Abstract
Despite the publication of several software tools for analysis of glycopeptide tandem mass spectra, there remains a lack of consensus regarding the most effective and appropriate methods. In part, this reflects problems with applying standard methods for proteomics database searching and false discovery rate calculation. While the analysis of small post-translational modifications (PTMs) may be regarded as an extension of proteomics database searching, glycosylation requires specialized approaches. This is because glycans are large and heterogeneous by nature causing glycopeptides to exist as multiple glycosylated variants. Thus, the mass of the peptide cannot be calculated directly from that of the intact glycopeptide. In addition, the chemical nature of the glycan strongly influences product ion patterns observed for glycopeptides. As a result, glycopeptidomics requires specialized bioinformatics methods. We summarize the recent progress towards a consensus for effective glycopeptide tandem mass spectrometric analysis.
2. Introduction
Analysis of intact glycopeptides entails analytical and informatics methods tailored to their hybrid nature. Unlike smaller PTMs, glycosylation is heterogeneous, making extension of traditional proteomics database searching problematic. In addition, glycosylation diminishes the strength of MS signals and results in ion suppression. As a result, investigators often enrich glycopeptides to eliminate competition from non-glycosylated peptides for ion signal [1].
The purpose of this review is to orient researchers from the proteomics or glycoscience fields who wish to use MS methods for glycopeptides. Readers interested in the use of proteomics for analysis of deglycosylated peptides are referred to a recent review [2]. In addition, a number of reviews of informatics methods for analysis of glycopeptide mass spectra have appeared [3–7].
We summarize database search methods in traditional proteomics and post-translational modification (PTM) proteomics, which shed light on the identification of glycopeptides. We then describe MS approaches for glycopeptide analysis and compare the effectiveness of tandem mass spectrometric dissociation methods. We finish with a summary of published and commercial tools and discuss approaches for false discovery rate calculation.
3. Summary of proteomics database search approaches
Accurate mass measurement does not suffice for assignment of glycopeptides even when the protein sequence is known [8]. This limitation arises from the multiplicity of chemical forms that result from the non-template driven biosynthetic processes that add glycosylation in the endoplasmic reticulum and the Golgi apparatus. Note that this complexity multiples when one considers other common modifications to peptides including oxidation, deamidation, dehydration, and partial proteolytic digestion. Thus, on the one hand, accurate mass measurement will not define glycopeptide composition adequately; on the other, the number of molecular forms results in a large search space for interpretation of glycopeptide tandem mass spectra.
In the early days of biomolecular mass spectrometry, the intent was to sequence proteins directly using fragmentation patterns generated from peptides [9,10]. During this time, the Edman degradation was more effective than mass spectrometry-based methods, particularly those based on liquid secondary ionization mass spectrometry/fast atom bombardment. With the development of electrospray ionization [11], it became much easier to interface liquid chromatography columns to mass spectrometers and the sensitivity for peptide tandem MS was improved by orders of magnitude. At the same time, researchers realized that it was not necessary to interpret peptide tandem mass spectra from first principles; rather, one could search the tandem mass spectra data against a list derived from genomic information [12,13]
Prior to the development of database search strategies, software tools for direct (de novo) interpretation of peptide tandem mass spectra were developed [14]. Such de novo methods calculate the peptide sequence from the mass shifts among product ions and approximate manual interpretation of tandem mass spectra. Modern software that build on de novo concepts include PepNovo [15], PEAKS [16] and Uninovo [17]. By contrast, database search methods calculate the most probable sequence from the tandem mass spectrum using in silico digested peptides from genomic information. The first database methods included extraction of sequence tags from tandem mass spectra for search against a database (PeptideSearch) [13] and used a cross-correlation function for automated database searches (SEQUEST) [18]. Other database searching methods have appeared, including ProteinProspector [19], Mascot [20], X!Tandem [21], OMSSA [22], MyriMatch [23], Andromeda [24] and Comet [25]. Recently, search engines designed specifically for high resolution tandem mass spectra have appeared including Morpheus [26] and MS Amanda [27]. Tools dedicated in integrating search engine results and managing search engines’ parameters have also become available [28,29].
4. Post-translational modifications and proteomics
The presence of a PTM multiplies the complexity of the proteome. Thus, the number of chemical forms increases as Xn where X = number of post-translationally modified forms and n = the number of modified amino acid residues. For a peptide with a single site of phosphorylation, there are two molecular forms. For a peptide with three sites of phosphorylation, there are 23 = 8 molecular forms. For a peptide with three sites of post-translational modification and three modified forms (for example unmodified, phosphorylated, or O-GlcNAcylated), the number of molecular forms is 33 = 27. Multiple PTM types and their preferred amino acid residues further complicate the situation. Despite this complexity, it is straightforward to calculate the molecular weight of the peptide using the precursor ion mass and the mass of the PTM group. Thus, traditional proteomics database search approaches are applicable for PTMs with defined molecular structure. These include phosphorylation, acetylation, methylation, ubiquitination, O-GlcNAcylation [30].
Complex glycosylation arises from a series of biosynthetic reactions that do not go to completion. Thus, a given peptide sequence will exist as a set of several glycoforms, multiplying the number of molecular forms by orders of magnitude. Therefore, it is computationally intractable to calculate the molecular weight of the peptide portion of a complex glycopeptide from the intact mass; however, such information can be extracted from a tandem mass spectrum.
5. Comparison of the effectiveness of collisional versus activated electron dissociation methods for glycopeptides
Ideally, we would like assign both the peptide sequence and glycan structure from a single tandem mass spectrum; however, this is not possible using present technology. One problem is that protonated glycoconjugates undergo rearrangements during collisional heating [31], rendering it possible to mis-interpret the data. Another problem is that detailed structural determination of branched glycans requires permethylation in order to define glycan topology [32,33]; this can only be accomplished on released glycans. Realistically, the goal of a glycopeptide tandem MS experiment using present technology should be to establish the peptide sequence, site of glycosylation, and glycan composition.
Commercial instruments offer collisional activated dissociation (CAD), also known as collision-induced dissociation, whereby ions are excited by collision with inert gas molecules, resulting in vibrational excitation. Collisional dissociation may occur by resonant excitation in a trapped ion instrument so as to dissociate a targeted precursor ion m/z window. The majority of productions have m/z values outside this window and are no longer vibrationally excited. The result is dissociation of the weakest precursor ion bonds with a low degree of subsequent dissociation. Thus, CAD of glycopeptides in a trapped ion instrument results in abundant ions from losses of monosaccharide units from the precursor. Non-resonant dissociation occurs when a beam of precursor ions collides with inert gas atoms, resulting in dissociation. The product ions may retain sufficient kinetic energy to undergo further dissociation. Thus, the extent of dissociation is higher for non-resonant beam-type dissociation such as occurs in triple quadrupole and quadrupole time-of-flight instruments. The so called higher energy collisional dissociation (HCD) term used with Thermo-Fisher™ instruments falls under non-resonant dissociation.
In glycopeptides the glycan dissociates at lower vibrational energies than does the peptide. As a result, a series of glycosidic bond dissociation events occurs before any peptide backbone dissociation in most cases. For many years, the general consensus in the field was that collisional dissociation of glycopeptides did not dissociate the peptide backbone to an appreciable degree. We now know that peptide bond dissociation occurs provided that sufficiently high collision energy is used [34–37]. Nonetheless, the relative abundances of such peptide backbone cleavage product ions resulting from collisional dissociation is relatively low.
Activated electron dissociation methods including electron capture dissociation (ECD) [38] and electron transfer dissociation (ETD) [39] favor dissociation of the peptide backbone, rather than the glycan, of glycopeptides [40]. Thus, the presence of glycopeptides can be identified by the production of low m/z product ions from HCD tandem mass spectra and used to trigger subsequent ETD [41]. Such experiments would represent the ideal for effective glycoproteomics were it possible to acquire ETD for all glycopeptides that produce oxonium ions detectable by HCD; however, the limitation of ETD is the precursor ion abundance required for efficient glycopeptide dissociation. Even with the most modern instruments, this abundance is considerably higher than that required for collisional dissociation of glycopeptide precursor ions.
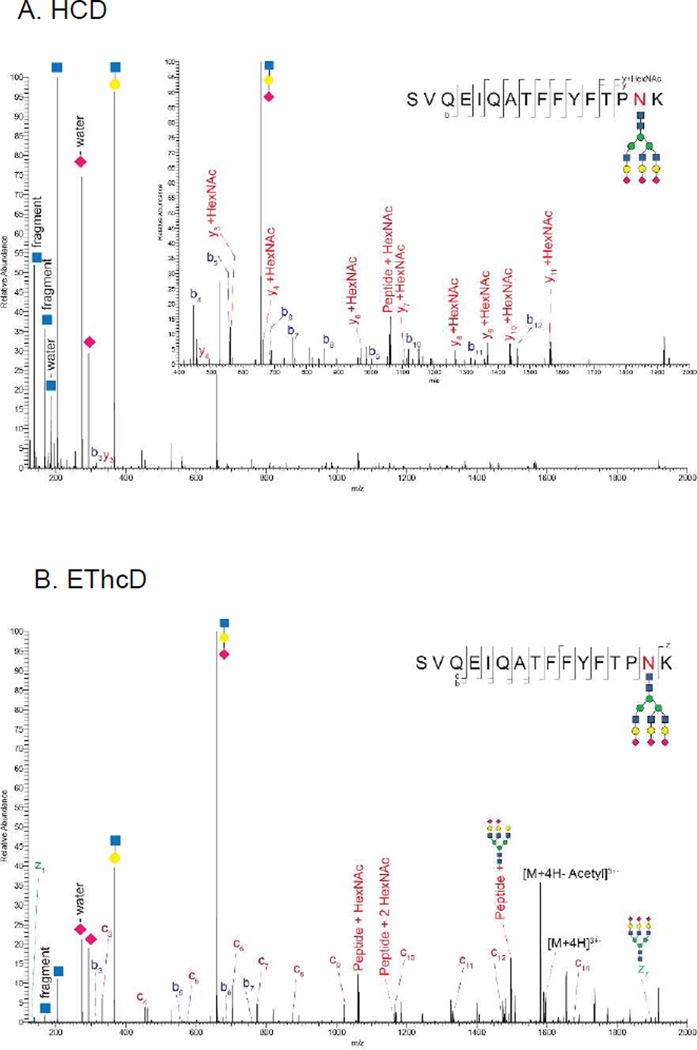
While ETD is essential for analysis of fragile PTMs including O-GlcNAcylation [42], it remains unclear the best approach for dissociation of complex glycopeptides. On the one hand, all modern MS instruments carry out collisional dissociation and can produce peptide backbone product ions, albeit in low relative abundances. On the other, while ExD product ion patterns favor peptide backbone dissociation and are more likely to define the sites of glycosylation, the technology is not mature, judging from the fact that the hardware configuration changes from one version of a given instrument to the next. A significant fraction of product ion abundance from ETD goes into formation of a charged reduced species that must be collisionally activated in order for product ions to be detected. Thus, a combination of activated electron dissociation and collisional excitation allows for improved product ion detection [43]. This principle has been used to generate mixed mode product ions for peptides using a combination of ETD and HCD (known as EThcD) [44]; as shown in Figure 1, this approach is useful for analysis of glycopeptides. Ultraviolet photo dissociation is also emerging as an alternative dissociation method for glycopeptides [45,46].
Figure 1.
Comparison of CAD (A) and electron transfer dissociation (B) for analysis of an influenza hemagglutinin glycopeptides. Data were acquired using a Thermo-Fisher Scientific Orbitrap XL mass spectrometer [97].
Figure 1 compares HCD and EThcD tandem mass spectra for an α1-acid glycoprotein glycopeptide. The CAD tandem mass spectrum (A) was acquired using ion trap dissociation, under which conditions product ions from dissociation of glycosidic bonds were abundant but peptide backbone product ions were not detected. It was possible to identify the peptide mass from the CAD tandem mass spectrum. The ETD tandem mass spectrum (B) enabled the sequencing of a 7 amino acid residue tag in the glycopeptide that unambiguously identified the peptide. Figure 2 shows a CAD tandem mass spectrum acquired using a Q-TOF mass spectrometer with higher dissociation energy than used in Figure 1. Under these conditions, it is possible to detect peptide backbone product ions for glycopeptides [34].
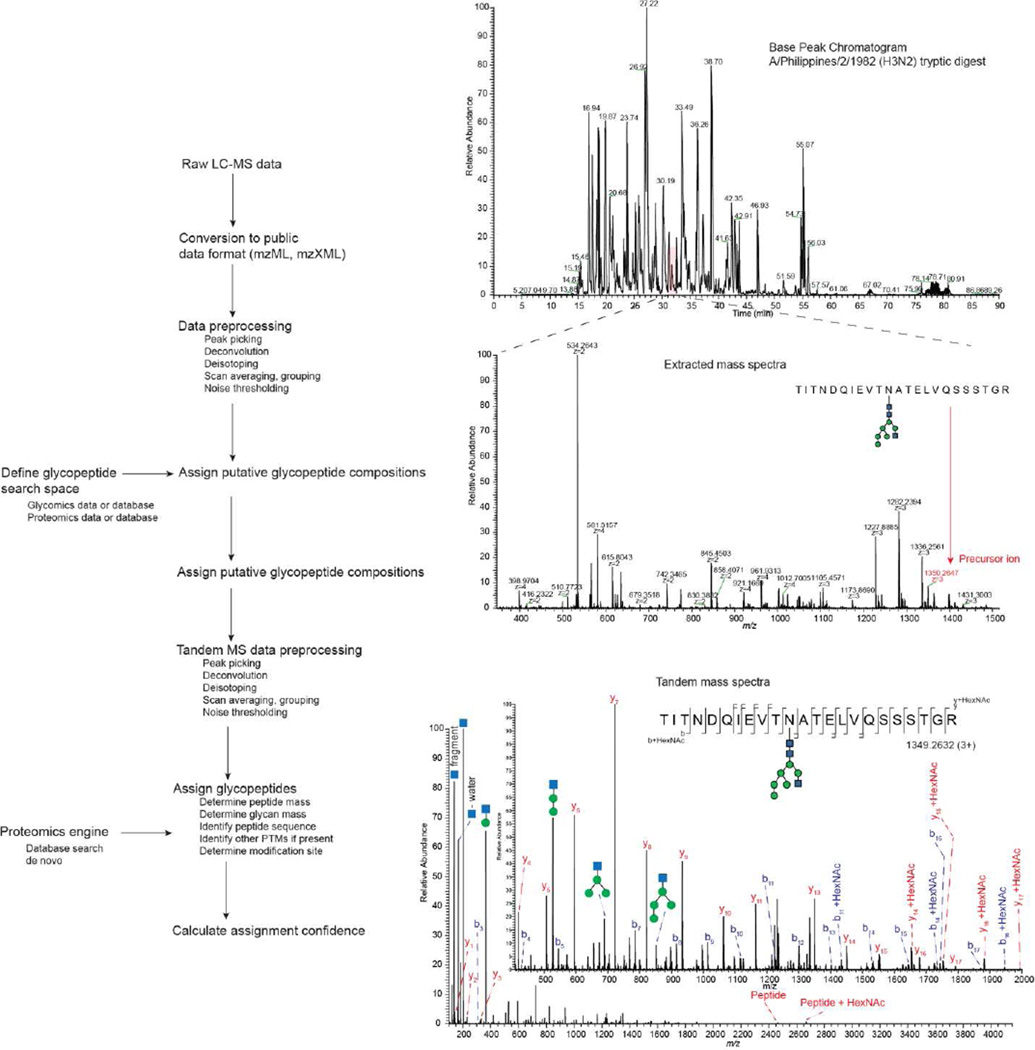
Figure 2.
Idealized workflow for assignment of glycoprotein site-specific N-glycosylation. Raw LC-MS data must first be converted to a public data format and then subjected to preprocessing steps including deconvolution and deisotoping. A glycopeptide search space corresponding to the theoretical glycopeptides is defined using a combination of proteomics and glycomics databases and experimental data. The compositions of glycopeptides detected in the MS dimension are assigned. Glycopeptide tandem mass spectra are assigned by determining the mass of the peptide and glycan portions of the glycopeptides and searching peptide backbone product ions using a proteomics engine. Scoring confidence for glycopeptides is calculated using rigorous bioinformatics principles.
6. Database search methods for glycoproteomics
Because glycopeptides are conjugates containing both peptide and glycan, it is necessary to make assumptions regarding the protein(s) and glycan(s) present in the sample. Thus, for a purified or recombinant glycoprotein, it is common practice to assume a single, known, protein sequence. With regard to the glycan, a list curated from a glycomics database is often used. The task then is to assign the peptide sequence and glycan composition from the data using these assumptions.
An idealized workflow for assigning site-specific glycoprotein glycosylation is shown in Figure 2. As with the most widely used proteomics workflows, the data are produced using LC-MS. In order to make the workflow vendor neutral, the data are converted into a public format using tools including ProteoWizard [47] or OpenMS [48]. Next, it is necessary to conduct MS preprocessing steps including deconvolution and deisotoping. For this purpose, the DeconTools program is publically available [49,50]. The user must also define glycopeptide search space that defines the range of theoretical glycopeptides. This search space can be based entirely on assumptions regarding the protein and its degree of purity and the range of glycans present in reference to a public database. A better choice is to refine the search space using proteomics and glycomics data acquired on the sample. The search space is used to assign the compositions of glycopeptides detected in the MS data. Even with exact masses, it is not possible to assign these compositions unambiguously [8]. Tandem mass spectral data must then be pre-processed and then assigned with reference to the glycopepide search space. To do this, the peptide mass is determined from the tandem mass spectral data and peptide backbone product ions are used to identify the peptide. Finally, the confidence of the glycopeptide assignment is calculated. At the present time, a consensus regarding the best methods for assigning confidence have yet to emerge.
The following is a detailed discussion of aspects of a glycoproteomics workflow:
-
a.
identification of the glycopeptides in a complex proteolytic peptide mixture
Fortunately, glycopeptides dissociate to form signature low m/z oxonium ions corresponding to mono-, di-, or oligosaccharides during collisional heating [51,52], enabling the use of precursor ion scans to identify glycopeptides in LC-MS data. Therefore, the presence of oxonium ions in a collisional tandem mass spectrum is diagnostic for a glycopeptide and can be used to trigger subsequent activated electron dissociation [41]. Another approach that leverages high resolution, high mass accuracy MS is to use a mass defect classifier to selectively differentiate glycopeptides from non-glycopeptides [53].
A significant subset of investigators prefer to perform glycopeptide analysis on tryptic digests without glycopeptide enrichment [54,55]. The advantage to this approach is the simplicity of the workup. The disadvantage is that unglycosylated tend to suppress ionization of glycosylated peptides; because data dependent dissociation algorithms favor analysis of the most abundant precursor ions, selection of glycopeptides will be disfavored. Several methods for enrichment of glycopeptides are available [1]. These include chromatographic or solid-phase enrichment based on increased size or hydrophilicity of glycopeptides relative to non-glycosylated peptides. Multiple lectin affinity chromatography has been used to enrich glycoproteins or glycopeptides for proteomics [56,57]. Enrichment provides the advantage that the majority of peptides detected are glycosylated, making use of data dependent tandem MS more straightforward.
-
b.
determination of the peptide mass from the tandem mass spectrum and identification of the possible peptide sequences
As mentioned above, the appearance of glycopeptide collisional tandem mass spectra depends on whether resonant or non-resonant dissociation is used. In either case, users can with care develop data acquisition methods in which a series of monosaccharide losses is observed from the precursor ion. This will result in product ions corresponding to a series of peptide + monosaccharide units, for which the peptide mass can be calculated reliably [58,37]. The peptide mass can then be used to limit the range of peptide sequences and facilitate database search. For ExD tandem mass spectra, abundant peptide backbone product ions are observed but the determination of the mass of the peptide versus glycan portions of the glycopeptide are not as straightforward as for collisional dissociation; typically there is a mass shift observed in the peptide sequence spanning the glycosylation site which in some cases be used to infer glycan mass.
-
c.
Calculation of corresponding glycan mass and monosaccharide composition
Once the mass of the peptide portion of the glycopeptide has been determined, calculation of glycan composition depends on assumptions regarding the class of glycosylation, organism, and database used. Because databases contain a large number of glycan compositions, investigators often reduce this number by manual curation based on knowledge of the biological system in question. Even with the relatively limited number of monosaccharides (compared with the 20 amino acids for proteins), and limited glycome size, ambiguous glycan compositions exist. It is common practice to cull a list of glycans from a glycomics database and manually curate according to the needs of the project in question. Databases in common use at the present time include Glycome DB [59], Unicarb [60–62] and GlyTouCan [63]
-
d.
Methods for evaluation of the statistical significance of proteomics data: lessons for glycoproteomics
As used in proteomics [64], false positive rate (FPR) is the probability that the score for a tandem mass spectrum exceeds the threshold to match that of a random peptide-spectrum match (PSM). False discovery rate (FDR) is the proportion of peptide sequence matches (PSMs) that are incorrect. In order to solve the problem that proteomics search engines return results for even unmatchable tandem mass spectra, the target-decoy approach (TDA) [65,66] was developed. Now the standard for validating database search results, this method distinguishes correct from incorrect identifications through the use of a decoy database, often constructed by reversing the target protein sequences, to estimate the likelihood of a spurious match. Essentially, TDA allows calculation of the hit number in the decoy versus target database. Thus, the more peptides in the dataset, the better the estimation of false discovery rate; as the number of peptides in the dataset decreases, so too does the accuracy of the estimation of the FDR [64].
Pevzner et al argue against use of TDA for calculation of FDR [67,64]. TDA is an approximation of a permutation test as a simulation of tandem mass spectra, used to test for spurious matches in high throughput experiments based on the number of permutations that are consistent with the observed case. It makes assumptions about the probability of false positives under a particular score from a given test. This black-box approach leads to concern about inappropriate use of TDA with scoring functions that violate the assumptions of TDA. The problem is that TDA does not determine the statistical significance of individual PSMs, thus it is possible for a PSM with a poor score to fall above the threshold for true positives. PSM-level false positive rates (FPRs) do not suffer from such limitations of TDA-based estimation of FDR.
Thus, TDA is not appropriate for evaluating individual peptide identifications and this is a problem given the need to characterize individual tandem mass spectra. The use of these more granular FPRs to calculate FDR is more rigorous.
In proteomics, peptide identification statistics are used to evaluate confidence in the discovery of individual peptides. Often these results are presented in the form of a p-value or probability under a null hypothesis. The less likely an outcome under the null hypothesis, the more significant the result; however, significant results are important if and only if the null hypothesis is well designed and appropriate to the experiment performed. It is still possible for a seemingly significant result to be true under the null hypothesis by random chance. At present, there are several schemes for formulating a null hypothesis in proteomics; however, not all are universally appropriate or rigorously defined.
In statistics, the FDR is usually estimated as a function of the number of false positives compared to the total number of positive outcomes. Despite the fact that in proteomics the term FDR appears to have become synonymous with TDA, the use of p-values would improve the extent to which biologically significant results could be extracted from proteomics data sets. The q-value calculation method developed by Storey [68] produces an alternative statistic based on the p-value distribution of an experiment and the false positive rate that describes the minimum false discovery rate for significance of a given result. This is the standard approach used in genomics. It is also the foundation of the proteomics tools MS-GF [67,69] and Percolator [70,17]. Unfortunately, these methods require modification in order to be applicable to glycoproteomics.
The use of FPR for glycoproteomics would allow calculation of p-values for individual glycopeptide tandem mass spectra, giving rise to a rigorous basis for differentiating true and false positive identifications. The use of p-value would allow direct assessment of the data validity in a manner that does not depend on the definition of a decoy database. Each glycopeptide glycoform in some ways resembles the existence of rare PTM-modified peptides in that each tandem mass spectrum requires individual evaluation that is best accomplished by calculating spectrum-level p-values. Unfortunately, some of the most widely used proteomics search engines (for example Sequest and Mascot) do not allow calculation of such statistics [64].
-
e.
Statistical methods used by glycoproteomics search algorithms
It is important to consider carefully the assumptions made by glycoproteomics algorithms. While the need for statistical rigor may not be apparent immediately, the ability to define confidence of assignments in a consistent manner is lacking in the glycoproteomics field. Consider the assumptions regarding the number of proteins/glycoproteins present in a sample and the glycans present. Such samples should be considered as proteomes that contain a range of common protein modifications resulting from glycosylation, other PTMs, oxidations, deamidations and others. Processing such samples consisting of populations of proteoforms results in proteolytic peptide variants. Thus, the sample complexity is higher than often assumed, requiring statistical rigor. With regard to the glycan portion, the glycome distribution is extracted from a glycomics database. The question regarding the reasonable glycans to consider often relies on manual curation based on expert knowledge and/or preferences. In direct analogy to the methods used for smaller PTMs, glycopeptide tandem mass spectra can be scored using standard proteomics database searching algorithms by extracting the unmodified peptide mass and removing the ions corresponding to losses of saccharides from the precursor ion.
-
f.
Summary of glycopeptide tandem MS analysis algorithms
The following is a summary of software tools for interpretation of intact glycopeptide mass spectral data. Approaches that analyze deglycosylated peptides using proteomics approaches are not included.
Software tools that calculate glycopeptide composition from MS-only data will not be summarized in detail. These include: GlycoMod [71], GlycoX [72], Glyco peakfinder [73], GlycoPep DB [74], GlycoMiner [75], and GlycoSpectrumScan [76],
Software tools that interpret glycopeptide tandem mass spectra
Sweet substitute[77] creates theoretical deconvoluted and deisotoped collisional dissociation glycopeptide QTOF-type tandem mass spectra against which actual data may be compared. The authors acknowledged that peak height modeling for ions produced by glycan dissociation of glycopeptide precursor ions is challenging due to the difficulty in standardizing acquisition parameters necessary for reproducible product ion abundances.
GlyDB[78] assigns glycopeptides using low resolution CID tandem mass spectra. A glycan database is converted into a linear notation to allow searching of the tandem mass spectra using the Sequest proteomics search engine [12].
Peptoonist [79] uses a proteomics database engine to assign unmodified peptides in a digest and then assigns glycopeptides by fitting the stable isotope cluster and/or tandem mass spectra. The algorithm recalibrates the MS data in order to achieve optimal mass accuracy. Glycopeptide tandem mass spectra are scored against a glycopeptide search space constructed from the assumed peptide sequence and a set of biosynthetically reasonable N-glycans. The authors state that while it would be ideal to have a scoring system based on rigorous statistical principles using known glycopeptides, this was not possible due to the paucity of high quality validated data. In lieu of this, they used an informal procedure whereby envelope quality and mass match were transformed into a score using a logistic function.
Medicel N-glycopeptide library [80,81] is intended to assign glycopeptides from complex biological mixtures using deconvoluted glycopeptide tandem mass spectra as the input. The algorithm calculates theoretical glycopeptides from the UniProt [82] database, then groups the glycopeptide tandem mass spectra, and matches them against the theoretical glycopeptide library. The algorithm specifies that tandem mass spectra must contain a peptide + HexNAc ion so as to calculate the respective masses of the peptide and glycan. The algorithm then calculates theoretical glycan compositions and attempts to match theoretical versus observed glycan fragmentation, assuming there is no peptide fragmentation. The algorithm uses a target decoy database generated using reversed peptide sequences that contain the N-glycosylation sequon. The algorithm calculates the probability that a random set of product ions matches the measured tandem mass spectrum in comparison to the product ions calculated for a given candidate glycopeptide. This scoring method resembles the Ascore used to describe confidence in localization of post-translational modifications in proteomics [83].
GlyPID [84,85] clusters glycopeptides in a reversed phase LC-MS dataset based on observed series of masses differing by monosaccharide units in MS data. It then scores tandem mass spectra on these ions. The algorithm assigns glycopeptide monoisotopic ions and charge states using a method to filter isotopic clusters for consistency. The algorithm scores each cluster of co-eluting glycopeptides using MS and tandem MS data. This relative significance score is converted into a probability score (P-value) according to a normal distribution.
GlycoPeptideSearch [86] assigns glycopeptide structure from collisional dissociation tandem mass spectra. The algorithm searches tandem mass spectra for oxonium ions, then checks for product ions that correspond to peptide with up to three monosaccharide residues attached. The mass of the glycan calculated from the tandem mass spectra is then searched against a set of glycans extracted from GlycomeDB [59]. They calculate FDR using the ratio of possible matches for a given spectrum against structures in the database.
GlycoPep grader [87] calculates theoretical glycopeptide compositions from the target glycoprotein sequences and a set of theoretical glycan compositions. Glycopeptide matches were verified against manual assignments. The user inputs candidate glycopeptide compositions generated using GlycoMod [71] or GlycoPeb DB [74] and the algorithm scores these against the tandem MS data. The algorithm generates an FDR value using a decoy database consisting of a set of glycopeptides with neutral mass values within 50 ppm of the measured accurate masses.
GlycoPep detector [88] assigns glycopeptide structure based on ETD tandem mass spectra. Users generate candidate glycopeptides in the same manner as described for GlycoPep grader. The algorithm calculates the theoretical m/z values for c-, z- and y-ions for each candidate glycopeptide, and searches against the ETD tandem mass spectra and assigns a final score. False discovery rate is calculated using a decoy database in the manner described for GlycoPep detector.
GlycoPep Evaluator [89] was developed to address the problem that target-decoy analysis is not appropriate for relatively small datasets, such as those for many glycopeptide samples. This algorithms generates decoy glycopeptides to facilitate determination of FDR. The program calculates 20 mock glycopeptide compositions with masses isobaric to the true glycopeptide and uses them to estimate the false discovery rate for ETD tandem mass spectra.
GlycoFragwork [90] combines label free quantification and glycopeptide identification. The algorithm uses collisional dissociation data to identify the glycan portion and ETD data to identify the peptide. The algorithm uses a TDA approach for the peptide identification.
Sweet-Heart [91] accepts low resolution, low mass accuracy ion-trap tandem mass spectra. Results are used to drive subsequent rounds of MS3 dissociation to determine the peptide backbone. Reasoning that peptide backbone product ions would be more abundant, these investigators have developed a workflow that includes an MS3 step of the peptide+HexNAc (Y1) ion [92]. This workflow utilizes the ability of the Thermo-Fisher™ Fusion™ Tribrid to use HCD data to trigger CID in the ion trap and ETD. The ion trap tandem mass spectra were analyzed using Sweet-Heart modified the identification of the peptide+HexNAc (Y1) ion. The HCD and ETD data were processed using Byonic (see below).
GP Finder [55] identifies glycopeptides from deconvoluted, deisotoped collisional dissociation tandem mass spectra acquired for glycopeptides generated using non-specific proteases. The algorithm uses the deconvoluted mass list and filters for diagnostic oxonium ions according to self-consistency among results. The algorithm was used with a decoy strategy in which an 11 Da residue was added to each theoretical glycan composition, thereby preserving the true peptide sequences so as to model effectively matches to peptide and peptide+monosaccharide peaks. Target and decoy libraries were searched against fabricated tandem mass spectra generated by adding 11 Da to each of the product ions. Scores are generated by calculating a base score reflecting how well a candidate glycopeptide matches the tandem mass spectrum, followed by a boost score from self-consistency in the data and adjusting for target-decoy bias.
SweetSEQer [93] uses dynamic programming to build sequentially a path for the peptide sequence and a directed subgraph for the glycan of glycopeptides. Essentially, the algorithm automates the process of assigning product ions resulting from glycan dissociation, allowing users to more rapidly identify glycopeptides from sets of tandem mass spectra.
GlycoMaster DB [94] searches a protein sequence database and a glycan database to identify the peptide-glycan pair that best matches the input tandem mass spectra generated by collisional or activated electron dissociation or mixed dissociation modes. Users provide a list of sequences for the proteins in the sample (glycosylated and unglycosylated) to be considered by the algorithm. The algorithm first filters the tandem mass spectra based on presence of oxonium ions and high m/z ladders from monosaccharide losses to eliminate non-glycosylated peptide tandem mass spectra. The algorithm then assigns the best matching glycan to the observed tandem mass spectrum from a glycan database. The glycan-spectrum match works similarly to those used for peptide identification in which matches to theoretical product ion m/z values are assigned an award or penalty value. The product ions are scored for glycan sequence matches and peptide sequence matches to produce a raw score reflecting the sum of the individual peak scores. If ETD tandem mass spectra are available, the algorithm identifies the peptide from the product ion pattern. If ETD data are not available, the algorithm does not identify the peptide sequence. The investigators used manual interpretation to identify an empirical cutoff for glycan and peptide sequence matches, respectively.
The MAGIC algorithm [37] automates the analysis of glycopeptide tandem mass spectra generated on beam-type (QTOF) instruments using collisional dissociation. The algorithm requires neither a glycosylated protein sequences nor a glycan database. It calculates an in silico deglycoysylated peptide tandem mass spectrum to facilitate use of a proteomics database search to identify the peptide. The algorithm generates a new mascot generic format (MGF) corresponding to the deglycosylated peptide that is used to search a proteomics database. The glycan composition is then assigned using the calculated glycan mass and a lookup table of theoretical compositions.
Commercial software
SimGlycan™ (Premier Biosoft) [95] predicts glycan structure from tandem mass spectra using a database of theoretical fragmentation. The algorithm can be applied to tandem mass spectra for glycopeptides in which the peptide mass is known.
Byonic™ (Protein Metrics, Inc) [96] is a proteomics search engine for discovery research that allows the user to define an unlimited number of modification types. The algorithm is designed to identify glycopeptides without prior knowledge of glycan mass or glycosylation sites. The published version allowed one glycosylation per peptide and uses internal tables of glycans to analyze the tandem mass spectra.
7. Conclusions
While progress has been made in developing approaches for analysis of glycopeptide tandem mass spectra, the methods are still maturing. Even in the more mature field of proteomics, statistically rigorous scoring methods have yet to be adopted widely. As a result, there remains a degree of uncertainty regarding the correctness of glycopeptide assignments that appear in the literature. This uncertainty arises from the underlying complexity of glycoprotein samples. In the first place, it is important to establish sample purity rigorously; when contaminant proteins are present, these should be taken into consideration in the data analysis. Secondly, the complexity of glycosylation is multiplied by the presence of other modifications, including oxidations, dehydrations, deamidations, and missed proteolytic cleavage sites. These modifications should also be considered in analysis of glycoproteomics data. Thirdly, the field needs to move towards statistically rigorous methods for estimating FPR and FDR; the datasets differ fundamentally from those used in proteomics and are likely to require tailored approaches.
Acknowledgements
The authors are supported by NIH grant P41GM104603. Thermo-Fisher Scientific provided instrument time for the acquisition of tandem mass spectra shown in Figure 1.
References cited
- 1.Wuhrer M, Deelder AM, Hokke CH. Protein glycosylation analysis by liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;825(2):124–133. doi: 10.1016/j.jchromb.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 2.Pan S, Chen R, Aebersold R, Brentnall TA. Mass spectrometry based glycoproteomics--from a proteomics perspective. Molecular & cellular proteomics : MCP. 2011;10(1) doi: 10.1074/mcp.R110.003251. R110 003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodin CL, Maxon M, Desaire H. Software for automated interpretation of mass spectrometry data from glycans and glycopeptides. Analyst. 2013;138(10):2793–2803. doi: 10.1039/c2an36042j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leymarie N, Zaia J. Effective use of mass spectrometry for glycan and glycopeptide structural analysis. Anal. Chem. 2012;84(7):3040–3048. doi: 10.1021/ac3000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dallas DC, Martin WF, Hua S, German JB. Automated glycopeptide analysis--review of current state and future directions. Briefings in bioinformatics. 2013;14(3):361–374. doi: 10.1093/bib/bbs045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F, Glinskii OV, Glinsky VV. Glycobioinformatics: Current strategies and tools for data mining in MS-based glycoproteomics. Proteomics. 2013;13(2):341–354. doi: 10.1002/pmic.201200149. [DOI] [PubMed] [Google Scholar]
- 7.Tang H, Mayampurath A, Yu CY, Mechref Y. Bioinformatics protocols in glycomics and glycoproteomics. Curr Protoc Protein Sci. 2014;76 doi: 10.1002/0471140864.ps0215s76. 2 15 11-17. [DOI] [PubMed] [Google Scholar]
- 8.Desaire H, Hua D. When can glycopeptides be assigned based solely on high-resolution mass spectrometry data? Int J Mass Spectrom. 2009;287(1–3):21–26. [Google Scholar]
- 9.Biemann K, Gapp F, Seibl J. Application of mass spectrometry to structure problems. I. Amino acid sequence in peptides. J. Am. Chem. Soc. 1959;81:2274. [Google Scholar]
- 10.Biemann K, Martin SA. Mass spectrometric determination of the amino acid sequence of peptides and proteins. Mass Spectrom Rev. 1987;6(1):1–76. [Google Scholar]
- 11.Meng CK, Mann M, Fenn JB. Of protons or proteins. Z. Phys. D. 1988;10:361–368. [Google Scholar]
- 12.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 13.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 1994;66(24):4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 14.Johnson RS, Biemann K. Computer program (SEQPEP) to aid in the interpretation of high-energy collision tandem mass spectra of peptides. Biomed Environ Mass Spectrom. 1989;18(11):945–957. doi: 10.1002/bms.1200181102. [DOI] [PubMed] [Google Scholar]
- 15.Frank A, Pevzner P. PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 2005;77(4):964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, Ma B. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics. 2012;11(4) doi: 10.1074/mcp.M111.010587. M111 010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong K, Kim S, Pevzner PA. UniNovo: a universal tool for de novo peptide sequencing. Bioinformatics. 2013;29(16):1953–1962. doi: 10.1093/bioinformatics/btt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore RE, Young MK, Lee TD. Protein identification using a quadrupole ion trap mass spectrometer and SEQUEST database matching. Curr Protoc Protein Sci. 2001 doi: 10.1002/0471140864.ps1610s22. Chapter 16, Unit 16 10. [DOI] [PubMed] [Google Scholar]
- 19.Clauser KR, Baker P, Burlingame AL. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 1999;71(14):2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 20.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 22.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open Mass Spectrometry Search Algorithm. J. Proteome Res. 2004;3(5):958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 23.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6(2):654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 25.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13(1):22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 26.Wenger CD, Coon JJ. A proteomics search algorithm specifically designed for high-resolution tandem mass spectra. J Proteome Res. 2013;12(3):1377–1386. doi: 10.1021/pr301024c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorfer V, Pichler P, Stranzl T, Stadlmann J, Taus T, Winkler S, Mechtler K. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res. 2014;13(8):3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10(12) doi: 10.1074/mcp.M111.007690. M111 007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaudel M, Barsnes H, Berven FS, Sickmann A, Martens L. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics. 2011;11(5):996–999. doi: 10.1002/pmic.201000595. [DOI] [PubMed] [Google Scholar]
- 30.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11(6):427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 31.Wuhrer M, Deelder AM, van der Burgt YE. Mass spectrometric glycan rearrangements. Mass Spectrom Rev. 2011;30(4):664–680. doi: 10.1002/mas.20337. [DOI] [PubMed] [Google Scholar]
- 32.Reinhold VN, Sheeley DM. Detailed characterization of carbohydrate linkage and sequence in an ion trap mass spectrometer: glycosphingolipids. Anal Biochem . 1998;259(1):28–33. doi: 10.1006/abio.1998.2619. [DOI] [PubMed] [Google Scholar]
- 33.Sheeley DM, Reinhold VN. Structural characterization of carbohydrate sequence linkage, and branching in a quadrupole Ion trap mass spectrometer: neutral oligosaccharides and N-linked glycans. Anal. Chem. 1998;70(14):3053–3059. doi: 10.1021/ac9713058. [DOI] [PubMed] [Google Scholar]
- 34.Khatri K, Staples GO, Leymarie N, Leon DR, Turiák L, Huang Y, Yip S, Hu H, Heckendorf CF, Zaia J. Confident Assignment of Site-Specific Glycosylation in Complex Glycoproteins in a Single Step. J. Proteome Res. 2014;13(10):4347–4355. doi: 10.1021/pr500506z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An Y, Cipollo JF. An unbiased approach for analysis of protein glycosylation and application to influenza vaccine hemagglutinin. Anal Biochem. 2011;415(1):67–80. doi: 10.1016/j.ab.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 36.An Y, Rininger JA, Jarvis DL, Jing X, Ye Z, Aumiller JJ, Eichelberger M, Cipollo JF. Comparative glycomics analysis of influenza Hemagglutinin (H5N1) produced in vaccine relevant cell platforms. J. Proteome Res. 2013;12(8):3707–3720. doi: 10.1021/pr400329k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynn KS, Chen CC, Lih TM, Cheng CW, Su WC, Chang CH, Cheng CY, Hsu WL, Chen YJ, Sung TY. MAGIC: an automated N-linked glycoprotein identification tool using a Y1-ion pattern matching algorithm and in silico MS(2) approach. Anal. Chem. 2015;87(4):2466–2473. doi: 10.1021/ac5044829. [DOI] [PubMed] [Google Scholar]
- 38.Håkansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Electron capture dissociation and Infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptide to yield complementary sequence information. Anal. Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 39.Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J Proteome Res. 2005;4(2):628–632. doi: 10.1021/pr049770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mechref Y. Use of CID/ETD mass spectrometry to analyze glycopeptides. Curr Protoc Protein Sci. 2012 doi: 10.1002/0471140864.ps1211s68. Chapter 12, Unit 12 11 11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh C, Zampronio CG, Creese AJ, Cooper HJ. Higher energy collision dissociation (HCD) product ion-triggered electron transfer dissociation (ETD) mass spectrometry for the analysis of N-linked glycoproteins. J Proteome Res. 2012;11(9):4517–4525. doi: 10.1021/pr300257c. [DOI] [PubMed] [Google Scholar]
- 42.Myers SA, Daou S, Affar el B, Burlingame A. Electron transfer dissociation (ETD): the mass spectrometric breakthrough essential for O-GlcNAc protein site assignments-a study of the O-GlcNAcylated protein host cell factor C1. Proteomics. 2013;13(6):982–991. doi: 10.1002/pmic.201200332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horn DM, Ge Y, McLafferty FW. Activated ion electron capture dissociation for mass spectral sequencing of larger (42 kDa) proteins. Anal. Chem. 2000;72(20):4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 44.Frese CK, Altelaar AF, van den Toorn H, Nolting D, Griep-Raming J, Heck AJ, Mohammed S. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 2012;84(22):9668–9673. doi: 10.1021/ac3025366. [DOI] [PubMed] [Google Scholar]
- 45.Madsen JA, Ko BJ, Xu H, Iwashkiw JA, Robotham SA, Shaw JB, Feldman MF, Brodbelt JS. Concurrent automated sequencing of the glycan and peptide portions of O-linked glycopeptide anions by ultraviolet photodissociation mass spectrometry. Anal. Chem. 2013;85(19):9253–9261. doi: 10.1021/ac4021177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ko BJ, Brodbelt JS. Comparison of Glycopeptide Fragmentation by Collision Induced Dissociation and Ultraviolet Photodissociation. Int J Mass Spectrom. 2015;377(1):385–392. doi: 10.1016/j.ijms.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinert K, Kohlbacher O. OpenMS and TOPP: open source software for LC-MS data analysis. Methods Mol Biol. 2010;604:201–211. doi: 10.1007/978-1-60761-444-9_14. [DOI] [PubMed] [Google Scholar]
- 49.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics. 2009;10:87. doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slysz GW, Baker ES, Shah AR, Jaitly N, Anderson GA, Smith RD. The DeconTools Framework: an Application Programming Interface Enabling Flexibility in Accurate Mass and Time Tag Workflows for Proteomics and Metabolomics. Proc. 58th ASMS Conf. Mass Spectrom. Allied Topics. 2010 [Google Scholar]
- 51.Huddleston MJ, Bean MF, Carr SA. Collisional fragmentation of glycopeptides by electrospray ionization LC/MS and LC/MS/MS: methods for selective detection of glycopeptides in protein digests. Anal. Chem. 1993;65(7):877–884. doi: 10.1021/ac00055a009. [DOI] [PubMed] [Google Scholar]
- 52.Carr SA, Huddleston MJ, Bean MF. Selective identification and differentiation of N-and O-linked oligosaccharides in glycoproteins by liquid chromatography-mass spectrometry. Protein Sci. 1993;2(2):183–196. doi: 10.1002/pro.5560020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Froehlich JW, Dodds ED, Wilhelm M, Serang O, Steen JA, Lee RS. A classifier based on accurate mass measurements to aid large scale, unbiased glycoproteomics. Mol Cell Proteomics. 2013;12(4):1017–1025. doi: 10.1074/mcp.M112.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Go EP, Liao HX, Alam SM, Hua D, Haynes BF, Desaire H. Characterization of host-cell line specific glycosylation profiles of early transmitted/founder HIV-1 gp120 envelope proteins. J. Proteome Res. 2013;12(3):1223–1234. doi: 10.1021/pr300870t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strum JS, Nwosu CC, Hua S, Kronewitter SR, Seipert RR, Bachelor RJ, An HJ, Lebrilla CB. Automated Assignments of N- and O-Site Specific Glycosylation with Extensive Glycan Heterogeneity of Glycoprotein Mixtures. Anal. Chem. 2013;85(12):5666–5675. doi: 10.1021/ac4006556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plavina T, Wakshull E, Hancock WS, Hincapie M. Combination of abundant protein depletion and multi-lectin affinity chromatography (M-LAC) for plasma protein biomarker discovery. J. Proteome Res. 2007;6(2):662–671. doi: 10.1021/pr060413k. [DOI] [PubMed] [Google Scholar]
- 57.Madera M, Mechref Y, Novotny MV. Combining lectin microcolumns with high-resolution separation techniques for enrichment of glycoproteins and glycopeptides. Anal. Chem. 2005;77(13):4081–4090. doi: 10.1021/ac050222l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng K, Chen R, Seebun D, Ye M, Figeys D, Zou H. Large-scale characterization of intact N-glycopeptides using an automated glycoproteomic method. J Proteomics. 2014;110:145–154. doi: 10.1016/j.jprot.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Ranzinger R, Frank M, von der Lieth CW, Herget S. Glycome-DB.org: a portal for querying across the digital world of carbohydrate sequences. Glycobiology. 2009;19(12):1563–1567. doi: 10.1093/glycob/cwp137. [DOI] [PubMed] [Google Scholar]
- 60.Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, Lisacek F, Packer NH. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Res. 2014;42(1):D215–D221. doi: 10.1093/nar/gkt1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayes CA, Karlsson NG, Struwe WB, Lisacek F, Rudd PM, Packer NH, Campbell MP. UniCarb-DB: a database resource for glycomic discovery. Bioinformatics. 2011;27(9):1343–1344. doi: 10.1093/bioinformatics/btr137. [DOI] [PubMed] [Google Scholar]
- 62.Campbell MP, Hayes CA, Struwe WB, Wilkins MR, Aoki-Kinoshita KF, Harvey DJ, Rudd PM, Kolarich D, Lisacek F, Karlsson NG, Packer NH. UniCarbKB: putting the pieces together for glycomics research. Proteomics. 2011;11(21):4117–4121. doi: 10.1002/pmic.201100302. [DOI] [PubMed] [Google Scholar]
- 63.Aoki-Kinoshita K. GlyTouCan: the glycan repository. 2015 https://www.glytoucan.org/
- 64.Gupta N, Bandeira N, Keich U, Pevzner PA. Target-decoy approach and false discovery rate: when things may go wrong. J. Am. Soc. Mass Spectrom. 2011;22(7):1111–1120. doi: 10.1007/s13361-011-0139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 66.Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods in molecular biology. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 2008;7(8):3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Storey JD. A direct approach to false discovery rates. J. Roy. Stat. Soc. Ser. B. (Stat. Method.) 2002;64(3):479–498. [Google Scholar]
- 69.Kim S, Mischerikow N, Bandeira N, Navarro JD, Wich L, Mohammed S, Heck AJ, Pevzner PA. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol Cell Proteomics. 2010;9(12):2840–2852. doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kall L, Storey JD, Noble WS. Non-parametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics. 2008;24(16):42–48. doi: 10.1093/bioinformatics/btn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cooper CA, Gasteiger E, Packer NH. GlycoMod--a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1(2):340–349. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 72.An HJ, Tillinghast JS, Woodruff DL, Rocke DM, Lebrilla CB. A new computer program (GlycoX) to determine simultaneously the glycosylation sites and oligosaccharide heterogeneity of glycoproteins. J Proteome Res. 2006;5(10):2800–2808. doi: 10.1021/pr0602949. [DOI] [PubMed] [Google Scholar]
- 73.Maass K, Ranzinger R, Geyer H, von der Lieth CW, Geyer R. “Glyco-peakfinder”--de novo composition analysis of glycoconjugates. Proteomics. 2007;7(24):4435–4444. doi: 10.1002/pmic.200700253. [DOI] [PubMed] [Google Scholar]
- 74.Go EP, Rebecchi KR, Dalpathado DS, Bandu ML, Zhang Y, Desaire H. GlycoPep DB: a tool for glycopeptide analysis using a “Smart Search”. Anal. Chem. 2007;79(4):1708–1713. doi: 10.1021/ac061548c. [DOI] [PubMed] [Google Scholar]
- 75.Ozohanics O, Krenyacz J, Ludanyi K, Pollreisz F, Vekey K, Drahos L. GlycoMiner: a new software tool to elucidate glycopeptide composition. Rapid Commun Mass Spectrom. 2008;22(20):3245–3254. doi: 10.1002/rcm.3731. [DOI] [PubMed] [Google Scholar]
- 76.Deshpande N, Jensen PH, Packer NH, Kolarich D. GlycoSpectrumScan: fishing glycopeptides from MS spectra of protease digests of human colostrum sIgA. J Proteome Res. 2010;9(2):1063–1075. doi: 10.1021/pr900956x. [DOI] [PubMed] [Google Scholar]
- 77.Clerens S, Van den Ende W, Verhaert P, Geenen L, Arckens L. Sweet Substitute: a software tool for in silico fragmentation of peptide-linked N-glycans. Proteomics. 2004;4(3):629–632. doi: 10.1002/pmic.200300572. [DOI] [PubMed] [Google Scholar]
- 78.Ren JM, Rejtar T, Li L, Karger BL. N-Glycan structure annotation of glycopeptides using a linearized glycan structure database (GlyDB) J Proteome Res. 2007;6(8):3162–3173. doi: 10.1021/pr070111y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goldberg D, Bern M, Parry S, Sutton-Smith M, Panico M, Morris HR, Dell A. Automated N-glycopeptide identification using a combination of single- and tandem-MS. J Proteome Res. 2007;6(10):3995–4005. doi: 10.1021/pr070239f. [DOI] [PubMed] [Google Scholar]
- 80.Joenvaara S, Ritamo I, Peltoniemi H, Renkonen R. N-glycoproteomics - an automated workflow approach. Glycobiology. 2008;18(4):339–349. doi: 10.1093/glycob/cwn013. [DOI] [PubMed] [Google Scholar]
- 81.Peltoniemi H, Joenväärä S, Renkonen R. De novo glycan structure search with the CID MS/MS spectra of native N-glycopeptides. Glycobiology. 2009;19(7):707–714. doi: 10.1093/glycob/cwp034. [DOI] [PubMed] [Google Scholar]
- 82.UniProt C. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37(Database issue):D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 84.Wu Y, Mechref Y, Klouckova I, Mayampurath A, Novotny MV, Tang H. Mapping site-specific protein N-glycosylations through liquid chromatography/mass spectrometry and targeted tandem mass spectrometry. Rapid Commun Mass Spectrom. 2010;24(7):965–972. doi: 10.1002/rcm.4474. [DOI] [PubMed] [Google Scholar]
- 85.Mayampurath AM, Wu Y, Segu ZM, Mechref Y, Tang H. Improving confidence in detection and characterization of protein N-glycosylation sites and microheterogeneity. Rapid communications in mass spectrometry : RCM. 2011;25(14):2007–2019. doi: 10.1002/rcm.5059. [DOI] [PubMed] [Google Scholar]
- 86.Pompach P, Chandler KB, Lan R, Edwards N, Goldman R. Semi-automated identification of N-Glycopeptides by hydrophilic interaction chromatography, nano-reverse-phase LC-MS/MS, and glycan database search. J. Proteome Res. 2012;11(3):1728–1740. doi: 10.1021/pr201183w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Woodin CL, Hua D, Maxon M, Rebecchi KR, Go EP, Desaire H. GlycoPep grader: a web-based utility for assigning the composition of N-linked glycopeptides. Anal. Chem. 2012;84(11):4821–4829. doi: 10.1021/ac300393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu Z, Hua D, Clark DF, Go EP, Desaire H. GlycoPep Detector: a tool for assigning mass spectrometry data of N-linked glycopeptides on the basis of their electron transfer dissociation spectra. Anal. Chem. 2013;85(10):5023–5032. doi: 10.1021/ac400287n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu Z, Su X, Go EP, Desaire H. New Glycoproteomics Software, GlycoPep Evaluator, Generates Decoy Glycopeptides de Novo and Enables Accurate False Discovery Rate Analysis for Small Data Sets. Anal. Chem. 2014;86(18):9212–9219. doi: 10.1021/ac502176n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mayampurath A, Yu CY, Song E, Balan J, Mechref Y, Tang H. Computational framework for identification of intact glycopeptides in complex samples. Anal. Chem. 2014;86(1):453–463. doi: 10.1021/ac402338u. [DOI] [PubMed] [Google Scholar]
- 91.Wu SW, Liang SY, Pu TH, Chang FY, Khoo KH. Sweet-Heart - an integrated suite of enabling computational tools for automated MS2/MS3 sequencing and identification of glycopeptides. J Proteomics. 2013;84:1–16. doi: 10.1016/j.jprot.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 92.Wu SW, Pu TH, Viner R, Khoo KH. Novel LC-MS(2) product dependent parallel data acquisition function and data analysis workflow for sequencing and identification of intact glycopeptides. Anal. Chem. 2014;86(11):5478–5486. doi: 10.1021/ac500945m. [DOI] [PubMed] [Google Scholar]
- 93.Serang O, Froehlich JW, Muntel J, McDowell G, Steen H, Lee RS, Steen JA. SweetSEQer, simple de novo filtering and annotation of glycoconjugate mass spectra. Molecular & cellular proteomics : MCP. 2013;12(6):1735–1740. doi: 10.1074/mcp.O112.025940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.He L, Xin L, Shan B, Lajoie GA, Ma B. GlycoMaster DB: software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry. J Proteome Res. 2014;13(9):3881–3895. doi: 10.1021/pr401115y. [DOI] [PubMed] [Google Scholar]
- 95.Apte A, Meitei NS. Bioinformatics in glycomics: glycan characterization with mass spectrometric data using SimGlycan. Methods Mol Biol. 2010;600:269–281. doi: 10.1007/978-1-60761-454-8_19. [DOI] [PubMed] [Google Scholar]
- 96.Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Current protocols in bioinformatics. 2012:13–20. doi: 10.1002/0471250953.bi1320s40. Chapter 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crouch E, Nikolaidis N, McCormack F, McDonald B, Allen K, Rynkiewicz M, Cafarella T, White M, Lewnard K, Leymarie N, Zaia J, Seaton B, Hartshorn K. Mutagenesis of SP-D informed by evolution and xray crystallography enhances defenses against Influenza A Virus in vivo. J Biol Chem. 2011;286(47):40681–40692. doi: 10.1074/jbc.M111.300673. [DOI] [PMC free article] [PubMed] [Google Scholar]