Abstract
Trypanosoma brucei causes African sleeping sickness for which no vaccine exists and available treatments are of limited use due to their high toxicity or lack of efficacy. T. brucei cultivated in the presence of deoxyadenosine accumulates high levels of dATP in an adenosine kinase-dependent process and dies within a few hours. Here we show that T. brucei treated with 1 mm deoxyadenosine accumulates higher dATP levels than mammalian cells but that this effect diminishes quickly as the concentration of the deoxynucleoside decreases. Radioactive tracer studies showed that the parasites are partially protected against lower concentrations of deoxyadenosine by the ability to cleave it and use the adenine for ATP synthesis. T. brucei methylthioadenosine phosphorylase (TbMTAP) was found to be responsible for the cleavage as indicated by the phosphate dependence of deoxyadenosine cleavage in T. brucei cell extracts and increased deoxyadenosine sensitivity in TbMTAP knockdown cells. Recombinant TbMTAP exhibited higher turnover number (kcat) and Km values for deoxyadenosine than for the regular substrate, methylthioadenosine. One of the reaction products, adenine, inhibited the enzyme, which might explain why TbMTAP-mediated protection is less efficient at higher deoxyadenosine concentrations. Consequently, T. brucei grown in the presence of adenine demonstrated increased sensitivity to deoxyadenosine. For deoxyadenosine/adenosine analogues to remain intact and be active against the parasite, they need to either be resistant to TbMTAP-mediated cleavage, which is the case with the three known antitrypanosomal agents adenine arabinoside, tubercidin, and cordycepin, or they need to be combined with TbMTAP inhibitors.
Keywords: nucleoside/nucleotide analogue, nucleoside/nucleotide metabolism, parasite, Trypanosoma brucei, trypanosome, methylthioadenosine phosphorylase (MTAP)
Introduction
African sleeping sickness (1) is a fatal disease that progresses through two stages and is caused by two subspecies of the parasite Trypanosoma brucei, T. brucei gambiense in Western and Central Africa and T. brucei rhodesiense in Eastern and parts of Southern Africa. Treatment of the disease is particularly difficult in the second stage in which the parasites leave the blood and lymph, enter the central nervous system, and eventually cause the patient to fall into a comatose state. T. brucei and related species also cause nagana, a disease in cattle that has a significant impact on socioeconomic development in many parts of rural Africa (2).
Many nucleoside analogues have been successfully used as drugs in the fields of cancer, virology, and to some extent parasitology. One advantage of using nucleoside analogues against second stage sleeping sickness is that a large proportion of them can cross the blood-brain barrier via transporters used for the uptake of nucleosides needed by the brain (3). Nucleoside analogues need to be phosphorylated inside the cells into their nucleotide forms to inhibit transcription, replication, or other nucleotide-dependent processes. In contrast to mammalian cells, which have several kinases to phosphorylate nucleosides and deoxynucleosides, T. brucei has only two: thymidine kinase primarily phosphorylates thymidine and deoxyuridine (4, 5), and adenosine kinase phosphorylates adenosine and to some extent deoxyadenosine (6).
T. brucei lacks de novo purine biosynthesis, and adenosine kinase is part of the efficient salvage pathways that enable the parasite to utilize nucleosides and bases from the host (6, 7). Adenosine is one of the major purine sources in human blood where the concentration is 2 μm (8). It is taken up by T. brucei via efficient transporters (9, 10) and phosphorylated by adenosine kinase into AMP and subsequently by other kinases into ADP and ATP (7). An alternative pathway that adenosine shares with other purine nucleosides is that the ribose moiety can be first cleaved off and the base salvaged instead. The adenosine cleavage reaction is catalyzed by inosine-adenosine-guanosine-nucleoside hydrolase (IAG-NH)3 (11). Although the relative contribution of each pathway for adenosine salvage is not known, the much higher affinity of adenosine kinase than IAG-NH for this substrate suggests that it represents the major route (6). As a side activity, T. brucei adenosine kinase is also able to phosphorylate deoxyadenosine (6). This reaction is detrimental to the parasites because addition of 0.5 mm deoxyadenosine to the culture medium causes the parasites to die within 15 h with grossly elevated dATP levels and a concomitant decrease in ATP, which is consumed in the phosphorylation of the deoxynucleoside (12).
Many adenosine analogues are metabolized in a similar manner as adenosine into their nucleotide forms and affect transcription and other ATP-dependent processes. Cordycepin (3′-deoxyadenosine) and tubercidin (7-deazaadenosine) are two such analogues that inhibit T. brucei growth with IC50 values in the nanomolar range (13). Cordycepin has been shown to cure both stages of the disease in T. brucei-infected mice when given together with deoxycoformycin, which protects the drug from deamination by serum adenosine deaminase (14). Promising results have also been obtained with 2-fluorocordycepin, which is resistant to adenosine deaminase and can be used as a single agent (15). So far, the development of antiparasitic nucleoside derivatives has focused mainly on ribonucleoside analogues such as cordycepin and tubercidin, whereas 2′-deoxynucleoside- and arabinoside-containing analogues are the ones most frequently used in anticancer and antiviral therapies. Arabinoside analogues can be considered equivalent to deoxynucleoside analogues because they are generally phosphorylated by the same kinases as deoxynucleosides and primarily inhibit DNA synthesis and other deoxynucleotide-dependent reactions. Adenine arabinoside (Ara-A) is an example of one such analogue that in combination with deoxycoformycin inhibits T. brucei DNA synthesis and cures T. brucei-infected mice (6).
The sensitivity of T. brucei to deoxyadenosine seems surprising in light of the fact that this is a naturally occurring nucleoside. In the current work, we show that T. brucei methylthioadenosine phosphorylase (TbMTAP) normally protects T. brucei cells against deoxyadenosine, but product inhibition by adenine sets an upper level on the capacity of the enzyme. Knowledge of the TbMTAP-mediated protection system is relevant for drug discovery because adenosine kinase substrate analogues need to be resistant to cleavage by TbMTAP (and TbIAG-NH) or be combined with cleavage enzyme inhibitors to be active against the parasite. Consequently, we found that cordycepin, tubercidin, and Ara-A were all resistant to TbMTAP activity and that the presence of the TbMTAP inhibitor adenine in the culture medium could increase deoxyadenosine-induced growth inhibition of T. brucei bloodstream forms (BSFs).
Experimental Procedures
Cultivation of T. brucei and Mammalian Cells
T. brucei BSFs were cultivated at 37 °C in HMI-9 medium (16) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Gibco) and grown in a humidified atmosphere containing 5% CO2. T. brucei procyclic forms were cultivated with similar atmospheric conditions at 27 °C in SDM-79 medium (Gibco) containing 10% (v/v) heat-inactivated fetal bovine serum and 1 μg/ml hemin (Sigma-Aldrich) (17). Madin-Darby bovine kidney cells (ATCC number CCL-22) and mouse Balb/3T3 fibroblasts (ATCC number CCL-163) were cultivated as monolayers in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with l-glutamine (0.584 g/liter), 10% (v/v) heat-inactivated fetal bovine serum, and 10 ml/liter 100× penicillin-streptomycin (Gibco) at 37 °C in a humidified atmosphere containing 7% CO2. Suspensions of human promyelocytic leukemia cells (HL-60 cells, ATCC number CCL-240) were cultivated in RPMI 1640 medium (Gibco) supplemented with 10% (v/v) heat-inactivated fetal bovine serum. Growth conditions and additional supplements (glutamine, penicillin, and streptomycin) were the same as for the Balb/3T3 and Madin-Darby bovine kidney cells.
Nucleotide Pool Measurements
NTP and dNTP pools from T. brucei and mammalian cells were extracted as described previously (12) and quantified by PolyWAX A (PolyLC, Columbia, MD) chromatography (18). A change compared with the previous analyses (12) is that here we used the regular T. brucei HMI-9 medium and did not exclude thymidine. The connection of the HPLC equipment to a flow scintillation analyzer (Radiomatic 150TR, PerkinElmer Life Sciences) enabled the detection of radiolabeled metabolites in experiments where the cells were treated with [2,8-3H] deoxyadenosine (Moravek Biochemicals, Brea, CA).
Preparation of T. brucei Cell Extracts
T. brucei TC221 cells were grown to a cell density of 2 × 106 cells/ml (50–100 ml) and centrifuged at 3,000 × g for 10 min at 4 °C. The pellet was washed with phosphate-buffered saline (PBS), recentrifuged, and resuspended in 200 μl of 10 mm Tris-HCl, pH 7.6. The resulting suspension was vortexed with zirconium beads to lyse the cells and recentrifuged. The supernatant was flash frozen in liquid nitrogen and stored at −80 °C. The protein concentration was determined by the Bio-Rad protein assay using bovine serum albumin as the reference. Enzyme assays were performed in the same manner as with the recombinant TbMTAP (see below) except that the phosphate concentration was decreased to 5 mm because cleavage of adenosine was inhibited at higher concentrations in the cell extracts (most likely due to inhibition of IAG-NH).
Measurement of IC50 Values in T. brucei
T. brucei cells were seeded in 96-well microtiter plates (5,000 cells/well for BSFs and 20,000 cells/well for procyclics) containing 200 μl of culture medium with various concentrations of deoxyadenosine or nucleoside analogues (in combination with 2 μm deoxycoformycin). After 48 h, the plates were incubated for 24 h with 20 μl of Alamar Blue dye (Invitrogen) and quantified by fluorescence (540-nm excitation and 590-nm emission) using an Infinite M200 microplate reader (Tecan Group, Männedorf, Switzerland). The low number of cells (5,000 cells/well) for testing deoxyadenosine sensitivity of BSFs was an adjustment because adenine (formed from the cleavage of deoxyadenosine) became detectable in the growth medium at significant levels if the number of cells was higher. The IC50 values were determined by fitting the data to a log[inhibitor] versus response curve (variable slope, four parameters) using GraphPad Prism 5.04 software (GraphPad Software, La Jolla, CA).
TbMTAP RNA Interference
Approximately 400 bp of the TbMTAP gene was amplified from T. brucei TC221 genomic DNA by using the primers 5′-GTC ACC TCG AGT GCC AAC TTC CGG AAG C-3′ (forward primer; the XhoI site is underlined) and 5′-AGC ACA AGC TTC TTT GTA ATG GCC TCT GGT TT-3′ (reverse primer; the HindIII site is underlined). The PCR product was digested with XhoI and HindIII restriction enzymes and ligated into pZJM (19). The resulting pZJM-TbMTAP construct was linearized with NotI and transfected into New York single marker T7NAP/TetR (NYSM) T. brucei BSFs (20) by electroporation (19), and positive clones were selected in the presence of 0.5 μg/ml phleomycin and 1 μg/ml G418 (20). Expression of the TbMTAP RNAi construct was induced with 1 μg/ml tetracycline for 7 days.
Western Blotting Analysis
Zirconium bead-homogenized T. brucei cell extracts (see above) were analyzed by Western blotting using an anti-TbMTAP immune serum, which was produced by Agrisera AB (Vännäs, Sweden) using recombinantly expressed and purified TbMTAP (see below) as the antigen. Western blotting was carried out using the antibody diluted 10,000-fold in Tris-buffered saline/Tween 20 containing 2% (w/v) skim milk. After incubation overnight, the membrane was washed and probed with an anti-rabbit secondary antibody for 1 h (GE Healthcare), developed with the ECL system (GE Healthcare), and imaged with the ChemiDocTM Touch Imaging System (Bio-Rad). For the determination of the loading control, the membrane was washed and incubated for 2 h with a 10,000-fold dilution of an antibody that recognizes Binding Protein (BiP), a member of the hsp70 heat shock protein family. The membrane was probed with secondary antibody and analyzed by ECL detection as described above. The anti-BiP antibody was a kind gift from J. Bang's laboratory at the University of Wisconsin Medical School.
Expression and Purification of Recombinant TbMTAP
The TbMTAP gene was amplified from T. brucei TC221 genomic DNA by using the primers 5′-AAC TGC TCA TGA TGT ACA CGA GTC CCC ACG-3′ (TbMTAP forward primer; the BspHI site is underlined) and 5′-ACG GGG TAC CTA TTA CGG AGC GAA TAT GGG ATA TTT-3′ (TbMTAP reverse primer; the Acc65I site is underlined). The PCR product was digested with BspHI and Acc65I and subcloned into a pETM-20 vector (European Molecular Biology Laboratory) that had been digested with NcoI (an isoschizomer of BspHI) and Acc65I. DNA sequencing from both directions confirmed the inserted gene to be correct. The resulting plasmid (pETM-20-TbMTAP) encodes a fusion construct with TbMTAP connected to an N-terminal His-tagged thioredoxin A (TrxA) protein via a linker containing a tobacco etch virus protease recognition site. The vector pETM20-TbMTAP was transformed into Escherichia coli BL21(DE3) pLysS cells (Merck), grown at 37 °C in 1 liter of Luria-Bertani broth supplemented with 60 μg/ml carbenicillin and 27.2 μg/ml chloramphenicol, induced at an A595 of 0.5 with 1 mm isopropyl β-d-thiogalactopyranoside, and harvested 3 h later. The bacteria were centrifuged, and the pellet was resuspended in 25 ml of buffer A (0.3 m NaCl and 20 mm Tris-HCl, pH 7.8). After freezing, thawing, and ultracentrifugation at 150,000 × g for 45 min, the supernatant was loaded onto a 1-ml nickel-NTA Superflow column (Qiagen, Hilden, Germany) equilibrated with buffer A. The column was subsequently washed with 15 ml of 5 mm and then 10 ml of 10 mm imidazole in buffer A. TrxA-tagged TbMTAP protein was finally eluted with 25 ml of 100 mm imidazole in buffer A. The eluted fractions were pooled and concentrated by two different methods. When adenine-free protein was required, the solution was precipitated with ammonium sulfate at 80% saturation at 4 °C (nominally 0.53 g/ml) and centrifuged, and the pellet was dissolved in 600 μl of buffer B (50 mm Tris-HCl, pH 7.6, and 0.1 mm DTT). When yield and stability were of higher importance (general protocol), the protein was concentrated in a 20-ml Vivaspin concentrator with a 30-kDa-molecular mass cutoff (Sartorius AG, Goettingen, Germany) to a final volume of 600 μl. The concentrated protein (using either of the two concentration methods) was buffer-exchanged into buffer B with a PD-10 Sephadex G-25 gel filtration column (GE Healthcare). TrxA-TbMTAP protein in 7.5 ml of buffer B was mixed with tobacco etch virus protease (2:1 ratio in milligrams) and incubated overnight at 4 °C. The digested protein mixture was loaded onto a 1-ml nickel-NTA Superflow column equilibrated with buffer B, and TbMTAP-containing fractions collected in the flow-through were subsequently pooled, concentrated by using a 30-kDa-cutoff 20-ml Vivaspin concentrator, frozen in liquid nitrogen, and stored at −80 °C. The Bio-Rad protein assay was used to assess the protein concentration with bovine serum albumin as the reference. The typical yield was 13 mg of TbMTAP from 1 liter of bacterial culture using the general protocol. In enzyme assays using methylthioadenosine (MTA), adenosine, or deoxyadenosine as substrates, the effect of adenine from the TbMTAP stock was negligible because strongly diluted protein solutions were used. With guanosine, inosine, and other low affinity purines and pyrimidines, much more enzyme was needed, and it was then crucial to use protein purified with the extra ammonium sulfate precipitation step.
TbMTAP Assays
A buffer consisting of 50 mm potassium acetate, 50 mm Tris-HCl, pH 7.4, and 0.05% (v/v) Tween 20 was used to dilute the enzyme to an appropriate concentration (0.05–1000 ng/μl depending on the substrate) prior to use in enzyme assays. The diluted enzyme (generally 2 μl) was mixed with a solution containing the nucleoside substrate in a total volume of 100 μl of 50 mm KH2PO4, pH 7.4 (pH adjusted with KOH). In assays where the reverse reaction was studied (nucleoside synthesis), the phosphate buffer was replaced by 50 mm HEPES-KOH, pH 7.4. The enzyme assay was incubated for 30 min at 37 °C. The reaction was terminated by incubation at 100 °C for 2 min in a heating block, and the reaction product was quantified by HPLC using an ACE UltraCore 2.5 Super C18 50 × 2.1-mm column (Advanced Chromatography Technologies, Aberdeen, UK). The column was run isocratically at 0.4 ml/min using a mobile phase consisting of 30 mm ammonium acetate, pH 5.8, and 5% (v/v) methanol. This procedure was sufficient to separate adenine (natural or modified) from each nucleoside substrate. Assays with guanine, hypoxanthine, cytosine, uracil, or thymine as reaction products were analyzed at 1 ml/min on an ACE 3 AQ 50 × 4.6-mm column (Advanced Chromatography Technologies) to enable the use of a methanol-free mobile phase (40 mm ammonium acetate, pH 5.8). All enzyme assays were performed within the linear range with respect to time and protein concentration.
Results
dATP Accumulates in Deoxyadenosine-treated T. brucei BSFs and Mammalian Cells
Generally, the levels of dNTPs are much lower than NTPs in T. brucei cells (12). However, when cultivated in the presence of 1 mm deoxyadenosine in the growth medium for 1 h, dATP became the most abundant nucleotide in the cell (Fig. 1A). Similarly to what was observed previously (12), the level of ATP, which is needed for the phosphorylation of deoxyadenosine, decreased significantly under these conditions (p < 0.001, unpaired t test). The level of dCTP increased, and dGTP decreased (Fig. 1A, inset) as a result of the effect of dATP as a specificity regulator of ribonucleotide reductase (12). Unlike in mammalian cells (21), dATP does not act as an overall activity inhibitor of the T. brucei ribonucleotide reductase (12, 22), and there was, therefore, no general decrease of the dNTP levels in Fig. 1A. The dTTP levels were much higher here than reported previously (12) due to the fact that we used regular HMI-9 growth medium, which contains 0.16 mm thymidine (16). The effect of deoxyadenosine on dNTP pools is likely to cause inhibition of DNA synthesis and an increased number of replication errors (23), whereas ATP depletion might cause more acute cytotoxicity.
FIGURE 1.
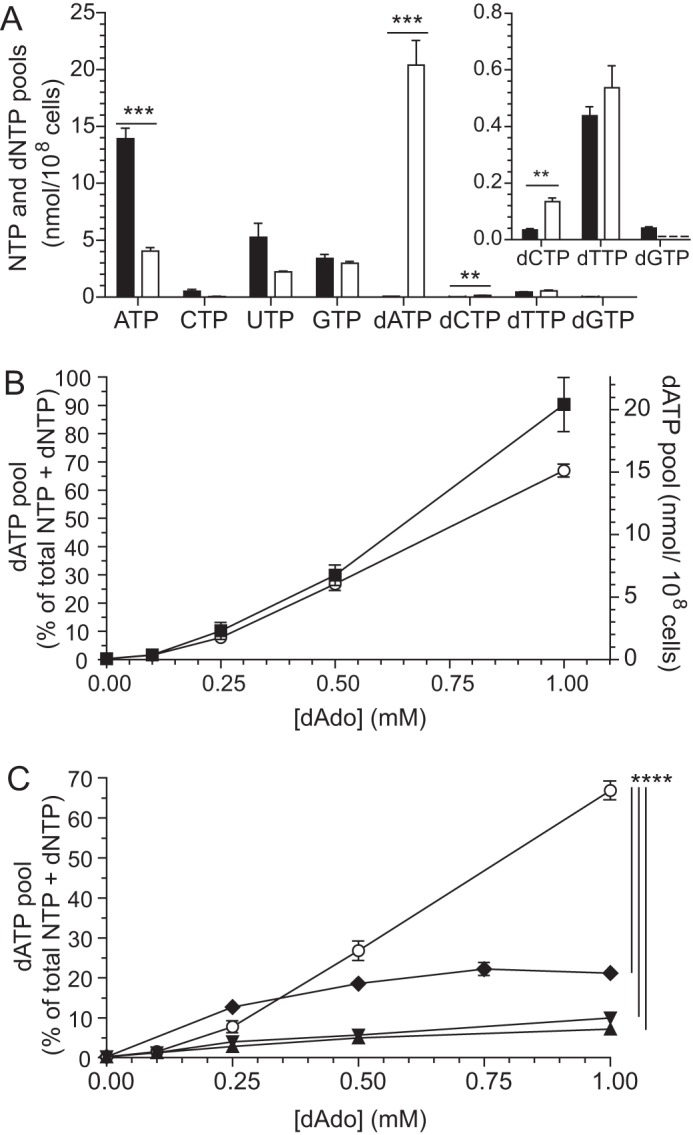
Deoxyadenosine salvage in T. brucei TC221 and mammalian cells. A, nucleotide pools in T. brucei bloodstream forms cultivated in the absence (black bars) or presence of 1 mm deoxyadenosine (white bars). The inset shows a magnification of the dCTP, dTTP, and dGTP pools. In the deoxyadenosine-treated sample, the dGTP pool is below the detection limit (indicated by a dashed line). B, accumulation of dATP in T. brucei cultivated in the presence of various concentrations of deoxyadenosine. The results are plotted as a percentage of the total nucleotide pool (○; left y axis) and as nmol/108 cells (■; right y axis). C, accumulation of dATP in deoxyadenosine-treated T. brucei (○), Madin-Darby bovine kidney cells (♦), Balb/3T3 fibroblasts (▾), and human promyelocytic leukemia cells (HL-60) (▴). All cell culture media used in A–C contained 2 μm deoxycoformycin to protect the deoxyadenosine from deamination in all experiments. The error bars in A and B show the S.E. from three independent experiments with p values indicated as follows: **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
In Fig. 1B, the dATP accumulation in T. brucei is shown as a function of the deoxyadenosine concentration in the growth medium. The data are plotted in two different ways. Presenting the data as a percentage of the total nucleotide pool (NTPs + dNTPs) has many advantages, including smaller standard errors, less variation depending on who is performing the experiment, and the possibility of comparing cell types with different volumes and thereby different total nucleotide levels. Because the increased dATP pool is to a large extent compensated for by a decrease in the other nucleotides, the two curves in Fig. 1B have fairly similar shapes. It is only when dATP becomes the major nucleotide in the cell (the 1 mm data point) that the total nucleotide pool increases significantly and creates a slight deviation between the two curve shapes.
The deoxyadenosine salvage efficiency of T. brucei was compared with mammalian cells selected from different species and cell types to make the results more generalizable (Fig. 1C). The data are shown as percentages of the total nucleotide pool to be able to compare the different cell types. T. brucei incubated with 1 mm deoxyadenosine accumulated a much higher percentage of dATP than the three mammalian cell types that were tested. This difference between T. brucei and mammalian cells is in fact slightly larger than in Fig. 1C if the deviation of the total nucleotide pool described in Fig. 1B is taken into account in the normalization procedure. The efficient deoxyadenosine salvage is consistent with the extraordinarily high affinity of all T. brucei adenosine transporters for deoxyadenosine (9, 24, 25). Despite the unusually efficient transport, the cellular percentage of dATP in T. brucei decreased very sharply at lower concentrations of deoxyadenosine and was then in the same range as in the mammalian cells, which all showed a more gradual relationship between the deoxyadenosine concentration and dATP accumulation. The comparatively low level of deoxyadenosine salvage at ≤0.25 mm in T. brucei suggested that the parasite has a protective system against deoxyadenosine, although this activity seems limited by an additional factor that prevents it from working efficiently at higher concentrations of the deoxynucleoside.
Deoxyadenosine Is Cleaved by T. brucei Cells
To investigate possible protection mechanisms against deoxyadenosine, T. brucei BSFs were incubated with different concentrations of [2,8-3H]deoxyadenosine, and the distribution of radioactive label in their ATP and dATP pools was analyzed (Table 1). There was no label detected in GTP, dGTP, or pyrimidine nucleotides. At the lower [3H]deoxyadenosine concentrations tested, most of the label appeared in the ATP pool, and this suggested that the nucleoside was cleaved and the labeled adenine salvaged into ATP, presumably by adenine phosphoribosyltransferase (APRT) (7, 26). A gradual decrease in radioactivity in the ATP pool at higher concentrations of [3H]deoxyadenosine in Table 1 indicated that the cleavage activity could be saturated and that the label went into dATP instead.
TABLE 1.
Accumulation of radiolabeled ATP and dATP in T. brucei TC221 bloodstream forms cultivated in the presence of different concentrations of [3H]deoxyadenosine combined with 2 μm deoxycoformycin
The results are based on three independent experiments with S.E. indicated for dATP (the remaining percentage is for ATP).
| [[3H]dAdo] | [[3H]dATP] | [[3H]ATP] |
|---|---|---|
| mm | % of total | % of total |
| 0.01 | 2.6 ± 0.1 | 97.4 |
| 0.1 | 8.8 ± 0.5 | 91.2 |
| 0.5 | 38 ± 3 | 62 |
| 1.0 | 86 ± 4 | 14 |
It should also be noted that a minor fraction of the radioactivity in dATP can come from ADP that is used as a substrate for ribonucleotide reductase. However, this cross-talk is inhibited by dATP, which acts as a specificity regulator of ribonucleotide reductase in T. brucei (12), and the cross-talk is therefore primarily relevant for the first data point.
Deoxyadenosine Cleavage in T. brucei Cell Extracts Is Phosphate-dependent
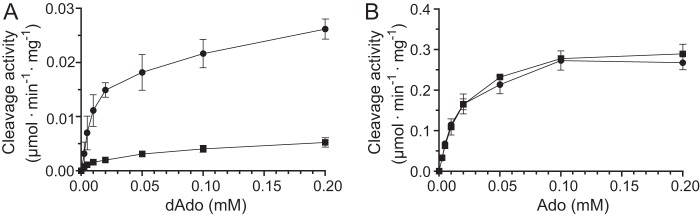
The two known adenosine/deoxyadenosine cleavage activities in T. brucei, TbMTAP and TbIAG-NH, are phosphate-dependent and phosphate-independent, respectively (11, 27). Analysis of deoxyadenosine cleavage in T. brucei cell extracts showed that the reaction was dependent on phosphate ions, suggesting cleavage by TbMTAP (Fig. 2A), whereas the corresponding reaction with adenosine was phosphate-independent, suggesting cleavage by TbIAG-NH (Fig. 2B). It is also clear that the total adenosine cleavage activity is much higher than with deoxyadenosine. If there is any TbMTAP-mediated adenosine cleavage, it is therefore likely to be overshadowed by that of TbIAG-NH.
FIGURE 2.
Cleavage of deoxyadenosine and adenosine by T. brucei cell extracts. A, cleavage of deoxyadenosine in the absence (■) or presence (●) of 5 mm potassium phosphate, pH 7.4. HEPES-KOH (50 mm) was used as an additional buffering agent in all samples. B, similar experiments as in A but with adenosine as the substrate. The same symbols were used as in A to indicate the absence and presence of phosphate. Three independent experiments were performed with S.E. indicated by error bars.
TbMTAP Is Able to Protect T. brucei from Deoxyadenosine
To investigate whether or not TbMTAP is able to protect T. brucei from deoxyadenosine, we created a tetracycline-inducible TbMTAP RNA interference construct and selected for its expression in T. brucei NYSM BSFs. Tetracycline induction of the knockdown construct for 7 days resulted in greatly reduced TbMTAP levels (Fig. 3A). Comparing the wild-type T. brucei lane (TC221 cells) in the immunoblot with the left lanes containing different amounts of recombinant protein showed that the cellular sample contained ∼3 ng of TbMTAP and, consequently, that the cellular level of this protein was ∼0.1% of the total protein (3.6 μg). A similar comparison of the other cellular extract lanes in Fig. 3A with the recombinant protein shows that the level of TbMTAP decreases at least 2-fold in the non-induced cells and ∼16-fold in the induced cells as compared with the NYSM cells. The small reduction of TbMTAP levels observed in the non-induced cells could be due to some RNAi expression leakage in the absence of tetracycline. The expression leakage in the non-induced cells had a tendency to increase over time, making it important to minimize the number of cellular generations. A second probing carried out with anti-BiP antibody confirmed that the loading was fairly equal in the four cellular extract lanes in Fig. 3A (the recombinant protein in the left lanes does not contain this protein). The protective role of TbMTAP against deoxyadenosine was demonstrated by IC50 value measurements, which showed a gradually increased sensitivity to deoxyadenosine among NYSM, non-induced, and induced cells (Fig. 3B). Note that the decrease in TbMTAP levels in the non-induced cells as compared with the NYSM cells led to a proportional increase in deoxyadenosine sensitivity but that the effect of further reduction of TbMTAP levels was quickly saturated. Consequently, the induced cells with much lower TbMTAP levels only had a 2-fold increased deoxyadenosine sensitivity as compared with the non-induced cells.
FIGURE 3.
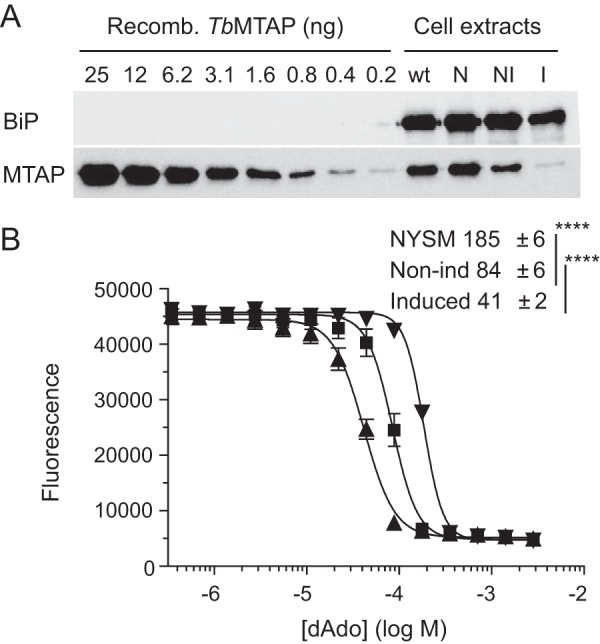
TbMTAP knockdown experiments. A, Western blotting analysis of a 2-fold dilution series of recombinant (Recomb.) TbMTAP protein and cell extracts from wild-type and knockdown T. brucei bloodstream forms. The cell extracts were prepared from the TC221 strain (wt), the NYSM parent strain (N), non-induced TbMTAP knockdown cells (NI), and tetracycline-induced knockdown cells (I). The membrane was blotted with α-TbMTAP and α-BiP antibodies (loading control for cell extracts). B, deoxyadenosine inhibition of proliferation in tetracycline-induced TbMTAP knockdown T. brucei (▴), non-induced (Non-ind) cells (■), and the NYSM strain (▾). The graph shows the average results with S.E. (error bars) from six independent experiments, and the table shows the calculated IC50 values in μm (**** indicates p < 0.0001). The cells in B were grown in the presence of 2 μm deoxycoformycin.
Recombinant TbMTAP Expression and Purification
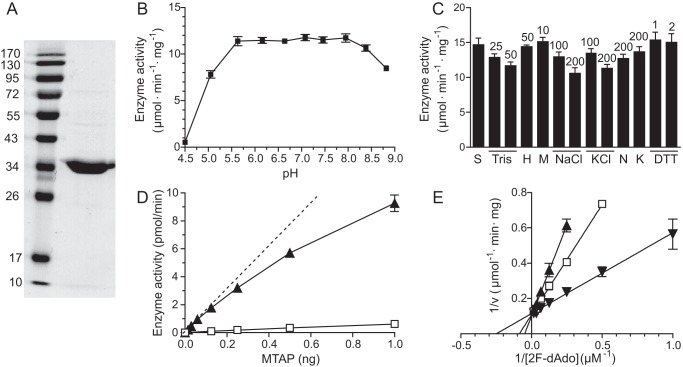
TbMTAP was expressed in E. coli as a fusion construct with an N-terminal His-tagged TrxA partner, which enabled the purification by nickel-NTA-agarose chromatography. SDS-PAGE analysis of the protein after removal of the fusion partner showed that it was pure and migrated according to its theoretical molecular mass of 34 kDa (Fig. 4A).
FIGURE 4.
Purification and characterization of recombinant TbMTAP. A, SDS-PAGE analysis of purified TbMTAP with a molecular mass marker to the left (PagerulerTM Prestained Protein ladder, Fermentas Life Sciences). B, influence of pH on TbMTAP-mediated cleavage of 100 μm adenosine. The solution was buffered by a mixture of 25 mm acetic acid, 50 mm KH2PO4, and 10 mm Tris base that was pH-adjusted with KOH. C, adenosine cleavage (100 μm) activity under standard conditions (S) or in the presence of Tris-HCl (Tris), HEPES-KOH (H), MgCl2 (M), NaCl, KCl, sodium acetate (N), potassium acetate (K), or DTT with concentrations in mm indicated on top of the bars. D, TbMTAP-mediated cleavage of 100 μm adenosine (▴) and the reverse reaction where adenosine is synthesized from 100 μm adenine and 100 μm ribose 1-phosphate (□). E, Lineweaver-Burk plot showing TbMTAP-mediated cleavage of various concentrations of 2-fluoro-2′-deoxyadenosine in the absence (▾) or presence of 0.75 (□) or 1.5 μm (▴) adenine. The results represent three independent experiments with S.E. (error bars) shown.
TbMTAP Is Inhibited by Adenine
Optimization of the enzyme assay conditions for TbMTAP showed that the protein had a broad pH optimum and was not dependent on common supplements such as Na+, K+, Mg2+, or DTT (Fig. 4, B and C). Most supplements had no obvious effect on enzyme activity except Tris-HCl and high concentrations of chloride salts (200 mm), which were slightly inhibitory. Similarly to other nucleoside phosphorylases, it could catalyze the enzymatic reaction in both directions, although the cleavage reaction (phosphorolysis) was more efficient than the synthesis reaction (Fig. 4D). By studying the enzyme activity in the presence of 50 mm phosphate (Pi), a concentration nearly 100 times above the Km value for Pi (Table 2), the synthesis reaction became undetectable, and we could selectively study the cleavage reaction. The linear range of the TbMTAP cleavage activity with respect to protein concentration was very limited (Fig. 4D), and it needed to be carefully optimized for each substrate. Deviation from linearity starts to be apparent already at 2 pmol/min in the figure, which corresponds to 0.6% substrate conversion during the 30-min incubation time. The limited linear range can be explained by the observation that the enzyme activity was strongly inhibited by adenine, a product in the reaction, which acted as a competitive inhibitor (Fig. 4E). The Ki value was calculated to be 0.56 ± 0.07 μm from the corresponding Michaelis-Menten graphs by non-linear regression using GraphPad Prism software. 2-Fluoro-2′-deoxyadenosine was used as the substrate in the inhibition studies to be able to get a product (2-fluoroadenine) that can be separated from the inhibitor (adenine). The catalytic parameters are fairly similar with this substrate as with deoxyadenosine (Table 2).
TABLE 2.
TbMTAP substrate selectivity
Kinetic parameters shown are Km, kcat (per polypeptide), and catalytic efficiency (kcat/Km) calculated by fitting Michaelis-Menten diagrams to hyperbolas using the GraphPad Prism software with S.E. of the regression analysis indicated. 2F-dAdo, 2-fluoro-2′-deoxyadenosine; 2Cl-dAdo, 2-chloro-2′-deoxyadenosine.
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | s−1 × μm−1 | |
| MTA | 0.09 ± 0.01 | 2.13 ± 0.06 | 24 |
| Ado | 13 ± 1 | 8.4 ± 0.2 | 0.65 |
| dAdo | 8.3 ± 0.7 | 9.6 ± 0.3 | 1.16 |
| 2F-dAdo | 5.0 ± 0.7 | 5.4 ± 0.2 | 1.08 |
| 2Cl-dAdo | 32 ± 6 | 6.4 ± 0.4 | 0.20 |
| Cordycepin | 260 ± 30 | 2.5 ± 0.1 | 0.0096 |
| Ara-Aa | 1210 ± 80 | 0.019 ± 0.001 | 0.000016 |
| Tubercidin | <0.003b | ||
| Guo | 150 ± 30 | 0.031 ± 0.002 | 0.00021 |
| dGuo | 150 ± 10 | 0.058 ± 0.002 | 0.00039 |
| Ino | 140 ± 20 | 0.079 ± 0.004 | 0.00056 |
| dIno | 180 ± 30 | 0.128 ± 0.006 | 0.00071 |
| Urd | 61 ± 10 | 0.052 ± 0.002 | 0.00086 |
| dUrd | 77 ± 12 | 0.036 ± 0.002 | 0.00047 |
| Cyd, dCyd, Thd | <0.003b | ||
| Pi (with Ado)c | 690 ± 30 | 9.0 ± 0.2 |
a Not tested up to the kcat plateau due to the solubility limit of Ara-A (∼1.8 mm).
b Measured with 100 μm substrate.
c Experiment performed under Pi-limiting conditions (100 μm Ado).
TbMTAP Substrate Analysis
TbMTAP was highly selective for adenine-containing nucleosides as compared with other natural purines and pyrimidines (Table 2). It had good activity with MTA, adenosine, and deoxyadenosine although with very different kinetic parameters for the three substrates. The much lower Km value (and higher catalytic efficiency) for MTA indicates that this is a first priority substrate. In contrast, deoxyadenosine and adenosine are high Km substrates with high turnover numbers (kcat), indicating that the enzyme is only able to cleave the nucleosides efficiently when they are present at high concentrations relative to MTA. Cleavage reactions with adenosine as the substrate were kinetically similar to those with deoxyadenosine (less than a 2-fold difference in kcat, Km, and catalytic efficiency). Interestingly, the Km values for deoxyadenosine and adenosine were an order of magnitude higher than the Ki value of adenine, indicating the strength of this inhibitor.
TbMTAP and Adenosine Analogues
Enzyme assays with some known antitrypanosomal adenosine analogues showed that tubercidin, Ara-A, and cordycepin were all poor substrates (Table 2). There was no measurable activity with tubercidin and very low catalytic efficiency with Ara-A. The kcat value with cordycepin was in the same range as the main substrates of the enzyme, but the Km value was very high, leading to a low catalytic efficiency for this substrate as well. A general trend seems, therefore, to be that known antitrypanosomal agents can circumvent TbMTAP-based cleavage, which would explain their efficiency. Halogenation of the 2-position of the base is a common way to make adenosine analogues resistant to deamination and thereby stable in the blood. We therefore tested the 2-fluorinated and 2-chlorinated analogues of deoxyadenosine (Table 2). Both were good substrates, and these modifications seemed to only have a modest effect on TbMTAP cleavage.
Adenine Promotes the Antitrypanosomal Activity of Deoxyadenosine
T. brucei BSFs showed enhanced sensitivity to deoxyadenosine in the presence of non-toxic concentrations of adenine (Table 3), supporting our conclusion that TbMTAP is inhibited by adenine. The concentration of adenine used here was confirmed to not affect growth by itself prior to the experiment. Similar experiments with the adenosine analogues Ara-A and cordycepin showed that the sensitivities to these compounds were not enhanced in the presence of adenine. With Ara-A, there was no adenine effect at all, and with cordycepin the sensitivity was slightly decreased instead of increased. The IC50 value with Ara-A was ∼3-fold higher than reported previously (6), and this deviation might be explained by the fact that here we used regular HMI-9 medium (with thymidine) and a different incubation time. Our conclusion about the role of adenine in TbMTAP inhibition was also confirmed in knock-out T. brucei procyclic cells, which lack the two genes for APRT (26). Lacking APRT makes these cells unable to use the adenine from deoxyadenosine cleavage for AMP biosynthesis, leading to adenine inhibition of TbMTAP and increased deoxyadenosine sensitivity (Table 3).
TABLE 3.
Growth inhibition of T. brucei bloodstream forms (TC221) and procyclic cells (S427) by deoxyadenosine, Ara-A, or cordycepin in the presence of 0, 10, or 100 μm adenine
The growth medium also contained 2 μm deoxycoformycin. The results show the average of three or four independent experiments with standard errors and p values as compared with the indicated controls (ctrl): ns, not significant; *, p < 0.1, **, p < 0.01, ****, p < 0.0001.
| Cell type | Condition | IC50 |
|---|---|---|
| μm | ||
| TC221 | Only dAdo | 205 ± 9 (ctrl) |
| + 10 μm Ade | 109 ± 4**** | |
| + 100 μm Ade | 52 ± 3**** | |
| TC221 | Only Ara-A | 0.28 ± 0.04 (ctrl) |
| + 100 μm Ade | 0.26 ± 0.01 (ns) | |
| TC221 | Only cordycepin | 0.0012 ± 0.0002 |
| + 100 μm Ade | 0.0022 ± 0.0002* | |
| S427 | Only dAdo | 70 ± 10 (ctrl) |
| S427 (APRT−/−) | Only dAdo | 33 ± 3** |
Discussion
The most well known protection mechanism against deoxyadenosine is via adenosine deaminase. Deficiency of this enzyme in humans leads to high dATP levels in lymphocytes, which results in lymphocytopenia and, consequently, severe combined immunodeficiency (28). It has been unclear, however, what level of protection T. brucei has against deoxyadenosine. Adenosine deaminase activity was initially reported from T. brucei cell extracts (29), but this finding could not be confirmed by other laboratories (30). The adenosine deaminase-like sequences reported from the genomes of T. brucei and related species in GenBankTM (e.g. EAN78471.1) are homologous to tRNA-editing enzymes and are thus not likely to work on the nucleoside level. Here, we have found that TbMTAP instead has the role of protecting the parasites against deoxyadenosine by cleaving it into adenine and deoxyribose 1-phosphate.
Characterization of the recombinant TbMTAP sheds light onto how its enzymatic function is optimized for a dual role in the cell (Fig. 5). First, it needs to cleave MTA efficiently to keep the methionine cycle and polyamine metabolism going. Thus the enzyme has a low Km (and a high catalytic efficiency) for MTA as a substrate compared with adenosine and deoxyadenosine. It is known from a previous study that knocking down TbMTAP leads to slightly slower growth and an increased proportion of anucleate and multinucleate T. brucei cells (31). The effect on the DNA content could possibly be a result of defective metabolism of polyamines, which are needed to stabilize DNA during replication. Second, TbMTAP needs to cleave deoxyadenosine to prevent any buildup of dATP. A comparatively high Km value for this substrate prevents it from competing for MTA cleavage. The deoxyadenosine concentrations used here are much higher than what can be expected to occur in the natural environment of the parasites, but it should be remembered that the HMI-9 growth medium contains hypoxanthine as the only purine source, and there is no selective pressure for the parasites to keep the adenosine salvage systems up-regulated. It is, for example, common that in vitro cultivated parasites lose almost all of their P2 nucleoside transporter activity, which transports adenine, adenosine, and deoxyadenosine (32); apparently the transporter is down-regulated under standard culture conditions. Although this does not eliminate all deoxyadenosine uptake, which can also occur by the P1 nucleoside transporter (9), the fact that T. brucei transporters can be differentially expressed under different conditions (33) suggests that in vitro IC50 values may not necessarily reflect in vivo sensitivities. In the human host, the concentration of deoxyadenosine is usually below the detection limit in the blood due to the action of adenosine deaminase. Nevertheless, it is important for the parasite to also have a protection system of its own; in studies on Trypanosoma evansi-infected rats it was found that the infection leads to down-regulation of the adenosine deaminase in the blood and some parts of the brain (34, 35). An even more unpredictable situation can be imagined to occur for the trypanosomes in the midgut of the tsetse fly where the deoxyadenosine concentration will be a direct consequence of what the insect feeds on.
FIGURE 5.
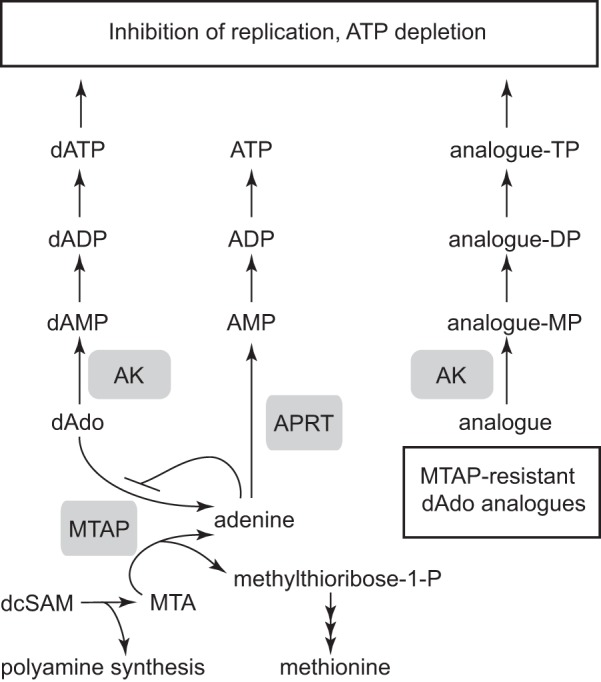
Metabolism of deoxyadenosine (left), MTA (bottom left) and TbMTAP-resistant deoxyadenosine analogues (right). MTA is a byproduct when decarboxylated S-adenosylmethionine (dcSAM) is used in polyamine biosynthesis. The recycling of methionine back into decarboxylated S-adenosylmethionine is not included in the figure. AK, adenosine kinase; MP, monophosphate; DP, diphosphate; TP, triphosphate.
In a previous report, using a partially purified preparation of TbMTAP, it was found that the enzyme has a broader substrate specificity than the mammalian enzyme and can cleave MTA along with adenosine, deoxyadenosine, and cordycepin (27). However, the enzyme activity measurements alone can give the false impression that these other substrates can never compete with MTA for which the enzyme has a much lower Km value and higher catalytic efficiency. Here, we have studied both the isolated enzyme and deoxyadenosine metabolism in living T. brucei cells. Experiments with radiolabeled deoxyadenosine demonstrated that the reaction is efficient in T. brucei cells despite the much lower catalytic efficiency with this substrate as compared with MTA. When present at a concentration of 10 μm in the growth medium, most of the deoxyadenosine taken up by the cells was cleaved, and 97% of the label was incorporated into ATP instead of dATP. No label was detected in GTP, dGTP, or other NTPs or dNTPs. The presence of the 3H label in ATP indicates that a major metabolic route for adenine is phosphoribosylation to AMP (and subsequent phosphorylation to ADP and ATP). If the adenine were deaminated prior to salvage, the label would be expected to occur in both ATP and GTP, which was not the case. The deoxyadenosine protection mechanism was confirmed in TbMTAP knockdown T. brucei cells, which, upon induction of the RNAi construct, became 5 times more sensitive to deoxyadenosine. The deoxyadenosine cleavage reaction seems, therefore, to occur efficiently in the cell despite the much higher Km for this substrate as compared with MTA. A possible explanation could be that MTA is immediately metabolized as soon as it is produced and does not accumulate to levels high enough to outcompete deoxyadenosine cleavage.
The TbMTAP-mediated protection system against deoxyadenosine in T. brucei seems to have an upper limit of how much deoxyadenosine it can process; the reaction is very efficient with 10 μm [3H]deoxyadenosine in the growth medium but gradually loses capacity at higher concentration of the nucleoside, leading to dATP pool expansion and an increased ratio of radiolabel in dATP as compared with ATP. As illustrated in Fig. 5, the cellular adenine concentration contributes to this limit of the protection system when the production of adenine exceeds the only metabolic use by APRT. T. brucei BSFs grown in the presence of adenine thus showed an enhanced sensitivity to deoxyadenosine, and characterization of the recombinant TbMTAP showed that adenine was a strong competitive inhibitor of deoxyadenosine cleavage (Ki = 0.56 μm). The low Ki value means that adenine easily outcompetes deoxyadenosine (Km = 8.3 μm) but has a much lesser effect on enzyme activity with the high affinity substrate MTA (Km = 0.09 μm). We believe that a major reason why the protection system fails at high concentrations of deoxyadenosine is because APRT does not metabolize the adenine efficiently enough. The role of APRT in this pathway was supported by our observation that knock-out cells lacking this enzyme showed enhanced sensitivity to deoxyadenosine.
The discovery of the deoxyadenosine protection system is very important for the rational development of adenosine and deoxyadenosine analogues against African sleeping sickness. Many of these analogues are only active against T. brucei in their nucleoside form, and then they need to be resistant to cleavage by both TbMTAP and TbIAG-NH. For 2′-deoxyadenosine analogues, TbMTAP is the more important of the two enzymes to consider (Fig. 5). As shown in Table 2, most nucleosides are to varying degrees substrates of the TbMTAP reaction, but a majority of them gave very low activity, including nearly all natural nucleosides except those containing adenine. Interestingly, the known antitrypanosomal nucleoside analogues tested were all very poor substrates in the reaction; tubercidin gave no detectable enzyme activity, and the catalytic efficiencies with cordycepin and Ara-A were 100 and 105 times lower, respectively, than with deoxyadenosine, showing that the N7 and 3′-OH moieties must be important for substrate binding and/or catalysis, whereas the effect of the 2′-OH group is strongly dependent on its orientation. Similar conclusions were drawn from our experiments on cultivated trypanosomes, which became more sensitive to deoxyadenosine but not to cordycepin and Ara-A when incubated in the presence of the TbMTAP inhibitor adenine. A prerequisite for these nucleoside analogues to be efficient as antitrypanosomal agents is that they remain intact and are not cleaved. Nucleoside analogues that are TbMTAP substrates need to be combined with TbMTAP inhibitors such as adenine or, preferably, a stable adenine analogue that is not an APRT substrate but an equally effective TbMTAP inhibitor. A possible alternative might be to create prodrug analogues that only become cytotoxic after cleavage and that need the selective activation by TbMTAP to target the parasites.
Author Contributions
The experiments were designed, performed, and analyzed by M. V. and F. R. (Figs. 1 and 3), F. R. and A. P. (Fig. 2), A. H. and M. V. (Fig. 4), M. V. (Table 1), A. H. and F. R. (Table 2), and F. R. (Table 3). H. P. d. K. contributed with ideas, expertise in nucleoside transporters, and writing. A. H. and M. V. designed the study and wrote the paper.
This work was supported by Swedish Research Council Grant 2012-1932, Swedish International Development Cooperation Agency Grant 2008-069, and the Kempe Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
- IAG-NH
- inosine-adenosine-guanosine-nucleoside hydrolase
- Ara-A
- adenine arabinoside
- MTAP
- methylthioadenosine phosphorylase
- Tb
- T. brucei
- BSF
- bloodstream form
- BiP
- binding protein
- NTA
- nitrilotriacetic acid
- MTA
- methylthioadenosine
- APRT
- adenine phosphoribosyltransferase
- NYSM
- New York single marker T7NAP/TetR
- TrxA
- thioredoxin A.
References
- 1. Malvy D., and Chappuis F. (2011) Sleeping sickness. Clin. Microbiol. Infect. 17, 986–995 [DOI] [PubMed] [Google Scholar]
- 2. Holmes P. (2013) Tsetse-transmitted trypanosomes—their biology, disease impact and control. J. Invertebr. Pathol. 112, (suppl.) S11–S14 [DOI] [PubMed] [Google Scholar]
- 3. Parkinson F. E., Damaraju V. L., Graham K., Yao S. Y., Baldwin S. A., Cass C. E., and Young J. D. (2011) Molecular biology of nucleoside transporters and their distributions and functions in the brain. Curr. Top. Med. Chem. 11, 948–972 [DOI] [PubMed] [Google Scholar]
- 4. Ranjbarian F., Vodnala M., Vodnala S. M., Rofougaran R., Thelander L., and Hofer A. (2012) Trypanosoma brucei thymidine kinase is tandem protein consisting of two homologous parts, which together enable efficient substrate binding. J. Biol. Chem. 287, 17628–17636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chello P. L., and Jaffe J. J. (1972) Comparative properties of trypanosomal and mammalian thymidine kinases. Comp. Biochem. Physiol. B 43, 543–562 [DOI] [PubMed] [Google Scholar]
- 6. Vodnala M., Fijolek A., Rofougaran R., Mosimann M., Mäser P., and Hofer A. (2008) Adenosine kinase mediates high affinity adenosine salvage in Trypanosoma brucei. J. Biol. Chem. 283, 5380–5388 [DOI] [PubMed] [Google Scholar]
- 7. el Kouni M. H. (2003) Potential chemotherapeutic targets in the purine metabolism of parasites. Pharmacol. Ther. 99, 283–309 [DOI] [PubMed] [Google Scholar]
- 8. Slowiaczek P., and Tattersall M. H. (1982) The determination of purine levels in human and mouse plasma. Anal. Biochem. 125, 6–12 [DOI] [PubMed] [Google Scholar]
- 9. de Koning H. P., and Jarvis S. M. (1999) Adenosine transporters in bloodstream forms of Trypanosoma brucei brucei: substrate recognition motifs and affinity for trypanocidal drugs. Mol. Pharmacol. 56, 1162–1170 [DOI] [PubMed] [Google Scholar]
- 10. de Koning H. P., Bridges D. J., and Burchmore R. J. (2005) Purine and pyrimidine transport in pathogenic protozoa: from biology to therapy. FEMS Microbiol. Rev. 29, 987–1020 [DOI] [PubMed] [Google Scholar]
- 11. Pellé R., Schramm V. L., and Parkin D. W. (1998) Molecular cloning and expression of a purine-specific N-ribohydrolase from Trypanosoma brucei brucei. Sequence, expression, and molecular analysis. J. Biol. Chem. 273, 2118–2126 [DOI] [PubMed] [Google Scholar]
- 12. Hofer A., Ekanem J. T., and Thelander L. (1998) Allosteric regulation of Trypanosoma brucei ribonucleotide reductase studied in vitro and in vivo. J. Biol. Chem. 273, 34098–34104 [DOI] [PubMed] [Google Scholar]
- 13. Geiser F., Lüscher A., de Koning H. P., Seebeck T., and Mäser P. (2005) Molecular pharmacology of adenosine transport in Trypanosoma brucei: P1/P2 revisited. Mol. Pharmacol. 68, 589–595 [DOI] [PubMed] [Google Scholar]
- 14. Rottenberg M. E., Masocha W., Ferella M., Petitto-Assis F., Goto H., Kristensson K., McCaffrey R., and Wigzell H. (2005) Treatment of African trypanosomiasis with cordycepin and adenosine deaminase inhibitors in a mouse model. J. Infect. Dis. 192, 1658–1665 [DOI] [PubMed] [Google Scholar]
- 15. Vodnala S. K., Lundbäck T., Yeheskieli E., Sjöberg B., Gustavsson A. L., Svensson R., Olivera G. C., Eze A. A., de Koning H. P., Hammarström L. G., and Rottenberg M. E. (2013) Structure-activity relationships of synthetic cordycepin analogues as experimental therapeutics for African trypanosomiasis. J. Med. Chem. 56, 9861–9873 [DOI] [PubMed] [Google Scholar]
- 16. Hirumi H., and Hirumi K. (1989) Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75, 985–989 [PubMed] [Google Scholar]
- 17. Brun R., and Schönenberger M. (1979) Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36, 289–292 [PubMed] [Google Scholar]
- 18. Håkansson P., Hofer A., and Thelander L. (2006) Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J. Biol. Chem. 281, 7834–7841 [DOI] [PubMed] [Google Scholar]
- 19. Wang Z., Morris J. C., Drew M. E., and Englund P. T. (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275, 40174–40179 [DOI] [PubMed] [Google Scholar]
- 20. Wirtz E., Leal S., Ochatt C., and Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 21. Hofer A., Crona M., Logan D. T., and Sjöberg B.-M. (2012) DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hofer A., Schmidt P. P., Gräslund A., and Thelander L. (1997) Cloning and characterization of the R1 and R2 subunits of ribonucleotide reductase from Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 94, 6959–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar D., Abdulovic A. L., Viberg J., Nilsson A. K., Kunkel T. A., and Chabes A. (2011) Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 39, 1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Al-Salabi M. I., Wallace L. J., Lüscher A., Mäser P., Candlish D., Rodenko B., Gould M. K., Jabeen I., Ajith S. N., and de Koning H. P. (2007) Molecular interactions underlying the unusually high adenosine affinity of a novel Trypanosoma brucei nucleoside transporter. Mol. Pharmacol. 71, 921–929 [DOI] [PubMed] [Google Scholar]
- 25. de Koning H. P., Watson C. J., and Jarvis S. M. (1998) Characterization of a nucleoside/proton symporter in procyclic Trypanosoma brucei brucei. J. Biol. Chem. 273, 9486–9494 [DOI] [PubMed] [Google Scholar]
- 26. Lüscher A., Lamprea-Burgunder E., Graf F. E., de Koning H. P., and Mäser P. (2014) Trypanosoma brucei adenine-phosphoribosyltransferases mediate adenine salvage and aminopurinol susceptibility but not adenine toxicity. Int. J. Parasitol. Drugs Drug Resist. 4, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ghoda L. Y., Savarese T. M., Northup C. H., Parks R. E. Jr., Garofalo J., Katz L., Ellenbogen B. B., and Bacchi C. J. (1988) Substrate specificities of 5′-deoxy-5′-methylthioadenosine phosphorylase from Trypanosoma brucei brucei and mammalian cells. Mol. Biochem. Parasitol. 27, 109–118 [DOI] [PubMed] [Google Scholar]
- 28. Blackburn M. R., and Kellems R. E. (2005) Adenosine deaminase deficiency: metabolic basis of immune deficiency and pulmonary inflammation. Adv. Immunol. 86, 1–41 [DOI] [PubMed] [Google Scholar]
- 29. Davies M. J., Ross A. M., and Gutteridge W. E. (1983) The enzymes of purine salvage in Trypanosoma cruzi, Trypanosoma brucei and Leishmania mexicana. Parasitology 87, 211–217 [DOI] [PubMed] [Google Scholar]
- 30. Ogbunude P. O., Ikediobi C. O., and Ukoha A. I. (1985) Adenosine cycle in African trypanosomes. Ann. Trop. Med. Parasitol. 79, 7–11 [DOI] [PubMed] [Google Scholar]
- 31. Berg M., Kohl L., Van der Veken P., Joossens J., Al-Salabi M. I., Castagna V., Giannese F., Cos P., Versées W., Steyaert J., Grellier P., Haemers A., Degano M., Maes L., de Koning H. P., et al. (2010) Evaluation of nucleoside hydrolase inhibitors for treatment of African trypanosomiasis. Antimicrob. Agents Chemother. 54, 1900–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ward C. P., Wong P. E., Burchmore R. J., de Koning H. P., and Barrett M. P. (2011) Trypanocidal furamidine analogues: influence of pyridine nitrogens on trypanocidal activity, transport kinetics, and resistance patterns. Antimicrob. Agents Chemother. 55, 2352–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Koning H. P., Watson C. J., Sutcliffe L., and Jarvis S. M. (2000) Differential regulation of nucleoside and nucleobase transporters in Crithidia fasciculata and Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 106, 93–107 [DOI] [PubMed] [Google Scholar]
- 34. da Silva A. S., Bellé L. P., Bitencourt P. E., Souza V. C., Costa M. M., Oliveira C. B., Jaques J. A., Leal D. B., Moretto M. B., Mazzanti C. M., Lopes S. T., and Monteiro S. G. (2011) Activity of the enzyme adenosine deaminase in serum, erythrocytes and lymphocytes of rats infected with Trypanosoma evansi. Parasitology 138, 201–208 [DOI] [PubMed] [Google Scholar]
- 35. Da Silva A. S., Bellé L. P., Bitencourt P. E., Perez H. A., Thomé G. R., Costa M. M., Oliveira C. B., Teixeira M. M., Moretto M. B., Mazzanti C. M., Lopes S. T., and Monteiro S. G. (2011) Trypanosoma evansi: adenosine deaminase activity in the brain of infected rats. Exp. Parasitol. 127, 173–177 [DOI] [PubMed] [Google Scholar]