Abstract
Much evidence points to a role of Na,K-ATPase in ouabain-dependent signal transduction. Based on experiments with different cell lines and native tissue membranes, a current hypothesis postulates direct interactions between the Na,K-ATPase and Src kinase (non-receptor tyrosine kinase). Na,K-ATPase is proposed to bind Src kinase and inhibit its activity, whereas ouabain, the specific Na,K-ATPase inhibitor, binds and stabilizes the E2 conformation, thus exposing the Src kinase domain and its active site Tyr-418 for activation. Ouabain-dependent signaling is thought to be mediated within caveolae by a complex consisting of Na,K-ATPase, caveolin, and Src kinase. In the current work, we have looked for direct interactions utilizing purified recombinant Na,K-ATPase (human α1β1FXYD1 or porcine α1D369Nβ1FXYD1) and purified human Src kinase and human caveolin 1 or interactions between these proteins in native membrane vesicles isolated from rabbit kidney. By several independent criteria and techniques, no stable interactions were detected between Na,K-ATPase and purified Src kinase. Na,K-ATPase was found to be a substrate for Src kinase phosphorylation at Tyr-144. Clear evidence for a direct interaction between purified human Na,K-ATPase and human caveolin was obtained, albeit with a low molar stoichiometry (1:15–30 caveolin 1/Na,K-ATPase). In native renal membranes, a specific caveolin 14–5 oligomer (95 kDa) was found to be in direct interaction with Na,K-ATPase. We inferred that a small fraction of the renal Na,K-ATPase molecules is in a ∼1:1 complex with a caveolin 14–5 oligomer. Thus, overall, whereas a direct caveolin 1/Na,K-ATPase interaction is confirmed, the lack of direct Src kinase/Na,K-ATPase binding requires reassessment of the mechanism of ouabain-dependent signaling.
Keywords: caveolin, Na+/K+-ATPase, protein-protein interaction, signaling, Src
Introduction
The major function of the Na,K-ATPase is as an active cation pump that maintains the normal trans-membrane gradients of sodium and potassium ions. Cardiac glycosides such as ouabain and digoxin are the classical inhibitors of the pump. In addition to the classical role of the Na,K-pump, there is now a large body of literature describing a cellular signaling function whereby binding of cardiac glycosides, such as ouabain, induce signal transduction cascades and increased cell-cell contact, gene transcription, cell proliferation, and hypertrophy of organs (1–4). Evidence for endogenous cardiac glycosides and the role of the Na,K-ATPase as the receptor of endogenous cardiac glycosides has been demonstrated in several studies (5)1.
The initial evidence for Na,K-ATPase involvement in signaling events came from observations that ouabain induces activation of the non-receptor tyrosine kinase Src kinase and the MAPK pathway in cardiac myocytes and various other cell types (3, 6) in a dose- and time-dependent manner (1–3). Activation of gene transcription was associated with tyrosine phosphorylation on Src kinase, EGF receptor, and activation of the p42/44 MAPK pathway (3). Src kinase was also reported to be immunoprecipitated with Na+/K+ ATPase (7), and, over a number of years, a variety of additional observations accumulated, leading to the proposal that there is a direct interaction between the Na,K-ATPase and Src kinase (2, 8–11). These findings fitted the assumption that the sodium pump mediates the activation of MAPKs pathway by ouabain and suggested the role of Src kinase as the first kinase that is activated after ouabain binding (8, 10). Subsequently, evidence accumulated that ouabain-induced signal events are initiated in caveolae microdomains (12, 13), the Na+/K+ ATPase in these microdomains being referred to as the “signaling complex.” In caveolae, signaling proteins are concentrated into a small surface area, hence increasing the probability of productive activation by specific ligands. Caveolin, the primary marker protein of caveolae, was found to immunoprecipitate with the pump (14). At the sequence level, the Na,K-ATPase contains caveolin binding motifs located on transmembrane segments M1 and M10 in the α subunit (ΦXXΦXXXXΦXXΦXXXXΦ for TM1 and ΦXΦXXXXΦXXXXΦXΦ for TM10, where Φ is an aromatic amino acid and X is any amino acid). There is also some evidence, so far uncorroborated, that the Na,K-ATPase located in microdomains, which mediates ouabain-induced signaling, is inactive as a cation pump (15).
More recently, other studies have challenged the model of direct interaction between Src kinase and Na,K-ATPase (14, 16, 17, 18) (see “Discussion”). Although several observations raise doubts about the direct interaction model, they do not rule out indirect interactions mediated by other proteins that might be involved in the signaling events.
This most detailed mechanistic hypothesis for ouabain-induced activation of Src kinase was proposed (8, 10). According to this hypothesis, the Src SH22 domain binds to the A domain of the α subunit, independently of the Na,K-ATPase conformation, whereas the Src kinase domain is associated with the N domain in a weak interaction, dependent on the Na,K-ATPase conformation state (11). In the E1 conformation, Src is bound on both A and N domains, and the Src kinase activity is inhibited. Upon binding of ouabain, the Na,K-ATPase is stabilized in the E2-P conformation state, exposing the Src kinase domain, and the active site Tyr-418 can then be autophosphorylated to activate the Src kinase. The Src kinase then phosphorylates downstream proteins, thus initiating activation of the MAPK pathway.
The hypothesis that Src kinase binds directly to the Na,K-ATPase, in a way dependent on E1 and E2 conformations, implies that determination of specific Src kinase binding should differ in media containing E1- or E2-stabilizing ligands. As a necessary corollary, Src kinase binding should also affect the E1-E2 conformational equilibria and, thus, the Na,K-ATPase activity.
A more general concern with the use of co-immunoprecipitation reactions to show protein/protein interactions is that they may be indirect and non-quantitative. Thus, there is a need for independent demonstrations of the specificity and determination of the stoichiometry of the interaction using purified proteins. Using both purified recombinant human Na,K-ATPase (α1β1FXYD1), which has been extensively characterized (19–21), and native rabbit kidney membrane vesicles enriched with Na,K-ATPase (α1β1FXYD2), we have tested the major assumption that Src kinase binds directly to the Na,K-ATPase α subunit in a conformation-dependent manner (11). In addition, we have then tested and characterized the interaction between caveolin 1 and Na,K-ATPase with recombinant proteins as well as in native tissue.
The results do not support direct binding of Src kinase to Na,K-ATPase but do show the existence of a complex of renal Na,K-ATPase with a caveolin4–5 oligomer, consistent with a distinct population of Na,K-pumps within caveolae. The work raises the question as to the detailed mechanism of ouabain-induced signaling.
Experimental Procedures
Materials
n-Dodecyl-β-maltoside (DDM) was purchased from Anatrace, and BD-Talon metal affinity resin was from Clontech. 1,2-Dioleoyl-sn-glycero-3-phospho-l-serine was purchased from Avanti Polar Lipids, as was sphingomyelin from bovine brain. AcTEVTM protease was from Life Technologies, Inc. ATP (disodium salt, special quality) was obtained from Roche. The following antibodies were used: caveolin 1 antibodies (BD Transduction, 610080), (Abcam, ab2910), phosphotyrosine antibody (Abcam, ab9319), Src antibody (Abcam ab47405), Src 418YP (MBL, AT-7135), and 2,2′-azinbis(3-ethylbenzothiazoline-6-sulfonic acid) (Sigma-Aldrich, A1888). Poly(Glu4-Tyr) (Sigma-Aldrich, P0275), 96-well plates (high binding capacity) (Corning, CI-9018), blue native marker (GE Healthcare high molecular weight Native Marker kit, 17044501), Metrizamide (Sigma, 860506), and OptiPrep (Sigma, D1556) were also used. All other reagents were purchased from Merck or Sigma-Aldrich at the highest quality level available.
Methods
Expression and Purification of Na+/K+ ATPase Human α1β1His10FXYD1
For expression of human Na+,K+-ATPase pump in Pichia pastoris SMD1165, pHIL-D2 (α/His10-β1) vector containing cDNAs encoding human α1 (Swiss-Prot accession numbers α1, P05023) and human His10-β1 (accession number P05026) was transformed into the yeast strain (21, 22). Expression of FXYD1 in Escherichia coli was done as described (23). Growth of the yeast cells, preparation of membranes, solubilization of membranes in DDM, binding to BD-Talon beads, washing and elution of purified Na,K-ATPase, purification of FXYD1 from E. coli, and reconstitution of the α1β1 complex with purified FXYD1 have all been described in detail (19, 21, 22). In standard conditions, beads were washed twice with 20 mm imidazole and eluted with 200 mm imidazole; 100 mm NaCl; 20 mm Tricine·HCl, pH 7.4; either 0.1 mg/ml C12E8 or 0.07 mg/ml SOPS; either 0.01 mg/ml cholesterol, or 0.3 mg/ml C12E8; 0.17 mg/ml SOPS; 0.05 mg/ml cholesterol; and 10% glycerol. In some cases, beads were washed twice with 20 mm imidazole and eluted with 200 mm imidazole, 100 mm NaCl, 20 mm Tricine·HCl, pH 7.4, 0.3 mg/ml C12E8, 0.1 mg/ml sphingomyelin from bovine brain, 0.07 mg/ml SOPS, 0.05 mg/ml cholesterol, 10% glycerol.
Na,K-ATPase Activity
Na,K-ATPase activity of the purified α1β1FXYD1 complex was measured as described (23). In short, activity was measured in triplicate for a single time or as single samples over 2, 4, 6, and 8 min. In the latter case, activity was calculated from the regression line ± S.E. (r > 0.98). Activity was expressed as μmol of Pi/min/mg of protein.
Preparation of Microsomes and Right-side Out Vesicles from Rabbit Kidney Outer Medulla
Rabbit kidney outer medulla microsomes were prepared as described (24). For preparation of right-side out membrane vesicles (25), microsomes were suspended in solution containing 250 mm sucrose, 25 mm histidine, pH 7.2, 1 mm EDTA (Tris), and 1 mm metrizamide was added. 1 ml of crude microsomes, ∼10 mg of protein, was layered onto 9 ml of 15% (w/v) metrizamide on 1 ml of 30% metrizamide (w/v) and centrifuged at 35,000 rpm for 2 h in a Beckman SW-41 swing-out rotor. Right-side out vesicles were collected on the layer of 15% metrizamide. The samples were stored at −80 °C with 20 mm histidine, pH 7.5, 20% glycerol. Unmasking was performed with deoxycholate at a 1:1 ratio (protein/detergent) for 30 min at room temperature.
Preparation of Caveolae from Rabbit Kidney Microsomes
A rabbit kidney caveolae fraction was prepared as described (26). In short, microsomal fractions were lysed by passages through a 21-gauge needle or sonicated with a setting of 1 min 10 s on, 10 s off at 30% amplitude, mixed with 50% OptiPrep, and overlaid on a linear 10–20% OptiPrep gradient. After ultracentrifugation, the top 4 ml were collected as the caveolae fraction. The sample was centrifuged again and was suspended in 20 mm histidine, pH 7.5, 20% glycerol and kept at −80 °C.
Expression and Purification of Src Kinase and Caveolin 1 in E. coli
Human Src kinase (P12931) and caveolin 1 (BAG70098) genes were incorporated into pET28-TevH vector, which contains a His6 tag at the N terminus and a TEV cleavage site for removal of the tag. A restriction-free procedure was used to incorporate the Src kinase gene into pET28-TevH vector (27). The plasmids were transformed into E. coli BL21(DE3) cells.
Src Kinase
For Src kinase expression, transformed BL21(DE3) cells were grown in LB medium with 1% glucose to A600 nm of 0.6–0.8. Induction was performed overnight with 0.5 mm IPTG and 0.1% glucose at 15 °C. Cells were homogenized and lysed in buffer that contained 50 mm Tricine, Tris, pH 7.4, 150 mm NaCl, 1 mm MgCl2, and 10 mm imidazole with protease inhibitor mixture (Sigma, P8340). The cells were lysed with a cooled Stansted French press at 10,000 p.s.i. and then centrifuged at 10,000 × g for 15 min at 4 °C. The supernatant fraction was incubated with Co2+ beads and washed twice with 10 mm imidazole. Elution was performed with 200 mm imidazole. 6 g of BL21(DE3) cells yield about 200 μg of purified Src kinase. The eluted fraction was further verified as full-length human Src kinase using LC-MS/MS (Biological Services/Mass Spectrometry Unit). Purifications and gels of the purified Src kinase were carried out five times. Figs. 1, 3, and 4 show representative experiments, and the figure legends denote the exact number of experiments performed (n = x, etc.).
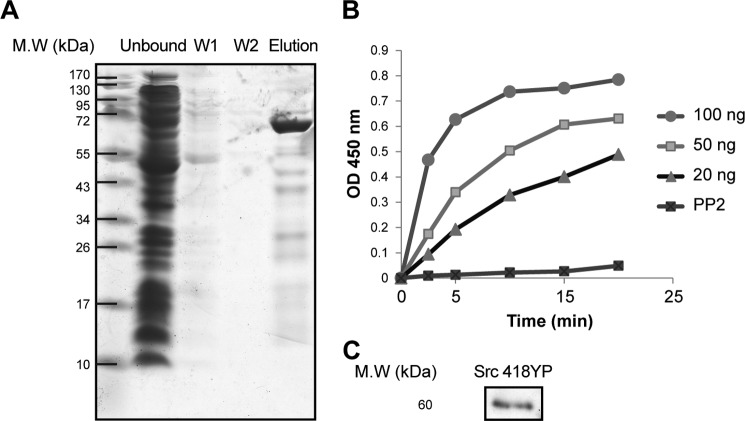
FIGURE 1.
Purification and properties of human recombinant Src tyrosine kinase. SDS gel separation showing different Src kinase purification steps (right to left): unbound, wash 1 (W1), wash 2 (W2), and elution fractions (n = 6). B, Src kinase activity with 20, 50, and 100 ng of Src kinase. With 20 ng of Src kinase, the rate of color development was linear over 20 min. The linear regression line, y = 0.0239x + 0.0416 (r2 = 0.97), gave a calculated activity of 1.195 ± 0.322 A450 nm/min/μg Src (n = 6). C, immunoblot of 20 ng of purified Src kinase with phosphorylated Tyr-418 antibody (n = 2).
FIGURE 3.
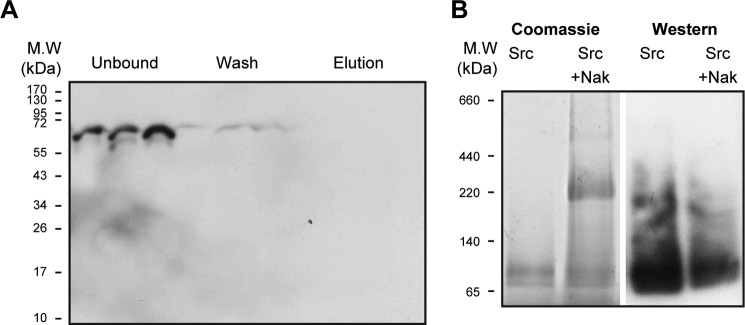
Tests for a direct interaction between purified human Src kinase and purified human Na,K-ATPase. A, pull-down assay. The Na,K-ATPase (1.5 μg diluted 10-fold to reduce imidazole to 20 mm) was incubated with TEV-treated Src kinase at molar ratios of 1:2, 1:4, and 1:8 (Na,K-ATPase/Src) as described under “Methods.” Unbound, wash, and elution fractions were loaded on an SDS gel. Western blot was probed with anti-Src (n = 2). B, BN gel separation. Lanes 1 and 2, Coomassie stain. Lanes 3 and 4, Western blots probed with anti-Src. Lane 1, Src (8 μg). Lane 2, Src (8 μg) premixed with purified human Na,K-ATPase (α1β1FXYD1-NaK) (10 μg). Lane 3, Src (8 μg). Lane 4, Src plus Na,K-ATPase (α1β1FXYD1-NaK) (10 μg) (n = 4).
FIGURE 4.
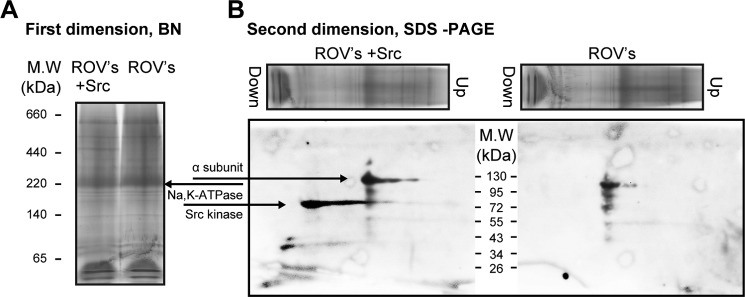
Blue native gel. A test for direct interaction between purified human Src kinase and native renal Na,K-ATPase is shown. A, blue native gel, first dimension, Coomassie-stained. Lane 1, marker proteins; lane 2, native rabbit kidney membranes, ROV (100 μg) plus Src (8 μg); lane 3, ROV (100 μg) (n = 4). B, SDS-PAGE, second dimension, Western blotting. Lane 2, probed with anti-α and anti-Src; lane 3, probed with anti-α (n = 2).
Caveolin 1
In short, E. coli BL21(DE3) cells harboring the plasmid pET28-TevH with the caveolin 1 gene were grown at 37 °C to an A600 nm of 0.6–0.8. Expression of caveolin 1 was induced with 0.5 mm IPTG. After induction, cells were grown for an additional 4 h and harvested at 10,000 × g for 15 min at 4 °C. Cells were then lysed with an emulsifier, passed through three times at 15,000 p.s.i. The nuclear fraction was removed by centrifugation at 10,000 × g for 15 min at 4 °C. The cloudy supernatant fraction (5 mg/ml protein) was dissolved to clarity at 20 °C in a buffer containing 8 m urea and 10 mg/ml DDM, diluted 2-fold, and centrifuged at 100,000 × g for 1 h, and the clear supernatant was incubated with Co2+ beads (BD-Talon) (500 μl per 10 mg of membranes). Beads were washed twice with 0.5 m NaCl, 50 mm Tris-Tricine, pH 7.4, 10% glycerol, 4 m urea, 5 mg/ml DDM, 10 mm imidazole. Elution was performed with the same buffer containing 200 mm imidazole. For caveolin purification with sphingomyelin, wash and elution buffer contained also 0.1 mg/ml sphingomyelin and 0.05 mg/ml cholesterol. To remove imidazole and 4 m urea, the eluted protein was dialyzed twice for 2 h at room temperature against 1000 volumes of a solution containing 0.5 m NaCl, 50 mm Tris-Tricine, pH 7.4, 10% glycerol. Purification, SDS-PAGE, and Western blotting of the purified caveolin 1 were carried out at least 10 times. Figs. 6–10 show representative experiments, and the figure legends denote the exact number of experiments done (n = x, etc.).
FIGURE 6.
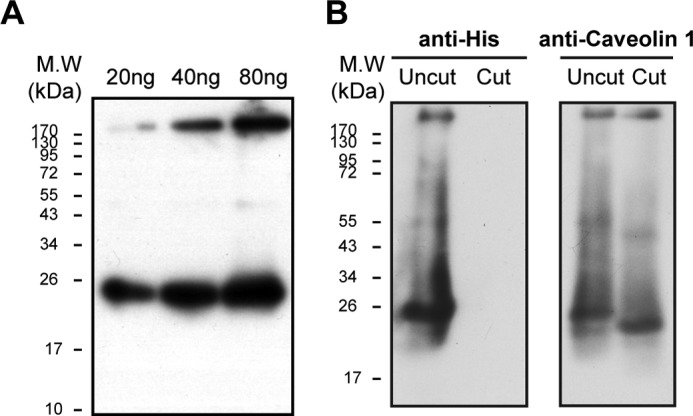
Purification and TEV cleavage of caveolin. A, Western blotting of 20, 40, and 80 ng of purified caveolin, probed with anti-caveolin 1 (n = 3). B, caveolin 1 before and after TEV treatment (see “Methods”). Western blotting was probed with anti-His (left) or anti-caveolin 1 (right)(n = 2).
FIGURE 7.
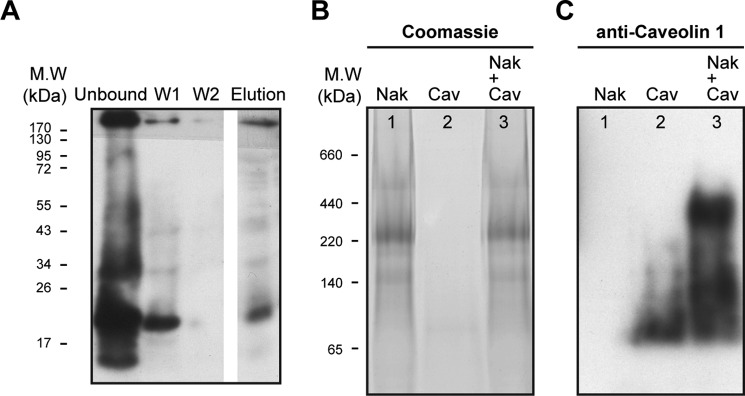
Tests for a direct interaction between purified human caveolin 1 and purified human Na,K-ATPase. A, pull-down assay. Western blotting of unbound, wash I and II, and elution fractions probed with anti-caveolin 1 (see “Methods”) (n = 2). B and C, blue native gel. B, Coomassie-stained. C, Western blot probed with anti-caveolin 1. Lane 1, markers; lane 2, 5 μg of purified human Na,K-ATPase plus purified caveolin (3 μg), a molar ratio of 3:1 (caveolin/Na,K-ATPase). C, as in lanes 1–3 of B, probed with anti-caveolin 1 antibody (n = 6).
FIGURE 8.
Interaction of purified Na,K-ATPase-caveolin 1 is not altered by the protein conformation or presence of sphingomyelin/cholesterol. A, lane 1, markers; lanes 1 and 2, Coomassie-stained; lane 3, 5 μg of purified human Na,K-ATPase preincubated with 100 μg of oligomycin, 1 mm ATP, 1 mm MgCl2, 100 mm NaCl at room temperature for 30 min, was mixed with 3 μg of purified caveolin; lane 4, purified human Na,K-ATPase preincubated with 2 mm ouabain, 1 mm ATP, 1 mm MgCl2, 100 mm NaCl at room temperature for 30 min was mixed with 3 μg of purified caveolin; lanes 3 and 4, like lanes 2 and 3, Western blots probed with anti-caveolin 1 (n = 4). B, lanes 1–4, Coomassie-stained. Lane 1, markers; lane 2, caveolin 1 (3 μg); lane 3, 5 μg of purified Na,K-ATPase prepared in standard conditions plus caveolin 1 (3 μg); lane 4, 5 μg of purified Na,K-ATPase prepared with sphingomyelin/cholesterol plus (3 μg) caveolin 1 that also was eluted with sphingomyelin and cholesterol; lanes 5–7, as in lanes 2–4, Western blots using anti-caveolin 1 (n = 4).
FIGURE 9.
Blue native gel separation of Na,K-ATPase with bound caveolin in rabbit renal caveolae membranes. A, BN gel. Lane 1, markers; lane 2, caveolae membranes (100 μg), Coomassie stain. Arrows, points of cutting the gel into six 1-mm slices below and above the main band at ∼250 kDa (n = 2). B, SDS-PAGE; Western blotting of second dimension separation of six gel pieces probed with Na,K-ATPase anti-α antibody (n = 2). C, SDS-PAGE; Western blotting of second dimension separation of six gel pieces probed with anti-caveolin 1 antibody (n = 2). D, SDS-PAGE; Western blotting. Lane 1, purified recombinant caveolin shows caveolin bands at 26 kDa and between stacking and separating gels. Lanes 2 and 3, native rabbit renal caveolin, lane 2 probed with anti-caveolin 1 (Ab1, BD Transduction, catalogue no. 610080) and lane 3 probed with anti-caveolin 1 (Ab2, Abcam, catalogue no. ab2910) (n = 2).
FIGURE 10.
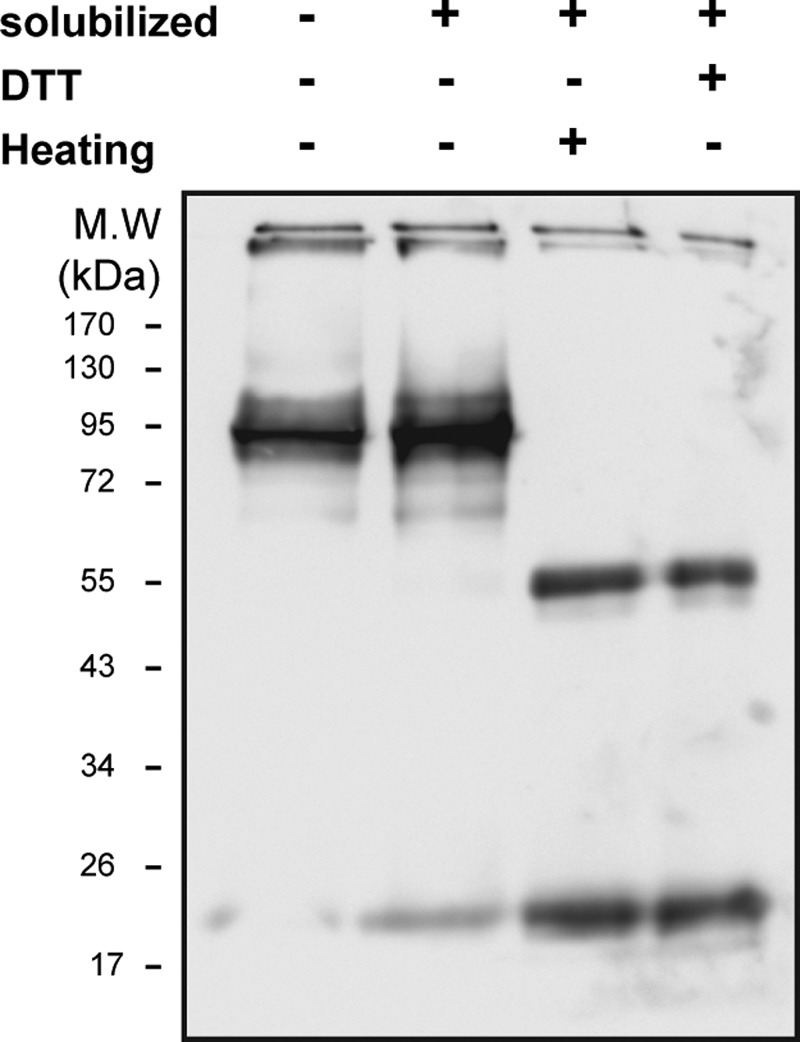
Dissociation of the 95-kDa caveolin4–5 oligomer to monomers and dimers. Shown are 10 μg of ROV samples solubilized at different conditions; Western blotting with anti-caveolin antibody. Lane 1, ROVs with no treatment; lane 2, ROVs solubilized with 5 mg/ml urea and 4 mg/ml DDM incubated for 30 min at room temperature; lane 3, ROVs solubilized with 5 mg/ml urea and 4 mg/ml DDM heated for 30 min at 65 °C; lane 4, ROVs solubilized with 5 mg/ml urea and 4 mg/ml DDM incubated with 10 mm DTT for 15 min at room temperature and then with 20 mm iodoacetamide for 15 min (n = 4).
TEV Protease Treatment of Src Kinase and Caveolin 1
Src kinase (0.8 μg/μl) or caveolin (0.3 μg/μl) was centrifuged at 200,000 × g for 30 min to remove aggregates, and 140 μl of the supernatant were dialyzed twice against 250 ml of 50 mm Tricine-Tris, pH 7.4, 150 mm NaCl, 10% glycerol. Samples were then centrifuged again, and the supernatants were incubated with TEV protease at a ratio of 1:10 (TEV/protein) for 9 h at 4 °C with rotation. 25 μl of cobalt beads were added and incubated for 1 h to remove any uncleaved protein and the tag. The samples were run on SDS-PAGE, and blots were probed with anti-His tag, anti-Src kinase, or anti-caveolin antibodies.
Src Kinase Activity Assay (28, 29)
Poly(Glu4-Tyr) was coated onto 96-well plates (Maxisorp) by adding 125 μl of 0.1 mg/ml poly(Glu4-Tyr) in PBS to each well. Plates were sealed and incubated for 16 h at 37 °C, washed once with TBST (Tris-buffered saline with 0.5% Tween 20), dried for 2–3 h at 37 °C, and stored at 4 °C. 20–50 ng of purified recombinant Src kinase protein in 20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 100 mm NaCl or choline chloride were added to each well with or without 1% BSA, as indicated in Fig. 2. Phosphotyrosine kinase activity was initiated by the addition of 1 mm of ATP and was terminated by the addition of EDTA to a final concentration of 200 mm. Plates were washed twice with TBST, and a third wash was done with low fat (1%) milk diluted 1:20 in TBST. Plates were incubated with anti-phosphotyrosine antibody for 1 h at room temperature, washed three times, and incubated with secondary antibody conjugated to peroxidase for 45 min. Detection was performed after three washes with TBST, using a color reagent (2,2′-azinbis(3-ethylbenzothiazoline-6-sulfonic acid)) in citrate-phosphate buffer, pH 4.0, that contains 0.004% H2O2. A450 nm was measured 10 min after the addition of the color reagent. Assays were run as single samples at time points (e.g. 5, 10, 5, and 20 min). The rate was calculated from the regression lines of the linear phase (r > 0.98) and expressed as A450 nm/min/μg of Src kinase.
FIGURE 2.
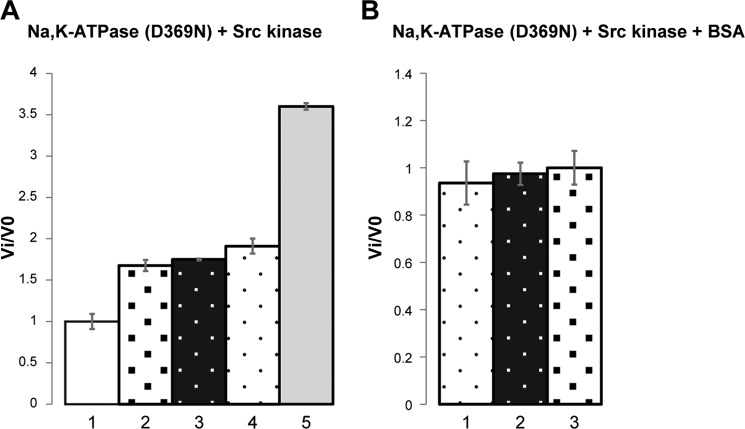
Effects of purified Na,K-ATPase (α1D369Nβ1FXYD1) and BSA on Src kinase activity. The specific Src kinase activity of the control sample was 1.195 A450 nm/min/μg. In the different conditions, Src kinase activity is presented as the ratio Vi/V0. A, purified porcine Na,K-ATPase (α1D369Nβ1FXYD1) was mixed with Src kinase at a molar ratio of 3:1 (Na,K-ATPase/Src). The Na,K-ATPase was pre-equilibrated with ouabain or oligomycin + 100 mm NaCl, 2 mm ATP, and 2 mm MgCl2 for 30 min at room temperature to stabilize E2-P·ouabain or E1–3Na·oligomycin or after denaturation (heating for 80 °C for 1 h). Where indicated, BSA was added at 1% (w/v) (∼150 μm). The values ± S.E. represent averages from 3 experiments/condition. A, column 1, Src kinase; column 2, Src kinase/Na,K-ATPase/ouabain; column 3, Src kinase/Na,K-ATPase/oligomycin; column 4, Src kinase/BSA; column 5, Src kinase/denatured Na,K-ATPase. B, column 1, Src kinase/BSA; column 2, Src kinase/BSA/Na,K-ATPase/oligomycin; column 3, Src kinase/BSA/Na,K-ATPase/ouabain. A, p values for the differences in columns 3, 4, and 5 versus control in column 1 (control) are in the range from 0.001 to 0.003. The p value for the difference between the values in column 5 (denatured Na,K-ATPase) and columns 3 and 4 (native Na,K-ATPase) is 0.0001.
Phosphorylation of the Na,K-ATPase by Src Kinase
Unmasked rabbit right-side out vesicles (ROVs) or recombinant purified human α1β1FXYD1 was incubated with recombinant purified human Src kinase at a molar ratio of 1:6 (Na,K-ATPase/Src). The reaction buffer contained 20 mm Tris, pH 7.4, 150 mm NaCl, 3 mm ATP, 50 μg/ml oligomycin, 1 mm ouabain, 10 mm MgCl2. The samples were incubated at room temperature for 2 h with rotation. Samples were run on SDS-PAGE, and phosphorylation of the Na,K-ATPase was detected by Western blotting using an anti-phosphotyrosine antibody (Abcam).
LC-Electrospray Ionization-MS/MS Analysis of Phosphotyrosine Src Kinase Digestion and Protein Identification
Protein bands were excised from an SDS gel stained with Coomassie Brilliant Blue. The protein bands were subsequently reduced, alkylated, and in-gel-digested with bovine trypsin (Promega) at a concentration of 12.5 ng/μl in 50 mm ammonium bicarbonate at 37 ºC, as described (30).
LC-MS/MS was performed using a 15-cm reversed-phase fused silica capillary column (inner diameter, 75 μm) made in house and packed with 3 μm ReproSil-Pur C18AQ medium (Dr. Maisch GmbH). The LC system, an UltiMate 3000 (Dionex), was used in conjunction with an LTQ Orbitrap XL (Thermo Fisher Scientific) operated in the positive ion mode and equipped with a nanoelectrospray ion source. Raw data files were searched with MASCOT (Matrix Science) against a Swiss-Prot database.
Blue Native Gels/SDS-PAGE
First Dimension
BN-PAGE for studying native protein complexes was carried out essentially as described (31). In brief, 100–200 μg of membranes were centrifuged at 70,000 rpm for 20 min and were solubilized in a buffer containing 30 mm imidazole, pH 7.00, 22 mm NaCl, 0.4 m 6-aminocaproic acid, 1.3 mm EDTA, 10% glycerol, and DDM at a 1:2 ratio to protein. The sample was incubated for 15 min on ice and centrifuged for 30 min at 200,000 × g to remove undissolved protein, and 0.1% Coomassie G-250 was added. Running conditions were as described before (31) except for the cathode buffer that contained 0.002% Coomassie G-250. The gel was either stained or blotted. For blotting of BN gels in the first dimension, the gel was incubated in 25 mm Tris, 192 mm glycine, 1% SDS for 1 h, followed by a 30-min incubation in 25 mm Tris, 192 mm glycine, 0.1% SDS. Semidry transfer was done in 25 mm Tris, 192 mm glycine, 0.03% SDS, 15% methanol.
Second Dimension, SDS Gel, Western Blotting
BN gels (whole lane or lanes cut into sections of 1.5 mm) were incubated in equilibration buffer containing 50 mm Tris, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, bromphenol blue trace with 100 mg/ml DTT for 1 h and then transferred to the same solution but with 0.2 g/ml iodoacetamide for another 1 h. The gel lanes or pieces were loaded onto SDS-PAGE. After electrophoresis, proteins were transferred to nitrocellulose paper, and the resolved polypeptides were identified by immunoblotting (e.g. with anti-caveolin antibodies).
All BN gels were run 4–6 times. The figures depict representative experiments, and figure legends denote the exact number of experiments done (n = x, etc.).
Western Blotting
For all Western blots comparing different conditions (e.g. Figs. 3, 5 (B and C), 6B, 7C, 8 (A and B), and 10), equal total amounts of protein were applied to the gels. All Western blots were repeated 2–6 times. The figures depict representative experiments, and figure legends denote the exact number of experiments done (n = x, etc.).
FIGURE 5.
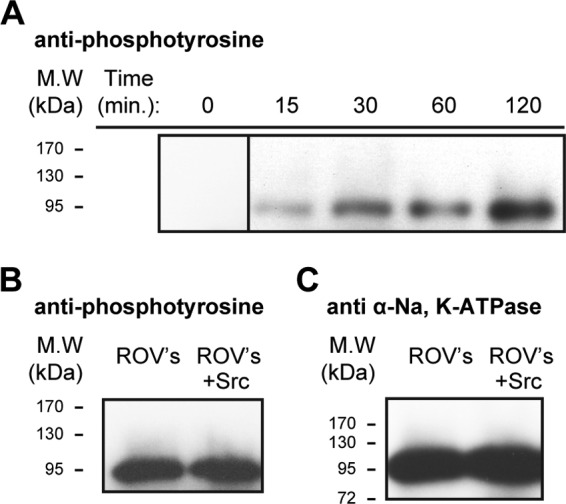
Phosphorylation of Na,K-ATPase α subunit by Src kinase. A, time-dependent phosphorylation of purified human Na,K-ATPase (α1β1FXYD1) by purified Src kinase condition. 2 μg of Src kinase were mixed with 15 μg of Na,K-ATPase with the addition of 1 mm ouabain, 2 mm ATP, 2 mm MgCl, 100 mm NaCl, and 50 mm Tris, pH 7.00. The sample was rotated at room temperature, and 2 μl were taken at each time point, mixed in gel buffer, and loaded on the gel (n = 3). B, right-side out rabbit kidney vesicles. Lane 2, untreated ROVs. Lane 3, after incubation of 10 μg of unmasked ROVs in a medium containing 1 mm ouabain, 2 mm ATP, 2 mm MgCl, 100 mm NaCl, and 50 mm Tris, pH 7.00, 2 μg of Src kinase were added. The sample was rotated at room temperature, and 2 μl were taken at each time point, mixed with gel buffer, and loaded on the gel. Western blots were probed with anti-phophotyrosine (n = 3). C, as in B but probed with Na,K-ATPase anti-α antibody (n = 3).
BN Gels of Purified Na,K-ATPase·Caveolin Complex
Purified dialyzed caveolin was centrifuged for 15 min at 10 °C at 50,000 rpm to remove aggregates. The supernatant (0.2 μg/μl protein) was mixed with purified Na,K-ATPase (5 μg) in the standard elution and incubated on ice for 15 min. The samples were centrifuged again, mixed with Coomassie G-250, and loaded on the blue native gel. The molar excess of caveolin over Na,K-ATPase in standard conditions is 3:1. The mixed sample, control samples of caveolin with elution buffer, and Na,K-ATPase with dialyzed caveolin elution buffer were loaded on the gel. The BN gel was then stained with standard staining solution for at least 2 h (see “Blue Native Gels/SDS-PAGE”) and destained overnight after scan, and the gel was transferred to a nitrocellulose membrane blot and incubated with anti-caveolin antibody.
BN Gels of ROV Vesicles
ROVs were dissolved with DDM at a ratio of (1:2) in a solution containing 30 mm imidazole, pH 7.00, 22 mm NaCl, 0.4 m 6-aminocaproic acid, 1.3 mm EDTA, 10% glycerol. The sample was incubated for 15 min on ice and centrifuged for 30 min at 200,000 × g to remove undissolved protein. Where indicated, BN gels were cut into sections of 1.5 mm, and each piece of gel was incubated in equilibration buffer containing 50 mm Tris, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, bromphenol blue trace with 100 mg/ml DTT for 1 h and then with 0.2 g/ml iodoacetamide for another 1 h. The pieces of gel were loaded onto SDS-PAGE. After electrophoresis, proteins were transferred to nitrocellulose paper, and the resolved polypeptides were identified by immunoblotting using anti-caveolin antibodies. Where indicated, the Na,K-ATPase and caveolin were mixed as in previous experiments, and also Src kinase (0.2 μg/μl) in 20 mm Tris, pH 7.00, 100 mm NaCl, 15% glycerol, 200 mm imidazole was added at different molar ratios.
Pull-down Assays
TEV-cleaved Src Kinase
TEV-cleaved Src kinase was mixed at molar ratios of 1:2, 1:4, and 1:8) (Na,K-ATPase/Src) with 1.5 μg of purified Na,K-ATPase, diluted 1:10 with standard elution buffer (so that the final concentration of imidazole was 20 mm), and rotated at 4 °C for 2 h. 30 μl of cobalt beads equilibrated with bead wash buffer were added to each tube, and the samples were incubated for 1.5 h. The samples were centrifuged, and the unbound fraction was removed. The beads were washed three times with 500 μl of wash buffer and eluted with 50 μl of elution buffer (containing 200 mm imidazole). The different samples were loaded on the gel and blotted with anti-Src kinase antibody.
TEV-cleaved Caveolin
TEV-cleaved caveolin was added to cobalt beads bound with the α1β1 from 50 mg of yeast membranes and reconstituted with FXYD1 to yield the bound α1β1FXYD1 complex, at an approximate molar ratio of 30:1 (caveolin/Na,K-ATPase). The sample was incubated overnight with 25 μl of beads at 4 °C and centrifuged, and the unbound fraction was removed. The beads were washed twice with 500 μl of the standard wash buffer. Elution was performed with 50 μl of the same buffer with an excess of 200 mm imidazole.
All pull-down assays were run at least twice. The figures depict representative experiments, and figure legends denote the exact number of experiments done (n = x, etc.).
Results
Binding of Src Kinase to Na,K-ATPase?
As a first step, we established the experimental systems for expression and purification of Src kinase and a convenient plate assay for Src kinase activity. Human Src kinase was expressed in E. coli BL21(DE3) cells and purified by affinity chromatography on BD-Talon (cobalt beads) via the His6 tag (see “Methods” and Fig. 1A). The purified recombinant Src kinase was phosphorylated on Tyr-418, suggesting that it is an active enzyme (Fig. 1C). The Src kinase activity was measured with an ELISA plate assay as described under “Methods.” In short, Src kinase was incubated with ATP and MgCl2 in a 96-well plate prebound with the substrate, poly(Glu4-Tyr), and Src kinase activity was detected using the anti-phosphotyrosine antibody. Fig. 1B shows a typical experiment of the development of the color with increasing amounts of added Src kinase and as seen, with 20 ng, the reaction was linear with time over 20 min. The specific activity estimated by linear regression was 1.195 ± 0.322 A450 nm/min/μg of protein. By determining the initial rate of color production at varying ATP concentrations, a saturation curve was obtained from which the best fit Km for ATP was found to be 30 ± 5.5 μm (not shown). This value is close to that reported for commercial GST-Src (32). It should be noted that although this assay is convenient, it does not provide an estimate of the absolute specific activity (in μmol of phospho-Tyr/min/mg of protein) because the total amount of bound substrate (poly(Glu4-Tyr)) and phosphorylated tyrosine is not known. Nevertheless, by ensuring that the initial rate of the color production was being measured at saturating ATP concentration (1 mm), the effects of different conditions on the reaction rate can be compared. For example, Fig. 1B also shows that activity can be fully inhibited with PP2, a known Src kinase inhibitor.
The concept of Src activation as a consequence of ouabain binding was tested by assaying Src kinase activity in the absence or presence of a 10-fold molar excess of Na,K-ATPase, stabilized either in an E1 (Na/oligomycin) or in an E2 (Mg/Pi/ouabain) conformation, conditions in which the Src kinase should be inactive or active, respectively. Because it was necessary to add sodium ions to the medium, containing also ATP and magnesium, an obvious confounding effect could be that the Na,K-ATPase might hydrolyze the ATP, and ouabain or oligomycin would inhibit the hydrolysis. Initial experiments showed indeed that the addition of the Na,K-ATPase in a medium containing Na/ATP/Mg, and especially Na/K/ATP/Mg, inhibited Src kinase activity, and either ouabain or oligomycin prevented the effect. This was due only to hydrolysis of the ATP.
In order to eliminate the effect of ATP consumption by the pump, Src kinase activity was then tested with an inactive Na,K-ATPase (Fig. 2). The active site aspartate Asp-369 of the pig α1 subunit was mutated to Asn, and the recombinant pig D369N mutant was expressed and purified (33). As described previously, the D369N mutant cannot be phosphorylated or hydrolyze ATP but binds ouabain and can undergo E1-E2 conformational changes normally. Assuming that the D369N mutant binds the Src kinase, the Src kinase activity should now be inhibited in the E1(3Na)oligomycin and active in the E2P·ouabain conformation, respectively3 (8). The result of this experiment (Fig. 2A) is that the addition of the D369N mutant Na,K-ATPase increased Src kinase activity by ∼60–70% (columns 2 and 3). In these conditions, the D369N mutant is expected to be stabilized primarily as E1(3Na). However, no difference was observed when the Na,K-ATPase was stabilized in either the E1(3Na)oligomycin or E2P·ouabain conformation. Because these effects are not the predicted ones, we have enquired whether the native states of the Na,K-ATPase are responsible for activation of Src kinase. As seen in Fig. 2A (column 5), the fully denatured Na,K-ATPase increased Src kinase activity to a much greater extent, almost 400%, showing that this is an effect of the unfolded Na,K-ATPase. This is a surprising observation. As a test for whether this effect is selective for sequences on the Na,K-ATPase, we added BSA to the medium and observed a similar degree of stimulation of Src kinase (70–80%) as was seen with the native Na,K-ATPase (Fig. 2A, column 4). Furthermore, upon the addition of the Na,K-ATPase D369N mutant, stabilized in either the E1(3Na)oligomycin or E2P·ouabain conformation, together with the BSA, no further stimulation occurred (Fig. 2B, columns 1–3). In other words, the stimulation of the Src kinase activity is a non-Na,K-ATPase-specific effect, which depends on the unfolding of the Na,K-ATPase, and is not dependent on a native conformation. In short, these experiments on the Src kinase activity provide no evidence for a specific direct interaction with the native Na,K-ATPase or the predicted effect of ouabain (see “Discussion”).
If, as proposed (8), bound Src kinase poises the conformation equilibrium of the Na,K-ATPase toward E1, Src kinase should reduce the Na,K-ATPase activity and could also affect the K0.5Na or K0.5K for activation of Na,K-ATPase. Thus, Na,K-ATPase activity was measured in reaction media with different concentrations of sodium and potassium, without and with Src kinase at a large molar excess of Src/Na,K (15–60:1). Table 1 shows that Src kinase had no detectable effect on Na,K-ATPase activity, either in Vmax conditions (130 mm sodium, 20 mm potassium) or at limiting sodium (7.5 mm sodium, 20 mm potassium) and potassium concentrations (130 mm sodium, 2 mm potassium) ions, or in conditions of excess potassium over sodium (10 mm sodium, 100 mm potassium). Thus, the experiment provides no indication that the Src kinase differentially stabilized E1 or E2 conformations.
TABLE 1.
Lack of Effect of Src Kinase on Na,K-ATPase Activity
Na,K-ATPase activity of the purified human α1β1FXYD1 complex was evaluated with the addition of purified recombinant Src kinase at ratios of 1:15–60 (Na,K/Src). In all conditions, the ionic strength (150 mm) was maintained constant using choline chloride. The data represent average values ±S.E. from 4–6 experiments in each condition.
| Condition | Na,K-ATPase activity without Src kinase ± S.E. | Na,K-ATPase activity with Src kinase ± S.E. |
|---|---|---|
| μmol/mg/min | μmol/mg/min | |
| 130 mm NaCl, 20 mm KCl | 18.95 ± 1.5 | 20.43 ± 1.9 |
| 130 mmNaCl, 2 mm KCl | 11.8 ± 1.1 | 12.71 ± 1.46 |
| 7.5 mm NaCl, 20 mm KCl | 12.05 ± 1.01 | 12.69 ± 1.14 |
| 10 mm NaCl, 100 mm KCl | 5.05 ± 0.49 | 5.36 ± 1.5 |
As a test for a direct Src-Na,K-ATPase interaction that might not be detected in an activity assay, a pull-down experiment was performed in which immobilized human Na,K-ATPase (α1β1FXYD1) serves as bait for Src kinase. The Na,K-ATPase was bound to cobalt beads via the β-His10 tag; TEV-cleaved Src was added in molar excess of 2:1, 4:1, and 8:1 over the Na,K-ATPase; and, after three washes, protein bound to the beads was eluted with 200 mm imidazole. The different fractions were loaded on a gel and analyzed for the presence of Src kinase (Fig. 3A). As seen in the figure, Src kinase is identified in the unbound and wash fractions but not in the eluted fraction. Although this pull-down experiment does not reveal an interaction between purified Src kinase and purified Na,K-ATPase, it does not exclude the existence of a weak interaction and the possibility that bound Src kinase dissociates during the washes. In order to exclude this possibility, we have carried out further experiments utilizing blue native gel technology (Fig. 3B). The BN technique is used widely to detect complexes of membrane proteins, including weak complexes, and is not subject to the same caveat as the pull-down experiment due to the fact that the samples are hardly diluted during the electrophoretic run. Purified Src kinase and purified human Na,K-ATPase were incubated for 30 min at 4 °C at a molar ratio of 10:1, and samples of Src or Src plus Na,K-ATPase were loaded onto the BN gel. After the electrophoretic run, the gel was either stained to reveal clearly all of the complexes (lanes 1 and 2), or proteins were transferred to a nitrocellulose membrane for Western blotting analysis (lanes 3 and 4). Lane 1 reveals the Coomassie-stained Src kinase itself (apparent molecular mass ∼70 kDa), and lane 2 shows Src kinase and the Na,K-ATPase, the latter running slightly above the position of the 220 kDa marker protein (i.e. with an apparent molecular mass of ∼250 kDa). Western blotting reveals Src kinase at its expected molecular weight as a monomer (lane 3), but there was no indication of a direct interaction of the Src kinase with Na,K-ATPase in the sample containing both proteins (lane 4).
Although the pull-down assay and BN gels using the purified proteins give a clear cut negative answer to the question of a direct Src/Na,K-ATPase interaction, they do not exclude indirect interactions of Src kinase with the Na,K-ATPase via the mediation of other proteins, lipids, or unknown factors. We have examined this possibility by looking for the hypothetical Src kinase/mediator/Na,K-ATPase interaction in native membranes prepared from rabbit kidney, assuming that they contain the full complement of the mediators. ROVs enriched with Na,K-ATPase were prepared from rabbit kidney outer medulla as described under “Methods.” A preliminary experiment showed that native Src kinase is present only at a very low molar ratio compared with Na,K-ATPase in the ROVs and cannot be present in a significant Src-Na,K-ATPase complex. Thus, the experiment utilized the purified Src kinase and looked for specific association with the native rabbit Na,K-ATPase by BN gel electrophoresis. The renal Na,K-ATPase (specific activity 120–180 μmol/h/mg) represents 5–10% of the total protein in ROVs and should be easily visible on a BN gel. The ROVs were solubilized/unmasked with deoxycholate at a ratio of 1:1 (protein/detergent) and incubated with purified Src kinase for 30 min at 4 °C (at a calculated molar ratio of 10:1 Src/Na,K-ATPase). The samples with or without added Src kinase were then loaded on the BN gels and visualized by Coomassie staining in a first dimension or loaded onto an SDS gel in a second dimension and then analyzed by Western blotting (Fig. 4). The first dimension gel shows the Na,K-ATPase band clearly with little or no change in the presence of Src kinase. The second dimension gel separation of the Na,K-ATPase complex from the native gel is shown in Fig. 4B. The purified Src kinase can be detected, but it is clearly separated from the Na,K-ATPase complex (i.e. there is no accumulation of Src kinase in the bandwith of Na,K-ATPase as would be expected if there was a specific Src·Na,K-ATPase complex). The conclusion is, once again, that Src is not in direct interaction with the Na,K-ATPase, confirming the results of the experiments in (Figs. 2 and 3) using the purified Na,K-ATPase complex and Src kinase.
Phosphorylation of Na,K-ATPase by Src Kinase
Although, by several criteria, Src kinase does not seem to interact stably with the Na,K-ATPase, one could still ask whether individual tyrosine residues are accessible to the Src kinase and can be phosphorylated. Furthermore, because phosphotyrosine residues are receptors for SH2 domains of signaling molecules, such as Src kinase, could it be that binding of Src kinase to Na,K-ATPase requires it to be phosphorylated on tyrosine residues? The results in Fig. 5 show that, indeed, the purified recombinant Na,K-ATPase is a substrate of Src kinase activity. Purified Na,K-ATPase was incubated with purified Src kinase, ATP, MgCl2, and oligomycin/ouabain (see “Methods”), and as seen in Fig. 5A, Src kinase phosphorylated the Na,K-ATPase in a time-dependent manner. The phosphorylated Na,K-ATPase was separated on an SDS gel, and the α1 band was cut out, dissolved, and cleaved with trypsin. The phosphotyrosine peptides were purified by affinity chromatography and were enriched with titanium and subjected to LC-electrospray ionization-MS/MS analysis (see “Methods”). This analysis showed that the phosphorylated residue is Tyr-144, located on the M2 cytoplasmic stalk segment of the α subunit. An assay of the Na,K-ATPase activity after the long preincubation with the Src kinase did not demonstrate any affect of Tyr-144 phosphorylation on activity (result not shown). As another approach, native rabbit kidney ROVs were also examined. Fig. 5B shows that, unlike the recombinant Na,K-ATPase, the native renal Na,K-ATPase α subunit was phosphorylated, and, interestingly, incubation with Src kinase did not increase the band intensity, suggesting that Tyr-144 may be fully phosphorylated in the native Na,K-ATPase.
Caveolin/Na,K-ATPase Interaction
Purified Caveolin and Na,K-ATPase
Here we examine the possibility of a human caveolin 1 interaction with the purified recombinant Na,K-ATPase (α1β1FXYD1 complex) as proposed previously (12, 13). Human caveolin 1 was expressed in BL21(DE3) cells and purified using cobalt beads via a His tag. Initial purification experiments showed a tendency for caveolin to aggregate (data not shown). Subsequently, refolding after denaturation was found to be the optimal method for purification of caveolin. In Coomassie-stained SDS gels, contaminant proteins were not observed, and there was only a faint band corresponding to the purified caveolin, which is known to stain poorly with different dyes (34). By contrast, the purified caveolin was easily visualized in immunoblots using anti-caveolin or anti-His antibodies, and as seen in Fig. 6, the detergent-soluble purified caveolin was detected as a mixture mostly of monomer (∼21 kDa) with some aggregated protein running between the stacking and separating gels. The gel shows the signals with increasing amounts of the purified caveolin (e.g. 20, 40, and 80 ng). In subsequent experiments, quantification of caveolin bound to either purified Na,K-ATPase or native renal Na,K-ATPase (e.g. see Figs. 7 and 8) was done using blots such as in Fig. 6A to calibrate the response of the anti-caveolin 1 in terms of ng of bound caveolin. Fig. 6B shows immunoblots using anti-His or anti-caveolin antibodies, before or after cleavage with TEV protease (referred to as Uncut or Cut). The TEV cleavage was quantitative, as shown by the disappearance of the anti-His staining band and reduction in size of the caveolin (to about 20 kDa) as detected with anti-caveolin in the cut samples.
Fig. 7 presents tests of caveolin/Na,K-ATPase interactions in pull-down (A) or BN gel experiments (B and C) using TEV-cleaved caveolin and purified human Na,K-ATPase (α1β1FXYD1). For the pull-down experiments (Fig. 7A), Na,K-ATPase was solubilized from the yeast membranes and bound to the BD-Talon beads. Purified caveolin was added at a molar ratio of ∼30:1 caveolin/Na,K-ATPase. After incubation overnight at 4 °C with rotation, the beads were washed twice, and proteins were finally eluted with imidazole (see “Methods”). The initial unbound, wash, and elution fractions were loaded on SDS-PAGE and analyzed in immunoblots probed with anti-caveolin 1 (Fig. 7A). Caveolin is clearly detected in the unbound, wash I, and elution fractions but not in the wash II fraction. This observation indicates the existence of a direct interaction between the purified Na,K-ATPase and purified caveolin. However, calculation of the molar ratio of bound caveolin/Na,K-ATPase, eluted from the beads, shows only a low stoichiometry of ∼1:16 (caveolin/Na,K-ATPase). Other experiments showed that increasing the initial ratio of caveolin/Na,K-ATPase above 30:1 did not increase the ratio of eluted caveolin/Na,K-ATPase.
For the BN gel in Fig. 7 (B and C), purified caveolin was mixed with the purified Na,K-ATPase (α1β1FXYD1)) at a molar ratio of 10:1. After preincubation for 15 min on ice, the caveolin/Na,K-ATPase mixture and, in addition, separate samples of Na,K-ATPase and caveolin were loaded onto the BN gel. Note that during electrophoresis, the sample is still at 4 °C, and caveolin and Na,K-ATPase in the mixed sample are in contact with each other for many hours. The BN gel was either stained with Coomassie (Fig. 7B) or analyzed by Western blotting using anti-caveolin 1 (C). The Coomassie stain shows the major band of Na,K-ATPase (lanes 1 and 3) with apparent molecular mass of ∼220–250 kDa, whereas caveolin itself was not detectable (lane 2). The immunoblotting detected caveolin (lane 2) at ∼65 kDa (corresponding to a trimer). In lane 3 with Na,K-ATPase, a clear cut band of caveolin was detected at ∼300–350 kDa in a position above, but overlapping, the major band of the Na,K-ATPase. Thus, the BN gel experiment supports the pull-down experiment showing spontaneous binding of the caveolin to the Na,K-ATPase. Another similarity with the pull-down experiment is that although caveolin was added in a large molar excess, the stoichiometry of caveolin/Na,K-ATPase was much less than 1:1. If the stoichiometry had been 1:1, the band of the Na,K-ATPase in the mixture would have run at the position of the caveolin.
Several different conditions of binding were tried in an attempt to increase the ratio of bound caveolin/Na,K-ATPase but without success (see the BN gels in Fig. 8). After incubating the Na,K-ATPase in sodium/oligomycin (E1-Na conformation) or sodium, ATP, MgCl2, and 1 mm ouabain (E2-P-ouabain conformation) or using Na,K-ATPase prepared with sphingomyelin/cholesterol that are naturally present in native caveolae, the bands were all clearly located, but differences in the relative amounts of caveolin in the 300–350 kDa band were not observed.
In summary, although clear evidence for a spontaneous caveolin/Na,K-ATPase interaction was obtained using the purified proteins, the stoichiometry of the complex was much lower than 1:1.
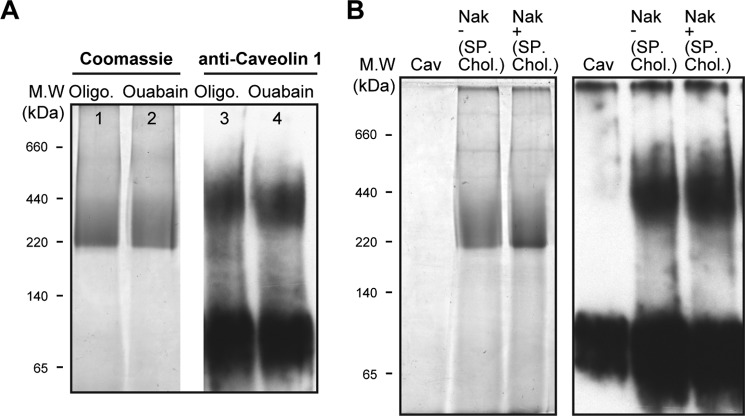
Native Renal Membranes
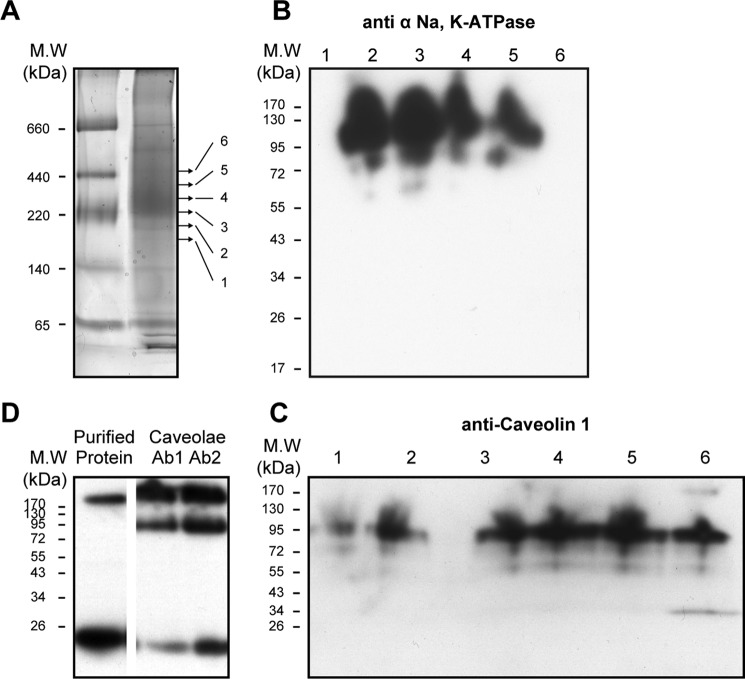
A final set of experiments investigated the caveolin·Na,K-ATPase complex in native rabbit renal caveolae vesicles (Fig. 9, A–D). The caveolae preparation was chosen because it seemed likely that a higher fraction of pumps would be complexed with caveolin 1 than in ROVs. The Coomassie-stained BN gel in Fig. 9A showed the expected 200–250 kDa band of Na,K-ATPase, but an immunoblot for caveolin (Fig. 9D) showed a clear difference from purified recombinant caveolin. Whereas the latter consists mainly of monomer and some aggregated caveolin, in the native renal membranes, caveolin appeared as new band with mass of about 95 kDa in addition to the higher aggregates and low amount of the monomer. The 95 kDa band recognized two separate anti-caveolin 1 antibodies (marked as Ab1 and Ab2), thus excluding the possibility that it represents an unrelated protein that happens to recognize the anti-caveolin 1. For quantification of the caveolin/Na,K-ATPase ratio in the native complex, the Na,K-ATPase band was cut into six narrow bands between 220 and 520 kDa (Table 2). The gel pieces were incubated with SDS-PAGE equilibration buffer, and proteins were separated by SDS-PAGE and then analyzed by immunoblotting, using both anti-α-Na,K-ATPase and anti-caveolin antibodies. It is striking that the caveolin detected in the complex consisted almost exclusively of the 95 kDa band, with very small amounts of lower molecular mass bands. Because the molecular mass of caveolin monomers is ∼21 kDa, one could infer that caveolin molecules are specifically associated in an SDS-resistant complex of 4–5 subunits (Fig. 9D). The relative amounts of Na,K-ATPase α subunit and caveolin were then approximately quantified using the purified human Na,K-ATPase and caveolin to calibrate the antibody responses (see Table 2). The α subunit was distributed in four fractions (fractions 2–5) with the majority in fractions 2 and 3. The caveolin overlapped the α subunit and was detected in all fractions, but the highest intensity was observed in slices 4 and 5. As an example of the quantification, in slice 5 (∼−370–440 kDa), the caveolin band intensity is equivalent to ∼60 ng of purified caveolin, and the Na,K-ATPase band is comparable with ∼150 ng, giving a molar ratio of ∼3:1 (caveolin/Na,K-ATPase), and in the other slices, the calculated ratio is lower. Because the caveolin is present essentially only as the 95 kDa band, corresponding to a tetramer or pentamer, the above result implies that, in fraction 5, 60–75% of the Na,K-ATPase is in a complex with the tetrameric or pentameric caveolin. Of course, fraction 5 represents only a small fraction of the total Na,K-ATPase protein (about 15%) in the caveolae membranes (and presumably a lower fraction in the general population of membranes).
TABLE 2.
Approximate quantification of caveolin/Na,K-ATPase ratios in slices cut out of BN gels
| Slice number | Approximate mass | Caveolin | Na,K-ATPase | Approximate molar ratio (Na,K-ATPase/caveolin) |
|---|---|---|---|---|
| kDa | ng | ng | ||
| 1 | 180–220 | ∼10 | 0 | |
| 2 | 220–270 | ∼20 | ∼500 | 1:0.28 |
| 3 | 270–320 | ∼50 | ∼500 | 1:0.7 |
| 4 | 320–370 | ∼50 | ∼200 | 1:2 |
| 5 | 370–440 | ∼60 | ∼150 | 1:3 |
| 6 | 440–520 | ∼30 | 0 |
The experiment in Fig. 10 shows that when caveolae were dissolved with DDM in the presence of 4 m urea and then treated with DTT or heated to 60 °C, the 95-kDa complex dissociates to a mixture of the monomers and dimers. This demonstrates directly that the 95 kDa band is a complex of associated caveolin monomers. The complex is robust enough to tolerate solubilization in SDS, but the solubilization in SDS/urea and then heating or DTT disassembles the complex.
Discussion
We discuss here the evidence presented that relates to basic assumptions of the proposed model of the Na,K-ATPase as a signaling molecule (8); namely, do Src kinase and caveolin bind specifically to Na,K-ATPase?
Direct Src Kinase/Na,K-ATPase Interaction?
Na,K-ATPase Activity
Src kinase had no effect on Na,K-ATPase activity of the purified α1β1FXYD1 complex either in conditions of Vmax or at limiting concentrations of sodium and potassium ions (Table 1). This finding is clearly at odds with the prediction that Src kinase binds preferentially to the E1 conformation, because this should inhibit Na,K-ATPase activity and reduce K0.5 for activation by potassium ions.
Src Kinase Activity
Src kinase activity, was stimulated ∼60% in the presence of the purified D369N mutant of Na,K-ATPase (Fig. 3A). However, this increase in activity was not specific for the intact Na,K-ATPase molecule, because (a) denatured Na,K-ATPase produced a 4-fold larger increase (Fig. 2A), (b) a similar increase was produced by BSA (Fig. 2B), and (c) in the presence of 1% BSA, stabilizing the pump in the two conformations, E1Na oligomycin and E2P·ouabain did not affect Src kinase activity. All of these findings are at odds with the prediction of the model.
Although the protein-dependent activation of Src kinase seen in Fig. 2 seems to be unrelated to the ouabain-dependent signaling hypotheses, it is, nevertheless, remarkable that Na,K-ATPase and BSA activated Src kinase to the same extent. One common property of Na,K-ATPase and BSA is that they both contain a polyproline II helix motif that is known to bind the SH3 domain of Src (35). Src kinase is negatively regulated by a polyproline II helix in the linker to the SH2 domain and can be activated by a peptide containing the polyproline II helix that displaces the SH3 domain from its negative internal interaction (35) (see below).
Src Kinase Binding in Pull-down Assays and Blue Native Gels
In pull-down assays with purified Na,K-ATPase as bait and purified Src kinase and BN native gels run with a mixture of Na,K-ATPase and Src kinase, no evidence for a stable complex between Src kinase and Na,K-ATPase was detectable (Fig. 3).
Native Renal Membranes
Blue native gel analysis of native rabbit renal ROVs, enriched in Na,K-ATPase, did not reveal association of the added purified Src kinase with the native Na,K-ATPase complex, although the Src kinase was present in a large molar excess (10:1). Thus, the experiment failed to provide evidence for a hypothetical mediator protein (such as in a complex of Src kinase, mediator, and Na,K-ATPase). Of course, this does not exclude the possibility of a water-soluble cellular mediator protein that is lost during the preparation of the membranes.
Phosphorylation of the Na,K-ATPase
Phosphorylation of the α subunit shows that Src kinase and Na,K-ATPase interact, at least transiently, as an enzyme·substrate complex, and there are no serious barriers to an interaction with surface-exposed residues or sequences. The phosphorylation was time-dependent (Fig. 5), and MS/MS analysis shows that the phosphorylation was identified on Tyr-144 on the α1 subunit stalk region leading to TM2. Furthermore, the native renal Na,K-ATPase was also phosphorylated (Fig. 5). The identity of the endogenous kinase and phosphorylated residue(s) in the native Na,K-ATPase is unknown, but the absence of increased phosphorylation upon the addition of Src kinase suggests that Tyr-144 could be phosphorylated. In any case, lack of stable interaction of added Src kinase in the native membranes (Fig. 4) is not attributable to lack of phosphorylation of a critical residue as an SH2 domain receptor (e.g. phosphorylated Tyr-144).
The phosphorylated recombinant Na,K-ATPase did not show any change in activity. This is consistent with a previous finding that the molar turnover rate of the α1β1 expressed in yeast membranes (unphosphorylated) is close to that of native kidney membranes (phosphorylated) (21). Some reports in the literature have documented elevated activity of ∼30% upon tyrosine phosphorylation of Na,K-ATPase (7, 36). However, the samples examined were in native tissue and might involve additional factors (e.g. bound FXYD proteins).
Prior Reports on Src Kinase Binding to Na,K-ATPase
Because our results give such a clear cut negative answer to the question of the direct Src-Na,K-ATPase interaction, it seems worthwhile to consider briefly some of the data that have given rise to this concept.
One type of evidence concerns the selectivity of ouabain for activation of Src kinase. Purified Na,K-ATPase was incubated with 100 μm vanadate, with and without 10 μm ouabain, and it was observed that phosphorylation on Tyr-418 was induced by ouabain but not by vanadate (8). However, subsequently, it was reported that Src kinase is activated either by ouabain or by vanadate with the conclusion that Src kinase activation is dependent on the energetic state of Na,K-ATPase, in particular the ATP/ADP ratio (18). Similarly, it was reported recently that all three inhibitors (ouabain, vanadate, and oligomycin) activate Src Tyr-418 autophosphorylation, and it was concluded that Src kinase activation is due primarily to the ATP-sparing effects of the ATPase inhibitors (16). Src kinase activation was also reportedly induced by stabilization of purified renal Na,K-ATPase using either ouabain or BeF and AlF (11). BeF and AlF stabilize E2 conformations, equivalent to the ground state E2-P conformation (BeF) or the “E2-P” transition state (AlF), respectively (37). The latter is exactly equivalent to that induced by vanadate, which is inconsistent with the finding in Ref. 8. Thus, there is uncertainty as to the selectivity of Src kinase activation by ouabain.
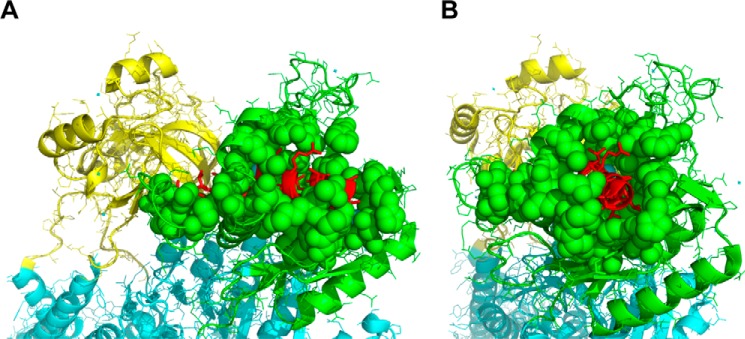
Two other sets of observations also raise questions. First, it was reported that purified isolated cytosolic domains of the α subunit interact directly with Src kinase; namely, one referred to as GST-CD2 (A domain) can bind Src kinase, whereas another, referred to as GST-CD3 (N domain), binds and inhibits the kinase activity of Src (8). However, there was no indication that isolated domains are folded in a native conformation, and the possibility cannot be excluded that the observed effects were nonspecific. Second, a 20-residue peptide derived from the N domain (NaKtide, 408SATWLALSRIAGLCNRAVFQ427; pig α1 numbering) was reported to “pull down” GST-Src and inhibit Src kinase activity (IC50 = 70 nm) (38), suggesting that, in the context of the native Na,K-ATPase α subunit, this sequence could bind the Src kinase. However, inspection of the structure of the Na,K-ATPase shows that the sequence corresponding to the NaKtide peptide is largely buried within the N domain of the Na,K-ATPase. Thus, it is not clear that it would be accessible to Src kinase in the native structure (39) (see Fig. 11). Most recently, a specific mutant A420P (Ala-413 in the pig α1 sequence) was shown to be incapable of interacting with NaKtide and regulating cellular Src kinase (40). Ala-413 is also largely hidden from access to the medium.
FIGURE 11.
NaKtide sequence is largely buried in the N domain of the α subunit. Shown is the crystal structure of the pig Na,K-ATPase in the E1 state (Protein Data Bank code 3WGV). The N domain is colored in green, and the A domain is colored in yellow. NaKtide is in the N domain, colored in red. In A and B, the N domain is seen from different angles.
Caveolin/Na,K-ATPase, Direct Interaction
Several studies have concluded that caveolae microdomains are a potential site for the signaling function of Na,K-ATPase (7, 13, 15). The Na,K-ATPase in these microdomains may not be an active cation pump (15). On the other hand, it has been shown recently that the specific Na,K-ATPase activity in caveolae membranes isolated from kidney is not different from that in the general population of kidney microsomal membranes (14) (although the caveolae were treated with SDS to partially purify the protein, a treatment that might have affected the sphingomyelin and cholesterol content).
Our findings clearly confirm that caveolin 1 spontaneously forms a complex with the Na,K-ATPase. Pull-down experiments with cobalt beads bound with Na,K-ATPase and BN gels provide clear cut evidence for a spontaneous complex, albeit with low stoichiometry (∼1:16–25 caveolin/ Na,K-ATPase) (Fig. 7). Because, in the BN gel, the caveolin·Na,K-ATPase complex runs with an apparent molecular mass of ∼350 kDa and the uncomplexed pump at 220 kDa (Fig. 7), one could speculate that each caveolin·Na,K-ATPase complex consists of 1 molecule of the Na,K-ATPase with 4–5 molecules of caveolin (4–5 separate monomers or as a tetramer-pentamer?). But why was the molar ratio of caveolin/Na,K-ATPase so low? The answer to this question came from studies on the caveolin·pump complex in the caveolae fraction isolated native rabbit kidney membranes (Fig. 9). The first important observation was that the native caveolin consists of an SDS-resistant 95-kDa complex (tetramer or pentamer) in addition to the 21-kDa monomer and higher aggregates. Furthermore, after separation on the BN gel and a second dimension separation on the SDS gel, essentially the only species of caveolin running in the complex with Na,K-ATPase was the 95-kDa tetramer or pentamer. The 95-kDa band was shown to be an SDS-resistant oligomer of caveolin because treatment with DTT or heating disassembled the aggregate to a monomer and dimers (Fig. 10). Most importantly, approximate quantification of the caveolin/pump ratio (Table 2) showed a maximal ratio of ∼1:3 Na,K-ATPase/caveolin 1 in fraction 5, which is close to a 1:1 molar ratio of purified caveolin 14–5/Na,K-ATPase. Thus, native Na,K-ATPase interacts almost exclusively with the tetramer-pentamer oligomer of caveolin 1. On the other hand, this caveolin 14–5·Na,K-ATPase complex represents only a small fraction of the total Na,K-ATPase (10–20%) in the rabbit renal caveolae fraction (i.e. the majority of the Na,K-ATPase is not complexed at all with caveolin). The fraction is likely to be <10% in the general renal membrane population. In the experiments with pure Na,K-ATPase and caveolin, we suspect that the molar ratio of caveolin/Na,K-ATPase was low, despite the large molar excess of caveolin present, because the purified caveolin was found primarily as monomers or high molecular mass aggregates and not as the 95-kDa tetramer-pentamer. Possibly, the initial exposure to 8 m urea dissociated the 95-kDa oligomer during the solubilization and purification process.
No functional effect of caveolin on the Na,K-ATPase activity was found. In the case of the purified recombinant protein, this result is not surprising because the fraction of bound caveolin to the Na,K-ATPase is low, and any functional effects should be undetectable. However, the additional finding that the ratio of caveolin bound to the pump was unaffected by the conformational state of the pump (Fig. 8) suggests that there are truly no conformational dependences on the interactions. In other words, the interaction between caveolin and the Na,K-ATPase may be purely structural.
Possible Mechanisms of Na,K-ATPase-mediated Cell Signaling?
Although the present findings using pure proteins and intact kidney membranes are clearly inconsistent with the hypothesis of direct ouabain-modified Src/Na,K-ATPase interactions proposed previously (8), the basic fact of ouabain-induced signal transduction is not in question (reviewed recently in Refs. 41 and 42). It is the molecular mechanism that remains elusive. As we have reported recently, the activity of recombinant Na,K-ATPase prepared with sphingomyelin and cholesterol is inhibited by 70–80%, due to a strong reduction in the molar turnover rate (20). Taken together, the findings of the Na,K-ATPase·caveolin complex and inhibition by sphingomyelin/cholesterol could be compatible with the presence of heterogeneous populations of the pump in the cell membrane. This would consist of (a) a majority of normal active cation pumps and (b) a minor fraction of inactive or poorly active pumps in the SM/cholesterol-rich caveolae that are complexed with caveolin4–5 oligomers and mediate ouabain-dependent signaling.
One central question regarding the mechanism is whether ouabain-induced Src kinase activation is the initial or a secondary step. A number of possibilities can be speculated upon as follows.
One interesting observation might link Src kinase to the Na,K-ATPase·caveolin complex. It has been shown that Src kinase can phosphorylate caveolin on Tyr-14, and the resulting phosphorylated caveolin binds the SH2 domain of the Src kinase, which is stabilized in the open, active conformation (43). Because Src kinase is known to interact with caveolin (44), conceivably the real signaling complex consists of Na,K-ATPase·caveolin with Src kinase binding transiently, or Src kinase stabilized in an active conformation by caveolin itself, when the pump binds ouabain. We tested for this possibility by mixing purified Na,K-ATPase with caveolin 1 and Src kinase. In a BN gel, we did not observe Src kinase in the higher band containing both Na,K-ATPase and caveolin 1, but this might be explained by the low stoichiometry of the caveolin·Na,K-ATPase complex.
The experiments have not excluded the possibility that, within the cellular environment, Src kinase interacts indirectly with the Na,K-ATPase via a soluble mediator protein, which is lost during membrane preparation. As a case in point, the cytoskeletal protein adducin is known to bind directly to the α subunit (45). Furthermore, an antihypertensive drug, rostafuroxin, exerts its antihypertensive and also antihypertrophic effects by antagonizing the Src-dependent signaling triggered by ouabain (46). In vivo, rostafuroxin acts as an antagonist of endogenous ouabain and has been shown to disrupt an interaction between Src kinase and adducin (47, 48). Thus, ouabain may affect the adducin and Src interaction.
With a stretch of the imagination, it is conceivable that in the cellular environment, the accessibility of the polyproline motif near the N terminus of the α subunit (PPPXXP), which was proposed above to bind to and activate Src kinase, may be regulated by the presence of ouabain (unlike the situation of the purified proteins).
The possibility that ouabain-dependent activation of Src kinase is a secondary event following inhibition of the pump and changes in the internal cation composition (sodium and calcium) would require that these events occur in a microdomain that is not in equilibrium with the general cytoplasmic space. This must be the case because the signaling pathway is activated by nanomolar concentrations (3), which could not inhibit a significant fraction of the pumps and indeed have been shown not to induce significant net changes in cytoplasmic sodium and calcium concentrations (49).
Author Contributions
E. Y. performed all of the experiments and wrote the paper; Y. P. produced the Src kinase and caveolin 1; T. M. performed mass spectroscopy; A. K. was responsible for setting up blue native gel technology; and S. J. D. K. planned the experiments, coordinated the study, and wrote the paper.
The authors declare that they have no conflicts of interest with the contents of this article.
Conceivably, also the Src·Na,K-ATPase complex might be hindered from accessing the poly(Glu4-Tyr) substrate on the plate, in which case the addition of the Na,K-ATPase would inhibit Src kinase activity in all conformations. As seen from the results in Fig. 2, in reality, this is not the observed phenomenon.
- SH2
- Src homology 2
- SH3
- Src homology 3
- BN
- blue native
- C12E8
- octaethylene glycol monodecyl ether
- DDM
- n-dodecyl-β-d-maltopyranoside
- ROV
- right-side out vesicle
- TEV
- tobacco etch virus
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- SOPS
- 1-stearoyl-2-oleoyl-sn-glycero-3-phospho-l-serine.
References
- 1. Kometiani P., Li J., Gnudi L., Kahn B. B., Askari A., and Xie Z. (1998) Multiple signal transduction pathways link Na+/K+-ATPase to growth-related genes in cardiac myocytes: the roles of Ras and mitogen-activated protein kinases. J. Biol. Chem. 273, 15249–15256 [DOI] [PubMed] [Google Scholar]
- 2. Haas M., Wang H., Tian J., and Xie Z. (2002) Src-mediated inter-receptor cross-talk between the Na+/K+-ATPase and the epidermal growth factor receptor relays the signal from ouabain to mitogen-activated protein kinases. J. Biol. Chem. 277, 18694–18702 [DOI] [PubMed] [Google Scholar]
- 3. Haas M., Askari A., and Xie Z. (2000) Involvement of Src and epidermal growth factor receptor in the signal-transducing function of Na+/K+-ATPase. J. Biol. Chem. 275, 27832–27837 [DOI] [PubMed] [Google Scholar]
- 4. Aperia A. (2007) New roles for an old enzyme: Na,K-ATPase emerges as an interesting drug target. J. Intern. Med. 261, 44–52 [DOI] [PubMed] [Google Scholar]
- 5. Schoner W. (2002) Endogenous cardiac glycosides, a new class of steroid hormones. Eur. J. Biochem. 269, 2440–2448 [DOI] [PubMed] [Google Scholar]
- 6. Aydemir-Koksoy A., Abramowitz J., and Allen J. C. (2001) Ouabain-induced signaling and vascular smooth muscle cell proliferation. J. Biol. Chem. 276, 46605–46611 [DOI] [PubMed] [Google Scholar]
- 7. Ferrandi M., Molinari I., Barassi P., Minotti E., Bianchi G., and Ferrari P. (2004) Organ hypertrophic signaling within caveolae membrane subdomains triggered by ouabain and antagonized by PST 2238. J. Biol. Chem. 279, 33306–33314 [DOI] [PubMed] [Google Scholar]
- 8. Tian J., Cai T., Yuan Z., Wang H., Liu L., Haas M., Maksimova E., Huang X. Y., and Xie Z. J. (2006) Binding of Src to Na+/K+-ATPase forms a functional signaling complex. Mol. Biol. Cell 17, 317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu J., Kesiry R., Periyasamy S. M., Malhotra D., Xie Z., and Shapiro J. I. (2004) Ouabain induces endocytosis of plasmalemmal Na/K-ATPase in LLC-PK1 cells by a clathrin-dependent mechanism. Kidney Int. 66, 227–241 [DOI] [PubMed] [Google Scholar]
- 10. Li Z., and Xie Z. (2009) The Na/K-ATPase/Src complex and cardiotonic steroid-activated protein kinase cascades. Pflugers Arch. 457, 635–644 [DOI] [PubMed] [Google Scholar]
- 11. Ye Q., Li Z., Tian J., Xie J. X., Liu L., and Xie Z. (2011) Identification of a potential receptor that couples ion transport to protein kinase activity. J. Biol. Chem. 286, 6225–6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang H., Haas M., Liang M., Cai T., Tian J., Li S., and Xie Z. (2004) Ouabain assembles signaling cascades through the caveolar Na+/K+-ATPase. J. Biol. Chem. 279, 17250–17259 [DOI] [PubMed] [Google Scholar]
- 13. Liu L., Mohammadi K., Aynafshar B., Wang H., Li D., Liu J., Ivanov A. V., Xie Z., and Askari A. (2003) Role of caveolae in signal-transducing function of cardiac Na+/K+-ATPase. Am. J. Physiol. Cell Physiol. 284, C1550–C1560 [DOI] [PubMed] [Google Scholar]
- 14. Liu L., Ivanov A. V., Gable M. E., Jolivel F., Morrill G. A., and Askari A. (2011) Comparative properties of caveolar and noncaveolar preparations of kidney Na+/K+-ATPase. Biochemistry 50, 8664–8673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang M., Tian J., Liu L., Pierre S., Liu J., Shapiro J., and Xie Z. J. (2007) Identification of a pool of non-pumping Na/K-ATPase. J. Biol. Chem. 282, 10585–10593 [DOI] [PubMed] [Google Scholar]
- 16. Gable M. E., Abdallah S. L., Najjar S. M., Liu L., and Askari A. (2014) Digitalis-induced cell signaling by the sodium pump: on the relation of Src to Na+/K+-ATPase. Biochem. Biophys. Res. Commun. 446, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 17. Clifford R. J., and Kaplan J. H. (2013) Human breast tumor cells are more resistant to cardiac glycoside toxicity than non-tumorigenic breast cells. PLoS One 8, e84306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weigand K. M., Swarts H. G., Fedosova N. U., Russel F. G., and Koenderink J. B. (2012) Na,K-ATPase activity modulates Src activation: a role for ATP/ADP ratio. Biochim. Biophys. Acta 1818, 1269–1273 [DOI] [PubMed] [Google Scholar]
- 19. Haviv H., Cohen E., Lifshitz Y., Tal D. M., Goldshleger R., and Karlish S. J. (2007) Stabilization of Na+,K+-ATPase purified from Pichia pastoris membranes by specific interactions with lipids. Biochemistry 46, 12855–12867 [DOI] [PubMed] [Google Scholar]
- 20. Habeck M., Haviv H., Katz A., Kapri-Pardes E., Ayciriex S., Shevchenko A., Ogawa H., Toyoshima C., and Karlish S. J. (2015) Stimulation, inhibition, or stabilization of Na,K-ATPase caused by specific lipid interactions at distinct sites. J. Biol. Chem. 290, 4829–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cohen E., Goldshleger R., Shainskaya A., Tal D. M., Ebel C., le Maire M., and Karlish S. J. (2005) Purification of Na+,K+-ATPase expressed in Pichia pastoris reveals an essential role of phospholipid-protein interactions. J. Biol. Chem. 280, 16610–16618 [DOI] [PubMed] [Google Scholar]
- 22. Strugatsky D., Gottschalk K. E., Goldshleger R., Bibi E., and Karlish S. J. (2003) Expression of Na+,K+-ATPase in Pichia pastoris: analysis of wild type and D369N mutant proteins by Fe2+-catalyzed oxidative cleavage and molecular modeling. J. Biol. Chem. 278, 46064–46073 [DOI] [PubMed] [Google Scholar]
- 23. Kapri-Pardes E., Katz A., Haviv H., Mahmmoud Y., Ilan M., Khalfin-Penigel I., Carmeli S., Yarden O., and Karlish S. J. (2011) Stabilization of the α2 isoform of Na,K-ATPase by mutations in a phospholipid binding pocket. J. Biol. Chem. 286, 42888–42899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jørgensen P. L. (1988) Purification of Na+,K+-ATPase: enzyme sources, preparative problems, and preparation from mammalian kidney. Methods Enzymol. 156, 29–43 [DOI] [PubMed] [Google Scholar]
- 25. Karlish S. J., Goldshleger R., and Jørgensen P. L. (1993) Location of Asn831 of the α chain of Na/K-ATPase at the cytoplasmic surface: implication for topological models. J. Biol. Chem. 268, 3471–3478 [PubMed] [Google Scholar]
- 26. Macdonald J. L., and Pike L. J. (2005) A simplified method for the preparation of detergent-free lipid rafts. J. Lipid Res. 46, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 27. Unger T., Jacobovitch Y., Dantes A., Bernheim R., and Peleg Y. (2010) Applications of the restriction free (RF) cloning procedure for molecular manipulations and protein expression. J. Struct. Biol. 172, 34–44 [DOI] [PubMed] [Google Scholar]
- 28. Lázaro I., Gonzalez M., Roy G., Villar L. M., and Gonzalez-Porqué P. (1991) Description of an enzyme-linked immunosorbent assay for the detection of protein tyrosine kinase. Anal. Biochem. 192, 257–261 [DOI] [PubMed] [Google Scholar]
- 29. Karni R., Mizrachi S., Reiss-Sklan E., Gazit A., Livnah O., and Levitzki A. (2003) The pp60c-Src inhibitor PP1 is non-competitive against ATP. FEBS Lett. 537, 47–52 [DOI] [PubMed] [Google Scholar]
- 30. Shevchenko A., Wilm M., Vorm O., and Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 31. Wittig I., Braun H. P., and Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 32. Susa M., Luong-Nguyen N. H., Crespo J., Maier R., Missbach M., and McMaster G. (2000) Active recombinant human tyrosine kinase c-Yes: expression in baculovirus system, purification, comparison to c-Src, and inhibition by a c-Src inhibitor. Protein Expr. Purif. 19, 99–106 [DOI] [PubMed] [Google Scholar]
- 33. Belogus T., Haviv H., and Karlish S. J. (2009) Neutralization of the charge on Asp 369 of Na+,K+-ATPase triggers E1 ↔ E2 conformational changes. J. Biol. Chem. 284, 31038–31051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li S., Song K. S., and Lisanti M. P. (1996) Expression and characterization of recombinant caveolin: purification by polyhistidine tagging and cholesterol-dependent incorporation into defined lipid membranes. J. Biol. Chem. 271, 568–573 [PubMed] [Google Scholar]
- 35. Moroco J. A., Craigo J. K., Iacob R. E., Wales T. E., Engen J. R., Smithgall T. E. (2014) Differential sensitivity of Src-family kinases to activation by SH3 domain displacement. PLoS One 9, e105629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bozulic L. D., Dean W. L., and Delamere N. A. (2005) The influence of SRC-family tyrosine kinases on Na,K-ATPase activity in lens epithelium. Invest. Ophthalmol. Vis. Sci. 46, 618–622 [DOI] [PubMed] [Google Scholar]
- 37. Cornelius F., Mahmmoud Y. A., and Toyoshima C. (2011) Metal fluoride complexes of Na,K-ATPase: characterization of fluoride-stabilized phosphoenzyme analogues and their interaction with cardiotonic steroids. J. Biol. Chem. 286, 29882–29892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Z., Cai T., Tian J., Xie J. X., Zhao X., Liu L., Shapiro J. I., and Xie Z. (2009) NaKtide, a Na/K-ATPase-derived peptide Src inhibitor, antagonizes ouabain-activated signal transduction in cultured cells. J. Biol. Chem. 284, 21066–21076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reinhard L., Tidow H., Clausen M. J., and Nissen P. (2013) Na+,K+-ATPase as a docking station: protein-protein complexes of the Na+,K+-ATPase. Cell. Mol. Life Sci. 70, 205–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lai F., Madan N., Ye Q., Duan Q., Li Z., Wang S., Si S., and Xie Z. (2013) Identification of a mutant α1 Na/K-ATPase that pumps but is defective in signal transduction. J. Biol. Chem. 288, 13295–13304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fontana J. M., Burlaka I., Khodus G., Brismar H., and Aperia A. (2013) Calcium oscillations triggered by cardiotonic steroids. FEBS J. 280, 5450–5455 [DOI] [PubMed] [Google Scholar]
- 42. Aperia A., Akkuratov E. E., Fontana J. M., and Brismar H. (2016) Na+,K+-ATPase, a new class of plasma membrane receptors. Am. J. Physiol. Cell Physiol. 310, C491–C495 [DOI] [PubMed] [Google Scholar]
- 43. Gottlieb-Abraham E., Shvartsman D. E., Donaldson J. C., Ehrlich M., Gutman O., Martin G. S., and Henis Y. I. (2013) Src-mediated caveolin-1 phosphorylation affects the targeting of active Src to specific membrane sites. Mol. Biol. Cell 24, 3881–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li S., Couet J., and Lisanti M. P. (1996) Src tyrosine kinases, Gα subunits, and H-Ras share a common membrane-anchored scaffolding protein, caveolin: caveolin binding negatively regulates the auto-activation of Src tyrosine kinases. J. Biol. Chem. 271, 29182–29190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ferrandi M., Salardi S., Tripodi G., Barassi P., Rivera R., Manunta P., Goldshleger R., Ferrari P., Bianchi G., Karlish S. J. (1999) Evidence for an interaction between adducin and Na+-K+-ATPase: relation to genetic hypertension. Am. J. Physiol. 277, H1338–H1349 [DOI] [PubMed] [Google Scholar]
- 46. Ferrari P., Ferrandi M., Valentini G., and Bianchi G. (2006) Rostafuroxin: an ouabain antagonist that corrects renal and vascular Na+-K+-ATPase alterations in ouabain and adducin-dependent hypertension. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R529–R535 [DOI] [PubMed] [Google Scholar]
- 47. Ferrandi M., Molinari I., Torielli L., Padoani G., Salardi S., Rastaldi M. P., Ferrari P., and Bianchi G. (2010) Adducin- and ouabain-related gene variants predict the antihypertensive activity of rostafuroxin, part 1: experimental studies. Sci. Transl. Med. 2, 59ra86. [DOI] [PubMed] [Google Scholar]
- 48. Ferrari P. (2010) Rostafuroxin: an ouabain-inhibitor counteracting specific forms of hypertension. Biochim. Biophys. Acta 1802, 1254–1258 [DOI] [PubMed] [Google Scholar]
- 49. Liu J., Tian J., Haas M., Shapiro J. I., Askari A., and Xie Z. (2000) Ouabain interaction with cardiac Na+/K+-ATPase initiates signal cascades independent of changes in intracellular Na+ and Ca2+ concentrations. J. Biol. Chem. 275, 27838–27844 [DOI] [PubMed] [Google Scholar]