Abstract
A critical function for skin is that when damaged it must simultaneously identify the nature of the injury, repair barrier function, and limit the intrusion of pathogenic organisms. These needs are carried out through the detection of damage-associated molecular patterns (DAMPs) and a response that includes secretion of cytokines, chemokines, growth factors, and antimicrobial peptides (AMPs). In this study, we analyzed how non-coding double-stranded RNA (dsRNAs) act as a DAMP in the skin and how the human cathelicidin AMP LL-37 might influence growth factor production in response to this DAMP. dsRNA alone significantly increased the expression of multiple growth factors in keratinocytes, endothelial cells, and fibroblasts. Furthermore, RNA sequencing transcriptome analysis found that multiple growth factors increase when cells are exposed to both LL-37 and dsRNA, a condition that mimics normal wounding. Quantitative PCR and/or ELISA validated that growth factors expressed by keratinocytes in these conditions included, but were not limited to, basic fibroblast growth factor (FGF2), heparin-binding EGF-like growth factor (HBEGF), vascular endothelial growth factor C (VEGFC), betacellulin (BTC), EGF, epiregulin (EREG), and other members of the transforming growth factor β superfamily. These results identify a novel role for DAMPs and AMPs in the stimulation of repair and highlight the complex interactions involved in the wound environment.
Keywords: antimicrobial peptide (AMP), double-stranded RNA (dsRNA), fibroblast growth factor (FGF), insulin-like growth factor (IGF), keratinocyte, vascular endothelial growth factor (VEGF), betacellulin, cathelicidin
Introduction
Epithelial tissues provide essential barriers to the external environment. The skin barrier, in particular, functions through control of moisture, ion transport, pH, lipid content, and antimicrobial actions (1). In humans, protection against invasive microorganisms is aided by the production of antimicrobial peptides (AMPs)2 produced by various epithelia including but not limited to defensins (2–4), cathelicidin (LL-37) (5), psoriasin (6), RNases (7, 8), and antimicrobial chemokines such as CXCL14 (9). Importantly, some AMPs such as LL-37 are induced upon wounding (10, 11) or ultraviolet (UV) injury (12, 13). Because intense UV radiation disrupts normal homeostasis in the skin epithelium, the production of AMPs is important to compensate for the loss of the physical barrier. Coincident with this response, UV or physical wounding also leads to cell death and the production of damage-associated molecular patterns (DAMPs) including double-stranded RNA (dsRNA) (14). These DAMPS can interact with pattern recognition receptors such as the Toll-like receptors (TLRs) to identify and respond to external and internal threats. Thus, AMPs and DAMPs are two critical elements of epithelial function and are essential for responding to injury. However, the mechanisms by which these molecules act in response to skin wounding are incompletely understood.
Production of AMPs is stimulated by several forms of injury including a low dosage of UV exposure (15, 16). LL-37 has broad antimicrobial properties but also directly influences host immune and repair responses including re-epithelialization of skin wounds (16, 17). Through its charge and amphipathic nature, LL-37 promiscuously binds multiple DAMP molecules such as single-stranded DNA (18), single-stranded RNA (19), dsRNA (20), and bacterial polysaccharides (21, 22). LL-37 is also an immunogenic effector molecule can that alter the signal response of TLRs to a LL-37·DAMP complex such as enhancing dsRNA sensing through TLR3 (23) or inhibiting LPS sensing through TLR4 (24).
Skin wounds normally contain both AMPs and DAMPs such as dsRNAs. Due to the ability of LL-37 and the synthetic dsRNA polyinosinic acid:polycytidylic acid (poly(I:C)) to individually stimulate epithelial repair and prior observations that these two molecules can interact, we hypothesized that the LL-37 and dsRNA may alter the production of growth factors important for wound repair. This investigation evaluated the response of multiple growth factors produced by primary cultures of human keratinocytes, endothelial cells, and fibroblasts. Our results suggest that LL-37 may alter wound repair by modifying the responses to dsRNAs and that this response is cell type-specific.
Experimental Procedures
Cell Culture and Stimuli
Normal human epidermal keratinocytes (NHEKs) were obtained from Life Technologies (catalogue number C-001-5C) and grown in serum-free EpiLife cell culture medium (Thermo Fisher) for up to seven passages under standard tissue culture conditions described previously (25). Cells were cultured until 60–80% confluence and then treated with 0.1 μg/ml poly(I:C) (Invivogen, San Diego, CA), 1 μg/ml U1 RNA (created via in vitro transcription), and/or LL-37 (1.75 or 4 μm; Genemed Synthesis Inc., San Antonio, TX) in 12-well flat bottom plates (Corning Life Sciences, Lowell, MA) for up to 24 h. After cell stimulation, 200 μl of supernatant was collected, and then RNA was extracted using TRIzol reagent (Invitrogen). RNA was stored at −80 °C until use.
Quantitative Real Time PCR
Total RNA was extracted from cultured keratinocytes using TRIzol reagent. Total RNA was quantified using a Nanodrop 2000/200c spectrophotometer (Thermo Fisher). Typically 500–1000 ng of total RNA was reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad). mRNA transcript levels were evaluated by TaqMan Gene Expression Assays using 6-carboxyfluorescein-labeled predeveloped probes (Thermo Fisher). As a control, mRNA transcripts were assessed using a factor V Leiden minor groove binder (VIC/MGB)-labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe described previously (25). The GAPDH probe was used as an internal control when quantifying interleukin 6 (IL-6), basic fibroblast growth factor (FGF2), heparin-binding EGF-like growth factor (HBEGF), transforming growth factor (TGF) α, and TGFβ mRNA transcription and an external control in evaluating epidermal growth factor (EGF), vascular endothelial growth factor C (VEGFC), betacellulin (BTC), amphiregulin (AREG), and epiregulin (EREG) expression. Analyses were performed with biological triplicates and were representative of two to five independent experiments. Quantitative PCR was analyzed with an ABI Prism 7300 Sequence Detection System (Thermo Fisher). -Fold induction relative to GAPDH was calculated using the ΔΔCt method. Results were considered to be significant if p < 0.05.
Growth Factor Enzyme-linked Immunoadsorbent Assay (ELISA) Analysis
Supernatant from cell cultures was collected prior to RNA isolation and stored at −80 °C until use. ELISA analysis was performed using a MILLIPLEX Human Angiogenesis/Growth Factor Magnetic Bead Panel (HAGP1MAG-12K, Millipore, Billerica, MA) kit following the manufacturer's instruction.
In Vitro Transcription of Self-noncoding RNA (ncRNA)
ncRNA was generated from template DNA using an Ampliscribe T7-Flash Transcription kit from Epicenter (an Illumina Co., Madison, WI). Templates used for reactions were gel-purified PCR products from primer pairs previously published (25).
Protein Extraction and Immunoblotting
NHEKs were lysed in a denaturing lysis buffer containing 20 mm HEPES, pH 7.4, 250 mm NaCl, 2 mm EDTA, and 1% SDS supplemented with Complete proteinase inhibitor mixture as well as 50 mm sodium fluoride, 5 mm N-ethylmaleimide, and 100 μm hemin chloride to maximally preserve protein post-translational modifications as described previously (26). Lysates were boiled for 3 min and then homogenized by sonication using a digital sonifier (Branson, Ferguson, MO) followed by centrifugation to remove DNA and cell debris. Protein concentrations were measured by BCA protein assay kit (Thermo Fisher). For immunoblotting, 20 μg of protein was separated on a 10% Tris-glycine precast gel (Bio-Rad) and transferred to PVDF membrane (Bio-Rad) followed by immunoblotting using the indicated primary antibodies followed by fluorescent secondary antibodies (LI-COR Biosciences, Lincoln, NE) and imaging using the Odyssey System (LI-COR Biosciences). Rabbit anti-phospho-p38, mouse anti-p38, rabbit anti-phospho-AKT, mouse anti-AKT, rabbit anti-IKK, rabbit anti-NF-κB, rabbit anti-phospho-EKR1/2, and mouse anti-ERK1/2 were from Cell Signaling Technology (Danvers, MA).
Drug Treatment/UV Irradiation of Human Biopsies
Skin punch biopsies of adults (male and female) ranging from 45 to 64 years old were suspended in a drop of EpiLife medium. For UVB irradiation, biopsies were dosed one time with 5 kJ/m2 UVB irradiation or no irradiation for control. Each 3-mm biopsy was transferred to an individual well in a 24-well plate (Corning) and suspended in EpiLife medium with the dermal side submerged. For drug-treated biopsies, 64 μm LL-37, 1.0 μg/ml poly(I:C), or both 64 μm LL-37 and 1.0 μg/ml poly(I:C) were added to the EpiLife medium. After a 24-h incubation, tissue was homogenized via bead beating in Trizol followed by isolation of the aqueous layer via phenol-chloroform extraction and RNA extraction and purification using a Purelink RNA Mini kit (Thermo Fisher). Replicates of tissue not used for RNA isolate were immediately frozen in optimum cutting temperature (O.C.T.) compound (VWR International, Radnor, PA) and stored at −80 °C until immunohistochemistry was performed.
Immunohistochemistry
Skin tissue sections were sliced using a Microm HM525 cryostat (Thermo Fisher) to ∼7 μm. Sections were adhered to Fisherbrand Superfrost Plus slides (Thermo Fisher) and left to air dry for 30 min before being placed in 4 °C for short term storage or −20 °C for long term storage. Unfixed slides were fixed and permeabilization via a 10-min incubation in −20 °C acetone, washed three times with phosphate-buffered saline (PBS), blocked for 1 h with 5% normal goat serum in PBS, pH 7.2, supplemented with 1% rehydrated powdered skim milk, 1% fish gelatin, 0.1% Triton X-100, 0.05% Tween 20, 0.05% sodium azide, and 0.3 m glycine. Slides were incubated overnight with primary antibodies normal rabbit IgG (sc-2027) or anti-FGF basic antibody (ab72316) (Abcam, Cambridge, MA), each with a dilution ratio of 1:300. Slides were washed four times in PBS with 0.05% Tween 20 (PBS-T), and then Alexa Fluor 568 secondary antibody (Thermo Fisher) was applied at a 1:500 dilution. Slides were incubated for 30 min at room temperature before being washed four times in PBS-T. Finally one drop of DAPI Fluoromount-G (SouthernBiotech, Birmingham, AL) was added, and the slide was allowed to dry overnight before imaging.
siRNA Knockdown
Control, TLR3, and MAVS Dharmacon siRNA were purchased from GE Healthcare. NHEK cells were trypsinized, split equally into two aliquots of ∼3 million cells, and subsequently transfected with 1 nmol of siRNA using an Amaxa Human Keratinocyte Nucleofector kit (Lonza, Basel, Switzerland) by electroporation via an Amaxa Nucleofector II device (Lonza) set to the T-18 high efficiency neonatal keratinocyte transfection setting. After transfection, aliquots were pooled together in a 10-cm plate. After 24 h, cells were washed with PBS, trypsinized, and plated into 12-well plates with a medium change 24 h after plating. Treatment with LL-37, poly(I:C), or LL-37/poly(I:C) co-treatment began 24 h later (72 h post-transfection). Cells were collected, and RNA was isolated after a 16-h treatment using a Purelink RNA isolation kit.
NHEK Inhibition
NHEKs were washed with PBS, trypsinized, and plated into 12-well plates, and medium was changed 24 h after plating. Cells were pretreated with various inhibitors for 30 min prior to the addition of LL-37, poly(I:C), or LL-37/poly(I:C) co-treatment 24 h after the previous medium change. Cells were collected, and RNA was isolated after a 16-h treatment using a Purelink RNA isolation kit. Final concentrations of inhibitors were 1 μm SB202190 (p38 MAPK inhibitor), 100 nm AG1478 (a selective inhibitor of epidermal growth factor receptor kinase (EGFR)), 10 nm bafilomycin A1 (blocks endosomal acidification), and 1 μm IKK-2 Inhibitor IV (inhibitor of IKK-2). SB202190 and AG1478 were purchased from Calbiochem EMD Millipore. IKK-2 Inhibitor IV was purchased from EMD Millipore. Bafilomycin A1 was purchased from Sigma.
RNA-seq Analysis
P5 NHEKs were treated with 1.75 μm LL-37, 0.1 μg/ml poly(I:C), or LL-37 and poly(I:C) co-treatment for 24 h before RNA was isolated by a Purelink RNA isolation kit. RNA samples were then submitted to the University of California San Diego Institute for Genomic Medicine for analysis by Tapestation bioanalyzer (Agilent, LA Jolla, CA), and samples with RNA integrity number values above 9.7 were submitted for Illumina library preparation (Illumina, San Diego, CA) and subsequent RNA sequencing using a high output run V4 platform (Illumina) in a single read 50-cycle run. Subsequent analysis was done on Partek flow software (Partek, St. Louis, MO) using the Tophat2 aligner to quantify relative to the transcriptome using hg19 RefSeq database version 15_05_07_v2.
Gene analysis used Partek Gene-Specific Analysis (GSA) run on the gene level with the following changes: lowest maximum coverage, 10; Poisson, true; and trimmed mean of M values (TMM) normalization. Gene filters used were p < 0.05 and-fold expression > 2. Hierarchical clustering and GO enrichment were performed using these filters utilizing Partek flow software.
Results
Growth Factor Expression Is Stimulated by dsRNA and LL-37
ncRNAs are released from mammalian cells after they are disrupted by injury, and some of those that are dsRNA have been shown to serve as endogenous signals for inflammation and differentiation (27, 28). In this study, we hypothesized that dsRNA might also induce growth factor production by keratinocytes, thus facilitating the repair process after injury. To test this hypothesis, we cultured NHEKs with poly(I:C), a synthetic dsRNA.
The human cathelicidin antimicrobial peptide LL-37 is abundantly released in normal skin after injury (29, 30). Production of the human cathelicidin gene CAMP is sensitive enough to be stimulated by a single dose of UVB irradiation (16); thus CAMP transcription can quickly respond to UVB exposure. LL-37 has been shown to induce re-epithelialization of skin wounds (17, 31, 32). Because LL-37 is an abundant component of the wound environment and has been shown to influence wound repair, we also investigated whether LL-37 could modify the production of growth factors induced by dsRNA.
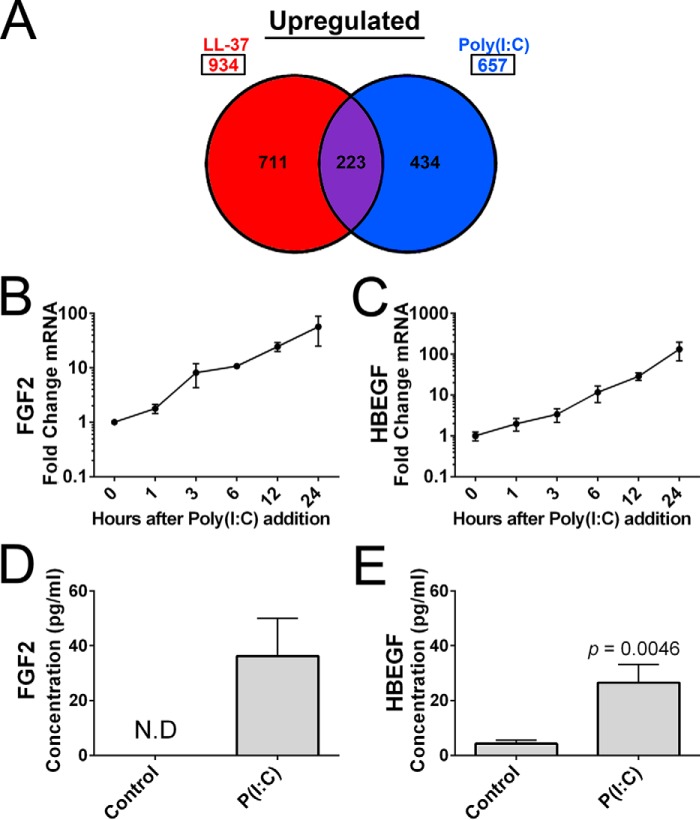
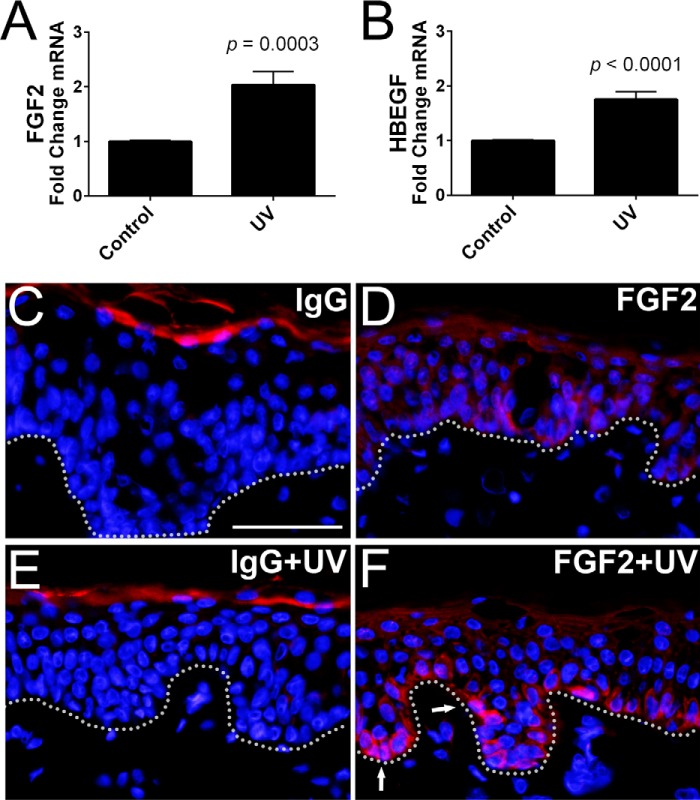
RNA-seq analysis of NHEKs incubated for 24 h with LL-37 or poly(I:C) revealed that LL-37 up-regulated 934 genes (≥2-fold, p < 0.05), whereas poly(I:C)-stimulated cells had 657 genes up-regulated (≥2-fold, p < 0.05) with 223 genes commonly up-regulated by either treatment (Fig. 1A). Heparin-binding growth factors FGF2 and HBEGF were two of the 223 commonly up-regulated genes, up-regulated 8.65- and 7.63-fold, respectively, by 0.1 μg/ml poly(I:C) and up-regulated 32.66- and 7.13-fold, respectively, by 1.75 μm LL-37 (Table 1). As expected, several other growth factors and cytokines were also observed to increase in transcript abundance after exposure to poly(I:C) or LL-37 (Table 1). Top enriched genes identified through gene ontology analysis revealed that LL-37 stimulation included genes involved in RNA binding, cell cycle, and metabolism, whereas poly(I:C) stimulation included genes involved immune system-based responses involving interferons and cytokine signaling and production (data not shown). Interestingly, LL-37 stimulation had greater enrichment scores for cellular growth processes or regulation than poly(I:C)-stimulated NHEKs. A time-dependent increase in mRNA for FGF2 and HBEGF was observed after poly(I:C) treatment, verifying the stimulatory effect of poly(I:C) (Fig. 1, B and C). An increase in FGF2 and HBEGF protein released into the culture supernatant was also detected by ELISA (Fig. 1, D and E). Human skin punch biopsies exposed to a single dose of UVB irradiation (5 kJ/m2) had increased mRNA abundance of FGF2 and HBEGF (Fig. 2, A and B). Because FGF2 was strongly up-regulated by poly(I:C) treatment, replicate punch biopsies used in Fig. 2, A and B, were immunostained with IgG control and anti-FGF2 antibodies (Fig. 2, C–F). After UVB irradiation (5 kJ/m2), basal layer keratinocytes had an increase in the expression of FGF2 compared with the non-irradiated control (Fig. 2, D and F). The increase in FGF2 expression in basal keratinocytes was also detectable at a lower dosage of UVB irradiation (2 kJ/m2) (data not shown).
FIGURE 1.
NHEKs have a growth factor response to UVB poly(I:C), or LL-37 stimulation. A, analysis of RNA-seq gene profiling for genes in NHEKs after a 24-h exposure to 0.1 μg/ml poly(I:C) or 4 μm LL-37. Up-regulated genes with a greater than 2-fold change and a significant p value (p < 0.05) are represented. B and C, qPCR measurements of FGF2 and HBEGF cDNA created from 1 μg of RNA isolated from NHEKs at the indicated time points after the addition of 0.1 μg/ml of poly(I:C) (P(I:C)). D and E, ELISA of supernatant of NHEKs treated with 0.1 μg/ml poly(I:C). p values were calculated by a two-tailed Student's t test. The time course qPCR and the ELISA data are mean ± S.D. of biological replicates, n = 3, and are representative data from at least three independent experiments. Samples not detected by ELISA are indicated by the abbreviation N.D. Error bars represent S.D.
TABLE 1.
RNA-seq analysis indicates multiple growth factor genes are up-regulated by poly(I:C) or LL-37
RNA-seq gene profiling was carried out for growth factors and select cytokines in NHEKs after a 24-h exposure to dsRNA mimic 0.1 μg/ml poly(I:C) or 1.75 μm LL-37. Represented genes were up-regulated a minimum of 2-fold or greater and had a significant p value (p < 0.05). Ligands along with their respective binding receptor(s) and calculated p values and -fold change when the treatment condition was compared with untreated control (Ctrl) NHEKs are listed.
| Growth factor ligand | Growth factor ligand gene | Binding receptor | p value | -Fold change (Ctrl vs. poly(I:C)) |
|---|---|---|---|---|
| Up-regulated by poly(I:C) | ||||
| Colony-stimulating factor 2 | CSF2 | CSF2R | 3.01E − 02 | 107.26 |
| Interleukin 6 | IL-6 | IL-6R | 2.75E − 02 | 27.51 |
| Inhibin, βA | INHBA | ACVR2A | 2.08E − 30 | 20.99 |
| Colony-stimulating factor 3 | CSF3 | CSF3R | 4.47E − 07 | 13.08 |
| Fibroblast growth factor 2 (basic) | FGF2 | FGFR1–4 | 1.57E − 11 | 8.65 |
| Heparin-binding EGF-like growth factor | HBEGF | ERBB1, -4 | 4.90E − 18 | 7.63 |
| Bone morphogenetic protein 2 | BMP2 | BMPR1, -2 | 8.74E − 06 | 6.82 |
| Colony-stimulating factor 1 | CSF1 | CSF1R | 2.51E − 02 | 3.95 |
| Vascular endothelial growth factor C | VEGFC | VEGFR2, -R3 | 7.27E − 21 | 3.26 |
| Vascular endothelial growth factor A | VEGFA | VEGFR1, -R2 | 2.79E − 19 | 2.53 |
| Transforming growth factor α | TGFα | ERBB1 | 1.78E − 17 | 2.43 |
| Up-regulated by LL-37 | ||||
| Interleukin 6 | IL-6 | IL-6R | 8.56E − 03 | 50.49 |
| Fibroblast growth factor 2 (basic) | FGF2 | FGFR1–4 | 9.20E − 30 | 32.66 |
| Growth differentiation factor 15 | GDF15 | ERBB2 | 2.02E − 06 | 19.16 |
| Inhibin, βA | INHBA | ACVR2A | 5.37E − 18 | 10.46 |
| Heparin-binding EGF-like growth factor | HBEGF | ERBB1, -4 | 8.06E − 17 | 7.13 |
| Bone morphogenetic protein 2 | BMP2 | BMPR1, -2 | 7.10E − 04 | 4.52 |
| Vascular endothelial growth factor A | VEGFA | VEGFR1, -R2 | 5.87E − 32 | 3.25 |
| Epiregulin | EREG | ERBB1, -4 | 1.47E − 21 | 3.02 |
FIGURE 2.
UVB irradiation leads to increased expression of FGF2 in basal keratinocytes. A and B, qPCR of FGF2 and HBEGF from whole skin biopsies after a 24-h incubation in EpiLife medium following either no UVB irradiation (Control) or a 5 kJ/m2 dose of UVB irradiation (UV). C–F, replica human punch biopsies from A and B were visualized via immunofluorescence of either non-irradiated human skin punch biopsies with IgG control (IgG), or a 5 kJ/m2 UV dose (IgG + UV) showed background staining in the stratum corneum but not in basal keratinocytes. Anti-FGF2 staining detected low levels of expression in non-irradiated basal keratinocytes (FGF2) and increased FGF2 expression after a 5 kJ/m2 UV dose (FGF2 + UV) (indicated by arrows). p values were calculated by a two-tailed Student's t test. Human biopsy qPCR data were from technical triplicates from n = 4 patients. The dotted line denotes the boundary between epidermal basal layer keratinocytes and the dermis. The scale bar represents 50 μm. Error bars represent S.E.
LL-37 Alters Growth Factor and Inflammatory Cytokine Responses to Poly(I:C)
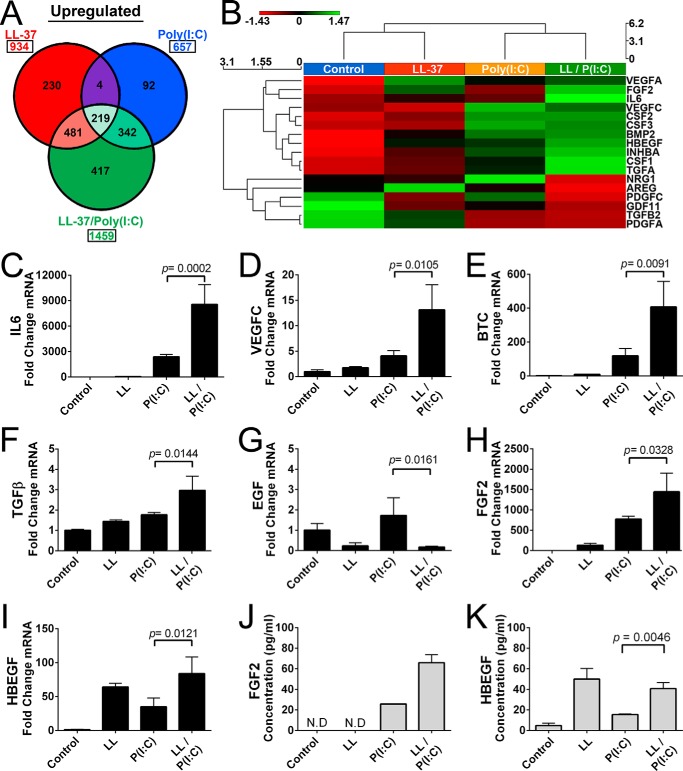
Co-stimulation of NHEKs with poly(I:C) and LL-37 greatly increased the number of up-regulated genes with a total of 1459 genes up-regulated (≥2-fold, p < 0.05) (Fig. 3A). Gene ontology analysis of NHEKs co-treated with poly(I:C) and LL-37 included many cell cycle, organelle, and cellular processes (data not shown). Hierarchical clustering reveals that poly(I:C) or LL-37 stimulation has different effects on growth factors, but co-stimulation with both LL-37 and poly(I:C) dramatically alters growth factor stimulation, usually having positive synergy (Fig. 3B). RNA-seq data indicated multiple cytokines and growth factors were up-regulated, and several genes were selected to verify the effect of poly(I:C) and/or LL-37 on mRNA abundance via qPCR. The cytokine IL-6 and growth factors VEGFC, BTC, and TGFβ1 were stimulated by either LL-37 or poly(I:C) alone (Fig. 3, C–F). When LL-37 was combined with poly(I:C), a further induction was observed in NHEKs for some factors (i.e. IL-6, VEGFC, BTC, and TGFβ). Thus co-stimulation of NHEKs with LL-37 and poly(I:C) had positive synergy (Fig. 3, C–F). Multiple other growth factors had a similar pattern to with significant p values such as TGFα (p = 0.0007), EREG (p < 0.0001), and AREG (p = 0.0045) (data not shown). However, not all tested genes had positive synergy. In NHEKs treated with LL-37 or co-treated with LL-37 and poly(I:C), there was a decrease in expression of EGF (Fig. 3G). Two growth factors with a strong response to LL-37 and poly(I:C) were selected to determine whether protein expression matched the increase in mRNA transcript. Similar to the response of the previously tested growth factors, exposure of NHEKs to poly(I:C) or LL-37 alone led to an increase in mRNA abundance of FGF2 and HBEGF (Fig. 3, H and I). Supernatant from treated NHEKs in Fig. 3, H and I, were subjected to ELISA, and an increase in secreted protein levels of FGF2 and HBEGF was detected that correlated to their previously measured relative mRNA abundance (Fig. 3, J and K). Furthermore, stimulatory and inhibitory responses of growth factors and cytokines were dose-dependent on their treatment with either LL-37 or poly(I:C) (data not shown).
FIGURE 3.
Growth factor response to poly(I:C) is enhanced by LL-37. A, analysis of RNA-seq gene profiling for genes in NHEKs after a 24-h exposure to dsRNA mimic 0.1 μg/ml poly(I:C) (P(I:C)), 4 μm LL-37 (LL), or 0.1 μg/ml poly(I:C) and 4 μm LL-37 (LL/P(I:C)) co-stimulation. Genes represented follow the guidelines established in Fig. 1. B, heat map generated from selected cytokines and growth factors from A. C–I, qPCR measurement of IL-6, VEGFC, BTC, EGF, FGF2, and HBEGF from NHEKs following protocols described in Fig. 1. J and K, ELISA of supernatant of NHEKs from H and I, respectively. p values were calculated by one-way analysis of variance. qPCR data are mean ± S.D. of biological replicates, n = 3, and are representative data from four independent experiments. Samples not detected by ELISA are indicated by the abbreviation N.D. Error bars represent S.D.
Growth Factor Expression by Dermal Endothelial Cells and Fibroblasts Is Enhanced by the dsRNA in the Presence of LL37
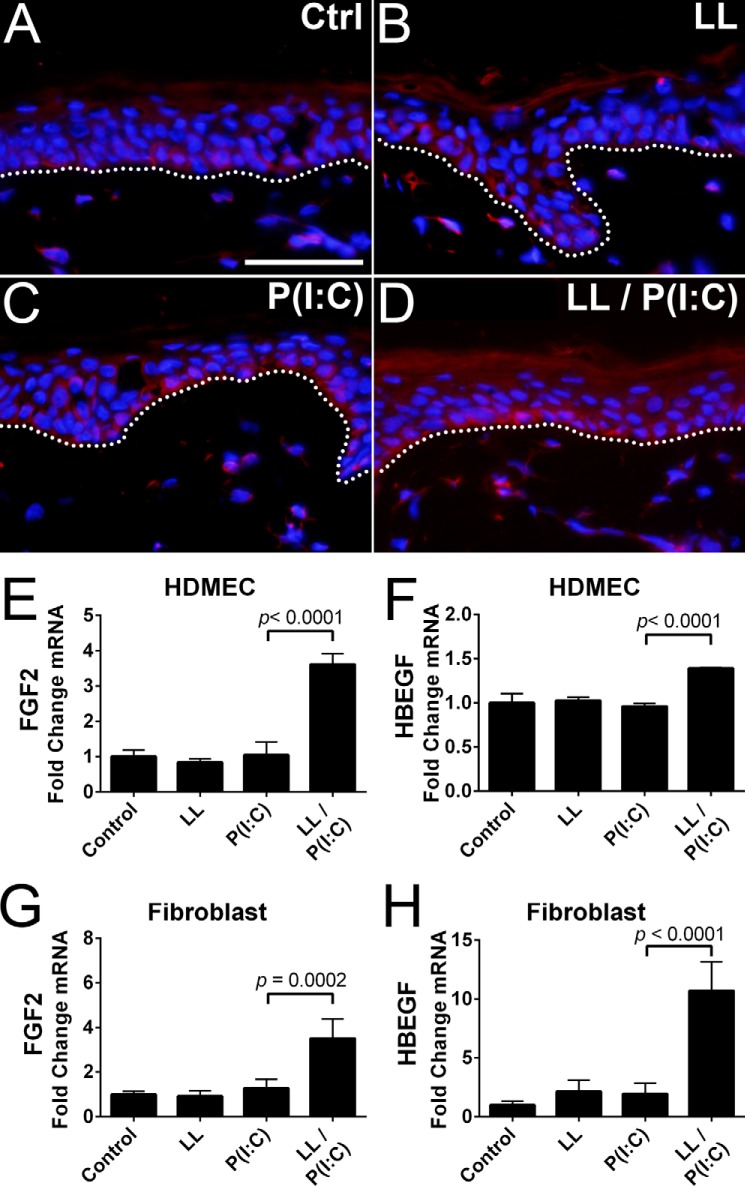
To determine whether these results occur in a more complex model, we treated human punch biopsies with 64 μm LL-37 or 1 μg/ml poly(I:C) or co-treated with LL-37 and poly(I:C) in EpiLife followed by FGF2 immunofluorescence staining. IgG-treated control had only nonspecific staining in the stratum corneum (data not shown), whereas control had slight expression of FGF2 mainly in the basal layer (Fig. 4A). Treatment with LL-37 or poly(I:C) showed an increase in FGF2 expression in basal keratinocytes compared with control (Fig. 4, B and C). Co-treatment with both LL-37 and poly(I:C) increased FGF2 expression compared with either poly(I:C) or LL-37 alone (Fig. 4D). These observations demonstrated that the keratinocyte growth factor response may be significantly modified in the physiologic wound environment where both AMPs and DAMPs are present.
FIGURE 4.
Poly(I:C) and LL-37 can stimulate FGF2 expression in basal layer keratinocytes. A–D, immunofluorescence staining of human skin punch biopsies with IgG control (IgG) antibodies indicated no background staining in basal keratinocytes. Anti-FGF2 antibodies detected low levels of FGF2 expression in basal keratinocytes of untreated control tissue (Ctrl). Tissue incubated with LL-37 (LL) or poly(I:C) (P(I:C)) had increased FGF2 expression in basal keratinocytes. Tissue co-incubated with poly(I:C) and LL-37 (LL/P(I:C)) had increased FGF2 expression compared with individual treatments. E and F, qPCR measurement of FGF2 and HBEGF from HDMECs using the treatment protocol described in Fig. 1. G and H, qPCR measurement of FGF2 and HBEGF from fibroblasts protocols described in E and F. Staining was performed on replicate punch biopsies of human skin from Fig. 2. The dotted line denotes the boundary between epidermal basal layer keratinocytes and the dermis. The scale bar represents 50 μm. p values were calculated by one-way analysis of variance. Data are mean ± S.D. of biological replicates, n = 3, and are representative data from at least three independent experiments. Error bars represent S.D.
The wound environment is a complex system that includes several cell types. To explore whether the production of growth factors after exposure to dsRNA and LL-37 is cell type-specific, we next examined mRNA abundance in cultured human dermal microvascular endothelial cells (HDMECs) and fibroblasts. Compared with NHEKs, the response of these cell types to either poly(I:C) or LL-37 was significantly weaker (Fig. 4, E–H). These cell types had distinct responses that were markedly different from NHEKs. Stimulation of HDMECs with either poly(I:C) or LL-37 alone had a minor effect on most growth factor and cytokine transcription. Co-stimulation with poly(I:C) and LL-37 led to a small but significant increase in FGF2 and HBEGF (Fig. 4, E and F).
For a majority of tested growth factors such as FGF2 (Fig. 4G), fibroblasts had a similar response as HDMECs. However, some growth factors varied significantly such as HBEGF where co-stimulation with poly(I:C) and LL-37 stimulation led to a significant increase in mRNA abundance in contrast to the weaker response of HDMECs (Fig. 4H). Co-stimulation of fibroblasts with LL-37 and poly(I:C) led to an increase in transcription of IL-6 and BTC with smaller increases in VEGFC, AREG, and TGFα (data not shown). In total, these observations demonstrate that the presence of LL-37 can significantly alter the production of growth factors in response to dsRNA and that these effects are specific for both the cell type and the growth factor studied.
Keratinocytes Respond to U1 ncRNA in the Presence of LL-37
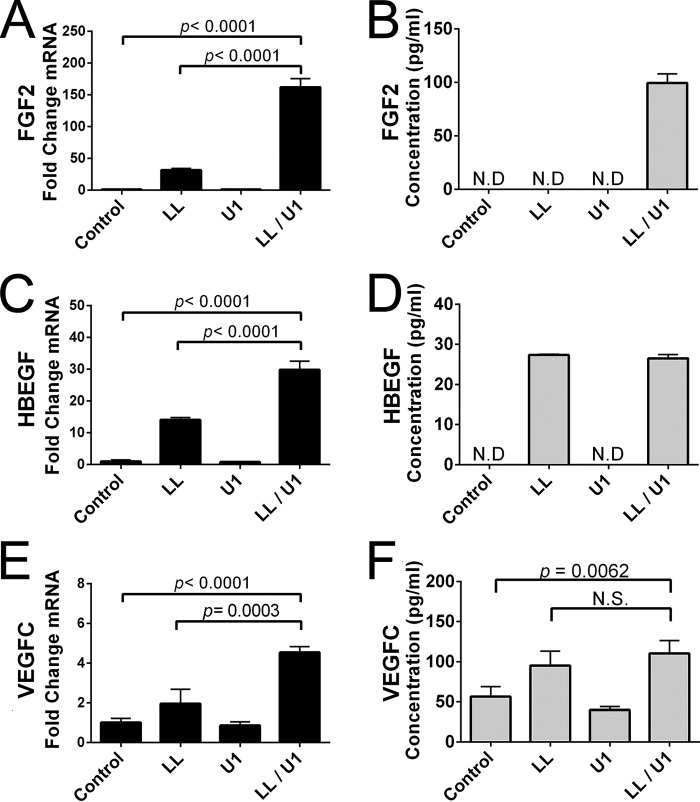
The self-noncoding RNA U1 (U1) is released following damaging UVB irradiation and is likely one of the major RNAs present in the physiologic environment after UV injury (14). This ncRNA forms dsRNA loop structures and activates TLR3 in a manner similar to poly(I:C) to stimulate increased production of inflammatory cytokines (14). To determine whether this endogenous mammalian ncRNA was also influenced by LL-37 and thus confirm that prior responses observed to poly(I:C) were reflective of endogenous responses to dsRNAs released after injury, we next evaluated the expression of FGF2, HBEGF, and VEGFC mRNA by NHEK after treatment with U1 and/or LL37. By itself, U1 was unable to induce transcript abundance of these growth factors (Fig. 5, A, C, and E). However, in the presence of LL-37, both FGF2 and HBEGF were greatly increased. Other growth factors and cytokines tested that had a similar response to U1 and LL37 include BTC (p < 0.0001), EREG (p < 0.0001), TGFα (p < 0.0001), TGFβ1 (p = 0.002), and IL-6 (p < 0.0001). Detection of growth factors in supernatant of NHEKs incubated for 24 h with LL-37, U1, or both LL-37 and U1 generally reflected the mRNA abundance of each condition (Fig. 5, B, D, and F). Interestingly, FGF2 was detected only during co-stimulation with LL-37 and U1, whereas incubation of NHEKs with LL-37 alone or with U1 led to detectable amounts of HBEGF and VEGFC in the supernatant (Fig. 5, B, D, and F). These data correlate with earlier NHEK experiments and implicate that LL-37 alone can alter the expression of HBEGF (Fig. 3, I and K, and data not shown). These observations confirmed that LL-37 and U1, which are both present in human skin following skin injury, can synergize to enhance growth factor production from primary cultures of normal human keratinocytes.
FIGURE 5.
Small ncRNAs induce growth factor expression. A, C, and E, qPCR measurement of FGF2, HBEGF, and VEGFC from cDNA created from 500 ng of RNA isolated from NHEK cells 24 h after the addition of 1.0 μg/ml ncRNA U1 (U1), 1.75 μm LL-37 (LL), or co-treatment with 1.0 μg/ml ncRNA U1 and 1.75 μm LL-37 (LL/U1). B, D, and E, ELISA of supernatant of fibroblasts treated in E and F. p values were calculated by one-way analysis of variance. Data are mean ± S.D. of biological replicates, n = 3, and are representative data from at least two independent experiments (ELISAs) or from four independent experiments (qPCR). Samples not detected by ELISA are indicated by the abbreviation N.D. Error bars represent S.D.
Activation of p38, NF-κB, or Endosomal Acidification Induces Growth Factors by Keratinocytes
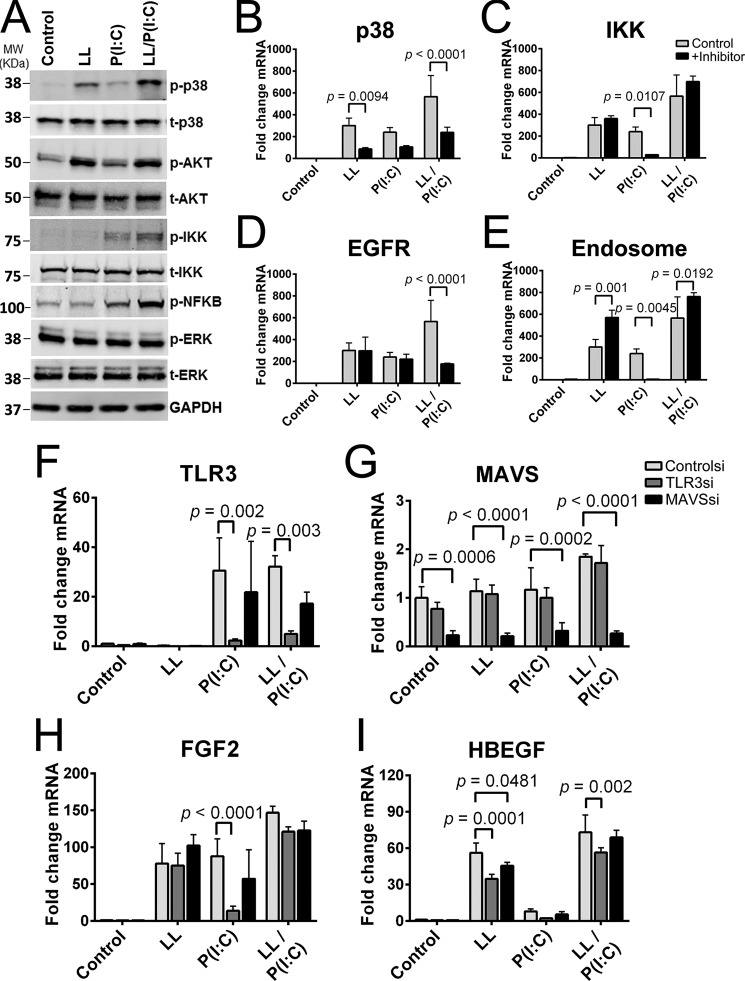
Western blotting comparing phosphorylated proteins with total protein was used to determine the signaling pathways involved in poly(I:C) and/or LL-37 stimulation of NHEKs (Fig. 6A). LL-37 strongly stimulated p38 mitogen-activated protein kinase and PI3K-AKT pathways, whereas poly(I:C) stimulated the NF-κB signaling pathway through phosphorylation of IKK and NF-κB. Co-stimulation with LL-37 and poly(I:C) led to a synergistic increase of the phosphorylation of p38 mitogen-activated protein kinase, IKK, and NF-κB (Fig. 6A).
FIGURE 6.
LL-37 affects multiple signal pathways and has synergy with poly(I:C) in stimulating the NF-κB pathway. A, keratinocytes were pretreated with LL-37 followed by stimulation with poly(I:C) for 2 h before cells were harvested for immunoblotting analyses using antibodies against phosphorylated and total forms of the following proteins: p38, AKT, IKK, NF-κB, and ERK. B–E, keratinocytes were preincubated with either p38, IKK, EGFR, or an endosomal inhibitor for 30 min prior to stimulation with 4 μm LL-37 (LL) and 0.1 μg/ml poly(I:C) (P(I:C)) for 16 h before RNA was isolated and FGF2 mRNA was quantified via qPCR. F–I, siRNA knockdown was performed on keratinocytes 72 h before stimulation with 4 μm LL-37 and 0.1 μg/ml poly(I:C) for 16 h before cells were collected, RNA was isolated, and qPCR measurements were performed. p values were calculated by two-way analysis of variance. Data are mean ± S.D. of biological replicates, n = 3, and are representative data from at least two or more independent experiments. Error bars represent S.D.
Inhibitors were next used to determine pathways required for the induction of FGF2. A p38 inhibitor reduced FGF2 mRNA induction by LL-37 (p = 0.0094) and LL-37 and poly(I:C) co-treatment (p < 0.0001) (Fig. 6B). An IKK-2 inhibitor, which prevents NF-κB signaling, was able to inhibit FGF2 induction by poly(I:C) stimulation (p = 0.0107), but combined LL-37 and poly(I:C) co-treatment was not (Fig. 6C). Use of a selective agent against EGFR did not have a significant effect on FGF2 expression induced either by LL-37 or poly(I:C) but did have a significant effect (p < 0.0001) on co-treatment with LL-37 and poly(I:C) (Fig. 6D). An inhibitor of endosomal acidification was able to significantly inhibit FGF2 induction by poly(I:C) (p = 0.0045) (Fig. 6E). However, endosomal acidification led to increased FGF2 induction by LL-37 and co-stimulation with LL-37 and poly(I:C).
We next performed siRNA-mediated knockdowns of TLR3 and MAVS to further test the mechanism of how LL-37 and poly(I:C) can induce FGF2 or HBEGF. These receptors were selected as knockdown targets because of their role in sensing dsRNA (33), including endogenous dsRNA (14), and the ability of nucleic acids to interact with LL-37 (23). 3 days postknockdown, NHEKs were stimulated with LL-37 and/or poly(I:C), and a partial silencing of TLR3 and MAVS was confirmed (Fig. 6, F and G). FGF2 induction by LL-37 was not significantly altered by TLR3 or MAVS knockdown (Fig. 6H). However, TLR3 knockdown did significantly reduce (p < 0.0001) the induction of FGF2 by poly(I:C) alone. A similar pattern of response was seen with HBEGF (Fig. 6I).
Discussion
A primary requirement for mammalian life is for the skin to repair itself after physical injury. Many redundant events are programmed into this essential process including inflammation, growth, and differentiation of many different cell types that reside in normal skin. It is recognized that the release of dsRNAs from necrotic cells can serve as an important signal of damage (34, 35) and thus is a DAMP. TLR3, which recognizes the presence of dsRNA, has classically been studied as an initiator of inflammation, and consistent with this role it has been shown that UV-induced inflammation is partly dependent on TLR3 recognition of dsRNA released by the irradiation event (14). In this study, we asked whether the response to dsRNA may be more than inflammation and evaluated whether cell types present in the skin might also produce growth factors important for wound repair. We report that the production of multiple growth factors is significantly increased by activation via TLR3. Furthermore, we demonstrate that the antimicrobial peptide LL-37 can stimulate production of certain growth factors and acts as an essential cofactor for mediation of this response to dsRNA signaling. Several pathways appear to be stimulated by LL-37 or poly(I:C) and lead to the induction of growth factors. Thus our findings uncover a previously unappreciated growth factor response by keratinocytes when in a wound environment containing dsRNA and LL-37.
The ability of LL-37 alone to modulate inflammatory responses was known to be dependent on the cell type and its TLR expression profile (1, 36, 37). Poly(I:C)-induced activation of macrophages, dendritic cells, and microglial cells was attenuated by LL-37 (1), whereas LL-37 and poly(I:C) co-stimulation synergistically up-regulate IFN-β in keratinocytes (38) and IL-8 and IL-6 in bronchial epithelial cells (39). The role of TLR3 was expanded when it was demonstrated that UVB irradiation altered a self-noncoding RNA U1 into an active form that was able to activate TLR3 (14). Utilizing poly(I:C), it also has been demonstrated that TLR3 activation leads to an increase in expression of genes involved in barrier repair and normal recovery after UV damage that is dependent on TLR3 signaling (25, 28). In a wounding model, TLR3−/− mice had a diminished IL-6 and TNFα response, indicating the role of TLR3 in inflammation after wounding (27). Wound healing was also shown to be delayed in TLR3−/− mice as opposed to wild-type mice where TLR3 mRNA and protein expression was up-regulated after wounding (40). When taken together with our work, these previous observations support our hypothesis that the enhanced growth factor response triggered by the combination of LL-37 and dsRNA is physiologically significant.
The responses observed in vitro by cultured primary cells were specific to the growth factor and cell type studied. Keratinocytes co-stimulated with poly(I:C) or ncRNA U1 and LL-37 had synergy, resulting in increased growth factor abundance detected through qPCR and/or ELISA. However, the synergistic effect of LL-37 and poly(I:C) on growth factor expression was not the same in fibroblasts. Fibroblasts treated with both poly(I:C) and LL-37 had positive synergy for some growth factors. In contrast, the effect of co-treatment with poly(I:C) and LL-37 on HDMECs was more muted compared with NHEKs or fibroblasts. Interestingly, LL-37 alone was sufficient to up-regulate some growth factors in NHEKs such as HBEGF. This observation is consistent with prior reports of accelerated re-epithelialization by addition of LL-37 alone (41) and that LL-37 transactivates EGFR (42).
The cause of altered signaling by the combination of dsRNA and LL-37 remains opaque. One hypothesis has been that LL-37 enhances the entry of nucleic acids into the endosome, thus facilitating TLR3 recognition of dsRNA (18). The presence of the cathelicidin precursor protein has also been found to facilitate nucleic acid associations with TLR in the endolysosome (43). LL-37 has been shown to bind dsRNA and traffic it to endosomes where it releases the dsRNA in a pH-dependent manner (44). Additionally, structural changes of LL-37·ligand complex were shown by electron microscopy illustrating that LL-37 and poly(I:C) individually formed globular structures, but a complex of the two formed filamentous structures (23). Thus, the LL-37 data presented here correlate with previous evidence supporting the role of LL-37 in facilitating the response to nucleic acids. Our findings provide a functional example of these ideas in action where dsRNA leads to the eventual induction of growth factors. The addition of LL-37 may enhance this induction through cooperative signaling or via an increase in dsRNA uptake.
The growth factor TGFα has been shown to induce AMP expression in keratinocytes (45), whereas HBEGF was shown to stimulate EGFR, leading to the induction of antimicrobial peptides (46). When coupled with our results of synergy between dsRNA and LL-37 leading to growth factor production, we can envision a positive feedback loop. In a wound scenario, synergy between basal levels of LL-37 and dsRNA produced from either UV injury or necrosis stimulates growth factor production. These growth factors appear to have multiple roles in wound repair as they are able to (i) stimulate proliferation to accelerate tissue regrowth and (ii) induce antimicrobial peptides to start a feedback loop. The importance of this feedback loop can be seen in human wound healing models using topical treatment with LL-37 (41) or poly(I:C) (47) resulting in an increased rate of wound closure. Therefore, future studies are justified to investigate the relationship between LL-37/dsRNA and growth factors that may form a feedback loop to promote wound healing. Inhibition of this feedback loop may assist in hyperproliferation-based diseases (e.g. psoriasis), whereas enhancement can lead to increased wound regeneration in pathological conditions of abnormal wound repair (e.g. diabetic ulcers).
Author Contributions
C. A. A performed a majority of the experiments and wrote the manuscript. A. W. B. conducted the initial research for the paper, gave technical assistance, and reviewed the manuscript. L-j. Z. performed phosphoblotting experiments, assisted with knockout and inhibitor experiments, gave technical assistance, and reviewed the manuscript. M. R. W. provided the ncRNA U1, gave technical assistance, and reviewed the manuscript. E. S. assisted with knockout experiments, gave technical assistance, and reviewed the manuscript. J. A. S. prepared RNA-seq samples and assisted with RNA-seq analysis, gave technical assistance, and reviewed the manuscript. R. L. G. supervised and designed experiments and wrote and prepared the manuscript. All authors reviewed and approved the final version of the manuscript.
Acknowledgment
We thank all the members of the Gallo laboratory for the input and stimulating conversations.
This work was supported by National Institutes of Health Grants R01 AR052728, R01 AR064781, R01 AI116576, R01 AI052453 (to R. L. G.) and T32 AR062496 (to C. A. A. and M. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Raw and processed data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE79343.
- AMP
- antimicrobial peptide
- RNA-seq
- RNA sequencing
- DAMP
- damage-associated molecular pattern
- dsRNA
- double-stranded RNA
- HBEGF
- heparin-binding EGF-like growth factor
- qPCR
- quantitative PCR
- BTC
- betacellulin
- EREG
- epiregulin
- TLR
- Toll-like receptor
- NHEK
- normal human epidermal keratinocyte
- AREG
- amphiregulin
- ncRNA
- self-noncoding RNA
- IKK
- IκB kinase
- MAVS
- mitochondrial antiviral signaling
- EGFR
- epidermal growth factor receptor kinase
- HDMEC
- human dermal microvascular endothelial cell
- U1
- self-noncoding RNA U1.
References
- 1. Hasan M., Ruksznis C., Wang Y., and Leifer C. A. (2011) Antimicrobial peptides inhibit polyinosinic-polycytidylic acid-induced immune responses. J. Immunol. 187, 5653–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fulton C., Anderson G. M., Zasloff M., Bull R., and Quinn A. G. (1997) Expression of natural peptide antibiotics in human skin. Lancet 350, 1750–1751 [DOI] [PubMed] [Google Scholar]
- 3. Harder J., Bartels J., Christophers E., and Schröder J. M. (1997) A peptide antibiotic from human skin. Nature 387, 861. [DOI] [PubMed] [Google Scholar]
- 4. Harder J., Bartels J., Christophers E., and Schroder J. M. (2001) Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276, 5707–5713 [DOI] [PubMed] [Google Scholar]
- 5. Frohm M., Agerberth B., Ahangari G., Stâhle-Bäckdahl M., Lidén S., Wigzell H., and Gudmundsson G. H. (1997) The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J. Biol. Chem. 272, 15258–15263 [DOI] [PubMed] [Google Scholar]
- 6. Gläser R., Harder J., Lange H., Bartels J., Christophers E., and Schröder J. M. (2005) Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat. Immunol. 6, 57–64 [DOI] [PubMed] [Google Scholar]
- 7. Harder J., and Schroder J. M. (2002) RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 277, 46779–46784 [DOI] [PubMed] [Google Scholar]
- 8. Fischer S., Nishio M., Dadkhahi S., Gansler J., Saffarzadeh M., Shibamiyama A., Kral N., Baal N., Koyama T., Deindl E., and Preissner K. T. (2011) Expression and localisation of vascular ribonucleases in endothelial cells. Thromb. Haemost. 105, 345–355 [DOI] [PubMed] [Google Scholar]
- 9. Maerki C., Meuter S., Liebi M., Mühlemann K., Frederick M. J., Yawalkar N., Moser B., and Wolf M. (2009) Potent and broad-spectrum antimicrobial activity of CXCL14 suggests an immediate role in skin infections. J. Immunol. 182, 507–514 [DOI] [PubMed] [Google Scholar]
- 10. Dorschner R. A., Pestonjamasp V. K., Tamakuwala S., Ohtake T., Rudisill J., Nizet V., Agerberth B., Gudmundsson G. H., and Gallo R. L. (2001) Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J. Invest. Dermatol. 117, 91–97 [DOI] [PubMed] [Google Scholar]
- 11. Schmid P., Grenet O., Medina J., Chibout S. D., Osborne C., and Cox D. A. (2001) An intrinsic antibiotic mechanism in wounds and tissue-engineered skin. J. Invest. Dermatol. 116, 471–472 [DOI] [PubMed] [Google Scholar]
- 12. Seo S. J., Ahn S. W., Hong C. K., and Ro B. I. (2001) Expressions of β-defensins in human keratinocyte cell lines. J. Dermatol. Sci. 27, 183–191 [DOI] [PubMed] [Google Scholar]
- 13. Kim J. E., Kim B. J., Jeong M. S., Seo S. J., Kim M. N., Hong C. K., and Ro B. I. (2005) Expression and modulation of LL-37 in normal human keratinocytes, HaCaT cells, and inflammatory skin diseases. J. Korean Med. Sci. 20, 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bernard J. J., Cowing-Zitron C., Nakatsuji T., Muehleisen B., Muto J., Borkowski A. W., Martinez L., Greidinger E. L., Yu B. D., and Gallo R. L. (2012) Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat. Med. 18, 1286–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hong S. P., Kim M. J., Jung M. Y., Jeon H., Goo J., Ahn S. K., Lee S. H., Elias P. M., and Choi E. H. (2008) Biopositive effects of low-dose UVB on epidermis: coordinate upregulation of antimicrobial peptides and permeability barrier reinforcement. J. Invest. Dermatol. 128, 2880–2887 [DOI] [PubMed] [Google Scholar]
- 16. Mallbris L., Edström D. W., Sundblad L., Granath F., and Stahle M. (2005) UVB upregulates the antimicrobial protein hCAP18 mRNA in human skin. J. Invest. Dermatol. 125, 1072–1074 [DOI] [PubMed] [Google Scholar]
- 17. Heilborn J. D., Nilsson M. F., Kratz G., Weber G., Sørensen O., Borregaard N., and Ståhle-Bäckdahl M. (2003) The cathelicidin anti-microbial peptide LL-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J. Invest. Dermatol. 120, 379–389 [DOI] [PubMed] [Google Scholar]
- 18. Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y. H., Homey B., Cao W., Wang Y. H., Su B., Nestle F. O., Zal T., Mellman I., Schröder J. M., Liu Y. J., and Gilliet M. (2007) Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449, 564–569 [DOI] [PubMed] [Google Scholar]
- 19. Ganguly D., Chamilos G., Lande R., Gregorio J., Meller S., Facchinetti V., Homey B., Barrat F. J., Zal T., and Gilliet M. (2009) Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J. Exp. Med. 206, 1983–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsukura S., Kokubu F., Kurokawa M., Kawaguchi M., Ieki K., Kuga H., Odaka M., Suzuki S., Watanabe S., Takeuchi H., Kasama T., and Adachi M. (2006) Synthetic double-stranded RNA induces multiple genes related to inflammation through Toll-like receptor 3 depending on NF-κB and/or IRF-3 in airway epithelial cells. Clin. Exp. Allergy 36, 1049–1062 [DOI] [PubMed] [Google Scholar]
- 21. Ciornei C. D., Sigurdardóttir T., Schmidtchen A., and Bodelsson M. (2005) Antimicrobial and chemoattractant activity, lipopolysaccharide neutralization, cytotoxicity, and inhibition by serum of analogs of human cathelicidin LL-37. Antimicrob. Agents Chemother. 49, 2845–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zughaier S. M., Svoboda P., Pohl J., Stephens D. S., and Shafer W. M. (2010) The human host defense peptide LL-37 interacts with Neisseria meningitidis capsular polysaccharides and inhibits inflammatory mediators release. PLoS One 5, e13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lai Y., Adhikarakunnathu S., Bhardwaj K., Ranjith-Kumar C. T., Wen Y., Jordan J. L., Wu L. H., Dragnea B., San Mateo L., and Kao C. C. (2011) LL37 and cationic peptides enhance TLR3 signaling by viral double-stranded RNAs. PLoS One 6, e26632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshioka M., Fukuishi N., Kubo Y., Yamanobe H., Ohsaki K., Kawasoe Y., Murata M., Ishizumi A., Nishii Y., Matsui N., and Akagi M. (2008) Human cathelicidin CAP18/LL-37 changes mast cell function toward innate immunity. Biol. Pharm. Bull. 31, 212–216 [DOI] [PubMed] [Google Scholar]
- 25. Borkowski A. W., Kuo I. H., Bernard J. J., Yoshida T., Williams M. R., Hung N. J., Yu B. D., Beck L. A., and Gallo R. L. (2015) Toll-like receptor 3 activation is required for normal skin barrier repair following UV damage. J. Invest. Dermatol. 135, 569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang L. J., Bhattacharya S., Leid M., Ganguli-Indra G., and Indra A. K. (2012) Ctip2 is a dynamic regulator of epidermal proliferation and differentiation by integrating EGFR and Notch signaling. J. Cell Sci. 125, 5733–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lai Y., Di Nardo A., Nakatsuji T., Leichtle A., Yang Y., Cogen A. L., Wu Z. R., Hooper L. V., Schmidt R. R., von Aulock S., Radek K. A., Huang C. M., Ryan A. F., and Gallo R. L. (2009) Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 15, 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borkowski A. W., Park K., Uchida Y., and Gallo R. L. (2013) Activation of TLR3 in keratinocytes increases expression of genes involved in formation of the epidermis, lipid accumulation, and epidermal organelles. J. Invest. Dermatol. 133, 2031–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koczulla R., von Degenfeld G., Kupatt C., Krötz F., Zahler S., Gloe T., Issbrücker K., Unterberger P., Zaiou M., Lebherz C., Karl A., Raake P., Pfosser A., Boekstegers P., Welsch U., Hiemstra P. S., Vogelmeier C., Gallo R. L., Clauss M., and Bals R. (2003) An angiogenic role for the human peptide antibiotic LL-37/hCAP-18. J. Clin. Investig. 111, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scott M. G., Davidson D. J., Gold M. R., Bowdish D., and Hancock R. E. (2002) The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J. Immunol. 169, 3883–3891 [DOI] [PubMed] [Google Scholar]
- 31. Carretero M., Escámez M. J., García M., Duarte B., Holguín A., Retamosa L., Jorcano J. L., Río M. D., and Larcher F. (2008) In vitro and in vivo wound healing-promoting activities of human cathelicidin LL-37. J. Invest. Dermatol. 128, 223–236 [DOI] [PubMed] [Google Scholar]
- 32. Bowdish D. M., Davidson D. J., and Hancock R. E. (2006) Immunomodulatory properties of defensins and cathelicidins. Curr. Top. Microbiol. Immunol. 306, 27–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alexopoulou L., Holt A. C., Medzhitov R., and Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 34. Seya T., Shime H., Takaki H., Azuma M., Oshiumi H., and Matsumoto M. (2012) TLR3/TICAM-1 signaling in tumor cell RIP3-dependent necroptosis. Oncoimmunology 1, 917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karikó K., Ni H., Capodici J., Lamphier M., and Weissman D. (2004) mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 279, 12542–12550 [DOI] [PubMed] [Google Scholar]
- 36. Kandler K., Shaykhiev R., Kleemann P., Klescz F., Lohoff M., Vogelmeier C., and Bals R. (2006) The anti-microbial peptide LL-37 inhibits the activation of dendritic cells by TLR ligands. Int. Immunol. 18, 1729–1736 [DOI] [PubMed] [Google Scholar]
- 37. Vandamme D., Landuyt B., Luyten W., and Schoofs L. (2012) A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell. Immunol. 280, 22–35 [DOI] [PubMed] [Google Scholar]
- 38. Takiguchi T., Morizane S., Yamamoto T., Kajita A., Ikeda K., and Iwatsuki K. (2014) Cathelicidin antimicrobial peptide LL-37 augments interferon-β expression and antiviral activity induced by double-stranded RNA in keratinocytes. Br. J. Dermatol. 171, 492–498 [DOI] [PubMed] [Google Scholar]
- 39. Filewod N. C., Pistolic J., and Hancock R. E. (2009) Low concentrations of LL-37 alter IL-8 production by keratinocytes and bronchial epithelial cells in response to proinflammatory stimuli. FEMS Immunol. Med. Microbiol. 56, 233–240 [DOI] [PubMed] [Google Scholar]
- 40. Lin Q., Fang D., Fang J., Ren X., Yang X., Wen F., and Su S. B. (2011) Impaired wound healing with defective expression of chemokines and recruitment of myeloid cells in TLR3-deficient mice. J. Immunol. 186, 3710–3717 [DOI] [PubMed] [Google Scholar]
- 41. Grönberg A., Mahlapuu M., Ståhle M., Whately-Smith C., and Rollman O. (2014) Treatment with LL-37 is safe and effective in enhancing healing of hard-to-heal venous leg ulcers: a randomized, placebo-controlled clinical trial. Wound Repair Regen. 22, 613–621 [DOI] [PubMed] [Google Scholar]
- 42. Tjabringa G. S., Aarbiou J., Ninaber D. K., Drijfhout J. W., Sørensen O. E., Borregaard N., Rabe K. F., and Hiemstra P. S. (2003) The antimicrobial peptide LL-37 activates innate immunity at the airway epithelial surface by transactivation of the epidermal growth factor receptor. J. Immunol. 171, 6690–6696 [DOI] [PubMed] [Google Scholar]
- 43. Nakagawa Y., and Gallo R. L. (2015) Endogenous intracellular cathelicidin enhances TLR9 activation in dendritic cells and macrophages. J. Immunol. 194, 1274–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh D., Vaughan R., and Kao C. C. (2014) LL-37 peptide enhancement of signal transduction by Toll-like receptor 3 is regulated by pH: identification of a peptide antagonist of LL-37. J. Biol. Chem. 289, 27614–27624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sørensen O. E., Cowland J. B., Theilgaard-Mönch K., Liu L., Ganz T., and Borregaard N. (2003) Wound healing and expression of antimicrobial peptides/polypeptides in human keratinocytes, a consequence of common growth factors. J. Immunol. 170, 5583–5589 [DOI] [PubMed] [Google Scholar]
- 46. Sørensen O. E., Thapa D. R., Roupé K. M., Valore E. V., Sjöbring U., Roberts A. A., Schmidtchen A., and Ganz T. (2006) Injury-induced innate immune response in human skin mediated by transactivation of the epidermal growth factor receptor. J. Clin. Investig. 116, 1878–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin Q., Wang L., Lin Y., Liu X., Ren X., Wen S., Du X., Lu T., Su S. Y., Yang X., Huang W., Zhou S., Wen F., and Su S. B. (2012) Toll-like receptor 3 ligand polyinosinic:polycytidylic acid promotes wound healing in human and murine skin. J. Invest. Dermatol. 132, 2085–2092 [DOI] [PubMed] [Google Scholar]