopowder, saturated aq NH4Cl/acetone, 25 °C, 20 min) and subsequent phenol silyl ether deprotection (Bu4NF, 25 °C, 20 min). A third analogue 92 shortened the linker of 6 by one carbon, matching the linker length of 5 but now containing an oxygen atom. Compound 92 was prepared by alkylation of the benzylic bromide, methyl 3-bromomethyl-4-nitrobenzoate, with 4-triisopropyloxyphenol (68%). Subsequent LiOH mediated hydrolysis of the methyl ester and concurrent silyl ether deprotection followed by coupling of the resulting carboxylic acid with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12 h) and a final nitro group reduction (Zn nanopowder, saturated aq NH4Cl/acetone, 25 °C, 20 min, 91% for 3 steps) provided 92.
Scheme 9.
A series of saturated all carbon linkers were explored that cover the range of (CH2)n where n = 0–3, including 5 (n = 2). Extension of the linker found in 5 by one carbon, providing an all carbon analogue 93 of 6, was prepared as shown in Scheme 10. Wittig reaction of methyl 3-formyl-4-nitrobenzoate with 2-((4-triisopropylsilyloxyphenyl)ethylene)triphenylphosphorane provided the substituted styrene (THF, 25 °C, 1 h, 66%, 3:1 Z:E). Methyl ester hydrolysis and concurrent silyl ether deprotection (LiOH, 25 °C, 1 h, 99%), coupling of the resulting carboxylic acid with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12 h, 86%), and final nitro group and alkene reduction (H2, Pd(OH)2, EtOAc, 25 °C, 81%) provided 93. The analogue 94 of 5 containing one less carbon in the linking chain was prepared by Suzuki coupling of 4-(triisopropylsilyloxy)phenyl boronic acid with methyl 3-bromomethyl-4-nitrobenzoate (cat. PdCl2, K2CO3, acetone/H2O, 25 °C, 1 h, 58%). LiOH mediated methyl ester hydrolysis and silyl ether deprotection, coupling of the resulting carboxylic acid with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12 h, 51%), and subsequent nitro group reduction (Zn nanopowder, saturated aqueous NH4Cl/acetone, 25 °C, 1 h, 80%) provided 94. The analogue 95 of 5, containing no linking chain and attached directly to the central residue (two carbons shorter), was prepared by Suzuki coupling of triflate 7 with 4-(triisopropylsilyloxy)phenyl boronic acid (cat. Pd(PPh3)4, K3PO4, DME/H2O,85 °C, 1.5 h, 98%). Methyl ester hydrolysis of the coupled product conducted with LiOH under conditions that did not induce phenol silyl ether deprotection (1.1 equiv LiOH, 1:1 t-BuOH/H2O, 25 °C, 2 h, 84%), coupling of the carboxylic acid with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12 h, 93%), nitro group reduction by hydrogenation (H2, Pd/C, EtOAc, 25 °C, 3 h, 98%), and a final silyl ether deprotection (Bu4NF, THF, 25 °C, 2 h, 63%) provided 95. In addition, the analogue 96 with a rigid alkyne two carbon linker was prepared by selective nitro group reduction of intermediate 10 employed for the synthesis of 5 (Zn nanopowder, saturated aqueous NH4Cl/acetone, 25 °C, 15 min, 62%).
Scheme 10.
Two additional analogues were prepared that contain a single atom heteroatom linker (Scheme 11). The first of these was the analogue 97 that contains a sulfur atom linker. This analogue is one that we expected to be an exceptional candidate since a sulfur atom is often regarded as an excellent isosteric replacement for the two carbons of a double bond. Room temperature aromatic nucleophilic substitution of 3-fluoro-4-nitrobenzoic acid with 4-(triisopropylsilyloxy)thiophenol (K2CO3, DMF, 25 °C, 12 h, 72%), followed by coupling of the carboxylic acid with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12 h, 76%), and final nitro group reduction (Zn nanopowder, saturated aqueous NH4Cl/acetone, 25 °C, 1 h, 97%) provided 97. An analogous approach was enlisted for the synthesis of 98 that contains a single nitrogen atom linker. Nucleophilic substitution of methyl 3-fluoro-4-nitrobenzoate with 4-benzyloxyaniline (DMSO, 110 °C, 6 h, 91%), methyl ester hydrolysis (LiOH, 93%), coupling of the carboxylic acid with L-HoPhe-OtBu (EDCI, HOAt, 2,6-lutidine, DMF, 25 °C, 12 h, 65%), and nitro group reduction and concurrent benzyl ether hydrogenolysis provided 98.
Scheme 11.
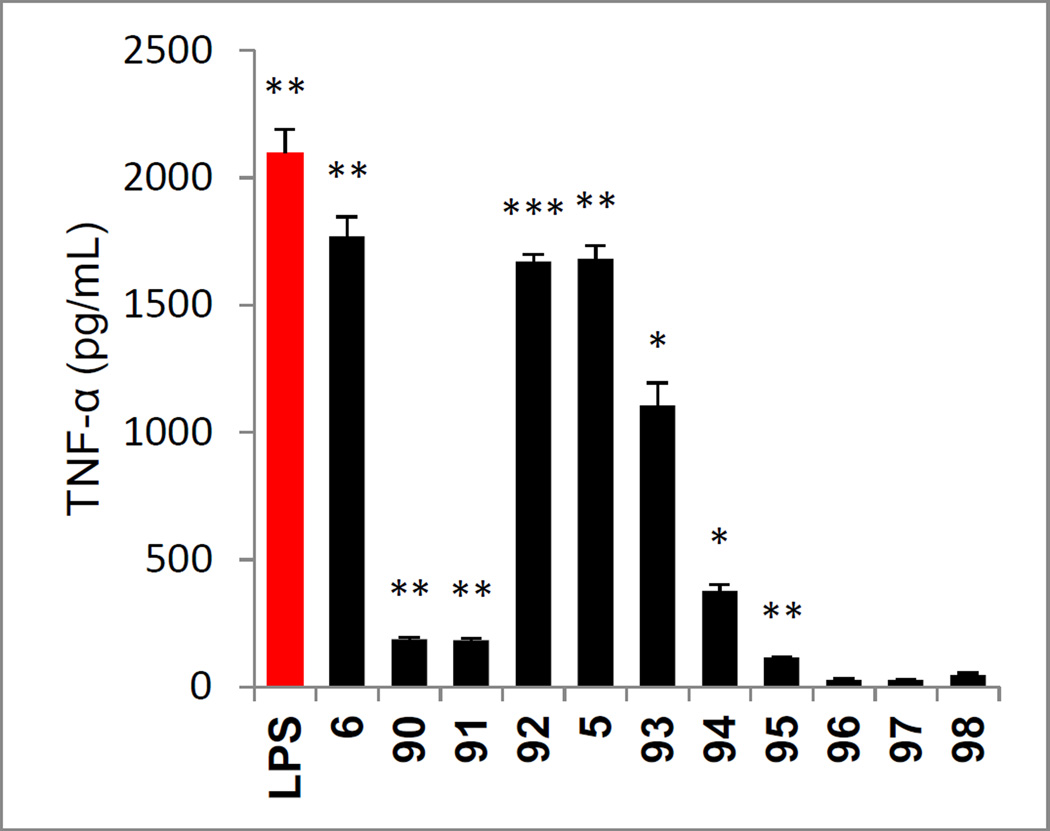
All analogues that contained saturated linkers composed of two or three carbons or oxygen were highly effective agonists and the results with analogues in this series exhibited well defined trends (Figure 12). The effective agonists included not only 5 and 6, but also 92 that incorporates an oxygen atom into the two atom linker of 5 and 93 that replaces the oxygen atom in the three atom linker of 6 with a carbon methylene. Extending the linker of 6 by even one or two more carbon atoms resulted in a near complete loss in agonist activity (analogues 90 and 91). Similarly shortening the two carbon atom linker of 5 by one carbon with 94 led to a substantial loss in activity, and removing the linker altogether with 95 led to essentially complete loss in activity. Additionally, replacement of the flexible two carbon saturated linker of 5 with the rigid two carbon alkyne linker in 96 resulted in a complete loss in activity. Similarly, replacement of the linker with a single nitrogen atom as found in 98 resulted in an inactive compound. Surprisingly, replacement of the linker with a sulfur atom as in 97 resulted in a complete loss in activity despite the fact that it is isosteric with two carbon atoms. The carboxylic acids derived from these series were also prepared by acid-catalyzed t-butyl ester deprotection and all were found to be inactive (not shown).
Figure 12.
Evaluation of analogues of 5 and 6 (Schemes 9–11) used to probe the linker between the central aromatic ring and the phenol assayed (50 µM) against mouse macrophages alongside LPS (1 ng/mL) for stimulated release of TNF-α. All data is the mean of triplicate assay versus vehicle control, error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001.
Adjuvant Activity of 5
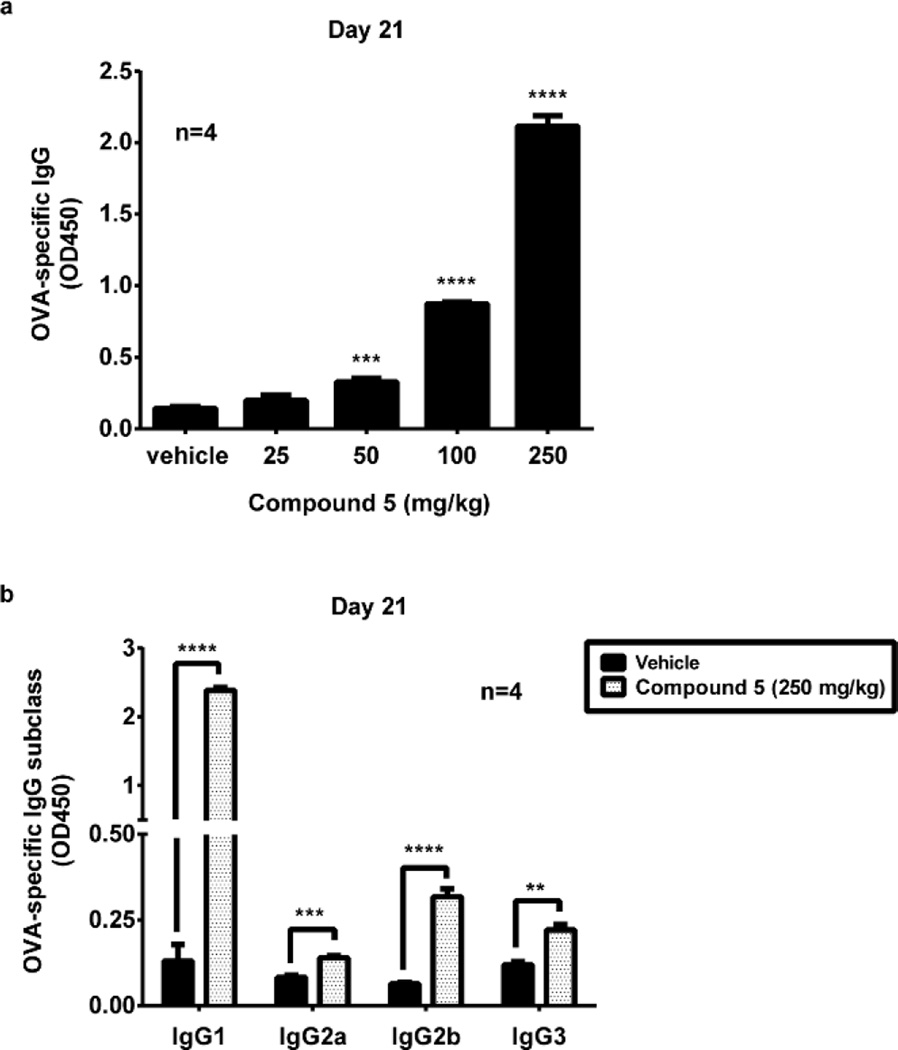
Extensive in vitro characterization of 5 as a mTLR4 agonist and its downstream signaling was disclosed elsewhere26 as summarized in the introduction, indicating it effectively elicits the innate immune response. Since TLR4 signaling plays an essential role in the generation of both innate and adaptive immune responses, we investigated whether 5 can also serve as an adjuvant in vivo in mice, indicating stimulation of the adaptive immune response. We analyzed IgG production against a protein antigen ovalbumin (OVA) in the absence or presence of escalating doses of 5. The data showed that 5 elicited a robust antibody response in a dose-dependent manner measured 21 days after immunization (Figure 13a). In mice, immunoglobulin G1 (IgG1) is associated with a Th2-like response, while a Th1 response is associated with the induction of IgG2a, IgG2b, and IgG3 antibodies. We further assessed the effect of 5 on OVA-specific IgG subclasses and found that 5 induced significant increases in OVA-specific IgG1, IgG2a, IgG2b and IgG3 (Fig.13b), suggesting it activated both Th1 and Th2-mediated immune responses. Taken together, these data indicate that 5 is an adjuvant in mouse and serves to stimulate the adaptive immune response. Just as significantly, the study indicates that compounds in this new class are of a caliber that exhibit in vivo activity, being active not only on its target proteins and in functional cellular assays, but in vivo in mice as well.
Figure 13.
Adjuvant activity of 5. (a) Dose-dependent adjuvant activity of 5. C57BL/6 mice (4 per group) were immunized i.m. by 100 µg OVA with vehicle (propylene glycol) or 5 in vehicle (propylene glycol) at indicated doses. After 21 days, serum titers of OVA-specific IgG were measured by ELISA. (b) Assessment of OVA-specific IgG subclasses. Mice (4 per group) were immunized with 100 µg of OVA mixed with 5 or vehicle alone. After 21 days, serum titers of OVA-specific IgG subclasses indicated were measured by ELISA. Error bars represent SEM. * = P ≤ 0.05; ** = P ≤ 0.01; *** = P ≤ 0.001; **** = P ≤ 0.0001.
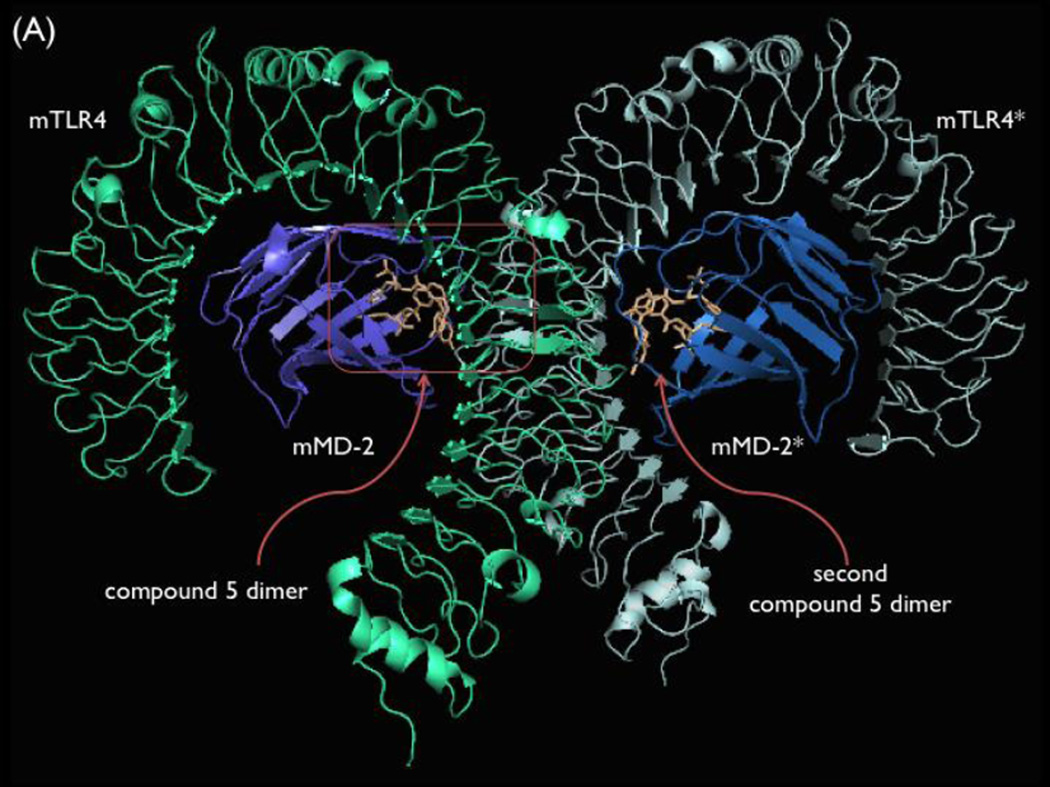
Retrospective Interpretation of the Structure–Activity Relationships Based on an X-ray Co-crystal Structure of Mouse TLR4/MD-2 with 5
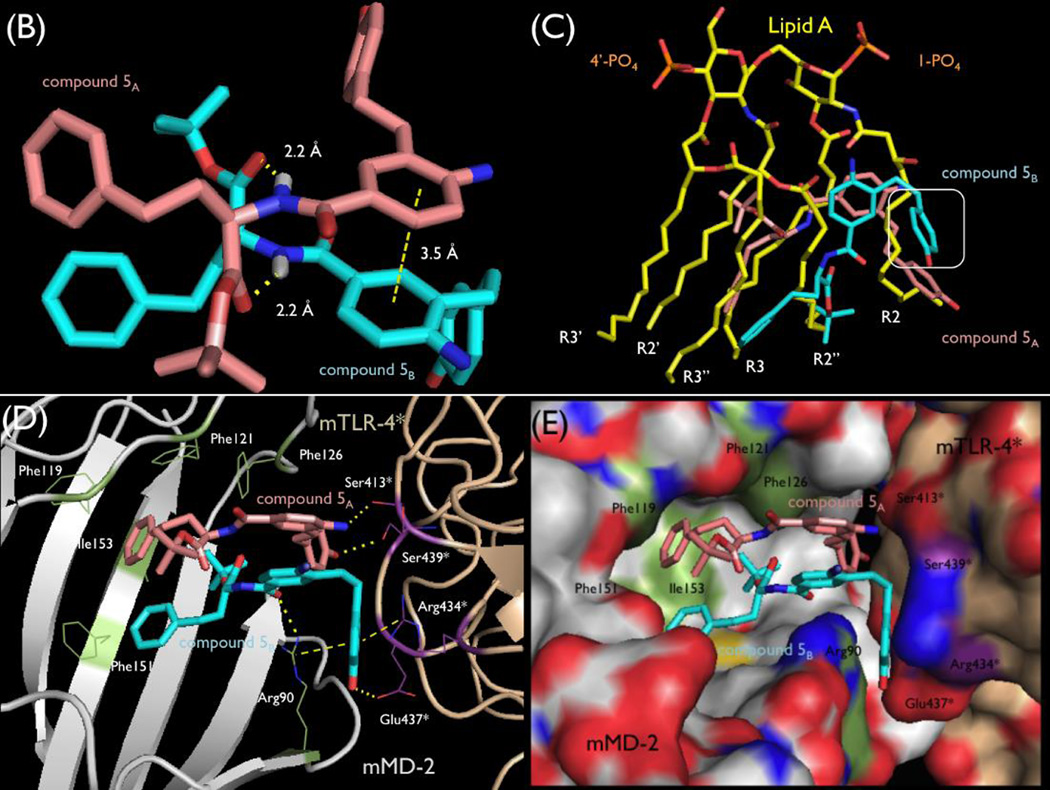
The crystal structure of mTLR4/mMD-2/5 recently reported by us revealed that 5 binds as an asymmetrical dimer within the hydrophobic pocket of mMD-2 (Figure 14).26 There it induces and stabilizes a dimerization interface between two mTLR4/mMD-2 complexes, similar to the active complex induced by Lipid A. However, 5 and Lipid A form overlapping but distinct molecular contacts with the receptor to achieve receptor dimerization and activation. Two molecules of 5 bind tightly to each other, being stabilized by π-stacking of the central aniline rings (π-stacking) and by two hydrogen bonds between the NH group of the amide bond of each molecule of 5 and the t-butyl ester carbonyl group of the other (2.2 Å each, Figure 14, panel B). Thus, it is not surprising that all modifications to the amide (71–73) resulted in complete loss in activity. Similarly, removal of hydrogen bond acceptor, the carbonyl of the t-butyl ester (43), results in complete loss of activity. What is unique is that these two hydrogen bonds represent key interactions, stabilizing the ligand dimer formation and not the interaction of 5 with the protein target. Additionally, a hydrogen bond between the side chain of mMD-2 residue Arg90 and one of the amide carbonyl groups stabilizes binding to the target.
Figure 14.
Panel A, Co-crystal structure of 5 with mTLR4/mMD-2 (pdb 5HG4).26 Panel B, Asymmetrical dimer of 5 bound at the mMD-2/mTLR4 interface. Panel C, Overlay of Lipid A (pdb 5HG6)26 and 5 as bound to mTLR4/mMD-2, highlighting their distinct activation interactions and differences in contact size. Boxed phenol of 5 has no overlay with Lipid A and represents a distinct site of inter-receptor interactions for 5. Panels D and E, Dimerization interface of the co-crystal structure of 5 with mTLR4/mMD-2, highlighting its binding at the entrance of the hydrophobic pocket of mMD-2 and key intra- and inter-receptor interactions.
One t-butyl group of the esters is bound deep in the hydrophobic pocket of mMD-2, mimicking one of the lipid chains of Lipid A (R2”; Figure 14, panel C), extending to the bottom of the pocket and contacting mMD-2 Phe126 stabilizing the Phe126 loop movement now characteristic of agonists. The second t-butyl group is found near the lip of the mMD-2 hydrophobic pocket interacting with both Phe121 and Phe119 and poised to extend into solvent. Thus, one t-butyl group contributes critical hydrophobic contacts deep in the hydrophobic pocket of mMD-2, whereas the other may provide stabilizing contacts at the lip of the hydrophobic pocket. Consistent with these roles, only the t-butyl ester provides potent agonist activity, the corresponding amides are inactive, and either smaller or more extended bulky esters lose activity (40–59). It is likely that the first of the t-butyl esters is essential (5B), whereas the second (5A) could tolerate significant changes.
The HoPhe side chains extend into the mMD-2 hydrophobic pocket, each mimicking a lipid chain of Lipid A, and they pack against one another similar to the Lipid A lipid chains (Figure 14, panels D and E). This is consistent with the requirement for an extended hydrophobic side chain (17–35), the accommodation of only hydrophobic phenyl substituents (63–69), and the remarkable accommodation of a cyclic saturated (70) versus aryl side chain. The two atom linker to the phenyl group for both molecules of 5 adopt gauche versus extended conformations, albeit different from one another. This permits the tip of the aryl rings to interact with either mMD-2 Phe151 or Ile153. A shorter linker to the aryl ring does not support receptor activation (23) because both aryl rings would fail to make these stabilizing contacts with mMD-2 and each other, whereas the more extended linkers (61 and 62) may not be effective because they would extend past and sterically clash with the side of the pocket. Likely this region will tolerate more extended hydrophobic substituents on the aryl ring, may tolerate a wide range of extended or branched hydrophobic groups in place of the aryl ring, and the linkers to each of the two phenyl groups may not need to be the same for each molecule in the dimer.
The central core aniline of 5 stabilizes ligand dimerization by π stacking interactions between the two aniline aromatic rings, which are separated by 3.5 Å (Figure 14, panel C). One of the amino groups of the aniline participates in a significant hydrogen bond with the recruited mTLR4* main chain Ser413* carbonyl, stabilizing active mTLR4 dimer formation. The other lies at the center of the entry to the mMD-2 binding pocket extending into solvent, making no contacts other than with solvent. Each amino group sterically orients the adjacent alkyl substituent extending to the aryl phenol, facilitating the participation of each phenol in two different and essential hydrogen bonds with the recruited mTLR4* and aromatic ring π stacking interactions at the dimer interface. Consistent with a significant role, removal of the aniline amino group (76 and 81), its replacement with a nitro group (16), its alkylation (77 and 78) or acylation (79), and its movement to the 6 position (75) results in loss in activity. Only its movement to the adjacent 5 position (74) maintains roughly half the agonist activity, presumably reducing but not necessarily completely losing the hydrogen bond to a mTLR4* backbone carbonyl. It is likely that only the first of the aniline amino groups is essential, whereas the second found at the entrance of the mMD-2 binding pocket could tolerate its removal or significant changes.
One of the most important features of 5 is the pendant phenol, the presence and position of its hydroxyl group, and the length and nature of the chain linking the phenol to the core central ring. The two phenol rings of each molecule of 5 do not bind in the hydrophobic pocket of mMD-2 but are buried at the dimerization interface with mTLR4*, providing the key interactions between mMD-2 and mTLR4*. The phenol hydroxyl group of 5A forms a specific hydrogen bond with the side chain of residue Ser439* of mTLR4* and is sufficiently strong and specific to anchor one ligand near the entrance of the hydrophobic pocket of mMD-2 for dimerization with mTLR4*. This phenol ring and its linker region overlap with the Lipid A lipid chain (R2; Figure 14, panel C) that has been suggested to be necessary for its agonist activity. The second phenol ring shows no overlap with bound Lipid A. This phenol is found sandwiched between the guanidinium groups of Arg434* of mTLR4* and Arg90 of mMD-2 and its hydroxyl group is hydrogen bonded to the side chain carbonyl of Glu437* of mTLR4*. Along with a hydrogen bond from the Arg90 side chain guanidine of mMD-2 with the amide carbonyl of the same molecule of 5, these interactions appear key to binding, dimerization, and adoption of the activated mTLR4/mMD-2 complex, creating a binding pocket that is not found in the activated Lipid A complex with mTLR4/mMD-2. Since both phenols are essential, it is not surprising that the removal of the phenol hydroxyl group (84 and 89), phenol O-methylation (85), hydroxyl movement to the 2-position on the aromatic ring (83), or benign substitution adjacent to the hydroxyl group (87 and 88) result in a loss in agonist activity. Only a shift of the hydroxyl group to the adjacent 3 versus 4 position (82 and 86) is tolerated. The chain linking the phenol to the central aromatic ring is flexible with each adopting different conformations (one extended, one gauche), and is optimal with a chain length of two or three atoms (5, 6, 92 and 93), permitting the placement of each phenol aromatic ring and hydroxyl group for key dimerization interactions with the mTLR4*. Shortening the linker (94, 95, 97 and 98), extending the linker beyond three atoms (90 and 91), or its rigidification (96) result in a loss of agonist activity.
It is worth highlighting that even bound as a dimer, 5 occupies less than 50% of the mMD-2 hydrophobic pocket, overlays with only three of six lipid chains of Lipid A (R2, R2”, and R3), and serves to promote mTLR4 dimerization and activation by enlisting many different stabilizing contacts than Lipid A at the dimerization interface.26 Finally, no amount of prospective modeling of the binding of 5 with mTLR4/mMD-2 could have predicted the requirement for dimer ligand binding.
Conclusions
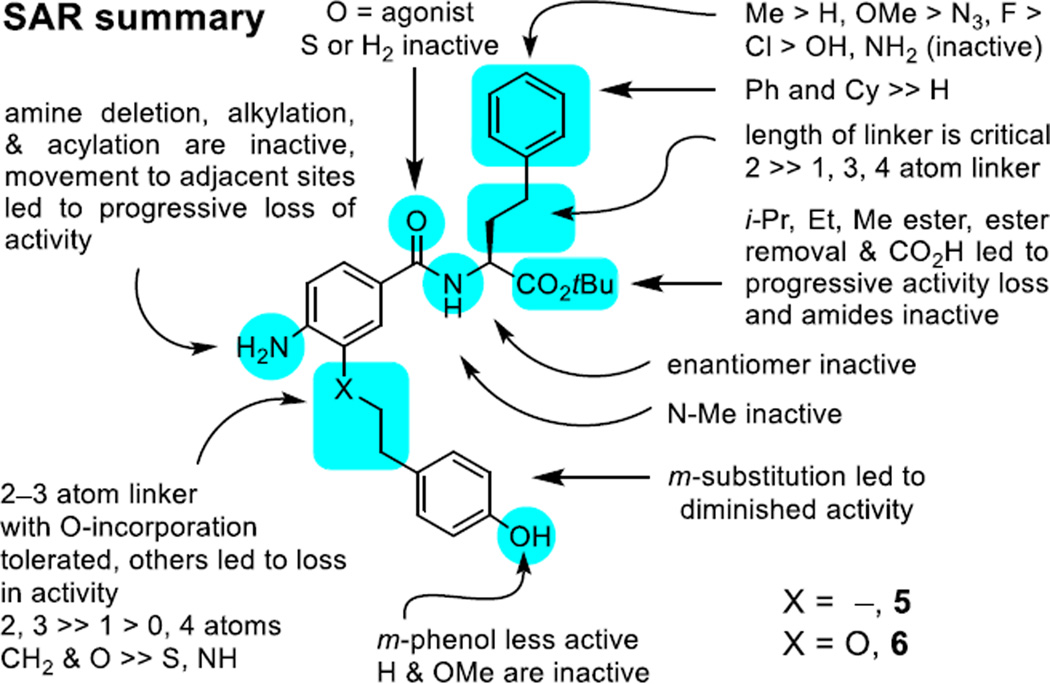
Although LPS and its biosynthetic precursor Lipid A have a well-defined target, the toxicity of these TLR4 ligands is considerable as is their instability in vivo and they are not easily modified. In work that defines a powerful paradigm for the discovery of new small molecule stimulators of the immune response, we report a remarkable class of small molecule mTLR4 agonists. The compound class referred to as the neoseptins emerged from a α-helix mimetic library, stimulate the immune response, act by a well-defined mechanism (mTLR4 agonist), are easy to produce and structurally manipulate, exhibit exquisite structure–activity relationships (Figure 15), are non-toxic, and elicit improved and qualitatively different responses than LPS even though they share the same receptor.26
Figure 15.
SAR summary.
Structural studies of compound 5 binding and mTLR4 activation disclosed elsewhere26 illustrated that it binds as a dimer within the mMD-2 hydrophobic pocket and promotes mouse TLR4/MD-2 dimerization with adoption of the same activated conformation induced by Lipid A, but that it does so by a different series of intra- and inter-receptor contacts than Lipid A. The requirement for agonist dimerization while also engaging this series of intricate intra- and inter-receptor contacts needed for receptor dimerization and activation accounts for the exquisitely sensitive SAR surrounding this class of agonists. It also defines a rational approach to further improvements in their potency and efficacy and for their modification to achieve human TLR4 activation that are underway. Finally, we are not aware of any prior reported example of ligand noncovalent dimerization at a target protein site, resulting in activation of receptor signaling (agonist vs antagonist/inhibitor) by ligand-induced receptor dimerization and adoption of an activated conformation. As such, the class of compounds represent the first ligands discovered and reported with structural features that simultaneously promote intermolecular ligand dimerization, target protein binding affinity, and induce receptor dimerization with adoption of an activated signaling conformation.
Experimental
General
Reagents and solvents were purchased reagent-grade and used without further purification. THF was freshly distilled from sodium benzophenone ketyl. All reactions were performed in oven-dried glassware under an Ar atmosphere. Evaporation and concentration in vacuo was performed at 20 °C. TLC was conducted using precoated SiO2 60 F254 glass plates from EMD with visualization by UV light (254 or 366 nm). Optical rotations were determined on a Rudolf Research Analytical Autopol III Automatic Polarimeter (λ = 589 nm, 25 °C). NMR (1 H or 13C) were recorded on Bruker DRX-500 and DRX-600 NMP spectrophotometers at 298K. Residual solvent peaks were used as an internal reference. Coupling constants (J) (H,H) are given in Hz. Coupling patterns are designated as singlet (s), doublet (d), triplet (t), quadruplet (q), multiplet (m), or broad singlet (br). IR spectra were recorded on a Thermo Scientific Nicolet 380 FT-IR spectrophotometer and measured neat. High resolution mass spectral data were acquired on an Agilent Technologies high resolution LC/MSD-TOF, and the detected masses are given as m/z with m representing the molecular ion. The purity of each tested and active compound (>95%) was determined on an Agilent 1100 LC/MS instrument using a ZORBAX SB-C18 column (3.5 mm, 4.6 mm × 50 mm, with a flow rate of 0.75 mL/min and detection at 220 and 254 nm) with a 10−98% acetonitrile/water/0.1% formic acid gradient (Supporting Information Figure S1).
Compound 5
Methyl 4-Nitro-3-(((trifluoromethyl)sulfonyl)oxy)benzoate (7). Methyl 3-hydroxy-4-nitrobenzoate (1.50 g, 7.61 mmol) was dissolved in anhydrous CH2Cl2 (40 mL) and cooled to 0 °C under a N2 atmosphere. Et3N (2.21 mL, 15.2 mmol, 2.00 equiv) was added slowly. After 15 min, trifluoromethanesulfonic anhydride (1.45 mL, 8.37 mmol, 1.10 equiv) was added dropwise. After 16 h at 25 °C, the reaction mixture was washed with saturated aqueous NH4Cl (30 mL) and H2O (40 mL). The aqueous phase was extracted with CH2Cl2 (2 × 20 mL), and the combined organic extracts were dried over Na2SO4 and concentrated in vacuo. Flash column chromatography (SiO2, 10–20% EtOAc/hexanes gradient) gave 1.98 g (79%) of 7 as a fluffy, off-white solid. IR (neat) vmax 1727, 1536, 1430, 1209, 975 cm−1; 1H NMR (300 MHz, CDCl3) δ 8.23 (t, J = 1.2 Hz, 1H), 8.12–8.07 (m, 2H), 4.02 (s, 3H); 13C NMR (125 MHz, CDCl3) δ 163.4, 141.3, 136.4, 130.0, 126.8, 125.3, 119.8, 117.3, 53.4. HRMS (ESI-TOF) m/z calcd for C9H6F3NO7SNa [M+Na]+ 351.9709, found 351.9707.
3-((4-Hydroxyphenyl)ethynyl)-4-nitrobenzoic Acid (9). A 100 mL two-neck round-bottom flask was charged with 7 (2.40 g, 7.29 mmol), PdCl2(PPh3)2 (510 mg, 0.73 mmol, 10 mol%), CuI (415 mg, 2.18 mmol, 30 mol%), and Bu4NI (8.06 g, 21.8 mmol, 3.0 equiv). The reagents were suspended in 5:1 DMF/Et3N (30 mL/6 mL), and the reaction mixture flask was submerged in a preheated oil bath at 70 °C. 4-(Triisopropylsiloxy)phenylacetylene (4.00 g, 14.6 mmol, 2.00 equiv) was added dropwise to the vigorously stirred reaction mixture. After 3 h, the mixture was cooled to room temperature, diluted with 1:1 EtOAc/hexanes (60 mL each) and washed with saturated aqueous NH4Cl (2 × 25 mL). The aqueous phase was extracted with 1:1 EtOAc/hexanes (3 × 30 mL), and combined organic phases were dried over Na2SO4 and concentrated to afford 8. The resultant dark oil was dissolved in 4:1:1 THF/MeOH/H2O (20 mL/5 mL/5 mL, respectively). LiOH•H2O (1.30 g, 31.0 mmol, 4.25 equiv) was added, and the suspension was stirred at room temperature overnight at 25 °C. After 16 h, the reaction mixture was cooled to 0 °C and aqueous 2 N HCl (approx. 8–10 mL) was added until a precipitate was observed. The mixture was diluted with EtOAc (100 mL) and washed with aqueous 2 N HCl until the aqueous phase remained pH ~2. The aqueous phase was extracted with EtOAc (2 × 30 mL), and the combined organic phases were dried over Na2SO4 and concentrated in vacuo. Flash chromatography (SiO2, 50:50:0.5 EtOAc/hexanes/HOAc) gave 1.69 g (81%) of 9 as a red solid. IR (neat) vmax 2207, 1683, 1597, 1241, 1170 cm−1; 1H NMR (500 MHz, CDCl3) δ 8.35 (d, J = 1.8 Hz, 1H), 8.10 (d, J = 8.5 Hz, 1H), 8.05 (dd, J = 8.6, 1.8 Hz, 1H), 7.50 (d, J = 8.6 Hz, 2H), 6.89 (d, J = 8.6 Hz, 2H), 4.19 (s, 1H). HRMS (ESI-TOF) m/z calcd for C15H10NO5 [M+H]+ 284.0553, found 284.0554.
(S)-tert-Butyl 2-(3-((4-Hydroxyphenyl)ethynyl)-4-nitrobenzamido)-4-phenylbutanoate (10). Compound 9 (320 mg, 1.13 mmol, 1.00 equiv), HoPhe-OtBu (266 mg, 1.13 mmol), and HOAt (170 mg, 1.24 mmol, 1.10 equiv) were combined in a 20 mL flask equipped with a stir bar. Anhydrous DMF (5 mL) and 2,6-lutidine (0.66 mL, 5.65 mmol, 5.00 equiv) were added, and the mixture stirred until complete dissolution of the reagents. EDCI•HCl (230 mg, 1.18 mmol, 1.05 equiv) was added, and the mixture was stirred at 25 °C for 18 h. The reaction mixture was diluted with EtOAc (30 mL), and washed with aqueous 0.1 N HCl (2 × 25 mL) and saturated aqueous NaCl (25 mL). The aqueous phase was extracted with EtOAc (2 × 10 mL), and combined organic phases were dried over Na2SO4 and concentrated. Flash column chromatography (SiO2, 0–30% EtOAc/hexanes gradient) afforded 555 mg (98%) of 10 as a red solid. 1H NMR (400 MHz, CDCl3) δ 8.07 (d, J = 8.5 Hz, 1H), 7.85 (d, J = 1.9 Hz, 1H), 7.64 (dd, J = 8.6, 2.0 Hz, 1H), 7.49 (d, J = 8.6 Hz, 2H), 7.35–7.28 (m, 2H), 7.25–7.17 (m, 3H), 6.86 (d, J = 8.6 Hz, 2H), 6.74 (d, J = 7.6 Hz, 1H), 4.80 (td, J = 7.0, 4.8 Hz, 1H), 2.76 (t, J = 7.7 Hz, 2H), 2.37 (dtd, J = 13.1, 8.1, 4.8 Hz, 1H), 2.26–2.15 (m, 1H), 1.56 (s, 9H). HRMS (ESI-TOF) m/z calcd for C29H29N2O6 [M+H]+ 501.2020, found 501.2018.
Compound 5. From 10: Compound 10 (530 mg, 1.06 mmol) was dissolved in EtOAc (10 mL) in a two-neck round-bottom flask equipped with a stir bar and three-way vacuum adapter. Pearlman’s catalyst (5 mol%, 20% Pd w/w on carbon) was suspended in the reaction solvent, and the solvent was sparged with N2 for 10 min. The reaction headspace was evacuated briefly until the solvent began to boil, and then back-filled with H2. This process was repeated 15–20 times to ensure maximum H2 atmosphere above the reaction mixture. After stirring for 16 h at 25 °C, the mixture was filtered through a plug of sand/Celite with a 2 mm top layer of SiO2, washing thoroughly with EtOAc. The solvent was removed in vacuo. Flash chromatography (SiO2, 30–50% EtOAc/hexanes gradient) gave 495 mg (98%) of 5 as a pale yellow solid. [α]+44 (c 0.10, CHCl3). Water solubility = 0.2 mg/mL or 700 µM. IR (neat) vmax 1714, 1621, 1494, 1365, 1229 cm−1. 1H NMR (500 MHz, CDCl3) δ 7.44 (dd, J = 8.3, 2.2 Hz, 1H), 7.34 (d, J = 2.2 Hz, 1H), 7.32–7.28 (m, 2H), 7.25–7.16 (m, 3H), 6.99 (d, J = 8.4 Hz, 2H), 6.78 (d, J = 8.4 Hz, 2H), 6.63 (d, J = 8.3 Hz, 1H), 6.49 (d, J = 7.8 Hz, 1H), 4.82–4.76 (m, 1H), 2.90–2.61 (m, 6H), 2.28 (dddd, J = 13.7, 10.2, 6.3, 5.1 Hz, 1H), 2.10 (m, 1H), 1.53 (s, 9H). 13C NMR (125 MHz, CDCl3) δ 172.1, 167.2, 154.5, 147.8, 141.2, 133.0, 129.5, 129.1, 128.49, 128.48, 128.37, 126.3, 126.1, 125.2, 123.4, 115.6, 114.7, 82.5, 53.0, 34.5, 34.2, 33.4, 31.6, 28.1. HRMS (ESI-TOF) m/z calcd for C29H35N2O4 [M+H]+ 475.2591, found 475.2592.
Alternative Synthesis of Compound 5
Methyl 3-(4-(Benzyloxy)styryl)-4-nitrobenzoate (13). (4-(Benzyloxy)benzyl)triphenyl-phosphonium bromide39 (1.98 g, 3.67 mmol, 1.10 equiv) was suspended in anhydrous THF (60 mL) and cooled to 0 °C. Sodium bis(trimethylsilyl)amide (5.50 mL, 1.5 equiv, 1.0 M solution in THF) was added slowly, forming an orange solution. The ice bath was removed and the mixture was stirred for 20 min at room temperature. The reaction mixture was cooled to 0 °C, and methyl 3-formyl-4-nitrobenzoate (0.700 g, 3.33 mmol, 1.00 equiv, solution in 15 mL anhydrous THF) was transferred via cannula to the reaction mixture. After stirring for 16 h at room temperature, saturated aqueous NH4Cl (50 mL) was added. The aqueous phase was extracted with EtOAc (2 × 50 mL). The organic phase was dried over Na2SO4 and concentrated. Flash column chromatography (SiO2, 15% EtOAc/hexanes) afforded 0.80 g (55%) of 13 as an inseparable 2:1 mixture of E/Z isomers. 1H NMR for the major isomer (500 MHz, CDCl3) δ 8.06 (s, 1H), 8.04 (d, J = 1.8 Hz, 1H), 8.00 (d, J = 1.9 Hz, 2H), 7.48–7.30 (m, 5H), 7.01 (d, J = 8.7 Hz, 1H), 6.98 (d, J = 8.7 Hz, 2H), 6.78 (d, J = 8.8 Hz, 2H), 6.74 (d, J = 4.8 Hz, 1H), 5.02 (s, 2H), 3.88 (s, 3H). HRMS (ESI-TOF) m/z calcd for C23H20NO5 [M+H]+ 390.1336, found 390.1348.
Compound 5. From 13: Compound 13 (288 mg, 0.740 mmol) was dissolved in THF/MeOH/H2O (2.0 mL, 0.5 mL, 0.5 mL, respectively) at room temperature. LiOH•H2O (156 mg, 3.70 mmol, 5 equiv) was added in one portion, and the mixture was stirred for 3 h at room temperature. Aqueous 1 N HCl was added (30 mL) and the aqueous phase was extracted with EtOAc (3 × 25 mL). The organic phase was dried over Na2SO4 and concentrated to give 254 mg (91%) of the benzoic acid, which was used without further purification. The benzoic acid (150 mg, 0.400 mmol), HoPhe-OtBu (94 mg, 0.40 mmol) and HOAt (60 mg, 0.44 mmol) were dissolved in anhydrous DMF (2 mL). 2,6-Lutidine (0.230 mL) was added, followed by EDCI•HCl (45 mg, 0.42 mmol). After 18 h at 25 °C, the mixture was diluted with EtOAc (50 mL) and washed with aqueous 1 N HCl (2 × 25 mL) and saturated aqueous NaCl (25 mL). The organic phase was dried over Na2SO4 and concentrated. Flash column chromatography (SiO2, 25% EtOAc/hexanes) gave 166 mg (70%) of the amide product. The amide product (146 mg, 0.246 mmol) was dissolved in EtOAc (1.5 mL) and Pearlman’s catalyst (50 mg) was added. The resulting suspension was stirred vigorously under H2 atmosphere for 12 h at 25 °C. The mixture was filtered through a 4 cm plug of Celite and sand with a 2 mm top layer of SiO2 to remove the catalyst. Removal of the solvent in vacuo provided 109 mg (93%) of 5, whose 1H NMR and HRMS data were identical with that reported from the samples obtained using the route in Scheme 1.
Compound 6
4-Nitro-3-(4-((triisopropylsilyl)oxy)phenethoxy)benzoic Acid (14). Sodium hydride (60%, 0.50 g, 12.4 mmol) was suspended in THF (10 mL) and 2-[4-(triisopropylsilyloxy)phenyl]ethanol (0.67 mL, 6.48 mmol) was added dropwise at 0 °C. The mixture was stirred at 0 °C for 15 min under an atmosphere of Ar before 3-fluoro-4-nitrobenzoic acid (1.0 g, 5.4 mmol) was added. The mixture was stirred at 0 °C for 5 min and room temperature for 2 h. The reaction mixture was quenched with the addition of saturated aqueous NH4Cl, diluted with EtOAc (50 mL), and washed with aqueous 0.1 N HCl (2 × 20 mL). The organic layer was collected, concentrated, and the product purified by flash chromatography (SiO2, 3:2:0.1 hexanes/Et2O/HOAc) to give 14 as a white solid (1.26 g, 85%). 1H NMR (600 MHz, DMSO-d6) δ 11.78 (s, 1H), 7.90 (d, J = 8.3 Hz, 1H), 7.83 (d, J = 6.1 Hz, 1H), 7.73 (dd, J = 8.3, 1.6 Hz, 1H), 7.24 (d, J = 8.5 Hz, 2H), 6.86 (d, J = 8.5 Hz, 2H), 4.45 (t, J = 6.7 Hz, 2H), 3.08 (t, J = 6.7 Hz, 2H), 1.34–1.22 (m, 3H), 1.11 (d, J = 7.2 Hz, 18H). HRMS (ESI-TOF) m/z calcd for C24H33NO6SiNa [M+Na]+ 482.1969, found 482.1964.
(S)-tert-Butyl 2-(4-Nitro-3-(4-((triisopropylsilyl)oxy)phenethoxy)benzamido)-4-phenylbutanoate (15). Compound 14 (95 mg, 0.20 mmol) was combined with HoPhe-OtBu (88 mg, 0.35 mmol) in DMF (0.75 mL). HOAt (61 mg, 0.45 mmol), EDCI•HCl (69 mg, 0.36 mmol), and 2,6-lutidine (0.17 mL, 1.5 mmol) were added. After stirring at room temperature for 12 h, the reaction mixture was poured into aqueous 1 N HCl (20 mL). The aqueous layer was extracted with EtOAc (2 × 20 mL). The combined organic extracts were washed with aqueous 1 N HCl, saturated aqueous NaHCO3 and saturated aqueous NaCl, dried over Na2SO4 and evaporated in vacuo. Flash chromatography (SiO2, 20% EtOAc/hexanes) yielded 15 (126 mg, 90%) as a white solid. 1H NMR (600 MHz, DMSO-d6) δ 7.80 (d, J = 8.3 Hz, 1H), 7.50 (s, 1H), 7.30–7.21 (m, 6H), 7.12 (d, J = 8.1 Hz, 2H), 6.85 (d, J = 7.3 Hz, 2H), 4.74–4.50 (m, 1H), 4.30 (t, J = 6.9 Hz, 2H), 3.10 (t, J = 6.7 Hz, 2H), 2.80–2.72 (m, 2H), 2.40–2.26 (m, 1H), 2.20–2.11 (m, 1H), 1.50 (s, 9H), 1.30–1.24 (m, 3H), 1.09 (d, J = 7.0 Hz, 18H). HRMS (ESI-TOF) m/z calcd for C38H53N2O7Si [M+H]+ 677.3616, found 677.3617.
(S)-tert-Butyl 2-(3-(4-Hydroxyphenethoxy)-4-nitrobenzamido)-4-phenylbutanoate (16). Compound 15 (250 mg, 350 mmol) was dissolved in anhydrous THF (8 mL) and treated with Bu4NF (0.43 mL, 1 M solution in THF, 1.20 equiv). The reaction mixture was stirred at room temperature for 1 h, before the solvent was removed in vacuo. Flash chromatography of the residue (SiO2, 25% EtOAc/hexanes) afforded 166 mg (91%) of 16 as a white solid. 1H NMR (600 MHz, DMSO-d6) δ 7.82 (d, J = 7.9 Hz, 1H), 7.50 (s, 1H), 7.40–7.11 (m, 8H), 6.85 (d, J = 7.3 Hz, 2H), 5.40 (brs, 1H), 4.80–4.65 (m, 1H), 4.38 (t, J = 7.0 Hz, 2H), 3.10 (t, J = 6.7 Hz, 2H), 2.80–2.72 (m, 2H), 2.50–2.30 (m, 1H), 2.25–2.10 (m, 1H), 1.50 (s, 9H). HRMS (ESI-TOF) m/z calcd for C29H33N2O7 [M+H]+ 521.2282, found 521.2283.
Compound 6. Compound 16 (120 mg, 0.23 mmol) was dissolved in acetone/saturated aqueous NH4Cl (1:1, 5 mL each). Zn nanopowder (151 mg, 2.30 mmol, 10 equiv) was added portionwise to the reaction mixture, which was stirred vigorously at room temperature for 1 h. The heterogeneous mixture was filtered through Celite to remove the Zn salts, and the filtrate was diluted with EtOAc (50 mL) and washed with H2O (50 mL). The organic phase was dried over Na2SO4 and concentrated to give 113 mg (99%) of 6 as a white solid. [α]+42 (c 0.10, CHCl3). IR (neat) vmax 1714, 1612, 1503, 1365, 1221 cm−1. 1H NMR (600 MHz, DMSO-d6) δ9.30 (s, 1H), 8.20 (d, J = 8.2 Hz, 1H), 7.50–7.30 (m, 7H), 7.20 (d, J = 7.1 Hz, 2H), 6.65 (d, J = 7.3 Hz, 2H), 5.25 (s, 2H), 4.80 (q, J = 6.0 Hz, 1H), 4.18 (t, J = 7.0 Hz, 2H), 3.00 (t, J = 6.7 Hz, 2H), 2.70–2.65 (m, 1H), 2.55–2.50 (m, 1H), 2.10–2.00 (m, 2H), 1.49 (s, 9H). 13C NMR (125 MHz, CDCl3) δ 172.0, 167.2, 155.0, 145.8, 141.1, 140.2, 129.9, 129.4, 128.5, 128.4, 126.1, 123.1, 120.0, 115.5, 113.3, 111.0, 82.5, 69.2, 53.0, 34.8, 34.5, 31.6, 28.0. HRMS (ESI-TOF) m/z calcd for C29H35N2O5 [M+H]+ 491.2540, found 491.2542.
(S)-2-(4-Amino-3-(4-hydroxyphenethyl)benzamido)-4-phenylbutanoic Acid (11)
Compound 5 (4.0 mg, 0.015 mmol, 1.0 equiv) was dissolved in 4 N HCl/dioxane (200 µL, 0.80 mmol, approx. 54 equiv) and allowed to stir for 8 h at 25 °C, after which the t-butyl ester was observed by LCMS to have been completely consumed. Concentration of the solvent provided 3.5 mg (99%) of 11 as a yellow solid. 1H NMR (500 MHz, methanol-d4) δ 8.06 (d, J = 2.1 Hz, 1H), 8.00 (dd, J = 8.3, 2.1 Hz, 1H), 7.62 (d, J = 8.3 Hz, 1H), 7.49–7.33 (m, 5H), 7.29 (d, J = 8.4 Hz, 2H), 6.92 (d, J = 8.4 Hz, 2H), 4.76 (dd, J = 9.6, 4.7 Hz, 1H), 3.25–3.17 (m, 2H), 3.16–3.09 (m, 2H), 2.96 (m, 2H), 2.48 (m, 1H), 2.37 (m, 1H). HRMS (ESI-TOF) m/z calcd for C25H27N2O4 [M+H]+ 419.1965, found 419.1968.
(S)-2-(4-Amino-3-(4-hydroxyphenethoxy)benzamido)-4-phenylbutanoic Acid (12)
Compound 6 (20 mg, 0.041 mmol) was dissolved in 4 N HCl/dioxane (1 mL), and the reaction mixture was stirred at 25 °C for 8 h. The solvent and excess HCl were evaporated under a stream of N2 to provide 17 mg (99%) of 12, which was not further purified. 1H NMR (500 MHz, methanol-d4) δ 7.63 (d, J = 1.7 Hz, 1H), 7.53 (dd, J = 8.1, 1.8 Hz, 1H), 7.41 (dd, J = 8.1 Hz, 1H), 7.28–7.12 (m, 5H), 7.21 (d, J = 8.5 Hz, 2H), 6.76 (d, J = 8.5 Hz, 2H), 4.57 (dd, J = 9.6, 4.7 Hz, 1H), 4.37 (t, J = 7.2 Hz, 2H), 3.11 (t, J = 7.2 Hz, 2H), 2.85–2.69 (m, 2H), 2.28 (m, 1H), 2.16 (m, 1H). HRMS (ESI-TOF) m/z calcd for C25H27N2O5 [M+H]+ 435.1914, found 435.1914.
Measurement of TNFα Release from Mouse Macrophages
Thioglycolate-elicited macrophages were recovered 4 d after i.p. injection of 2 mL BBL thioglycolate medium, brewer modified (4%; BD Biosciences) by peritoneal lavage with 5 mL phosphate buffered saline (PBS). The peritoneal macrophages were cultured in DMEM cell culture medium [DMEM containing 10% FBS (Gemini Bio Products), 1% penicillin and streptomycin (Life Technologies)] at 37 °C and 95% air/5% CO2. Cells were seeded onto 96-well plates at 1 × 105 cells per well and treated with 5, 6, or the candidate analogues (dissolved in DMSO, and final assay DMSO concentrations (≤0.5%) were kept constant in all experiments) and ultra-pure LPS (dissolved in H2O, Enzo Life Sciences) for 4 h and assessed alongside DMSO treated controls.40 Mouse TNF-α in the supernatants was measured by ELISA kits according to the manufacturer’s instructions (eBioscience and PBL Assay Science). All data is mean of triplicates and SD is within +10% of mean. Mouse cells were from wild type C57BL/6J mice.
Adjuvant Activity
C57BL/6 mice were purchased from The Jackson Laboratory. EndoFit™ ovalbumin (OVA) was purchased from Invivogen with 98% purity minimum (SDS-PAGE) and has endotoxin levels <1 EU/mg. Mice (4 per group) were immunized i.m. by 100 µg OVA with vehicle (propylene glycol) or 5 in vehicle (propylene glycol) at the indicated doses. After 21 days, serum titers of OVA-specific IgG or IgG subclasses were measured by ELISA. All experimental procedures using mice were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Texas Southwestern Medical Center, and were conducted in accordance with institutionally approved protocols and guidelines for animal care and use. All the mice were maintained at the University of Texas Southwestern Medical Center in accordance with institutionally approved protocols.
Supplementary Material
Acknowledgments
We gratefully acknowledge the financial support of the National Institutes of Health (CA042056, AI082657 D.L.B.; AI082657 B.B.; GM104496 H.Z.).
Abbreviations Used
- Abu
aminobutyric acid
- CD14
cluster of differentiation 14
- DAMP
damage-associated molecular pattern
- DME
dimethoxyethane
- DMF
dimethylformamide
- DMSO
dimethylsulfoxide
- DMAP
4-(dimethylamino)pyridine
- EDCI
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- ERK
extracellular signal-regulated kinase
- HOAt
1-hydroxy-7-azabenzotriazole
- HoPhe
homophenyl-alanine
- IKBKG
inhibitor of nuclear factor kappa-B kinase subunit gamma protein
- IFN
interferon
- IL
interleukin
- IRAK
interleukin-1 receptor-associated kinase
- JNK
c-Jun N-terminal kinase
- LPS
lipopolysaccharide
- LBP
LPS binding protein
- MAPK
mitogen-activated protein kinase
- MD-2
myeloid differentiation protein 2
- Met
methionine
- MPLA
monophosphoryl Lipid A
- MyD88
myeloid differentiation primary response gene 88 protein
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- OVA
ovalbumin
- PAMP
pathogen-associated molecular pattern
- Ra-Ni
Raney-nickel
- Tf
trifluoromethylsulfonyl
- TLR
Toll-like receptor
- TIPS
tri-isopropylsilyl
- TRIF
TIR-domain-containing adapter-inducing interferon-β
- TNF-α
tumor necrosis factor alpha
- SAR
structure–activity relationship
Footnotes
Associated Content
Full experimental details are provided. The Supporting Information is available free of charge on the ACS Publications web site at DOI:
Accession Codes
X-ray crystal structures have been previously published,26 are currently available, and have been deposited in the Protein Data Bank as follows: asymmetrical dimer of compound 5 bound to mTLR4/mMD-2, PDB code 5HG4; LPA bound to mTLR4/mMD-2, PDB code 5HG6.
References
- 1.Beutler B. Innate immunity: an overview. Mol. Immunol. 2004;40:845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Hoebe K, Janssen E, Beutler B. The interface between innate and adaptive immunity. Nat. Immunol. 2004;5:971–974. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- 3.(a) Moresco EMY, LaVine D, Beutler B. Toll-like receptors. Curr. Biol. 2011;21:R488–R493. doi: 10.1016/j.cub.2011.05.039. [DOI] [PubMed] [Google Scholar]; (b) Blasius AL, Beutler B. Intracellullar Toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]; (c) Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;13:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]; (d) Beutler B. TLRs and innate immunity. Blood. 2009;113:1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto C, Hudson KL, Anderson KV. The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell. 1988;52:269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- 5.(a) Poltorak A, He X, Smirnova I, Liu M-Y, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg MA, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]; (b) Beutler B, Du X, Poltorak A. Identification of Toll-like receptor 4 (TLR4) as the sole conduit for LPS signal transduction: genetic and evolutionary studies. J. Endotoxin Res. 2001;7:277–280. [PubMed] [Google Scholar]; (c) Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 7.(a) Vogel FR. Improving vaccine performance with adjuvants. Clin. Infect. Dis. 2000;30(Suppl 3):S266–S270. doi: 10.1086/313883. [DOI] [PubMed] [Google Scholar]; (b) Guy B. The perfect mix: recent progress in adjuvant research. Nat. Rev. Microbiol. 2007;5:505–517. doi: 10.1038/nrmicro1681. [DOI] [PubMed] [Google Scholar]; (c) Coffman RL, Sher A, Seder RA. Vaccine adjuvants: Putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Czarniecki M. Small molecule modulators of Toll-like receptors. J. Med. Chem. 2008;51:6621–6626. doi: 10.1021/jm800957k. [DOI] [PubMed] [Google Scholar]; (b) Peri F, Calabrese V. Toll-like receptor 4 (TLR4) modulation by synthetic and natural compounds: an update. J. Med. Chem. 2014;57:3612–3622. doi: 10.1021/jm401006s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Smith C, Yin H. Targeting Toll-like receptors with small molecule agents. Chem. Soc. Rev. 2013;42:4859–4866. doi: 10.1039/c3cs60039d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Hennessy EJ, Parker AE, O’Neill LAJ. Targeting Toll-like receptors: emerging therapeutics. Nat. Rev. Drug Discovery. 2010;9:293–301. doi: 10.1038/nrd3203. [DOI] [PubMed] [Google Scholar]; (b) Meyer T, Stockfleth E. Clincal investigations of Toll-like receptor agonists. Expert Opin. Invest. Drugs. 2008;17:1051–1065. doi: 10.1517/13543784.17.7.1051. [DOI] [PubMed] [Google Scholar]; (c) Hoebe K, Jiang Z, Georgel P, Tabeta K, Janssen E, Du X, Beutler B. TLR signaling pathways: Opportunities for activation and blockade in pursuit of therapy. Curr. Pharmaceut. Des. 2006;12:4123–4134. doi: 10.2174/138161206778743466. [DOI] [PubMed] [Google Scholar]
- 11.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonist and antagonists. Nat. Med. 2007;13:552–559. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- 12.Prins RM, Craft N, Bruhn KW, Khan-Farooqi H, Koya RC, Stripecke R, Miller JF, Liau LM. The TLR-7 agonist, imiquimod, enhances dendritic cell survival and promotes tumor antigen-specific T cell priming: relation to central nervous system antitumor immunity. J. Immunol. 2005;176:157–164. doi: 10.4049/jimmunol.176.1.157. [DOI] [PubMed] [Google Scholar]
- 13.Lee J, Chuang T-H, Redecke V, She L, Pitha PM, Carson DA, Raz E, Cottam HB. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of Toll-like receptor 7. Proc. Natl. Acad. Sci. USA. 2003;100:6646–6651. doi: 10.1073/pnas.0631696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Wu CCN, Lee KJ, Chuang T-H, Katakura K, Liu YT, Chan M, Tawatao R, Chung M, Shen C, Cottam HB, Lai MMC, Raz E, Carson DA. Activation of anti-hepatitis C virus responses via Toll-like receptor 7. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1828–1833. doi: 10.1073/pnas.0510801103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smits ELJM, Cools N, Lion E, Camp K, Ponsaerts P, Berneman ZN, Van Tendeloo VFI. The Toll-like receptor 7/8 agonist resiquimod greatly increases the immunostimulatory capacity of human acute myeloid leukemia cells. Cancer Immunol. Immunother. 2010;59:35–46. doi: 10.1007/s00262-009-0721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beesu M, Malladi SS, Fox LM, Jones CD, Dixit A, David SA. Human Toll-like receptor 8-selective agonistic activities in 1-alkyl-1H–benzimidazol-2-amines. J. Med. Chem. 2014;57:7325–7341. doi: 10.1021/jm500701q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu TYH, Singh M, Miller AT, De Gregorio E, Doro F, D’Oro U, Skibinski DAG, Mbow ML, Bufali S, Herman AE, Cortez A, Li Y, Nayak BP, Tritto E, Filippi CM, Otten GR, Brito LA, Monaci E, Li C, Aprea S, Valentini S, Calabro S, Laera D, Brunelli B, Caproni E, Malyala P, Panchal RG, Warren TK, Bavari S, O’Hagan DT, Cooke MP, Valiante NM. Rational design of small molecules as vaccine adjuvants. Sci. Transl. Med. 2014;6(263) doi: 10.1126/scitranslmed.3009980. 263ra160. [DOI] [PubMed] [Google Scholar]
- 18.(a) Guan Y, Omueti-Ayoade K, Mutha SK, Hergenrother PJ, Tapping RI. Identification of novel synthetic Toll-like receptor 2 agonists by high throughput screening. J. Biol. Chem. 2010;285:23755–23762. doi: 10.1074/jbc.M110.116046. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cheng K, Gao M, Godfroy JI, Brown PN, Kastelowitz N, Yin H. Specific activation of the TLR1-TLR2 heterodimer by small molecule agonists. Sci. Adv. 2015;1:e1400139. doi: 10.1126/sciadv.1400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Salunke DB, Connelly SW, Shukla NM, Hermanson AR, Fox LM, David SA. Design and development of stable, water-soluble, human Toll-like receptor 2 specific monoacyl lipopeptides as candidate vaccine adjuvants. J. Med. Chem. 2013;56:5885–5900. doi: 10.1021/jm400620g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Salunke DB, Shukla NM, Yoo E, Crall BM, Balakrishna R, Malladi SS, David SA. Structure-activity relationships in human Toll-like receptor 2-specific monoacyl lipopeptides. J. Med. Chem. 2012;55:3353–3363. doi: 10.1021/jm3000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schumann R, Leong S, Flaggs G, Gray P, Wright S, Mathison J, Tobias P, Ulevitch R. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 21.Wright S, Ramos R, Tobias P, Ulevitch R, Mathison J. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 22.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, A molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Jin MS, Lee JO. Structures of TLR-ligand complexes. Curr. Opin. Immunol. 2008;20:414–419. doi: 10.1016/j.coi.2008.06.002. [DOI] [PubMed] [Google Scholar]; (b) Kim HM, Park BS, Kim J, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo JO, Lee J. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]; (c) Park B, Song D, Kim H, Choi B, Lee H, Lee J. The structural basis of lipopolysaccharide recognition by the TLR4MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]; (d) Ohto U, Fukase K, Miyake K, Shimizu T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc. Natl. Acad. Sci. USA. 2012;109:7421–7426. doi: 10.1073/pnas.1201193109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Ohto U, Fukase K, Miyake K, Satow Y. Crystal structure of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 24.O‧ Connor CJ, Laraiaa L, Spring DR. Tutorial review chemical genetics. Chem. Soc. Rev. 2011;40:4332–4345. doi: 10.1039/c1cs15053g. [DOI] [PubMed] [Google Scholar]
- 25.(a) Whitby LR, Boger DL. Comprehensive peptidomimetic libraries targeting protein-protein interactions. Acc. Chem. Res. 2012;45:1698–1709. doi: 10.1021/ar300025n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Boger DL, Desharnais J, Capps K. Solution-phase combinatorial libraries: modulating cellular signaling by targeting protein-protein or protein-DNA interactions. Angew. Chem. Int. Ed. 2003;42:4138–4176. doi: 10.1002/anie.200300574. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Su L, Morin MD, Jones BT, Whitby LR, Surakattula MMRP, Huang H, Shi H, Choi JH, Wang K, Moresco EMY, Berger M, Zhan X, Zhang H, Boger DL, Beutler B. TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS. Proc. Natl. Acad. Sci. USA. 2016;113:E884–E893. doi: 10.1073/pnas.1525639113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaginian A, Whitby LR, Hong S, Hwang I, Faroogi B, Chen J, Searcey M, Vogt PK, Boger DL. Design, synthesis, and evaluation of an α-helix mimetic library targeting protein-protein interactions. J. Am. Chem. Soc. 2009;131:5564–5572. doi: 10.1021/ja810025g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitby LR, Ando Y, Setola V, Vogt PK, Roth BL, Boger DL. Design, synthesis, and evaluation of a β-turn mimetic library targeting protein-protein and peptide-receptor interactions. J. Am. Chem. Soc. 2011;133:10184–10194. doi: 10.1021/ja201878v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stover JS, Shi J, Jin W, Vogt PK, Boger DL. Discovery of inhibitors of aberrant gene transcription from libraries of DNA binding molecules: inhibition of LEF-1 mediated gene transcription and oncogenic transformation. J. Am. Chem. Soc. 2009;131:3342–3348. doi: 10.1021/ja809083d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otrubova K, Srinivasan V, Boger DL. Discovery libraries targeting the major enzyme classes: the serine hydrolases. Bioorg. Med. Chem. Lett. 2014;24:3807–3813. doi: 10.1016/j.bmcl.2014.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(a) Simon MM, Moresco EMY, Bull KR, Kumar S, Mallon AM, Beutler B, Potter PK. Current strategies for mutation detection in phenotype-driven screens utilizing next generation sequencing. Mammal. Gen. 2015;26:486–500. doi: 10.1007/s00335-015-9603-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Arnold CN, Barnes MJ, Berger M, Blasius AL, Brandl K, Croker B, Crozat K, Du X, Eidenschenk C, Georgel P, Hoebe K, Huang H, Jiang Z, Krebs P, La Vine D, Li X, Lyon S, Moresco EMY, Murray AR, Popkin DL, Rutschmann S, Siggs OM, Smart NG, Sun L, Tabeta K, Webster V, Tomisato W, Won S, Xia Y, Xiao N, Beutler B. ENU-induced phenovariance in mice: Inferences from 587 mutations. BMC Res. Notes. 2012;5:577. doi: 10.1186/1756-0500-5-577. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Beutler B, Du X, Xia Y. Precis on forward genetics in mice. Nat. Immunol. 2007;8:659–664. doi: 10.1038/ni0707-659. [DOI] [PubMed] [Google Scholar]; (d) Beutler B. The Toll-like receptors: analysis by forward genetic methods. Immunogen. 2005;57:385–392. doi: 10.1007/s00251-005-0011-3. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd S, Huber M, Kalis C, Keck S, Galanos C, Freudenberg M, Beutler B. CD14 is required for MyD88-independent LPS signaling. Nat. Immunol. 2005;6:565–570. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 33.(a) Mullarkey M, Rose JR, Bristol J, Kawata T, Kimura A, Kobayashi S, Przetak M, Chow J, Gusovsky F, Christ WJ, Rossignol DP. Inhibition of endotoxin response by e5564, a novel Toll-like receptor 4-directed endotoxin antagonist. J. Pharmacol. Exp. Ther. 2003;304:1093–1102. doi: 10.1124/jpet.102.044487. [DOI] [PubMed] [Google Scholar]; (b) Golenbock DT, Hampton RY, Qureshi N, Takayama K, Raetz CR. Lipid A-like molecules that antagonize the effects of endotoxins on human monocytes. J. Biol. Chem. 1991;266:19490–19498. [PubMed] [Google Scholar]
- 34.Okemoto K, Kawasaki K, Hanada K, Miura M, Nishijima MA. potent adjuvant monophosphoryl lipid A triggers various immune responses, but not secretion of IL-1 beta or activation of caspase-1. J. Immunol. 2006;176:1203–1208. doi: 10.4049/jimmunol.176.2.1203. [DOI] [PubMed] [Google Scholar]
- 35.Johnson DA. Synthetic TLR4-active glycolipids as vaccine adjuvants and stand-alone immunotherapeutics. Curr. Top. Med. Chem. 2008;8:64–79. doi: 10.2174/156802608783378882. [DOI] [PubMed] [Google Scholar]; (b) Casella CR, Mitchell TC. Putting endotoxin to work for us: monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cell Mol. Life Sci. 2008;65:3231–3240. doi: 10.1007/s00018-008-8228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Persing DH, Coler RN, Lacy MJ, Johnson DA, Baldridge JR, Hershberg RM, Reed SG. Taking toll: lipid A mimetics as adjuvants and immunomodulators. Trends Microbiol. 2002;10:S32–S37. doi: 10.1016/s0966-842x(02)02426-5. [DOI] [PubMed] [Google Scholar]
- 36.Neve JE, Wijesekera HP, Duffy S, Jenkins ID, Ripper JA, Teague SJ, Campitelli M, Garavelas A, Nikolakopoulos G, Le PV, de ALeone P, Pham NB, Shelton P, Fraser N, Carroll AR, Avery VM, McCrae C, Williams N, Quinn RJ. Euodenine A: a small molecule agonist of human TLR4. J. Med. Chem. 2014;57:1252–1275. doi: 10.1021/jm401321v. [DOI] [PubMed] [Google Scholar]
- 37.Chan M, Hayashi T, Mathewson RD, Nour A, Hayashi Y, Yao S, Tawatao RI, Crain B, Tsigelny IF, Kouznetsova VL, Messer K, Pu M, Corr M, Carson DA, Cottam HB. Identification of substituted pyrimido[5,4-b]indoles as selective Toll-like receptor 4 ligands. J. Med. Chem. 2013;56:4206–4223. doi: 10.1021/jm301694x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmer SM, Liu J, Clayton JL, Stephens DS, Snyder JP. Paclitaxel binding to human and murine MD-2. J. Biol. Chem. 2008;283:27916–27926. doi: 10.1074/jbc.M802826200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carreño MC, Des Mazery R, Urbano A, Colobert F, Solladié G. Reductive cyclization of hydroxysulfinyl ketones: enantioselective access to tetrahydropyran and tetrahydrofuran derivatives. J. Org. Chem. 2003;68:7779–7787. doi: 10.1021/jo034817x. [DOI] [PubMed] [Google Scholar]
- 40.1% DMSO or less does not induce TNF-α release from mouse macrophages, see Supporting Information Figure S1. It has been reported that DMSO (1%) enhances LPS induced IL-1β release, but has no effect on LPS-induced TNF-α and IL-6 release or NF-κB activation. See: Xing L, Remick DG. Mechanisms of dimethyl sulfoxide augmentation of IL-1β production. J. Immunol. 2005;174:6195–6202. doi: 10.4049/jimmunol.174.10.6195.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.