Abstract
Protein ubiquitination controls protein stability and subcellular localization of tyrosine kinase receptors, hence affecting signaling both quantitatively and qualitatively. In this report, we demonstrate that, after ligand stimulation, the PDGF β receptor (PDGFRβ) becomes ubiquitinated in a manner requiring both the c-Cbl and Cbl-b ubiquitin ligases. Simultaneous depletion of c-Cbl and Cbl-b resulted in reduced ligand-induced PDGFRβ clearance from the cell surface because of reduced endocytosis of the receptor. Cbl-b formed a complex with c-Cbl, as well as with the PDGFRβ, in response to PDGF-BB stimulation. We were unable to find a direct interaction between the receptor and c-Cbl, raising the possibility that Cbl-b is necessary for c-Cbl to interact with PDGFRβ. Phosphorylated Tyr-1021 in PDGFRβ was the primary interaction site for Cbl-b, with some contribution from Tyr-1009. Depletion of c-Cbl and Cbl-b led to an increased ligand-induced tyrosine phosphorylation of the receptor. Several tyrosine residues with elevated phosphorylation (i.e. Tyr-579, Tyr-581, Tyr-1009, and Tyr-1021) have previously been shown to interact with Src kinases and PLCγ. Indeed, in cells depleted of c-Cbl and Cbl-b, both Src and PLCγ phosphorylation were enhanced, whereas activation of other pathways, such as Erk1/2 MAP kinase and Akt, were not affected. In addition, Stat3 phosphorylation, which has been connected to Src activity, was also elevated in cells lacking c-Cbl and Cbl-b. Functionally, we found that cells depleted of c-Cbl and Cbl-b were more prone to migrate toward PDGF-BB, whereas no reproducible effect on cell proliferation could be observed. In conclusion, internalization as well as signaling via PDGFRβ are controlled by ubiquitination.
Keywords: CBLB, chemotaxis, E3 ubiquitin ligase, protein kinase, receptor tyrosine kinase, Src, STAT3, PLC
Introduction
The PDGF family consists of four polypeptide chains (PDGF-A, B, C, and D) that dimerize and form five biologically active growth factors (PDGF-AA, AB, BB, CC, and DD) (1). The PDGF isoforms stimulate cells of mesenchymal origin to proliferate, migrate, and survive by binding to two tyrosine kinase receptors: PDGFRα, which binds PDGF-A, B, and C chains, and PDGFRβ, which binds PDGF-B and D chains (1). Binding of the dimeric PDGF molecule causes PDGFR dimerization and, subsequently, autophosphorylation of tyrosine residues in the intracellular part of the receptor. The binding specificities of the PDGF isoforms allow for formation of both receptor homo- and heterodimers, i.e. PDGFRαα, ββ, and αβ (2). The phosphorylated tyrosine residues serve as docking sites for signaling proteins containing Src homology 2 domains and couple the activated receptor to intracellular signaling pathways (3).
The Cbl family of RING finger-containing E3 ubiquitin ligases has been implicated in ubiquitination of tyrosine kinase receptors, including PDGFRs. c-Cbl overexpression has been shown to lead to enhanced PDGFRα and β ubiquitination and degradation as well as reduced PDGF-dependent proliferation and survival (4, 5). The Cbl family consists of c-Cbl, Cbl-b, and Cbl-c (6). In their N termini, all Cbl family members have a four-helix bundle domain, a variant of an Src homology 2 domain, and a Ca2+-binding EF hand motif (6). The N-terminal domain is called tyrosine kinase binding domain, and it is critical in mediating interactions with tyrosine kinases that are Cbl substrates. The central parts of Cbl family members contain RING finger domains that control the ubiquitin ligase activity by interacting with E2-conjugating enzymes (7). In the C-terminal parts, c-Cbl and Cbl-b, but not Cbl-c, contain a proline-rich region followed by a stretch of amino acid residues with several serine and tyrosine phosphorylation sites and a ubiquitin association domain (6, 8). Notably, only the ubiquitin association domain of Cbl-b can interact with ubiquitin. Members of the Cbl family catalyze the monoubiquitination or polyubiquitination of target proteins (9, 10). These functions of the Cbl family members makes their roles complex because certain types of polyubiquitination have been associated with proteasomal degradation, whereas monoubiquitination facilitates interactions with ubiquitin association domain-containing proteins and regulates intracellular sorting (11). c-Cbl and Cbl-b have redundant as well as non-redundant functions, as observed in the immune system and the skeletons of c-Cbl and Cbl-b knockout mice (6). c-Cbl and Cbl-b contain several tyrosine residues that are phosphorylated and could possibly function as binding sites for Src homology 2 domain-containing proteins. Proteins that have been shown to interact with phosphorylated Cbl proteins include CrkL, Vav, and the p85 subunit of PI3K (6). Cbl family proteins are in many cases important negative regulators of receptor tyrosine kinase signaling, and their loss of function has been shown to lead to increased signaling, promoting tumorigenesis (12). The objective of this study was to clarify the involvement of different Cbl family members in PDGFRβ ubiquitination and investigate their roles in receptor signaling.
Experimental Procedures
Reagents
Recombinant human PDGF-BB was from RayBiotech, Inc. Antibodies against Erk1/2 (4695), phosphorylated Erk1/2 (9101), phosphorylated Akt (4060), PLCγ1 (5690), phosphorylated PLCγ1 (2821), Stat3 (4904), phosphorylated STAT3 (9145), Src (2109), phosphorylated Src (2101), c-Cbl (2747), Cbl-b (9498), and phosphorylated Tyr-857-PDGFRβ were from Cell Signaling Technology. Antibodies against Akt1/2/3 (sc-8312) and ubiquitin (sc-8017) were from Santa Cruz Biotechnology. The antibody against α-tubulin (T6074) was from Sigma-Aldrich. The rabbit polyclonal antibody against PDGFRβ was raised against a glutathione S-transferase fusion protein containing the C-terminal amino acid residues of PDGFRβ. PDGFRβ antibodies for co-immunoprecipitation (HPA028499) and for immunofluorescence staining (3169) were from Sigma-Aldrich and Cell Signaling Technology, respectively. Secondary antibodies for immunoblotting were peroxidase goat anti-mouse IgG (62-6520) and peroxidase goat anti-rabbit IgG (65-6120) from Invitrogen. Secondary antibodies for immunofluorescence staining were Alexa Fluor 594 donkey anti-rabbit (A21207) and Alexa Fluor 488 donkey anti-mouse (A21202) from Life Technologies. Pefabloc SC was from Roche. EasyTag EXPRESS35S protein labeling mixture was from PerkinElmer Life Sciences.
Cell Culture
Human foreskin fibroblasts, AG01523 (Coriell Cell Repositories) were cultured in Eagle's minimum essential medium and human BJ hTERT fibroblasts in Dulbecco's modified Eagle's medium, both supplemented with 2 mm l-glutamine and 10% FBS. For starvation, cells were washed once and incubated in the same medium containing 0.1% FBS for 20 h.
siRNA Knockdown
Down-regulation of Cbl-b and c-Cbl was performed by using 20 nm siRNA CBLBHSS101420 and 40 nm siRNA CBLHSS101418 from Invitrogen, respectively. Stealth RNAi Negative Control Duplex (12935-112) from Invitrogen was used as a control. Transfection of siRNA was done for 28 h with SiLentFect from Bio-Rad, and experiments were performed after an additional 20 h. Levels of knockdown were tested by immunoblotting.
Cell Lysis, Receptor Precipitation, and Immunoblotting
Subconfluent cells, transfected as indicated, were starved and stimulated with 20 ng/ml PDGF-BB for the indicated periods of time. Cells were washed with ice-cold PBS and lysed in 1% Nonidet, 0.5% sodium deoxycholate, 0.1% SDS, 20 mm Tris (pH 7.4), 150 mm NaCl, 1 mm Pefabloc, and 1 mm sodium orthovanadate for 20 min on ice. The lysates were cleared by centrifugation at 13,000 rpm for 15 min at 4 °C. PDGFRβ was precipitated by incubation with the indicated antibody overnight on ice, followed by 45-min incubation with Dynabeads protein A (Novex, Life Technologies) rotating at 4 °C. Beads were washed three times with ice-cold lysis buffer, and attached proteins were eluted by boiling for 5 min in reducing SDS sample buffer and then separated by SDS-PAGE. Immunoblotting was performed as described previously (13). For co-immunoprecipitation, starved cells were stimulated with 20 ng/ml PDGF-BB for the indicated periods of time and lysed in 1% Triton X-100, 20 mm Tris (pH 7.5), 150 mm NaCl, 1 mm Pefabloc, and 1 mm sodium orthovanadate and then subjected to immunoprecipitation followed by immunoblotting as described above. Densitometrical analysis of immunoblots was performed using advanced image data analyzer software (Fujifilm). The densitometrical data were normalized against unstimulated cells subjected to control treatment.
Inhibition of Proteasomal or Lysosomal Degradation
Serum-starved cells were incubated with the proteasomal inhibitor MG132 (10 μm) or the lysosomal inhibitor chloroquine (25 μm) (Sigma-Aldrich) for 4 h, followed by stimulation with 20 ng/ml PDGF-BB for the indicated periods of time. Cells were then lysed and subjected to SDS-PAGE and immunoblotting as described above.
Biotinylation and receptor Endocytosis
Serum-starved cells were incubated with 0.3 mg/ml Sulfo-NHS-SS-Biotin (Pierce) in PBS for 30 min at 4 °C to label cell surface proteins, followed by stimulation with 20 ng/ml PDGF-BB for different periods of time. Labeled receptors present on the cell surface were selectively stripped of biotin by three subsequent incubations for 15 min in ice-cold 100 mm 2-mercaptoethane sulfonic acid (Sigma-Aldrich) dissolved in 50 mm Tris (pH 8.8), 100 mm NaCl, 1 mm EDTA, and 0.2% BSA or just 50 mm Tris (pH 8.8), 100 mm NaCl, and 1 mm EDTA for control. Stripped cells were solubilized in lysis buffer (0.2% Triton X-100, 150 mm NaCl, 25 mm KCl, and 10 mm Tris (pH 7.4) supplemented with 0.1 mm EDTA, 1 mm Pefabloc, and 1 mg/ml iodoacetamide. Cleared lysates were incubated with streptavidin-Sepharose (GE Healthcare) on a rotator for 40 min at 4 °C. Protein-bound streptavidin beads were washed three times in lysis buffer, and biotinylated proteins were eluted by addition of reducing SDS sample buffer and subjected to SDS-PAGE, followed by immunoblotting.
Immunofluorescence Staining
Cells were fixed in 3.5% paraformaldehyde in PBS for 20 min at room temperature and then washed in PBS. The fixed cells were permeabilized in 0.3% Triton X-100 in PBS for 10 min, washed in PBS, and incubated in 1% BSA, 10 mm glycine in PBS for 1 h at room temperature. Primary and secondary antibodies were diluted in PBS containing 1% BSA. The cells were incubated with the primary antibodies overnight at 4 °C and with the secondary antibodies for 1 h at room temperature, followed by washing in PBS. The coverslips were mounted on object slides with Vectashield H-100 (Vector Laboratories). The cells were photographed under a fluorescence microscope (Axio Imager M2, Zeiss) with an AxioCam MRm digital camera, using a Plan-Apochromat ×63 objective lens (Zeiss). Images were analyzed with Zen software.
Pulse-Chase Experiment
After being starved for 2 h in methionine/cysteine-free MCDB 104 supplemented with 0.01% BSA, cells were pulsed with 150 μCi/ml of [35S]methionine/[35S]cysteine labeling mixture in methionine/cysteine-free MCDB 104, 0.01% BSA for 15 min at 37 °C. Cells were chased for the indicated periods of time by aspirating labeling medium and adding MCDB 104 and 0.01% BSA containing five times the labeling concentration of methionine/cysteine and 20 ng/ml PDGF-BB. Cells were washed with ice-cold PBS, followed by lysis in 1% Nonidet, 0.5% sodium deoxycholate, 0.1% SDS, 20 mm Tris (pH 7.4), 150 mm NaCl, and 1 mm Pefabloc. Samples were then subjected to immunoprecipitation as described above, followed by SDS/PAGE. The dried gel was exposed to film.
Cell Migration Assay
A ChemoTx disposable chemotaxis system (101-8, Neuro Probe Inc.) was used to study cell migration. Cells were transfected with siRNA as indicated. After serum starvation, cells were trypsinized and suspended in starvation medium containing aprotinin to inactivate trypsin. Cells were sedimented by centrifugation at 1000 rpm for 4 min and resuspended in medium with 0.1% BSA. Cells were seeded on a 96-filter ChemoTx frame (3 × 104 cells/filter) coated with 50 μg/ml fibronectin (BD Biosciences) in PBS. The filter frame was put on top of wells containing different concentration of PDGF-BB in quadruplicates, and the cells were allowed to migrate for 4 h at 37 °C. The chemotactic device was taken apart, and cells remaining on the upper surface of the filters were removed with a cell scraper, followed by rinsing in PBS. Cells on the lower side of the filters were fixed in 96% ethanol for 3 min, washed in three subsequent water baths, and stained by immersing the filter frame in undiluted Giemsa solution for 3 min, followed by three additional washes in water. After drying, the filters were analyzed in an Enspire multimode plate reader (PerkinElmer Life Sciences).
Results
Simultaneous Depletion of c-Cbl and Cbl-b Inhibits PDGF-BB-induced PDGFRβ Ubiquitination and Enhances Receptor Phosphorylation without Affecting Its Stability
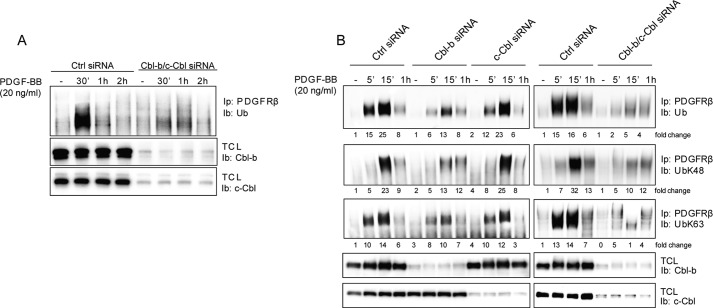
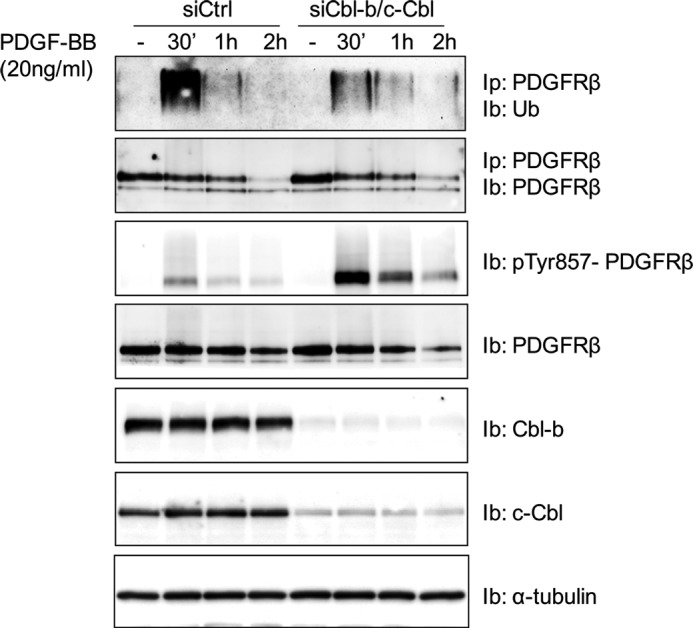
To explore the role of the Cbl family of ubiquitin ligases in the ubiquitination of PDGFR, we depleted primary fibroblasts (AG1523) of c-Cbl and Cbl-b expression using siRNAs. Cbl-c was not detected in these cells. Knockdown of either c-Cbl or Cbl-b only partially impacted PDGFRβ ubiquitination, but the combined depletion of both c-Cbl and Cbl-b robustly decreased PDGFRβ ubiquitination (Fig. 1, A and B). Using antibodies selective for Lys-48 or Lys-63 polyubiquitin chains, we found that depletion of both c-Cbl and Cbl-b or depletion of Cbl-b alone affected both types of ubiquitin chains, whereas depletion of c-Cbl only had minor effects and primarily on Lys-63 polyubiquitination chains (Fig. 1B). Thus, in contrast to studies with other cells and receptors, such as PDGFR, VEGFR2, EGF receptor, CSF-1R, c-KIT, and MET, in which loss of c-Cbl expression or binding was sufficient to decrease ubiquitination (reviewed in Ref. 6), our results showed that, in AG1523 fibroblasts, there is cooperation between c-Cbl and Cbl-b in promoting PDGFRβ ubiquitination, with Cbl-b having a dominant effect. We could not detect any ubiquitination of c-Cbl or Cbl-b in response to PDGF-BB stimulation (data not shown).
FIGURE 1.
Depletion of c-Cbl and Cbl-b results in decreased PDGFRβ ubiquitination. A, AG01523 fibroblasts were transfected with siRNAs targeting c-Cbl and Cbl-b and serum-starved overnight. Cells were then stimulated with 20 ng/ml PDGF-BB for the indicated periods of time, followed by lysis, PDGFRβ immunoprecipitation (Ip), SDS-PAGE and immunoblotting (Ib) for poly-ubiquitin (Ub, top panel). Total cell lysates (TCL) were also collected, and the efficiency of c-Cbl and Cbl-b depletion was analyzed (center and bottom panels). Ctrl, control. B, the ubiquitin chain type was analyzed using antibodies selective for Lys-48 or Lys-63 polyubiquitin chains. The experiments were repeated at least three times.
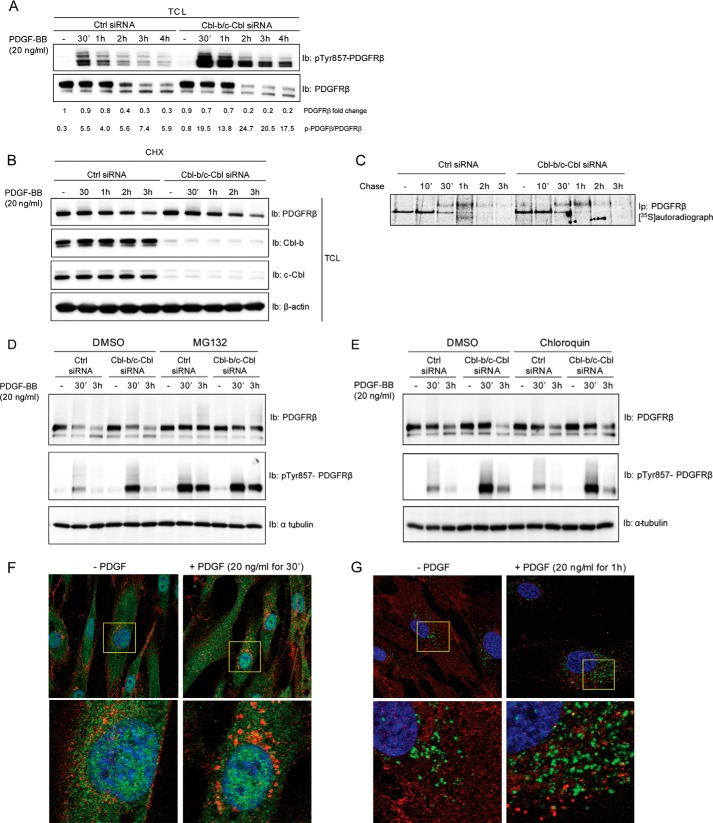
Next we investigated whether the decrease in PDGFRβ ubiquitination observed after depletion of c-Cbl and Cbl-b affected PDGF-BB-induced receptor phosphorylation. We found that, in the absence of c-Cbl and Cbl-b, there was an increased level of ligand-induced receptor phosphorylation on tyrosine residue 857 in the activation loop of the kinase domain (Fig. 2A). An increased phosphorylation of other tyrosine residues in PDGFRβ was also observed, including tyrosine residues located in the juxtamembrane region (Tyr-579/581), in the kinase insert (Tyr-751 and Tyr-771), and in the C-terminal tail of the receptor (Tyr-1009 and Tyr-1021), as determined by phosphospecific antibodies (data not shown).
FIGURE 2.
Depletion of c-Cbl and Cbl-b results in increased PDGFRβ phosphorylation but does not influence PDGF-BB-induced receptor degradation. A, AG01523 fibroblasts were transfected with siRNA targeting both c-Cbl and Cbl-b and serum-starved overnight. Cells were then stimulated with 20 ng/ml PDGF-BB for the indicated periods of time, followed by lysis. Total cell lysate (TCL) subjected to SDS-PAGE and immunoblotting (Ib) for phosphorylated Tyr-857. Ctrl, control. B, cells were treated with 20 μg/ml cycloheximide (CHX) for 30 min and stimulated for the indicated periods of time with 20 ng/ml PDGF-BB. Total extracts were prepared and separated by SDS-PAGE, and the level of PDGFRβ was analyzed by immunoblotting. C, cells were incubated for 4 h in the absence of Met and Cys and then pulsed with [35S]methionine/cysteine for 15 min, followed by chase in methionine/cysteine-containing medium for the indicated periods of time in the presence of 20 ng/ml PDGF-BB. Ip, immunoprecipitation. D and E, AG01523 fibroblasts were transfected with siRNAs targeting c-Cbl and Cbl-b and treated with a proteasomal inhibitor (MG132, D) or the lysosomal inhibitor chloroquine (E) for 4 h before stimulation with 20 ng/ml PDGF-BB for the indicated periods of time. Total extracts were prepared and separated by SDS-PAGE, and the level of PDGFRβ and phosphorylation was analyzed by immunoblotting. F and G, AG01523 fibroblasts were serum-starved overnight. Cells were then stimulated with 20 ng/ml PDGF-BB for the indicated periods of time. Subcellular localization of stimulated PDGFRβ was visualized using rabbit PDGFRβ antibodies, followed by tetramethylrhodamine isothiocyanate (TRITC)-conjugated anti-rabbit antibodies. Proteasomes were visualized using Alexa Fluor 488-conjugated antibodies against PSMD9 (F) and lysosomes using antibodies against LAMP1 (G). The bottom panels are enlargements of the cells shown in the top panels. Nuclei were stained blue with DAPI. Experiments were repeated at least three times.
Surprisingly, the rate of PDGFRβ degradation was not significantly affected by depletion of c-Cbl and Cbl-b, as shown by immunoblotting (Fig. 2B) or immunoprecipitation of the receptor from [35S]methionine/cysteine pulse-chase-labeled cells (Fig. 2C), after stimulation with PDGF-BB for different time periods. We investigated whether proteasomal inhibition influenced receptor stability because we could see clear effects on receptor ubiquitination upon depletion of c-Cbl and Cbl-b. To this end, we treated control cells and cells depleted of c-Cbl and Cbl-b with the proteasomal inhibitor MG132 and analyzed receptor stability. We found that both in the presence and absence of c-Cbl/Cbl-b, proteasomal inhibition stabilized the receptor (Fig. 2D). The almost complete inhibition of PDGFRβ degradation in the presence of MG132 suggests that the proteasome is a major degradation pathway for the receptor. In contrast, interference with lysosomal function using chloroquine did not result in stabilization of the ligand-activated receptor in cell expressing, or not expressing, c-Cbl and Cbl-b (Fig. 2E). This finding was further supported by immunofluorescence microscopy demonstrating a co-localization between PDGFRβ and proteasomes (using a PSMD9 marker) after 30 min of PDGF-BB stimulation (Fig. 2F). In contrast, we could not detect co-localization with a lysosomal marker (LAMP1) and PDGFRβ after 60 min of PDGF-BB stimulation (Fig. 2G). Also, other time periods of PDGF-BB stimulation were tested without detectable co-localization (data not shown).
Using the human fibroblast cell line BJ hTERT, we could reproduce the effect of c-Cbl/Cbl-b depletion on PDGFRβ ubiquitination, phosphorylation, and degradation (Fig. 3). In summary, c-Cbl and Cbl-b collaborate to ubiquitinate PDGFRβ, and loss of these two ubiquitin ligases leads to suppressed receptor ubiquitination and enhanced receptor phosphorylation but has no significant impact on receptor degradation, which primarily occurs by the proteasomes.
FIGURE 3.
Depletion of c-Cbl and Cbl-b in BJ hTERT fibroblast results in decreased PDGFRβ ubiquitination and increased receptor phosphorylation without affecting its degradation. BJ hTERT fibroblasts were transfected with siRNA targeting both c-Cbl and Cbl-b and serum-starved overnight. Cells were then stimulated with 20 ng/ml PDGF-BB for the indicated periods of time, followed by lysis, PDGFRβ immunoprecipitation (Ip), SDS-PAGE, and immunoblotting (Ib) for poly-ubiquitin, PDGFRβ, or phosphorylated Tyr-857 in PDGFRβ. Total cell lysates were also collected, and equal protein loading was verified by the efficiency of c-Cbl and Cbl-b depletion. α-Tubulin immunoblotting was used to verify equal protein loading. Experiments were repeated at least two times. Ctrl, control; Ub, ubiquitin.
Ligand-induced PDGFRβ Clearance from the Cell Surface Is Slower in Cells with Reduced c-Cbl and Cbl-b Expression
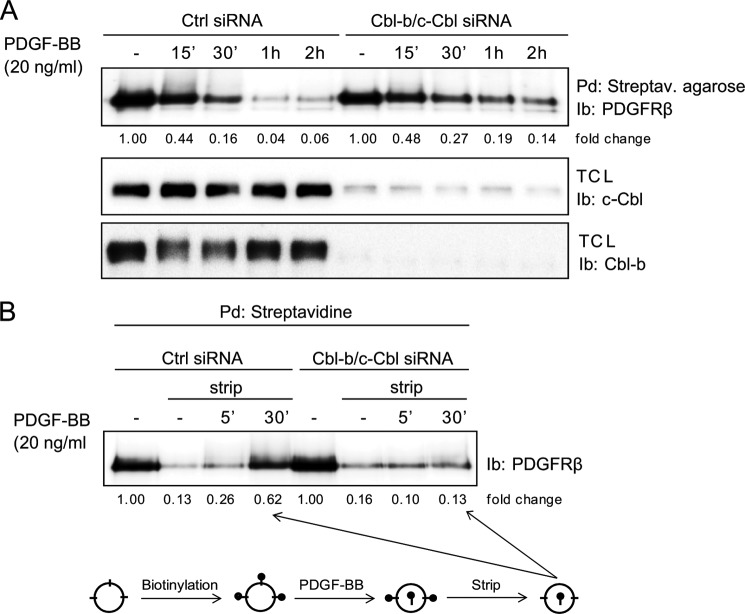
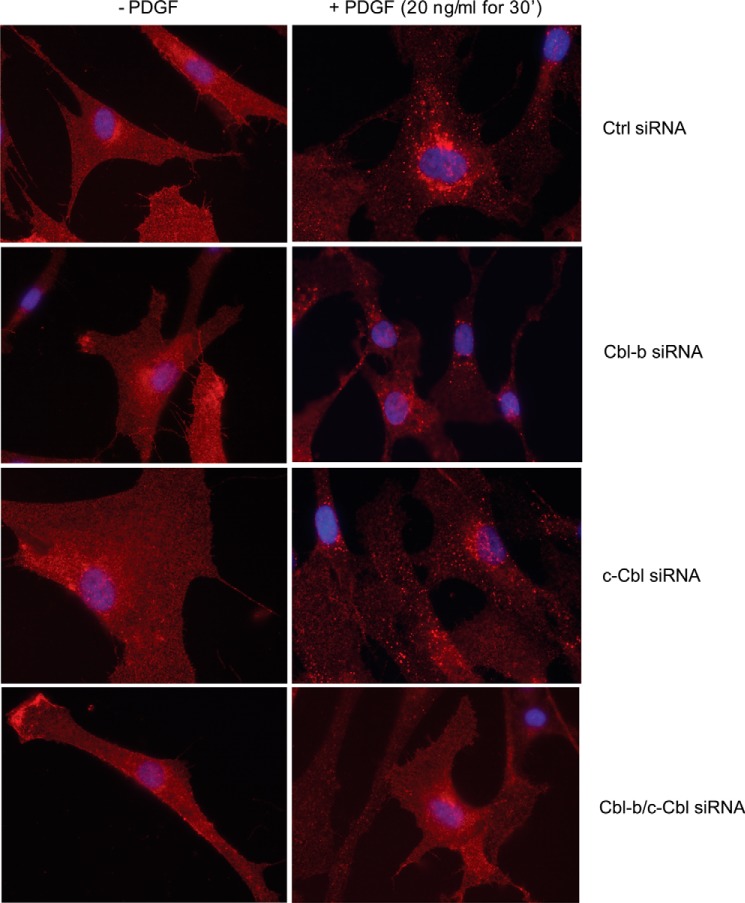
Ubiquitination has been linked to internalization and intracellular sorting of receptors (14). Therefore, we investigated whether the level of PDGFRβ ubiquitination affected the rate of cell surface clearance using biotinylation of extracellular parts of cell surface receptors. We found that, under conditions were c-Cbl and Cbl-b were depleted, the PDGF-BB-induced clearance of PDGFRβ from the cell surface was decreased compared with control conditions (Fig. 4A). We found that, in cells depleted of c-Cbl and Cbl-b, there was a clear inhibition of PDGF-BB-induced internalization of the receptor, seen as a diminished amount of biotinylated receptor accumulating in the cells (Fig. 4B). Moreover, punctuated immunofluorescence staining following PDGF-BB stimulation, which is interpreted as PDGFR accumulation in the endosomal compartment, was not observed when c-Cbl and Cbl-b were depleted, indicating an inability of the non-ubiquitinated receptor to enter the normal intracellular compartments (Fig. 5). Single c-Cbl or Cbl-b knockdown revealed that c-Cbl depletion by itself had a minimal effect, whereas a clear effect could be seen after Cbl-b knockdown (Fig. 5). This is consistent with a dominant role of Cbl-b in regulating PDGFRβ ubiquitination (Fig. 1B).
FIGURE 4.
Depletion of c-Cbl and Cbl-b results in a decreased rate of PDGF-induced PDGFRβ cell surface clearance. A, AG01523 fibroblasts were transfected with siRNA targeting both c-Cbl and Cbl-b and serum-starved overnight. Cells were treated for the indicated periods of time with 20 ng/ml PDGF-BB. After biotinylation of cell surface proteins, cells were lysed. Biotinylated proteins were precipitated with streptavidin (Streptav.)-agarose, separated on SDS-PAGE, and immunoblotted (Ib) for PDGFRβ to determine the level of receptor present at the cell surface. Ctrl, control; Pd, pulldown; TCL, total cell lysate. B, cells were biotinylated, followed by stimulation by 20 ng/ml PDGF-BB. Before lysis, cell surface biotin was removed by incubation with 20 mm sodium 2-mercaptoethanesulfonate in 50 mm Tris (pH 8.6) and 100 mm NaCl on ice. Residual sodium 2-mercaptoethanesulfonate was inactivated by incubation with 20 mm iodoacetic acid in PBS for 10 min on ice. Next, biotinylated proteins were precipitated with streptavidin-agarose, separated by SDS-PAGE, and immunoblotted for PDGFRβ. A schematic of the experiment is shown in the bottom panel. Experiments were repeated at least three times.
FIGURE 5.
Depletion of c-Cbl and Cbl-b results in defective intracellular sorting of activated PDGFRβ. AG01523 fibroblasts were transfected with siRNA targeting Cbl-b, c-Cbl, or a combination of both. After serum starvation, cells were stimulated with 20 ng/ml PDGF-BB for 30 min. Subcellular localization of stimulated PDGFRβ was visualized using rabbit PDGFRβ antibodies, followed by Alexa Fluor 594-conjugated anti-rabbit antibodies. Nuclei were stained blue with DAPI. Experiments were repeated at least three times. Ctrl, control.
c-Cbl and Cbl-b Interact with Each Other and with PDGFRβ
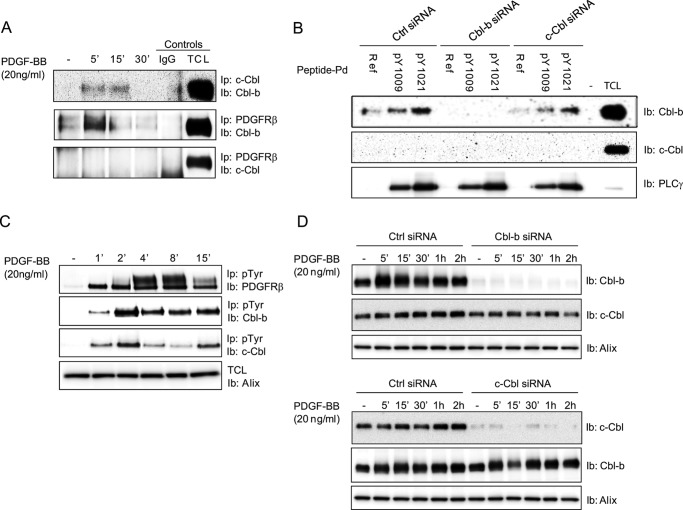
To further explore the connection between c-Cbl, Cbl-b, and PDGFRβ, we performed co-immunoprecipitation experiments. We found that c-Cbl and Cbl-b formed a transient complex with each other in response to PDGF-BB stimulation and that Cbl-b co-immunoprecipitated with the PDGFRβ after ligand stimulation (Fig. 6A). However, we were unable to observe any co-immunoprecipitation of PDGFRβ and c-Cbl. It is possible that the recruitment of c-Cbl to the activated receptor occurs indirectly through its interaction with Cbl-b or, alternatively, that the amount of c-Cbl associated with PDGFRβ is lower than the detection limit. To further map the binding site of Cbl-b and to explore the possibility of a weak interaction between c-Cbl and PDGFRβ, we used synthetic peptides corresponding to different autophosphorylation sites in the receptor. Cbl-b interacted in a phosphorylation-dependent manner primarily with Tyr-1021 in PDGFRβ and, to some extent, with Tyr-1009 (Fig. 6B), but not with Tyr-579/581, Tyr-716, Tyr-740, Tyr-751, Tyr-763, Tyr-771, Tyr-775, Tyr-857, and Tyr-1009 (data not shown). We were unable to detect any interaction between the phosphorylated Tyr-1021 peptide, or any other phosphorylated peptide, and c-Cbl. The well established interaction between PLCγ1 and phosphorylated Tyr-1009 and Tyr-1021 was used as a control (Fig. 6B). PDGF-BB stimulation induced tyrosine phosphorylation of Cbl-b (Fig. 6C). Consistent with the possibility that c-Cbl is brought into close proximity of PDGFRβ by its interaction with Cbl-b, we found that also c-Cbl was also efficiently and rapidly phosphorylated in response to PDGF-BB stimulation (Fig. 6C). Next we investigated whether c-Cbl and Cbl-b affect the stability of each other by depleting cells of either c-Cbl or Cbl-b, followed by immunoblotting for the other Cbl family member. We obtained no evidence that c-Cbl and Cbl-b regulate the stability of each other in PDGF-BB-stimulated cells (Fig. 6D). Furthermore, we were unable to detect any ubiquitination of c-Cbl or Cbl-b in response to PDGF-BB stimulation (data not shown).
FIGURE 6.
PDGF stimulation promotes formation of complexes between c-Cbl and Cbl-b and between Cbl-b and PDGFRβ. A, AG01523 fibroblasts were serum-starved overnight and treated for the indicated periods of time with 20 ng/ml PDGF-BB, followed by lysis. Lysates were subjected to immunoprecipitation (Ip) using antibodies against c-Cbl and PDGFRβ, followed by SDS-PAGE and immunoblotting (Ib) for Cbl-b (top and center panels) or c-Cbl (bottom panel). TCL, total cell lysate. B, synthetic peptides corresponding to the amino acid sequence surrounding Tyr-1009 and Tyr-1021 (phosphorylated or not) were used to investigate the interaction between c-Cbl and Cbl-b with PDGFRβ in vitro by performing pulldown (Pd) followed by immunoblotting for c-Cbl or Cbl-b. The interaction between these peptides and PLCγ was used as a positive control (Ctrl). Ref refers to the unphosphorylated peptide. C, AG01523 fibroblasts were serum-starved overnight and treated for the indicated periods of time with 20 ng/ml PDGF-BB, followed by lysis and immunoprecipitation by phospho-tyrosine antibodies. The extent of c-Cbl, Cbl-b, and PDGFRβ phosphorylation was determined by immunoblotting for these proteins. D, total cell lysate was prepared from cells transfected with either Cbl-b or c-Cbl (or control) siRNA and stimulated with 20 ng/ml PDGF-BB for the indicated periods of time. Protein extracts were separated by SDS-PAGE, and the effect on Cbl-b or c-Cbl stability was assayed by immunoblotting. Before immunoprecipitation, a small fraction of lysates was taken and analyzed for Alix expression as a loading control (C and D). Experiments were repeated at least three times.
Depletion of c-Cbl and Cbl-b Selectively Enhances PDGF-BB-induced Src, Stat3, and PLCγ Activation
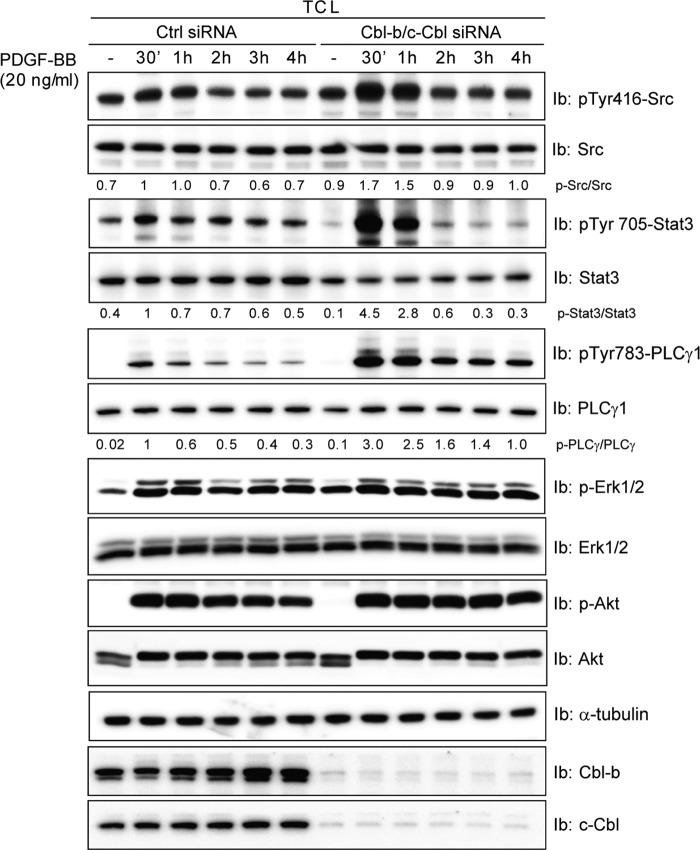
To assess whether the observed changes in PDGFRβ ubiquitination and phosphorylation after depletion of c-Cbl and Cbl-b lead to changes in PDGF-BB-induced signal transduction, we studied the phosphorylation of several well established signaling proteins downstream of PDGFRβ. We found that Src, Stat3, and PLCγ1 phosphorylation were elevated in the absence of c-Cbl and Cbl-b (Fig. 7), in contrast to the unaltered phosphorylation of Erk1/2 and Akt (Fig. 7), as well as SHP2 and p38 (data not shown). The observed changes in signaling in cells depleted of c-Cbl and Cbl-b are consistent with the increased level of phosphorylation of Tyr-579/581, which has been shown to promote recruitment of Src kinases (15), and of Tyr-1009 and Tyr-1021, which has been shown to promote PLCγ binding (16).
FIGURE 7.
c-Cbl and Cbl-b depletion results in increased Src, Stat3, and PLCγ phosphorylation. AG01523 fibroblasts were transfected with siRNA targeting both c-Cbl and Cbl-b, serum-starved overnight, and treated for the indicated periods of time with 20 ng/ml PDGF-BB. Total cell lysates (TCL) were prepared and separated by SDS-PAGE, and immunoblotting (Ib) was performed with antibodies against pPLCγ, PLCγ, pStat3, Stat3, α-tubulin, Cbl-b, c-Cbl, pSrc, or Src. For quantifications, control (Ctrl) cells stimulated with PDGF-BB for 30 min were set to 1. Experiments were repeated at least three times.
Depletion of c-Cbl and Cbl-b Enhances the Chemotactic Response toward PDGF-BB but Does Not Significantly Affect Proliferation
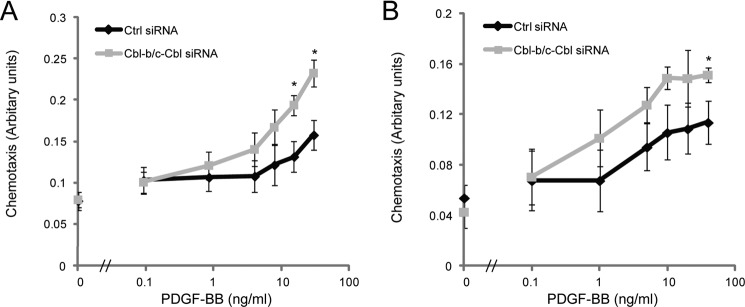
Src, Stat3, and PLCγ are central signaling proteins downstream of many receptors, including PDGFRβ, and have been connected to cell proliferation and migration (17, 18). Consistently, we found an elevated chemotactic response in cells lacking the Cbl ligases (Fig. 8). The increased chemotaxis upon combined c-Cbl and Cbl depletion was detected in both normal human fibroblasts (AG1523, Fig. 8A) and the human glioblastoma cell line U251-MG (Fig. 8B). However, we were unable to observe a significant effect on cell proliferation by depletion of c-Cbl and Cbl-b (data not shown).
FIGURE 8.
c-Cbl and Cbl-b depletion potentiates PDGF-BB-induced chemotaxis. AG01523 fibroblasts and U251MG glioblastoma cells were transfected with siRNAs targeting c-Cbl and Cbl-b, serum-starved overnight, and then plated on fibronectin-coated polycarbonate membranes (pore size, 8 μm) in 96-well microchemotaxis chambers. Cells were then allowed to migrate toward the indicated concentrations of PDGF-BB for 4 h. The membrane was then fixed in ethanol, and cells that had migrated through the membrane were stained with Giemsa. The intensity of the staining was measured using a charge-coupled device camera. Data are plotted as mean of quadruplicate samples with one standard deviation indicated. Experiments were repeated at least three times. Ctrl, control.
Discussion
The aim of this study was to elucidate the roles of the Cbl family of ubiquitin ligases in PDGFR down-regulation and signaling. We found that depletion of Cbl-b had a clear effect on PDGFRβ ubiquitination, whereas depletion of c-Cbl had only a minor impact. Depletion of both c-Cbl and Cbl-b caused a dramatic reduction of PDGFRβ ubiquitination. Furthermore, we found that Cbl-b promoted the formation of both Lys-48- and Lys-63-type polyubiquitin chains, whereas c-Cbl knockdown only led to a small decrease in Lys-63-type chains. We also observed that Lys-63 polyubiquitination occurred more rapidly than Lys-48 polyubiquitination. Collectively, these results suggest that PDGFRβ is ubiquitinated by different types of ubiquitin chains, with different kinetics, and involving at least two different ubiquitin ligases.
The decreased ubiquitination caused by the dual depletion of c-Cbl and Cbl-b correlated with a decreased rate of receptor internalization from the cell surface and to increased PDGF-BB-induced receptor phosphorylation. Consistently, an important role for ubiquitination in receptor internalization has been reported for IGF-1R (19), whereas, in contrast, studies on EGFR and FGFR1 suggested that their ubiquitination was not essential for internalization (20, 21). It is possible that the decreased internalization delays the deactivation mechanism that limits receptor activity, including exposure to phosphatases. Surprisingly, we did not observe any significant effect of Cbl-b/c-Cbl depletion on PDGF-induced receptor degradation. We could demonstrate that the dominant degradation pathway for PDGFRβ involves proteasomes. However, although the rapid Cbl-induced receptor ubiquitination was crucial for receptor internalization, it did not affect its degradation. The observation that PDGFRβ was also degraded by the proteasome when c-Cbl and Cbl-b were depleted, which led to a dramatic loss of receptor ubiquitination, was unexpected. However, there are reports showing that proteins containing disordered regions can be recognized by the proteasome without being ubiquitinated (22). Notably, 60–80% of signaling molecules have been described as intrinsically disordered (23). Alternatively, residual ubiquitination found on the receptor after c-Cbl/Cbl-b depletion, induced by low levels of remaining Cbl proteins or other ubiquitin ligases acting on the receptor, may be sufficient for proteasomal recognition. Earlier studies have shown that 125I-labeled PDGF is degraded in lysosomes (24), suggesting that the ligand and receptor are degraded by different routes. The exact mechanism and localization of PDGFRβ degradation remain to be elucidated.
The increased PDGFRβ phosphorylation translated into hyperactivation of several pathways downstream of the receptor, in particular the Src, Stat3, and PLCγ1 pathways. Our results show that Cbl-mediated ubiquitination dampens the signals emitted by the receptor, most likely by affecting its subcellular localization. A previous report has also shown increased PLCγ1 signaling in the absence of c-Cbl expression, and it was suggested that this was due to loss of competition for binding to PDGFRβ because both PLCγ1 and c-Cbl have been shown to bind phosphorylated Tyr-1021 in the receptor (5). We did not observe increased binding between endogenously expressed PDGFRβ and PLCγ1 in cells depleted of c-Cbl and Cbl-b (data not shown).
The possibility that c-Cbl directly binds to the PDGFRβ has been addressed previously but with unclear results. Studies using endogenously expressed c-Cbl have failed to detect a complex, whereas, upon overexpression of c-Cbl, an interaction between c-Cbl and PDGFRβ could be observed (4, 25). In addition, GST pulldown assays have suggested that c-Cbl may bind to the phosphorylated Tyr-1021 in PDGFRβ (5). We found that, in response to ligand stimulation, Cbl-b, but not c-Cbl, co-immunoprecipitated with the PDGFR in a transient manner consistent with the possibility that receptor ubiquitination occurs rapidly at the plasma membrane before the receptor is internalized (14). The observation that c-Cbl did not co-immunoprecipitate with PDGFRβ is also consistent with a previous report (26). Using an in vitro system, we were able to show that Cbl-b, but not c-Cbl, interacted with Tyr-1021 and, to some extent, with Tyr-1009, the most C-terminal autophosphorylation sites in PDGFRβ. We found that, in response to PDGF-BB treatment, c-Cbl and Cbl-b formed a complex, which suggests that Cbl-b primarily binds to PDGFRβ and that c-Cbl interacts indirectly with the receptor via binding to Cbl-b. We found that depletion of both c-Cbl and Cbl-b was necessary to drastically reduce receptor ubiquitination, suggesting that the functional ubiquitin ligase complex may contain both c-Cbl and Cbl-b. Individual depletion of c-Cbl or Cbl-b revealed that c-Cbl had a minor function in PDGFRβ ubiquitination, whereas Cbl-b had a dominant role.
We found that cells, both normal human fibroblasts (AG1523) and glioblastoma cells (U251MG) lacking c-Cbl and Cbl-b had a stronger chemotactic response toward PDGF compared with control cells, partially consistent with a previous study where a similar effect was seen by cells lacking only c-Cbl (5). Previous studies have shown that the effect of c-Cbl overexpression on PDGF-induced proliferation depends on the passage number of the clone (4), suggesting that the cell adapts to continued c-Cbl overexpression. To avoid the possibility of cell adaptation to changes in Cbl levels, we used primary AG1523 fibroblasts and performed transient siRNA-mediated depletion of c-Cbl and Cbl-b. In these cells, we did not observe any clear effect on cell proliferation. These results suggest that proliferation and migration depend on signals originating at different subcellular locations where prolonged cell surface exposure promotes the chemotactic, but not the proliferative, response. Consistent with the lack of effect on cell proliferation, Cbl depletion did not affect activation of the Erk1/2 MAP kinase and Akt pathways, which promote proliferation and survival.
To conclude, we found that both c-Cbl and Cbl-b contribute to polyubiquitination of PDGFRβ and that this modification is important for receptor internalization. The majority of the receptor is degraded in the proteasome, but, surprisingly, diminished ubiquitination as a consequence of Cbl knockdown did not appreciably influence the degradation rate. Functionally, depletion of c-Cbl and Cbl-b leads to increased signaling through certain pathways, i.e. Src, Stat3, and PLCγ, and increased chemotactic response.
Author Contributions
C. R. conducted most of the experiments and analyzed the results. M. T. performed the experiments and analyzed the data. C. H. H. conceived idea for the project, analyzed data, and wrote the paper together with J. L. J. L. conceived idea for the project, performed some experiments, analyzed the data, and wrote the paper together with C. H. H.
Footnotes
This work was supported by Swedish Cancer Society Grant 140332 and the Ludwig Institute for Cancer Research. The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1. Fredriksson L., Li H., and Eriksson U. (2004) The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 15, 197–204 [DOI] [PubMed] [Google Scholar]
- 2. Heldin C. H., and Westermark B. (1999) Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79, 1283–1316 [DOI] [PubMed] [Google Scholar]
- 3. Pawson T. (1995) Protein modules and signalling networks. Nature 373, 573–580 [DOI] [PubMed] [Google Scholar]
- 4. Miyake S., Mullane-Robinson K. P., Lill N. L., Douillard P., and Band H. (1999) Cbl-mediated negative regulation of platelet-derived growth factor receptor-dependent cell proliferation. A critical role for Cbl tyrosine kinase-binding domain. J. Biol. Chem. 274, 16619–16628 [DOI] [PubMed] [Google Scholar]
- 5. Reddi A. L., Ying G., Duan L., Chen G., Dimri M., Douillard P., Druker B. J., Naramura M., Band V., and Band H. (2007) Binding of Cbl to a phospholipase Cγ1-docking site on platelet-derived growth factor receptor β provides a dual mechanism of negative regulation. J. Biol. Chem. 282, 29336–29347 [DOI] [PubMed] [Google Scholar]
- 6. Mohapatra B., Ahmad G., Nadeau S., Zutshi N., An W., Scheffe S., Dong L., Feng D., Goetz B., Arya P., Bailey T. A., Palermo N., Borgstahl G. E., Natarajan A., Raja S. M., Naramura M., Band V., and Band H. (2013) Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl-family ubiquitin ligases. Biochim. Biophys. Acta 1833, 122–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Joazeiro C. A., Wing S. S., Huang H., Leverson J. D., Hunter T., and Liu Y. C. (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286, 309–312 [DOI] [PubMed] [Google Scholar]
- 8. Davies G. C., Ettenberg S. A., Coats A. O., Mussante M., Ravichandran S., Collins J., Nau M. M., and Lipkowitz S. (2004) Cbl-b interacts with ubiquitinated proteins; differential functions of the UBA domains of c-Cbl and Cbl-b. Oncogene 23, 7104–7115 [DOI] [PubMed] [Google Scholar]
- 9. Mosesson Y., Shtiegman K., Katz M., Zwang Y., Vereb G., Szollosi J., and Yarden Y. (2003) Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 278, 21323–21326 [DOI] [PubMed] [Google Scholar]
- 10. Thien C. B., Walker F., and Langdon W. Y. (2001) RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol. Cell 7, 355–365 [DOI] [PubMed] [Google Scholar]
- 11. Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., and Dikic I. (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5, 461–466 [DOI] [PubMed] [Google Scholar]
- 12. Kales S. C., Ryan P. E., Nau M. M., and Lipkowitz S. (2010) Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 70, 4789–4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lennartsson J., Ma H., Wardega P., Pelka K., Engström U., Hellberg C., and Heldin C. H. (2013) The Fer tyrosine kinase is important for platelet-derived growth factor-BB-induced signal transducer and activator of transcription 3 (STAT3) protein phosphorylation, colony formation in soft agar, and tumor growth in vivo. J. Biol. Chem. 288, 15736–15744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haglund K., and Dikic I. (2012) The role of ubiquitylation in receptor endocytosis and endosomal sorting. J. Cell Sci. 125, 265–275 [DOI] [PubMed] [Google Scholar]
- 15. Mori S., Rönnstrand L., Yokote K., Engström A., Courtneidge S. A., Claesson-Welsh L., and Heldin C. H. (1993) Identification of two juxtamembrane autophosphorylation sites in the PDGF β-receptor: involvement in the interaction with Src family tyrosine kinases. EMBO J. 12, 2257–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Valius M., Bazenet C., and Kazlauskas A. (1993) Tyrosines 1021 and 1009 are phosphorylation sites in the carboxy terminus of the platelet-derived growth factor receptor β subunit and are required for binding of phospholipase C γ and a 64-kilodalton protein, respectively. Mol. Cell. Biol. 13, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y. Z., Wharton W., Garcia R., Kraker A., Jove R., and Pledger W. J. (2000) Activation of Stat3 preassembled with platelet-derived growth factor β receptors requires Src kinase activity. Oncogene 19, 2075–2085 [DOI] [PubMed] [Google Scholar]
- 18. Bornfeldt K. E., Raines E. W., Graves L. M., Skinner M. P., Krebs E. G., and Ross R. (1995) Platelet-derived growth factor: distinct signal transduction pathways associated with migration versus proliferation. Ann. N.Y. Acad. Sci. 766, 416–430 [DOI] [PubMed] [Google Scholar]
- 19. Mao Y., Shang Y., Pham V. C., Ernst J. A., Lill J. R., Scales S. J., and Zha J. (2011) Polyubiquitination of insulin-like growth factor I receptor (IGF-IR) activation loop promotes antibody-induced receptor internalization and down-regulation. J. Biol. Chem. 286, 41852–41861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang F., Kirkpatrick D., Jiang X., Gygi S., and Sorkin A. (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 21. Haugsten E. M., Malecki J., Bjørklund S. M., Olsnes S., and Wesche J. (2008) Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol. Biol. Cell 19, 3390–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Erales J., and Coffino P. (2014) Ubiquitin-independent proteasomal degradation. Biochim. Biophys. Acta 1843, 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iakoucheva L. M., Brown C. J., Lawson J. D., Obradović Z., and Dunker A. K. (2002) Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 323, 573–584 [DOI] [PubMed] [Google Scholar]
- 24. Heldin C. H., Wasteson A., and Westermark B. (1982) Interaction of platelet-derived growth factor with its fibroblast receptor: demonstration of ligand degradation and receptor modulation. J. Biol. Chem. 257, 4216–4221 [PubMed] [Google Scholar]
- 25. Bonita D. P., Miyake S., Lupher M. L. Jr., Langdon W. Y., and Band H. (1997) Phosphotyrosine binding domain-dependent up-regulation of the platelet-derived growth factor receptor alpha signaling cascade by transforming mutants of Cbl: implications for Cbl's function and oncogenicity. Mol. Cell. Biol. 17, 4597–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galisteo M. L., Dikic I., Batzer A. G., Langdon W. Y., and Schlessinger J. (1995) Tyrosine phosphorylation of the c-cbl proto-oncogene protein product and association with epidermal growth factor (EGF) receptor upon EGF stimulation. J. Biol. Chem. 270, 20242–20245 [DOI] [PubMed] [Google Scholar]