Abstract
Macrophages are pleiotropic cells capable of performing a broad spectrum of functions. Macrophage phenotypes are classified along a continuum between the extremes of proinflammatory M1 macrophages and anti-inflammatory M2 macrophages. The seemingly opposing functions of M1 and M2 macrophages must be tightly regulated for an effective and proper response to foreign molecules or damaged tissue. Excessive activation of either M1 or M2 macrophages contributes to the pathology of many diseases. Emodin is a Chinese herb-derived compound and has shown potential to inhibit inflammation in various settings. In this study, we tested the ability of emodin to modulate the macrophage response to both M1 and M2 stimuli. Primary mouse macrophages were stimulated with LPS/IFNγ or IL4 with or without emodin, and the effects of emodin on gene transcription, cell signaling pathways, and histone modifications were examined by a variety of approaches, including microarray, quantitative real-time PCR, Western blotting, chromatin immunoprecipitation, and functional assays. We found that emodin bidirectionally tunes the induction of LPS/IFNγ- and IL4-responsive genes through inhibiting NFκB/IRF5/STAT1 signaling and IRF4/STAT6 signaling, respectively. Thereby, emodin modulates macrophage phagocytosis, migration, and NO production. Furthermore, emodin inhibited the removal of H3K27 trimethylation (H3K27m3) marks and the addition of H3K27 acetylation (H3K27ac) marks on genes required for M1 or M2 polarization of macrophages. In conclusion, our data suggest that emodin is uniquely able to suppress the excessive response of macrophages to both M1 and M2 stimuli and therefore has the potential to restore macrophage homeostasis in various pathologies.
Keywords: cell signaling, histone modification, inflammation, innate immunity, macrophage, emodin, macrophage memory, polarization
Introduction
Macrophages are a heterogeneous population of innate immune cells found in most tissues of the body (1, 2). They are capable of displaying a variety of functional phenotypes along a broad spectrum, at the extremes of which are classically activated M1 and alternatively activated M2 macrophages (3, 4). Macrophages are induced to a proinflammatory M1 state by Th1 cytokines (such as IFNγ and TNFα) and bacterial products (such as LPS). M1 macrophages play major roles in host defense against bacteria or tissue remodeling after injury through production of proinflammatory cytokines (such as IL12, TNFα, and IL1), reactive oxygen species and NO, and proteases (such as MMP 2 and 9). IFNγ and TNFα activate the JAK/STAT cascade and lead to STAT1 activation, whereas LPS activates the NFκB and MAPK cascade upon ligation with TLR4 (4, 5). A combination of stimuli, including Th2 cytokines (such as IL4, IL10, and IL13), growth factors (such as TGFβ and CSF1), glucocorticoids, and immune complexes, can polarize macrophages toward an anti-inflammatory M2 phenotype (6). M2 macrophages have major roles in tissue homeostasis and repair, inflammation resolution, and immune regulation. M2 macrophages are characterized by high expression of Arg1, YM1, Mrc1, and IL10 and low expression of proinflammatory cytokines (7). IL4/IL13 signal through the common IL4ra receptor and lead to STAT6 phosphorylation and activation, which, along with several other secondary transcription factors, including IRF4, PPARγ,2 and KLF4, fine-tunes transcriptional responses in the cells (5, 8). Macrophage are also able to retain a memory for the signals to which they have been exposed through epigenetic modification, which results in increased transcription (priming) or repressed transcription (tolerance) upon future exposure (9–11). Cytokines cause the addition of the positive histone modifications H3K4m3 or H3K27ac to gene promoters that lead to increased expression. The mechanisms of tolerance are incompletely understood but could involve the loss of positive histone modifications and/or an increase in negative histone modifications (such as H3K27m3) (11). The different macrophage functional phenotypes need to be tightly controlled for a proper response to environmental stimuli.
An imbalance in macrophage phenotypes has been shown to contribute to the pathology of a large number of diseases (12–14). Chronic M1 macrophage activation promotes tissue damage in neurodegenerative disorders, arthritis, and autoimmune diseases (12, 15, 16). Although necessary for the initial stages of tissue repair, excessive M1 activation inhibits the healing of damaged tissue through excessive matrix degradation and inhibition of tissue regeneration (17, 18). Chronic M1 activation has also been shown to promote the development of cancer (19). Increased inflammatory monocyte/macrophage infiltration has been shown to correlate with disease severity for patients with myocardial infarction, atherosclerosis, and metabolic disorders (20–22). Shifting the balance toward M2 macrophages has shown to be beneficial in experimental models; however, prolonged or excessive M2 macrophage activation has also been shown to be detrimental. M2 macrophages contribute to lung inflammation and damage in allergy and asthma (13, 14). They have also been shown to impair tissue functions through promoting fibrosis (23). M2 macrophage infiltration correlates with increased cancer growth and metastasis in multiple types of cancer (14, 24). M2 macrophages promote cancer growth and metastasis by supporting extracellular matrix remodeling, angiogenesis, and immune suppression. Targeting macrophage-driven inflammation has shown benefits in many experimental disease models; however, there have been many challenges in developing therapies for clinical use (25). If treatment is administered too late during the disease, irreversible tissue damage can occur; therefore, targeting the macrophage/inflammatory component of disease would likely require chronic therapies administered early during disease progression. Furthermore, most current treatments target one molecule or pathway, but there are many redundant and compensatory pathways built into the inflammatory response.
There is much interest in herb-derived compounds because they can modulate multiple inflammatory pathways, are inexpensive, and have low toxicity for chronic treatment (26). Many compounds have shown potential to inhibit macrophage activation; however, experimental studies with these compounds haven been focused on response to stimuli that induce a narrow range of activation (either M1 or M2 phenotypes but not both) (26–29). A compound that could inhibit a broad range of macrophage phenotypes through regulation of multiple signaling pathways would have great clinical potential. Emodin is a trihydroxy-anthraquinone that is found in several Chinese herbs, including Rheum palmatum and Polygonum multiflorum (30). Emodin has been shown to attenuate the severity of experimental disease models, including arthritis, liver damage, atherosclerosis, myocardial ischemia, and cancer (30). Studies in our laboratory and others have shown that emodin has the potential to regulate both Th1- and Th2-driven inflammation (31–34), which indicates that emodin is able to regulate a broad spectrum of macrophage phenotypes.
In this study, we tested the ability of emodin to regulate both M1 and M2 macrophage activation. We stimulated primary mouse macrophages with LPS/IFNγ as an M1 stimulus or IL4 as a M2 stimulus and investigated the mechanism of action of emodin using a whole-genome microarray. Emodin was able to inhibit the change in expression of a large percentage of both M1- and M2-associated genes. Emodin inhibited the NFκB, IRF5, and STAT1 pathways following LPS and IFNγ stimulation and the STAT6 and IRF4 signaling pathways following IL4 stimulation. We demonstrated the effects of emodin on macrophage functions such as phagocytosis, migration, and NO production. Finally, our data showed, for the first time, that emodin is able to regulate macrophage memory by inhibiting changes in H3K27m3 and H3K27ac at the promoter regions of several key genes. Taken together, our data show that emodin has the ability to bidirectionally modulate macrophage activation by targeting multiple pathways.
Experimental Procedures
Peritoneal Macrophage Isolation and Culture
Three milliliters of 4% thioglycolate solution was injected intraperitoneally into 8- to 12-week-old C57BL/6 mice. Three days later, macrophages were collected by lavaging the peritoneal cavity with PBS. The cells were then resuspended in DMEM containing 10% FBS. After 2 h, non-adherent cells were washed away with PBS, and the macrophages were cultured overnight in serum-free DMEM. The macrophages were then treated with DMEM containing IL4 (10 ng/ml, BioAbChem Inc., Ladson, SC) or LPS (100 ng/ml, Sigma-Aldrich) and IFNγ (20 ng/ml, BioAbChem Inc.) with or without emodin. Emodin was purchased from Nanjing Langze Medicine and Technology Co. Ltd. (Nanjing, China) and dissolved in DMSO at a concentration of 10 mg/ml as a stock solution.
For macrophage memory experiments, macrophages were stimulated overnight with IL4, IFNγ, and/or emodin. Then the cells were washed three times with PBS and cultured for 2 or 5 days in DMEM with 2% FBS. The medium was changed every 2 days. The cells were then restimulated with IL4 or LPS for 6 h.
Microarray Analysis
Microarray analysis was carried out as described previously (35) with a few alterations. Macrophages were stimulated with IL4 for 6 h or LPS + IFNγ for 24 h with or without emodin (50 μm). Samples were prepared in biological replicates of four. Cells were lysed with Qiazol, and RNA was extracted using the miRNeasy kit (Qiagen, Valencia, CA). Next, an Agilent 2100 Bioanalyzer was used to determine the quality and quantity of the RNA. All RNA samples had an RNA integrity number (RIN) of 9.2 or higher. The RNA was amplified and labeled with the Agilent Low Input Quick Amp labeling kit according to the instructions of the manufacturer. Labeled RNA was then purified using the Qiagen RNeasy mini kit, and dye incorporation and cRNA yield were assessed. Labeled samples were hybridized to Agilent whole mouse genome microarrays 8 × 60,000 using a gene expression hybridization kit (Agilent) according to the instructions of the manufacturer. Microarray analysis was performed using an Agilent DNA microarray scanner system (catalog no. G2565CA).
A heatmap of genes from relevant pathways identified by Ingenuity pathway analysis was generated using R function heatmap.2. A principle component analysis was performed using all genes differentially expressed in at least one of the treatment groups using the R function prcomp.
Phagocytosis Assay
Macrophages were seeded into a 96-well plate (2 × 105 cells/well) and stimulated with IL4 or LPS + IFNγ for 24 h with or without emodin. Then phagocytotic activity was measured using a Vybrant phagocytosis assay kit (Molecular Probes). The medium was removed, and the cells were washed with PBS. The fluorescent bioparticle suspension was added to the cells and incubated up to 6 h. After the indicated time, the bioparticle suspension was removed, any extracellular fluorescence was quenched with trypan blue, and intracellular fluorescence was detected using a Spectramax M5 microplate reader (Molecular Devices, Sunnyvale, CA).
Macrophage Migration Assay
Macrophages were stimulated with IL4 or LPS + IFNγ for 24 h with or without emodin. The media was then removed, and the cells were washed with PBS. The cells were then resuspended in DMEM with scraping, and 2 × 105 macrophages were seeded in triplicate into the top chamber of transwell inserts with 8-μm pores (Corning) and placed in 24-well plates. Serum-free DMEM with 20 ng/ml MCP1 was placed in the bottom of the wells. After 4 h, inserts were fixed in 4% paraformaldehyde for 20 min. The cells in the upper inserts were swabbed using cotton buds, and the cells left on the membrane were stained with DAPI (1 μg/ml) for 1 min. The inserts were then cut out, mounted onto slides, and imaged under an Eclipse NI-U fluorescence microscope (Nikon Inc., Melville, NY) at ×200 magnification (5 fields/insert). DAPI-stained cells were quantified using Nikon NIS-Elements software.
NO Production Assay
Macrophages were stimulated with LPS + IFNγ for 24 h with various concentrations of emodin (0–50 μm). The culture media were then collected, and the NO content was detected using a nitrite/nitrate colorimetric kit (Sigma-Aldrich) according to the instructions of the manufacturer. The assay was read using a Spectramax M5 microplate reader (Molecular Devices).
Western Blotting and Real-time PCR
Following stimulation with IL4 or LPS + IFNγ with or without emodin for varying periods, macrophages were lysed with cell signaling lysis buffer (Millipore) for whole cell lysates. Cytoplasmic and nuclear extracts were prepared using the Epiquik nuclear extraction kit (Epigentek, Farmingdale, NY). Total protein was separated on 4–20% Tris/glycine precast gels (Pierce) and transferred onto nitrocellulose membranes (Bio-Rad). Antibodies are shown in supplemental Table 1. The membranes were then probed with HRP-conjugated secondary antibodies, and signals were detected using Pierce ECL Western blotting substrate (Pierce).
For qPCR, macrophages were lysed with Qiazol, and RNA was extracted using a Zymo Research Direct-zol RNA isolation kit. cDNA was then made from 1 μg of RNA using the iScript cDNA synthesis kit (Bio-Rad). Primers are listed in supplemental Table 2. Run conditions were 95 °C for 10 s, 58 °C for 15 s, and 70 °C for 15 s. Samples were run in duplicate on a Bio-Rad CFX real-time thermocycler.
ChIP Assay
Macrophages were stimulated with IL4 or LPS +IFNγ for 24 h with or without emodin. They were then fixed in 1% formaldehyde. Excess formaldehyde was quenched with glycine, and the cells were collected in PBS by scraping. The cells were lysed (0.5% IGEPAL, 4 mm HEPES), and the nuclei were resuspended in nuclear lysis buffer (1% SDS, 10 mm EDTA, and 50 mm Tris (pH 8.1)). The DNA was sheared by sonication using a Diagnode Bioruptor Pico for 15 cycles of 30 s on/30 s off. Then 10 μg of chromatin was diluted 1:10 (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl (pH 8.1), and 167 mm NaCl), and 2% of the input was removed from each sample and saved for analysis. Anti-H3K27m3 or anti-H3K27ac (Abcam) was added to each sample along with 20 μl of protein A+G magnetic beads (Millipore), and the samples were incubated overnight at 4 °C. The beads were washed with low-salt, high-salt, LiCl, and Tris-EDTA wash buffers sequentially, and the DNA was eluted off the beads with proteinase K at 62 °C for 2 h (elution buffer: 200 mm NaCl, 1% SDS, and 50 mm Tris). The DNA was then analyzed by real-time PCR using primers listed in supplemental Table 3.
Statistical Analysis
For the microarray analysis, data were extracted from images with Feature Extractor software version 10.7.3.1 (Agilent). Background correction using detrending algorithms was performed. Subsequently, background-corrected data were uploaded into GeneSpring GX version 11.5.1 for analysis. In this process, data were log2-transformed, quantile-normalized, and baseline-transformed using the median of all samples. Then data were filtered by flags in a way that three of the four biological replicates had a “detected” flag in at least one of the three treatment groups. Differentially expressed genes were determined by analysis of the data using Mann-Whitney unpaired statistics. A cutoff p value of 0.05 and a -fold change cutoff value of 2.0 were used to filter the data. Pathway analysis was performed using Ingenuity Pathway Analysis software.
For all other experiments, data were presented as mean ± S.E. Statistical significance was calculated by Student's t test (two-group comparison) using the GraphPad Prism statistical program (GraphPad Software Inc., San Diego, CA). p ≤ 0.05 was considered significant.
Results
Emodin Affects the Activation of Different Transcriptional Programs Depending on the Nature of the Stimuli
LPS + IFNγ and IL4 induce macrophages to adopt opposing proinflammatory or anti-inflammatory phenotypes, respectively, through the activation of different, often mutually exclusive signaling cascades. Therefore, we tested the ability of emodin to inhibit both forms of macrophage activation.
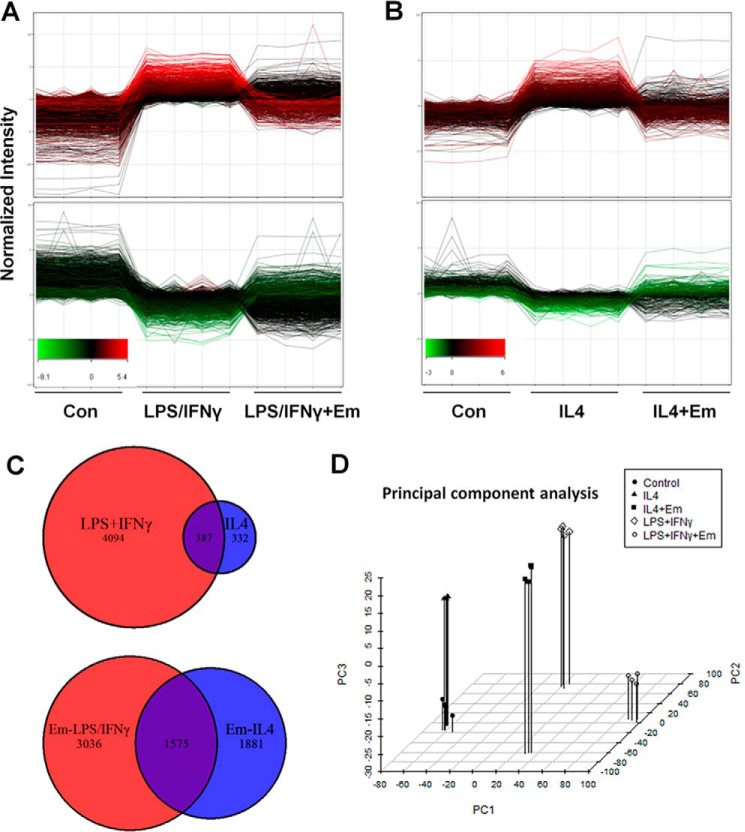
To comprehensively characterize the effects of emodin on macrophage activation and to determine the mechanism of action, gene expression was analyzed using a whole-genome microarray. Mouse peritoneal macrophages were stimulated with LPS/IFNγ or IL4 with/without emodin. We found that LPS/IFNγ stimulation changed the expression of over 4400 genes, and IL4 changed the expression of over 700 genes (≥2-fold, p ≤ 0.05) (GEO accession number GSE73311). Fig. 1A shows the effect of emodin treatment on the expression of genes that are significantly increased (top panel) or decreased (bottom panel) by LPS/IFNγ. Emodin treatment attenuated the LPS/IFNγ-induced changes in about 31% of the LPS/IFNγ-responsive genes. Similarly, emodin inhibited IL4-induced changes in almost 60% of IL4-responsive genes (Fig. 1B). These results indicate that emodin significantly inhibited the transcription programs induced by both M1 and M2 stimuli.
FIGURE 1.
Emodin inhibits LPS/IFNγ- and IL4-induced transcriptional changes in macrophages. A and B, mouse peritoneal macrophages were stimulated with LPS (100 ng/ml) and IFNγ (20 ng/ml) for 24 h (A) or IL4 (10 ng/ml) for 6 h (B) with or without emodin (Em, 50 μm). Gene expression was then detected using a whole-genome microarray. Genes significantly increased (top panel) or decreased (bottom panel) by LPS/IFNγ or IL4, respectively. The y axes correspond to normalized intensity values for gene expression and the x axes to treatments. Each line represents one gene, and the red and green colors mark high and low expression of genes, respectively, in the LPS/IFNγ or IL4 treatment groups. Con, control. C, Venn diagrams showing genes significantly changed by LPS/IFNγ or IL4 and by emodin under LPS/IFNγ or IL4 stimulation. D, principle component analysis of genes significantly changed in one of the treatment groups.
LPS/IFNγ and IL4 induce macrophage activation through competing signaling pathways (STAT1 versus STAT6, IRF4 versus IRF5), resulting in little overlap between the transcriptional programs induced by LPS/IFNγ or IL4. The majority of genes changed by LPS/IFNγ treatment and almost half of the genes changed by IL4 were unchanged in the other treatment group (Fig. 1C, top panel). The expression of almost 6500 genes was changed by emodin under at least one of the conditions; among them, only 1575 were changed in both groups (Fig. 1C, bottom panel). Therefore, emodin treatment predominately affected different transcriptional programs under the two different conditions. These results were confirmed with a principle component analysis of the genes that were significantly changed in at least one of the treatment groups (∼10,000 genes) (Fig. 1D). The samples within each group cluster near each other, with LPS/IFNγ-treated cells clustering much further from naive cells than IL4 treated cells, indicating that M1 activation involves much greater transcriptional changes than M2, in agreement with previous studies (36). Similarly, there is significant distance between the two emodin treatment groups, indicating that emodin treatment differentially affected the expression of transcriptional programs under the different conditions.
Emodin Inhibits the Induction of Signaling Pathways Associated with Macrophage Polarization and Function
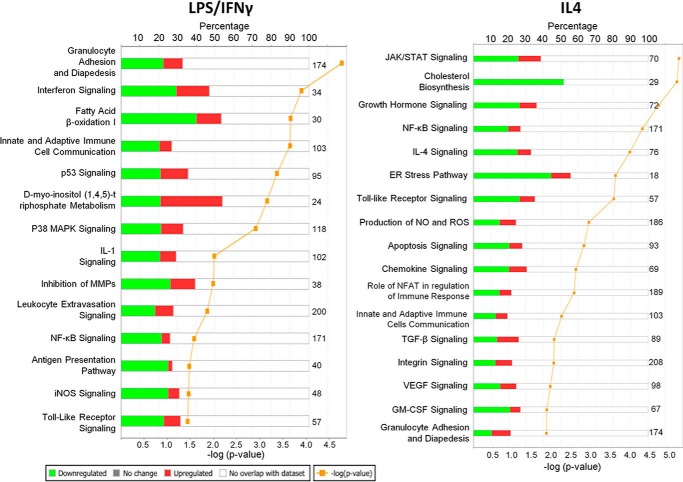
Next we investigated which cell signaling pathways might be targeted by emodin by performing a canonical signaling pathway analysis using Ingenuity IPA. The genes influenced by emodin treatment were enriched for genes associated with immune cell signaling, inflammation, cell adhesion, and metabolism. Fig. 2 shows a list of pathways with the highest significance. Several pathways were targeted by emodin under both conditions, including communication between immune cells, granulocyte adhesion, NFкB signaling, and Toll-like receptor signaling. However, most of the pathways targeted by emodin were different under the different conditions. The genes changed by emodin under LPS/IFNγ stimulation were enriched for M1-associated pathways (antigen presentation, IL1 signaling, and inducible nitric-oxide synthase (iNOS/NOS2) signaling), whereas the genes changed by emodin under IL4 stimulation were enriched for M2 associated pathways (IL4, JAK/STAT, TGFβ, and VEGF signaling).
FIGURE 2.
Emodin inhibits the induction of signaling pathways associated with macrophage polarization and function. Shown are the most significantly affected pathways relevant for macrophage activation as determined by Ingenuity IPA canonical pathway analyses.
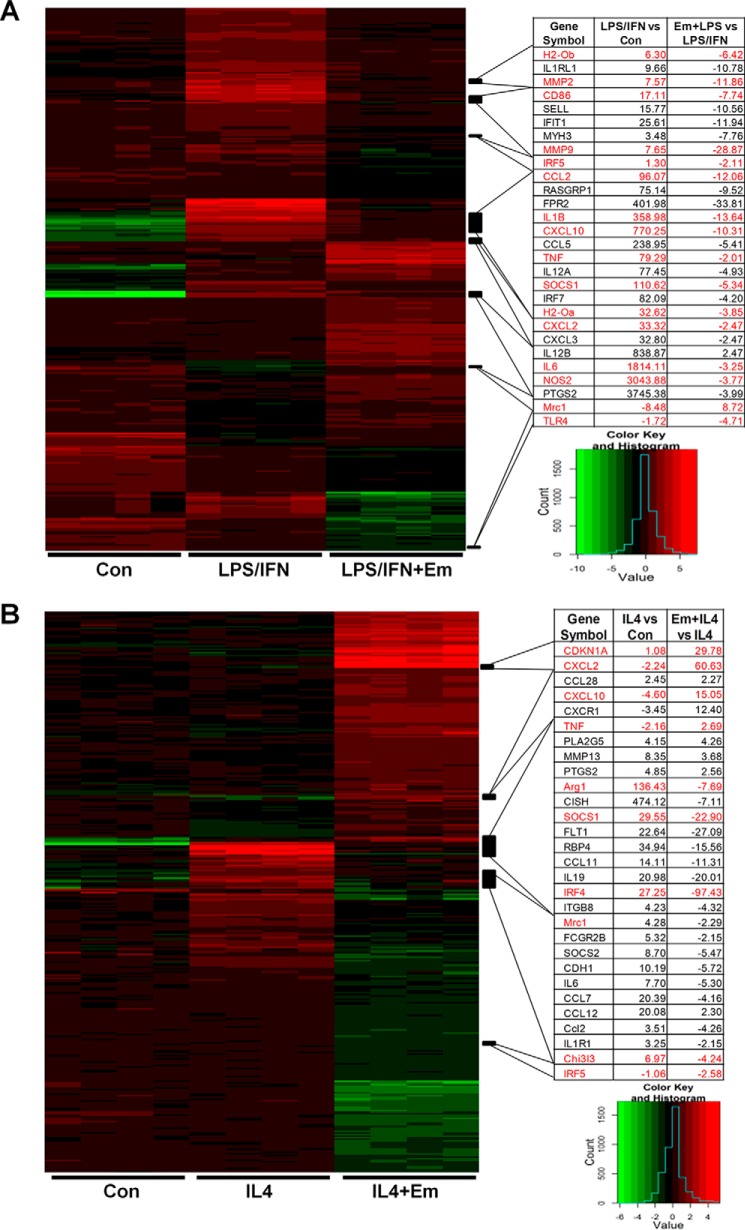
The expression of genes in these pathways is shown in Fig. 3. Emodin significantly attenuated the LPS/IFNγ-induced changes in a large number of genes, including canonical M1-associated genes: the proinflammatory cytokines IL1β, TNFα, and IL6 (13.64-, 2.01-, and 3.25-fold reduction, respectively); the proteases MMP2/9 (11.86- and 28.87-fold reduction, respectively); and the antigen presentation genes CD86 and H2-Oa/b (7.74-, 3.85-, and 6.42-fold reduction, respectively) (Fig. 3A). Similarly, emodin inhibited the IL4 induced expression of the canonical M2 genes Arg1, Mrc1, and Ch3l3 (7.69-, 2.29-, and 4.24-fold reduction, respectively) and the transcription factors SOCS1 and IRF4 (22.9- and 97.43-fold reduction, respectively), which have both shown to be necessary for M2 activation (8, 37, 38) (Fig. 3B). Emodin also increased the expression of CDKN1A (p21), which has been shown to inhibit macrophage proliferation and activation (39, 40).
FIGURE 3.
Effects of emodin on the expression of genes that regulate macrophage activation. A and B, heat maps showing the expression of genes associated with the most significantly affected macrophage canonical signaling pathways. Right panels, the -fold changes of genes caused by LPS/IFNγ or IL4 treatment compared with control (Con) and emodin (Em) treatment compared with LPS/IFN or IL4 for select macrophage activation genes.
Interestingly, we also found that emodin treatment had inverse effects on a subset of 86 genes under the two different stimuli (supplemental Table 4). For examples, emodin increased the expression of some proinflammatory genes, including TNFα, CXCL2, and CXCL10 (2.7-, 60.6-, and 15.5-fold, respectively) under IL4 stimulation while significantly decreasing them (−2.47-, −10.31-, and −2.01-fold, respectively) under LPS/IFNγ stimulation (Fig. 3). Similarly, emodin increased the expression of the M2 genes YM1 and Mrc1 under LPS/IFNγ stimulation (2.04- and 8.72-fold, respectively), while reducing them in IL4-stimulated cells (Fig. 3 and supplemental Table 4). These results show the ability of emodin to tune macrophage phenotype back toward the center between the extremes of the M1 or M2 activation states.
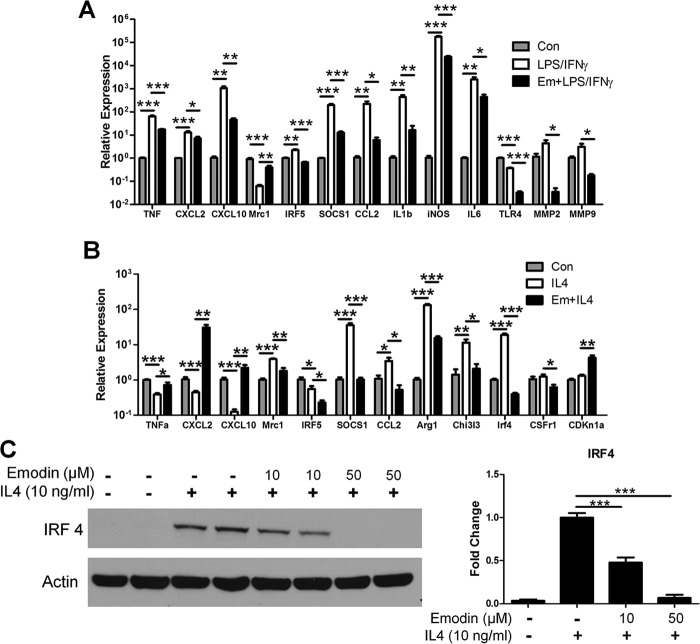
The expression of several important genes for macrophage activation was confirmed by qPCR (Fig. 4, A and B). Emodin inversely regulated the M1 genes TNFα, CXCL2, and CXCL10 and the M2 gene Mrc1 in the two settings. Interestingly, emodin inhibited the expression of the transcription factors IRF5 and SOCS1 and the chemoattractant CCL2 under both IL4 and LPS stimulation. In agreement with the microarray, emodin inhibited the expression of many proinflammatory mediators, including IL1β, iNOS, and IL6, as well as the proteases MMP2 and MMP9 under LPS/IFNγ stimulation and inhibited the M2 genes Arg1 and Chi3l3 under IL4 stimulation. Emodin also inhibited the expression of the receptors TLR4 and CSFr1 under LPS/IFNγ or IL4 stimulation, respectively, which could further inhibit macrophages from detecting activation signals in the environment. IRF4, a major regulator of M2 macrophage activation, was the most down-regulated gene by emodin under IL4 stimulation in the microarray dataset. We examined the production of IRF4 protein and found that emodin dose-dependently inhibited IRF4 production (Fig. 4C).
FIGURE 4.
Emodin inhibits the expression of M1 and M2 genes. A and B, qPCR was performed to verify the microarray results for select M1 and M2 genes. Error bars represent the mean ± S.E. For each treatment, n = 4. Con, control; Em, emodin. C, macrophages were stimulated with IL4 with or without emodin (0–50 μm) for 6 h. The cells were lysed, and IRF4 protein levels were detected by Western blotting. Results are shown as the mean ± S.E. for two independent experiments (n = 4). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
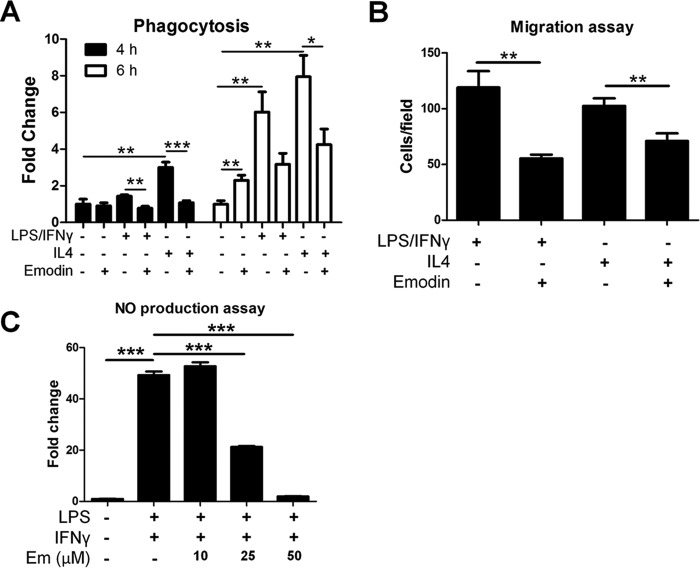
Emodin Inhibits Functions of Activated Macrophages
Next we investigated the effects of emodin on the functions of macrophages, which are predicted to be inhibited by emodin in one or both of the groups based on pathway analysis of the microarray results. We examined the phagocytic ability of activated macrophages. Macrophages were pretreated with LPS/IFNγ or IL4 with or without emodin for 24 h. Then the cells were incubated with FITC-labeled E. coli bioparticles. In agreement with previous studies (41, 42), emodin treatment alone was able to increase the phagocytic activity of naive cells at the 6-h time point. However, emodin decreased phagocytosis in IL4- or LPS/IFNγ-treated cells. Both IL4- and LPS/IFNγ-stimulated macrophages showed time-dependent increases in bioparticle uptake (6- 8-fold increase at 6 h). However, emodin significantly inhibited particle uptake under both conditions (by almost 2-fold) (Fig. 5A). Next, macrophages were stimulated with LPS/IFNγ or IL4 with or without emodin, and then their migration potential was detected. Macrophages were seeded into the top chamber of transwell inserts, and medium containing MCP1 (20 ng/ml) was placed in the bottom. Emodin significantly reduced macrophage migration under both conditions (Fig. 5B). We then verified the ability of emodin to inhibit M1 activation by detecting NO production. Macrophage production of NO was measured in the culture media following stimulation with LPS/IFNγ for 24 h. The result showed that emodin dose-dependently inhibited NO production (Fig. 5C).
FIGURE 5.
Emodin modulates the functions of activated macrophages. Mouse peritoneal macrophages were stimulated with LPS (100 ng/ml) and IFNγ (20 ng/ml) or IL4 (10 ng/ml) with or without emodin (50 μm) for 24 h. A, macrophages were washed, and the cells were incubated with FITC-labeled E. coli bioparticles for 4–6 h. Fluorescence was detected with a microplate reader as an indicator of phagocytosis. Results are shown as the mean ± S.E. (n = 4). B, macrophages were seeded into the top chamber of a transwell insert in DMEM, and DMEM with MCP1 (20 ng/ml) was placed in the bottom of the well. After 4 h, cells were fixed, stained with DAPI, and imaged with five fields of view at ×200 magnification per membrane. Results are shown as the mean ± S.E. for two independent experiments (n = 3). C, macrophages were incubated with LPS/IFNγ with emodin (Em) at various concentrations. After 24 h, the medium was collected, and the NO content was detected. Results are shown as the mean ± S.E. (n = 4).*, p < 0.05; **, p < 0.01; ***, p < 0.001.
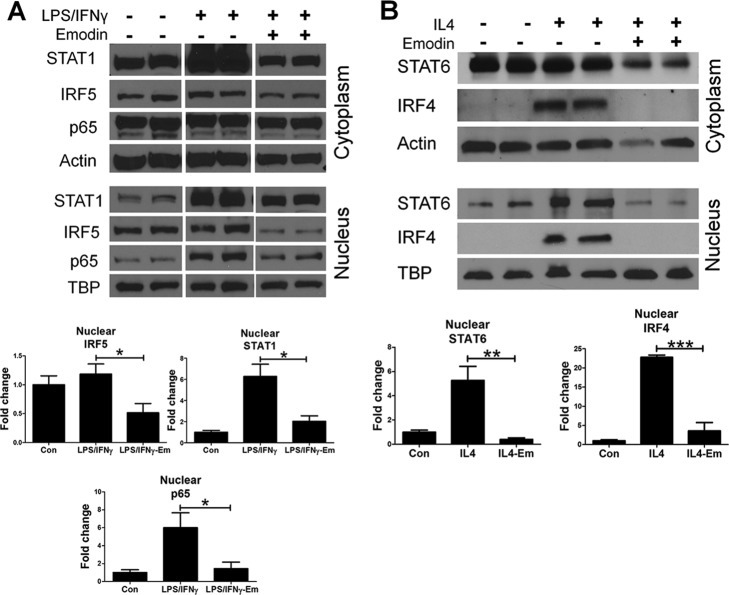
Emodin Modulates IL4- and LPS/IFNγ-induced Activation of Cell Signaling Pathways
We next attempted to identify the cell signaling pathways targeted by emodin under the different conditions. Macrophage polarization is controlled by multiple different signaling pathways, several of which have been shown to be antagonistic, promoting the exclusive nature of the M1 and M2 phenotypes (4, 5). Nuclear translocation of transcription factors was investigated by Western blotting using cytoplasmic and nuclear cell fractions. In agreement with results published previously (30, 31, 33), emodin was able to drastically inhibit NFκB p65 nuclear translocation in response to LPS/IFNγ stimulation (Fig. 6A) and STAT6 nuclear translocation in response to IL4 stimulation (Fig. 6B). Emodin also inhibited nuclear IRF4 (Fig. 6B), in agreement with our gene expression data. Furthermore, we found that emodin inhibited STAT1 and IRF5 nuclear translocation (Fig. 6A). These results indicate that emodin is able to inhibit the key signaling pathways necessary for macrophage polarization.
FIGURE 6.
Emodin inhibits LPS/IFNγ- and IL4-induced activation of signaling pathways. A and B, macrophages were stimulated with (A) LPS (100 ng/ml) and IFNγ (20 ng/ml) or (B) IL4 (10 ng/ml) with or without emodin (Em, 50 μm) for 24 h. Cells were lysed, and cytoplasmic and nuclear fractions were collected. Transcription factors were then detected in both fractions using Western blotting. Bottom panels, quantifications of blots of nuclear fractions normalized to the loading control (Con) TATA-binding protein (TBP). Results are shown as the mean ± S.E. for two independent experiments (n = 4). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
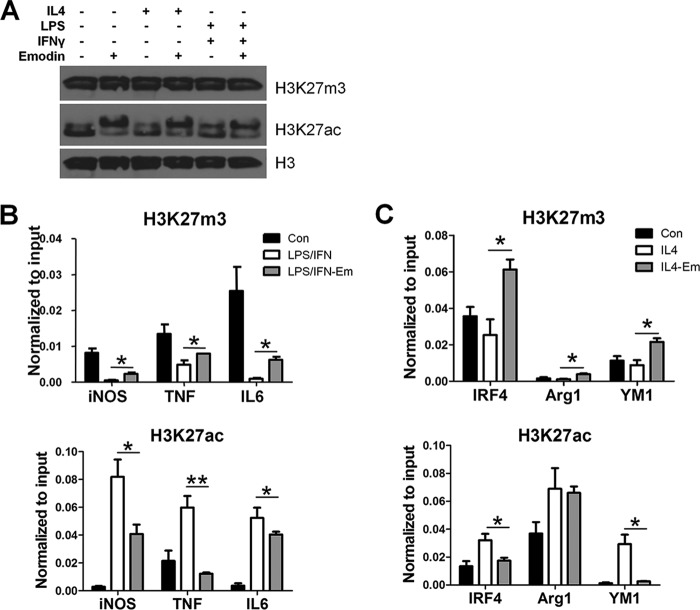
Emodin Inhibits IL4- and LPS/IFNγ-induced Changes in the Epigenetic Landscape in Macrophages
The microarray revealed that emodin changed the expression of several histone-modifying enzymes, including those that regulate H3K27 methylation and acetylation under both IL4 and LPS/IFNγ stimulation (supplemental Table 5). H3K27me3 attenuates M2 polarization by inhibiting the expression of IRF4; therefore, its removal by the demethylase JMJD3 promotes M2 activation (37, 43). H3K27ac has been shown to prime both M1 and M2 genes for expression upon subsequent stimulation (43, 44). Therefore, we next investigated the effects of emodin on these histone modifications in macrophages. We investigated the global expression of these histone modifications using Western blotting and found that emodin had no effect on the global expression of either H3K27m3 or H3K27ac (Fig. 7A). We then examined gene-specific changes in histone modifications using ChIP assays. We found that emodin attenuated the LPS/IFNγ-induced decrease of H3K27m3 in the promoter of the iNOS, TNFα, and IL6 genes and reversed the IL4-induced decrease of H3K27m3 in the promoter of the IRF4, Arg1, and YM1 genes. Emodin also suppressed the LPS/IFNγ-induced increase of H3K27ac in the promoters of the iNOS, TNFα, and IL6 genes and the IL4-induced increase of H3K27ac in the promoter of the IRF4 and YM1 genes (Fig. 7, B and C). These data, for the first time, show that emodin epigenetically regulates macrophage activation. Furthermore, unlike the effects of emodin on signaling pathways, the effects of emodin on histone modification are not stimulus-dependent but gene-specific.
FIGURE 7.
Emodin inhibits LPS/IFNγ- and IL4-induced histone modifications in macrophages. Macrophages were stimulated with LPS (100 ng/ml) and IFNγ (20 ng/ml) or IL4 (10 ng/ml) with or without emodin (50 μm) for 24 h. A, global histone modification levels were detected using Western blotting. The experiment was performed in triplicate. Con, control; Em, emodin. B and C, ChIP-PCR was used to detect gene-specific changes in histone modifications. Results are shown as the mean ± S.E. (n = 3). *, p ≤ 0.05; **, p ≤ 0.01.
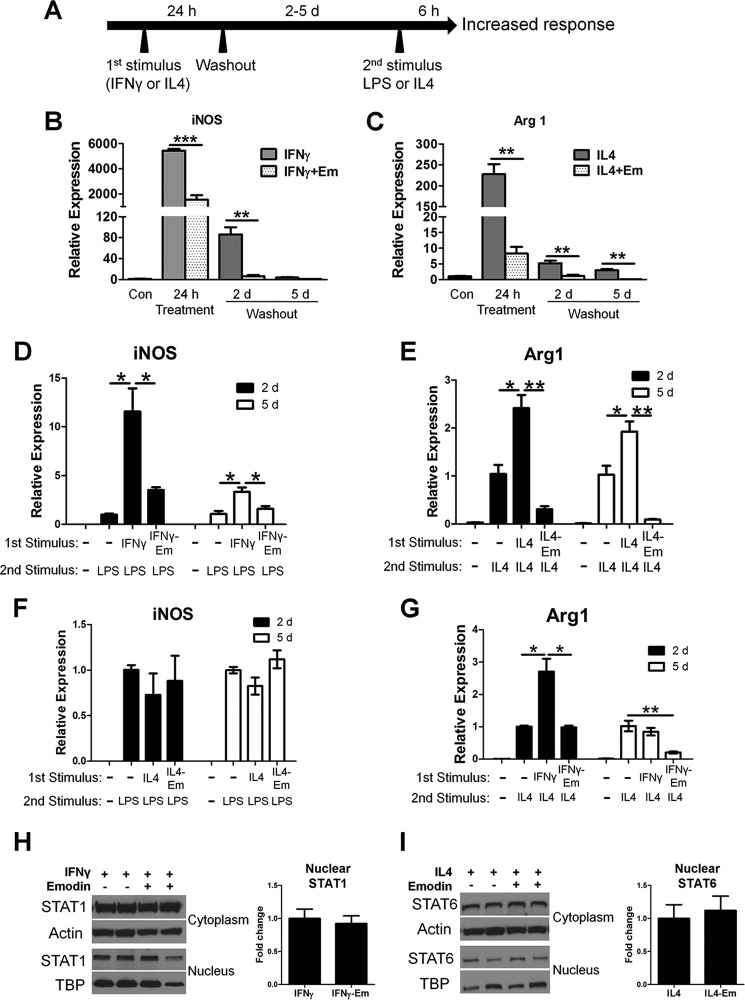
Macrophages are able to retain a memory of the environmental signals to which they have been exposed, allowing them to alter their response upon subsequent stimulation (9, 44, 45). Therefore, we next investigated the effects of emodin on macrophage memory. Macrophages were stimulated with IFNγ or IL4 for 24 h with or without emodin. Then they were cultured in fresh medium for 2–5 days before being stimulated again with LPS or IL4 for 6 h without emodin (Fig. 8A). The expression of iNOS returned to baseline levels by 5 days after stimulation with IFNγ, whereas the expression of Arg1 was still elevated 5 days after IL4 treatment (Fig. 8, B and C). IFNγ treatment significantly increased the response of the macrophages to a subsequent LPS treatment, with an 11.5- and 3.3-fold increase in iNOS expression after 2- and 5-day washout after the first IFNγ treatment, respectively, compared with cells without IFNγ treatment (Fig. 8D). Similarly, after 2- and 5-day washout after the first treatment with IL4, macrophages responded significantly more robustly to a second IL4 treatment, with 2.4- and 1.9-fold increases in Arg1 expression, respectively (Fig. 8E). However, when the cells were first concomitantly treated with emodin with IFNγ or IL4, their responses to the second LPS or IL-4 treatment were significantly diminished (Fig. 8, D and E). Interestingly, in a crossover experiment, IL4 pretreatment showed no effects on macrophage response to LPS 2 or 5 days after the pretreatment, and co-pretreatment with emodin did not have any effects (Fig. 8F). However, pretreatment with IFNγ increased the macrophage response to IL4 2 days after IFNγ treatment, but this effect was diminished within 5 days after IFNγ treatment, and emodin co-pretreatment decreased the response to the secondary IL-4 stimulation (Fig. 8G). After 5 days of the washout period, there was no significant difference in the nuclear STAT1 or STAT6 in IFNγ- or IL4-pretreated cells, respectively, regardless of whether the cells were also treated with emodin (Fig. 8, H and I). The lack of differences in the STAT1 and STAT6 signaling pathway prior to restimulation indicates that emodin may regulate macrophage memory through epigenetic modification of the key genes in macrophage activation.
FIGURE 8.
Emodin inhibits macrophage memory. A, macrophage treatments for testing the effects of emodin on macrophage memory. Macrophages were incubated with IFNγ (20 ng/ml) or IL4 (10 ng/ml) with or without emodin (50 μm) for 24 h. The cells were washed and incubated for 2 or 5 days (washout periods) and then stimulated with either IL4 or LPS for 6 h. B and C, gene expression was analyzed with qPCR after the first treatment with IFNγ or IL4 for 24 h and after the 2- or 5-day (d) washout period and compared with the baseline control (Con, set as 1). Em, emodin. D and E, gene expression was analyzed after the cells were restimulated with LPS or IL-4 for 6 h following the 2- or 5-day washout period. F and G, in crossover experiments, macrophages were first treated with IL4 or IFNγ for 24 h. After the 2- or 5-day washout period, the cells were further treated with LPS or IL4 for 6 h, respectively, and gene expression was analyzed. B–G, results are shown as the mean ± S.E. for two independent experiments (n = 3). H and I, macrophages were lysed after the 5-day rest period, and cytoplasmic and nuclear protein fractions were collected and analyzed via Western blotting. Results are shown as the mean ± S.E. for two independent experiments (n = 4). *, p ≤ 0.05; **, p ≤ 0.01; ***, p < 0.001.
To examine whether emodin has any effects on naive macrophages and whether emodin pretreatment alone may affect the subsequent response to polarization signals, we first treated mouse peritoneal macrophages with emodin at 25 or 50 μm for 24 h and found no effects of emodin on the expression of the macrophage polarization markers iNOS, Arg1, and YM1 (Fig. 9A). We then pretreated macrophages with 50 μm emodin alone for 24 h and, after a 2-day washout period, stimulated them with LPS or IL4 for 6 h. Emodin pretreatment had no effect on macrophage response to LPS but significantly reduced their response to IL4 (Fig. 9B). These data further suggest that, although emodin may not influence the basal phenotype of macrophages, it can change their response to subsequent polarization signals, particularly M2 polarization signals, possibly by altering the epigenetic landscape of the cells.
FIGURE 9.
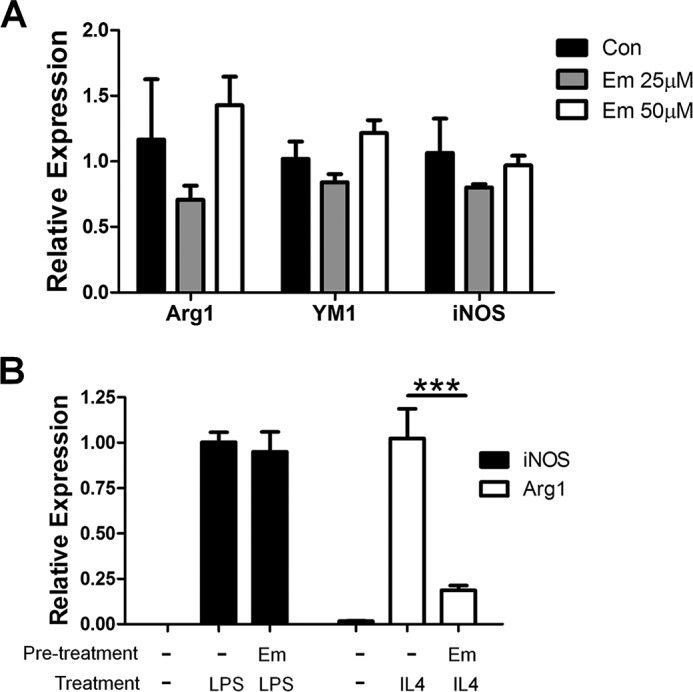
The effects of emodin on naive macrophages. A, macrophages were incubated with various concentrations of emodin (0–50 μm) for 24 h, and gene expression was analyzed using qPCR. Con, control. B, macrophages were incubated with or without emodin (50 μm) for 24 h. Then the cells were washed and incubated for 2 days and further stimulated with either IL4 or LPS for 6 h, and gene expression was analyzed with qPCR. Results are shown as the mean ± S.E. for two independent experiments (n = 3). *, p ≤ 0.05; **, p ≤ 0.01.
Discussion
This study reveals the ability of emodin to inhibit both M1 and M2 polarization of macrophages through transcriptional and epigenetic regulation. Gene expression analysis of emodin-treated macrophages revealed that emodin attenuated the transcriptional changes in 31% of genes altered by LPS/IFNγ and 60% of genes altered by IL4. Emodin mostly targeted different transcriptional networks in the two different stimulation settings, indicating the ability of emodin to differentially affect a broad spectrum of signaling pathways, depending on the microenvironment. Further analysis revealed that emodin inversely regulated a subset of genes, including Mrc1, YM1, TNFα, CXCL2, and CXCL10, in LPS/IFNγ- or IL4-treated macrophages. Emodin was able to inhibit numerous signaling pathways, including NFκB, STAT1, and IRF5, following LPS/IFNγ stimulation and STAT6 and IRF4 signaling triggered by IL4 stimulation. Finally, our data showed that emodin modulates macrophage training/memory. Taken together, our data show that emodin has great potential as a treatment for pathologies driven by an imbalance in macrophage activation and polarization.
It has been implied in the literature that emodin may have different effects on macrophages depending on the environment of the cells being studied. The majority of studies performed on emodin have focused on its ability to shift the Th1 inflammatory response to Th2 (30). Lin et al. (46) showed that emodin shifted the Th1/Th2 balance toward Th2 by decreasing IFNγ and IL2 and increasing IL4 levels in the serum of rats that received orthotopic liver transplantation. Emodin has also shown effectiveness in treating acute models of inflammation in the pancreas, lungs, kidneys, and liver at least partly through inhibiting NFκB signaling (47–50). Our laboratory has shown that emodin is able to attenuate liver inflammation in an experimental model of non-alcoholic fatty liver disease by inhibiting inflammatory cell infiltration in the liver (31). Emodin has also been shown to promote expression of the M2-associated molecules TGFβ and PPARγ (30, 51). On the other hand, there have been a few studies that have implied that emodin may also suppress Th2 responses. Emodin significantly inhibited the secretion of the Th2 cytokines IL4, IL5, and IL13 in the lungs of mice challenged with ovalbumin (34, 52). Furthermore, our laboratory has found that emodin inhibits breast cancer metastasis to the lungs in mice by reducing macrophage infiltration and M2 polarization through inhibiting STAT6 and CCAAT/enhancer-binding protein β signaling (33). But the interesting dual effects of emodin on macrophages have not been comprehensively studied further.
Our data suggest that emodin is a bidirectional regulator of macrophage activation by targeting multiple signaling pathways to return the macrophage phenotype to the homeostatic center between the extremes of M1 or M2 in various environmental settings. Our results showed that emodin inhibited proinflammatory cytokine/chemokines expression through blocking NFκB signaling as well as STAT1 signaling in response to LPS/IFNγ stimulation and also inhibited M2 activation markers through blocking STAT6 signaling. Emodin was found to regulate the IRF signaling pathways. IRF4 and IRF5 have been shown to be inversely regulated, pushing macrophages toward an M2 or M1 phenotype, respectively (4, 53). IRF5 is activated by TLR4 ligation and promotes the transcription of proinflammatory genes (e.g. IL12b) while suppressing the expression of anti-inflammatory genes (e.g. IL10) (5). IRF4 competitively binds to MyD88 and is a negative regulator of IRF5 signaling. IRF4 is regulated through the removal of H3K27m3 by the histone demethylase JMJD3 (37, 53). Our results show that emodin inhibits both IRF4 and IRF5 signaling. Taken together, these data indicate that emodin targets multiple signaling pathways to inhibit macrophage activation and return them to a homoeostatic state.
Emodin is a poorly selective tyrosine kinase inhibitor, and it has been reported to directly interact with a variety of proteins that are able to regulate macrophage activation (54). Emodin is a potent inhibitor of casein kinase 2 (CK2), which regulates JAK/STAT signaling (55). Furthermore, emodin has been shown to directly inhibit JAK activity (56). Emodin has also been reported to be an agonist for PPARγ (51). PPARγ activation can inhibit M1 macrophage activation. Yang et al. (57) showed that emodin could inhibit LPS-induced inflammatory responses by activating PPARγ in mouse epithelial cells. Emodin may also directly act on many other kinases, including activated protein kinase (AMPK), phosphoinositide 3-kinase (PI3K), Ca2+/calmodulin-dependent protein kinase (CAMK), and protein kinase D1 (54). Unfortunately, analysis of our microarray results using Ingenuity IPA was unable to narrow down what the direct target(s) of emodin might be. Most likely, the effects of emodin on macrophage polarization are due to direct interaction with a combination of these kinases/receptors, depending on the microenvironment to which the cells are exposed.
Cytokines such as IFNγ can prime genes for increased expression by the recruitment of transcription-promoting histone markers (such as H3K4m3, H3K9m3, and H3K27ac) to the promoter or enhancer regions (58). Similarly prolonged exposure to foreign molecular patterns could lead to increased or decreased immune responses (for examples, β-glucan or LPS, respectively) (44). Our microarray analysis revealed that emodin significantly changed the expression of several histone-modifying enzymes, particularly those responsible for regulating H3K27 trimethylation and acetylation. Emodin had no effects on genome-wide levels of H3K27ac and H3K27m3. Instead, it significantly increased H3K27m3 levels while decreasing H3K27ac levels on the promoters of many M1 genes following LPS/IFNγ treatment and M2 genes following IL4 treatment.
Our data further showed that emodin modulated macrophage memory. Prestimulation of macrophages with IFNγ resulted in an enhanced response to LPS up to 5 days later and to IL4 up to 2 days later. However, prestimulation with IL4 increased the macrophage response to subsequent IL4 stimulation while having no effect on LPS stimulation 2 or 5 days later. Emodin co-treatment during the prestimulation stage significantly diminished the exaggerated responses even though iNOS and Arg1 expression returned to near baseline levels prior to the second stimulation. Similarly, there was no difference in STAT1 or STAT6 activation prior to the second stimulation. Therefore, these data suggest that at least part of the effects of emodin on macrophage activation and memory could be attributed to gene-specific epigenetic modifications.
A few studies have shown that emodin delivered orally could attenuate the severity of some inflammatory diseases; however, the potential therapeutic benefits of emodin are limited by its low oral bioavailability (59). In humans, emodin acts as a laxative and reduces water absorption in the colon (60). Studies in rodents have shown that, following intravenous administration, the half-life of emodin is only 23 min (61). Emodin in the serum predominately exists as glucuronides/sulfates, whereas the free form is mainly found in organs such as the liver and kidneys (62). For emodin to be effectively developed into a clinical therapy, methods will need to be developed to improve its bioavailability.
In summary, emodin effectively inhibited macrophage activation in response to both M1 and M2 stimuli by suppressing the activation of multiple signaling pathways, including NFκB, IRF5, MAPK, STAT1 or STAT6, and IRF4, depending on the environmental settings. Emodin thus regulated a subset of genes depending on whether the cell was exposed to M1 or M2 stimuli, pushing the phenotype of the cell back toward the center of the two poles. This phenomenon opens the possibility that emodin may exert very different homeostasis-maintaining effects on macrophages in different locations and thus target two very different pathologies within the same individual. Our data showed overall more profound effects of emodin on M2 polarization, suggesting that emodin could be most beneficial for patients with M2 macrophage-driven diseases.
Author Contributions
D. F. conceived and coordinated the study and wrote the paper. S. I. and J. W. designed, performed, and analyzed the experiments and wrote the paper. D. A. performed the microarray experiments. Y. H. provided technical support. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grants HL116626 and AT003961–8455. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables 1–5.
- PPAR
- peroxisome proliferator-activated receptor
- m3
- trimethylation
- ac
- acetylation
- qPCR
- quantitative PCR
- IRF
- interferon regulatory factor
- iNOS
- inducible nitric-oxide synthase.
References
- 1. Mosser D. M., and Edwards J. P. (2008) Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chawla A. (2010) Control of macrophage activation and function by PPARs. Circ. Res. 106, 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sica A., and Mantovani A. (2012) Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou D., Huang C., Lin Z., Zhan S., Kong L., Fang C., and Li J. (2014) Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 26, 192–197 [DOI] [PubMed] [Google Scholar]
- 5. Lawrence T., and Natoli G. (2011) Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11, 750–761 [DOI] [PubMed] [Google Scholar]
- 6. Murray P. J., Allen J. E., Biswas S. K., Fisher E. A., Gilroy D. W., Goerdt S., Gordon S., Hamilton J. A., Ivashkiv L. B., Lawrence T., Locati M., Mantovani A., Martinez F. O., Mege J. L., Mosser D. M., Natoli G., Saeij J. P., Schultze J. L., Shirey K. A., Sica A., Suttles J., Udalova I., van Ginderachter J. A., Vogel S. N., and Wynn T. A. (2014) Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gordon S., and Martinez F. O. (2010) Alternative activation of macrophages: mechanism and functions. Immunity 32, 593–604 [DOI] [PubMed] [Google Scholar]
- 8. El Chartouni C., Schwarzfischer L., and Rehli M. (2010) Interleukin-4 induced interferon regulatory factor (Irf) 4 participates in the regulation of alternative macrophage priming. Immunobiology 215, 821–825 [DOI] [PubMed] [Google Scholar]
- 9. Deng H., Maitra U., Morris M., and Li L. (2013) Molecular mechanism responsible for the priming of macrophage activation. J. Biol. Chem. 288, 3897–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ifrim D. C., Quintin J., Joosten L. A., Jacobs C., Jansen T., Jacobs L., Gow N. A., Williams D. L., van der Meer J. W., and Netea M. G. (2014) Trained immunity or tolerance: opposing functional programs induced in human monocytes after engagement of various pattern recognition receptors. Clin. Vaccine Immunol. 21, 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ivashkiv L. B. (2013) Epigenetic regulation of macrophage polarization and function. Trends Immunol. 34, 216–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wynn T. A., Chawla A., and Pollard J. W. (2013) Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murray P. J., and Wynn T. A. (2011) Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pollard J. W. (2009) Trophic macrophages in development and disease. Nat. Rev. Immunol. 9, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shechter R., and Schwartz M. (2013) Harnessing monocyte-derived macrophages to control central nervous system pathologies: no longer “if” but “how”. J. Pathol. 229, 332–346 [DOI] [PubMed] [Google Scholar]
- 16. Karsdal M. A., Woodworth T., Henriksen K., Maksymowych W. P., Genant H., Vergnaud P., Christiansen C., Schubert T., Qvist P., Schett G., Platt A., and Bay-Jensen A. C. (2011) Biochemical markers of ongoing joint damage in rheumatoid arthritis: current and future applications, limitations and opportunities. Arthritis Res. Ther. 13, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wicks K., Torbica T., and Mace K. A. (2014) Myeloid cell dysfunction and the pathogenesis of the diabetic chronic wound. Semin. Immunol. 26, 341–353 [DOI] [PubMed] [Google Scholar]
- 18. Nahrendorf M., and Swirski F. K. (2013) Monocyte and macrophage heterogeneity in the heart. Circ. Res. 112, 1624–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kundu J. K., and Surh Y. J. (2008) Inflammation: gearing the journey to cancer. Mutat. Res. 659, 15–30 [DOI] [PubMed] [Google Scholar]
- 20. Hermansson C., Lundqvist A., Magnusson L. U., Ullstrom C., Bergström G., and Hultén L. M. (2014) Macrophage CD14 expression in human carotid plaques is associated with complicated lesions, correlates with thrombosis, and is reduced by angiotensin receptor blocker treatment. Int. Immunopharmacol. 22, 318–323 [DOI] [PubMed] [Google Scholar]
- 21. McNelis J. C., and Olefsky J. M. (2014) Macrophages, immunity, and metabolic disease. Immunity 41, 36–48 [DOI] [PubMed] [Google Scholar]
- 22. Lê K. A., Mahurkar S., Alderete T. L., Hasson R. E., Adam T. C., Kim J. S., Beale E., Xie C., Greenberg A. S., Allayee H., and Goran M. I. (2011) Subcutaneous adipose tissue macrophage infiltration is associated with hepatic and visceral fat deposition, hyperinsulinemia, and stimulation of NF-κB stress pathway. Diabetes 60, 2802–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xue J., Sharma V., Hsieh M. H., Chawla A., Murali R., Pandol S. J., and Habtezion A. (2015) Alternatively activated macrophages promote pancreatic fibrosis in chronic pancreatitis. Nat. Commun. 6, 7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Komohara Y., Jinushi M., and Takeya M. (2014) Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 105, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tabas I., and Glass C. K. (2013) Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science 339, 166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gupta S. C., Tyagi A. K., Deshmukh-Taskar P., Hinojosa M., Prasad S., and Aggarwal B. B. (2014) Downregulation of tumor necrosis factor and other proinflammatory biomarkers by polyphenols. Arch. Biochem. Biophys. 559, 91–99 [DOI] [PubMed] [Google Scholar]
- 27. Shehzad A., Rehman G., and Lee Y. S. (2013) Curcumin in inflammatory diseases. Biofactors 39, 69–77 [DOI] [PubMed] [Google Scholar]
- 28. Wang Q., and Li X. K. (2011) Immunosuppressive and anti-inflammatory activities of sinomenine. Int. Immunopharmacol. 11, 373–376 [DOI] [PubMed] [Google Scholar]
- 29. Liang Q., Wu Q., Jiang J., Duan J., Wang C., Smith M. D., Lu H., Wang Q., Nagarkatti P., and Fan D. (2011) Characterization of sparstolonin B, a Chinese herb-derived compound, as a selective Toll-like receptor antagonist with potent anti-inflammatory properties. J. Biol. Chem. 286, 26470–26479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shrimali D., Shanmugam M. K., Kumar A. P., Zhang J., Tan B. K., Ahn K. S., and Sethi G. (2013) Targeted abrogation of diverse signal transduction cascades by emodin for the treatment of inflammatory disorders and cancer. Cancer Lett. 341, 139–149 [DOI] [PubMed] [Google Scholar]
- 31. Jia X., Iwanowycz S., Wang J., Saaoud F., Yu F., Wang Y., Hu J., Chatterjee S., Wang Q., and Fan D. (2014) Emodin attenuates systemic and liver inflammation in hyperlipidemic mice administrated with lipopolysaccharides. Exp. Biol. Med. 239, 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tong H., Chen K., Chen H., Wu H., Lin H., Ni Z., and Lin S. (2011) Emodin prolongs recipient survival time after orthotopic liver transplantation in rats by polarizing the Th1/Th2 paradigm to Th2. Anat. Rec. 294, 445–452 [DOI] [PubMed] [Google Scholar]
- 33. Jia X., Yu F., Wang J., Iwanowycz S., Saaoud F., Wang Y., Hu J., Wang Q., and Fan D. (2014) Emodin suppresses pulmonary metastasis of breast cancer accompanied with decreased macrophage recruitment and M2 polarization in the lungs. Breast Cancer Res. Treat. 148, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chu X., Wei M., Yang X., Cao Q., Xie X., Guan M., Wang D., and Deng X. (2012) Effects of an anthraquinone derivative from Rheum officinale Baill, emodin, on airway responses in a murine model of asthma. Food Chem. Toxicol. 50, 2368–2375 [DOI] [PubMed] [Google Scholar]
- 35. Tomar S., Graves C. A., Altomare D., Kowli S., Kassler S., Sutkowski N., Gillespie M. B., Creek K. E., and Pirisi L. (June 15, 2015) Human papillomavirus status and gene expression profiles of oropharyngeal and oral cancers from European American and African American patients. Head Neck 10.1002/hed.24072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martinez F. O., Gordon S., Locati M., and Mantovani A. (2006) Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 [DOI] [PubMed] [Google Scholar]
- 37. Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., Honma K., Matsuyama T., Yui K., Tsujimura T., Standley D. M., Nakanishi K., Nakai K., and Akira S. (2010) The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 [DOI] [PubMed] [Google Scholar]
- 38. Whyte C. S., Bishop E. T., Rückerl D., Gaspar-Pereira S., Barker R. N., Allen J. E., Rees A. J., and Wilson H. M. (2011) Suppressor of cytokine signaling (SOCS)1 is a key determinant of differential macrophage activation and function. J. Leukocyte Biol. 90, 845–854 [DOI] [PubMed] [Google Scholar]
- 39. Xaus J., Cardó M., Valledor A. F., Soler C., Lloberas J., and Celada A. (1999) Interferon γ induces the expression of p21waf-1 and arrests macrophage cell cycle, preventing induction of apoptosis. Immunity 11, 103–113 [DOI] [PubMed] [Google Scholar]
- 40. Lloberas J., and Celada A. (2009) p21(waf1/CIP1), a CDK inhibitor and a negative feedback system that controls macrophage activation. Eur. J. Immunol. 39, 691–694 [DOI] [PubMed] [Google Scholar]
- 41. Chang Y. C., Lai T. Y., Yu C. S., Chen H. Y., Yang J. S., Chueh F. S., Lu C. C., Chiang J. H., Huang W. W., Ma C. Y., and Chung J. G. (2011) Emodin induces apoptotic death in murine myelomonocytic leukemia WEHI-3 cells in vitro and enhances phagocytosis in leukemia mice in vivo. Evid. Based Complement. Alternat. Med. 2011, 523596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ni Q., Sun K., Chen G., and Shang D. (2014) In vitro effects of emodin on peritoneal macrophages that express membrane-bound CD14 protein in a rat model of severe acute pancreatitis/systemic inflammatory response syndrome. Mol. Med. Rep. 9, 355–359 [DOI] [PubMed] [Google Scholar]
- 43. Ishii M., Wen H., Corsa C. A., Liu T., Coelho A. L., Allen R. M., Carson W. F. 4th, Cavassani K. A., Li X., Lukacs N. W., Hogaboam C. M., Dou Y., and Kunkel S. L. (2009) Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114, 3244–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saeed S., Quintin J., Kerstens H. H., Rao N. A., Aghajanirefah A., Matarese F., Cheng S. C., Ratter J., Berentsen K., van der Ent M. A., Sharifi N., Janssen-Megens E. M., Ter Huurne M., Mandoli A., van Schaik T., Ng A., Burden F., Downes K., Frontini M., Kumar V., Giamarellos-Bourboulis E. J., Ouwehand W. H., van der Meer J. W., Joosten L. A., Wijmenga C., Martens J. H., Xavier R. J., Logie C., Netea M. G., and Stunnenberg H. G. (2014) Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Netea M. G., Quintin J., and van der Meer J. W. (2011) Trained immunity: a memory for innate host defense. Cell Host Microbe 9, 355–361 [DOI] [PubMed] [Google Scholar]
- 46. Lin S. Z., Chen K. J., Tong H. F., Jing H., Li H., and Zheng S. S. (2010) Emodin attenuates acute rejection of liver allografts by inhibiting hepatocellular apoptosis and modulating the Th1/Th2 balance in rats. Clin. Exp. Pharmacol. Physiol. 37, 790–794 [DOI] [PubMed] [Google Scholar]
- 47. Ni Q., Zhang W., Sun K., Yin C., An J., and Shang D. (2014) In vitro effects of emodin on peritoneal macrophage intercellular adhesion molecule-3 in a rat model of severe acute pancreatitis/systemic inflammatory response syndrome. Biomed. Rep. 2, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiao M., Zhu T., Zhang W., Wang T., Shen Y. C., Wan Q. F., and Wen F. Q. (2014) Emodin ameliorates LPS-induced acute lung injury, involving the inactivation of NF-κB in mice. Int. J. Mol. Sci. 15, 19355–19368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Y., Xiong W., Yang J., Zhong J., Zhang L., Zheng J., Liu H., Zhang Q., Ouyang X., Lei L., and Yu X. (2015) Attenuation of inflammation by emodin in lipopolysaccharide-induced acute kidney injury via inhibition of Toll-like receptor 2 signal pathway. Iran. J. Kidney Dis. 9, 202–208 [PubMed] [Google Scholar]
- 50. Xue J., Chen F., Wang J., Wu S., Zheng M., Zhu H., Liu Y., He J., and Chen Z. (2015) Emodin protects against concanavalin A-induced hepatitis in mice through inhibiting activation of the p38 MAPK-NF-κB signaling pathway. Cell Physiol. Biochem. 35, 1557–1570 [DOI] [PubMed] [Google Scholar]
- 51. Fu X., Xu A. G., Yao M. Y., Guo L., and Zhao L. S. (2014) Emodin enhances cholesterol efflux by activating peroxisome proliferator-activated receptor-γ in oxidized low density lipoprotein-loaded THP1 macrophages. Clin. Exp. Pharmacol. Physiol. 41, 679–684 [DOI] [PubMed] [Google Scholar]
- 52. Wang T., Zhong X. G., Li Y. H., Jia X., Zhang S. J., Gao Y. S., Liu M., and Wu R. H. (2015) Protective effect of emodin against airway inflammation in the ovalbumin-induced mouse model. Chin. J. Integr. Med. 6, 431–437 [DOI] [PubMed] [Google Scholar]
- 53. Negishi H., Ohba Y., Yanai H., Takaoka A., Honma K., Yui K., Matsuyama T., Taniguchi T., and Honda K. (2005) Negative regulation of Toll-like-receptor signaling by IRF-4. Proc. Natl. Acad. Sci. U.S.A. 102, 15989–15994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sandholt I. S., Olsen B. B., Guerra B., and Issinger O. G. (2009) Resorufin: a lead for a new protein kinase CK2 inhibitor. Anticancer Drugs 20, 238–248 [DOI] [PubMed] [Google Scholar]
- 55. Zheng Y., Qin H., Frank S. J., Deng L., Litchfield D. W., Tefferi A., Pardanani A., Lin F. T., Li J., Sha B., and Benveniste E. N. (2011) A CK2-dependent mechanism for activation of the JAK-STAT signaling pathway. Blood 118, 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Muto A., Hori M., Sasaki Y., Saitoh A., Yasuda I., Maekawa T., Uchida T., Asakura K., Nakazato T., Kaneda T., Kizaki M., Ikeda Y., and Yoshida T. (2007) Emodin has a cytotoxic activity against human multiple myeloma as a Janus-activated kinase 2 inhibitor. Mol. Cancer Ther. 6, 987–994 [DOI] [PubMed] [Google Scholar]
- 57. Yang Z., Zhou E., Wei D., Li D., Wei Z., Zhang W., and Zhang X. (2014) Emodin inhibits LPS-induced inflammatory response by activating PPAR-γ in mouse mammary epithelial cells. Int. Immunopharmacol. 21, 354–360 [DOI] [PubMed] [Google Scholar]
- 58. Qiao Y., Giannopoulou E. G., Chan C. H., Park S. H., Gong S., Chen J., Hu X., Elemento O., and Ivashkiv L. B. (2013) Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and Toll-like receptor signaling. Immunity 39, 454–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang B., Chen L., Sun Y., Zhu Y., Sun Z., An T., Li Y., Lin Y., Fan D., and Wang Q. (2015) Development of phenylboronic acid-functionalized nanoparticles for emodin delivery. J. Mater Chem. B Mater. Biol. Med. 3, 3840–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Srinivas G., Babykutty S., Sathiadevan P. P., and Srinivas P. (2007) Molecular mechanism of emodin action: transition from laxative ingredient to an antitumor agent. Med. Res Rev. 27, 591–608 [DOI] [PubMed] [Google Scholar]
- 61. National Toxicology Program (2001) NTP Toxicology and carcinogenesis studies of EMODIN (CAS no. 518-82-1) feed studies in F344/N rats and B6C3F1 mice. Natl. Toxicol. Program Tech. Rep. Ser. 493, 1–278 [PubMed] [Google Scholar]
- 62. Shia C. S., Tsai S. Y., Lin J. C., Li M. L., Ko M. H., Chao P. D., Huang Y. C., and Hou Y. C. (2011) Steady-state pharmacokinetics and tissue distribution of anthraquinones of rhei rhizoma in rats. J. Ethnopharmacol. 137, 1388–1394 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.