Abstract
Degradation and remodeling of the extracellular matrix by matrix metalloproteinases (MMPs) plays important roles in normal development, inflammation, and cancer. MMP-9 efficiently degrades the extracellular matrix component gelatin, and the hemopexin domain of MMP-9 (PEX9) inhibits this degradation. To study the molecular basis of this inhibition, we generated GST fusion proteins containing PEX9 or truncated forms corresponding to specific structural blades (B1–B4) of PEX9. GST-PEX9 inhibited MMP-9-driven gelatin proteolysis, measured by gelatin zymography, FITC-gelatin conversion, and DQ-gelatin degradation assays. However, GST-PEX9 did not prevent the degradation of other MMP-9 substrates, such as a fluorogenic peptide, αB crystalline, or nonmuscular actin. Therefore, PEX9 may inhibit gelatin degradation by shielding gelatin and specifically preventing its binding to MMP-9. Accordingly, GST-PEX9 also abolished the degradation of gelatin by MMP-2, confirming that PEX9 is not an MMP-9 antagonist. Moreover, GST-B4 and, to a lesser extent, GST-B1 also inhibited gelatin degradation by MMP-9, indicating that these regions are responsible for the inhibitory activity of PEX9. Accordingly, ELISAs demonstrated that GST-B4 and GST-B1 specifically bound to gelatin. Our results establish new functions of PEX9 attributed to blades B4 and B1 and should help in designing specific inhibitors of gelatin degradation.
Keywords: catalysis, enzyme catalysis, inhibition mechanism, inhibitor, matrix metalloproteinase (MMP), hemopexin, PEX9 blades 1 and 4, enzyme antagonist, gelatin degradation
Introduction
The matrix metalloprotease (MMP)5 family comprises more than 25 Zn2+-dependent proteases that mainly degrade extracellular matrix components but also signaling molecules, membrane receptors, and intracellular and nuclear proteins (1–6). MMPs play roles in normal developmental processes (embryogenesis, wound healing, and cell mobilization), as well as in many pathological conditions, such as cancer and inflammatory reactions (7, 8). Indeed, the catalytic regions of MMPs have been tested as targets for cancer drugs, but most clinical trials failed (9). This was probably due to the high homology of these regions across MMPs, with the consequent low selectivity of the inhibitors and the induction of secondary effects of the drugs.
MMP-9, also known as gelatinase B, is one of the most complex members of the MMP family. In contrast to gelatinase A (MMP-2), which is constitutively expressed, MMP-9 is highly regulated by numerous agonists/antagonists (10). Like other MMPs, MMP-9 is a multidomain enzyme composed of a prodomain, a catalytic domain, and a carboxyl-terminal hemopexin domain. Both MMP-2 and MMP-9 possess a domain with three fibronectin type II homology repeats that yield high affinity binding to gelatins. Only MMP-9 contains a serine-, threonine-, and proline-rich O-glycosylated domain, which confers high flexibility to the enzyme (10).
The hemopexin domain of MMP-9 (PEX9) and other MMPs consists of four blade β-propeller structures (blades 1, 2, 3, and 4) (11). PEX9 is responsible for the interactions of MMP-9 with many molecules, including substrates, cell receptors, such as integrins and CD44, and tissue inhibitors of MMPs (12–15). PEX9 also contributes to MMP-9 trimerization (16). Because of these properties and the low homology with the hemopexin domains of other MMPs (25–30% identical residues) (12), PEX9 is revealed as an interesting target to increase the selectivity of MMP-9 inhibitors.
The main substrate of MMP-9 is gelatin, the product of denaturation or degradation of collagen by collagenases (MMP-1, MMP-8, and MMP-13). It was previously shown that the murine PEX9 binds to gelatin, and a claim was made that PEX9 acts as an antagonist of MMP-9 (17). This was based on the fact that PEX9 inhibited gelatinolysis in zymography assays, as well as invasion of melanoma and colorectal cancer cells (17, 18). In the present study, we have further investigated this, and we demonstrate that human PEX9 inhibits gelatin degradation by shielding gelatin and specifically preventing its interaction with active MMP-9. Moreover, we have identified the specific regions of PEX9 responsible for the inhibitory effect, and we show that PEX9 is not an antagonist of MMP-9 catalytic activity.
Experimental Procedures
MEC-1 Cells and Transfectants
The MEC-1 cell line, established from a chronic lymphocytic leukemia (CLL) patient, was purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). MEC-1 cells stably expressing MMP-9 (MMP-9–MEC-1 cells) were generated by lentiviral transfection exactly as described (19). The cells were maintained in Iscove's modified Dulbecco's medium (Lonza, Basel, Switzerland), 10% fetal bovine serum.
Antibodies, Reagents, Proteins, and Peptides
Rabbit polyclonal antibodies to GST (sc-459) and MMP-9 (sc-6841-R) were from Santa Cruz Biotechnology (Santa Cruz, CA). Recombinant full-length human pro-MMP-9 was prepared as reported (15). DQTM gelatin was purchased from Invitrogen, and DQTM collagen and fluorogenic peptide (DNPPro-Cha-Gly-Cys(Me)-His-Ala-Lys(N-Me-Abz)-NH2) were from Calbiochem. MMP-2 and FITC-gelatin were from Molecular Probes (Eugene, OR). Human nonmuscle actin and subunit αB-crystallin recombinant proteins were from Cytoskeleton, Inc. (Denver, CO) and Enzo Life Sciences (Farmingdale, NY), respectively. Pfu DNA polymerase was from Agilent Technologies (Waldbronn, Germany).
Construction of Plasmids
The full-length human MMP-9 cDNA (cloned in pEGFP-N1) was obtained from Dr. Santos Mañes (Centro Nacional de Biotecnología, Madrid, Spain) (20). The hemopexin domain cDNA was amplified by PCR using the following primers, custom-made by Sigma-Aldrich: forward, 5′-GAATTCCCCTTTGAGTCCGGTGGACG-3′ engineering an internal EcoRI site (underlined in Table 1); and reverse, 5′-CTCGAGCTAGTCCTCAGGGCACTGCA-3′ containing an internal XhoI restriction site. Amplification of DNAs was performed with the use of cloned Pfu DNA polymerase, and the resultant PCR fragments were inserted into the pGEX4T3 vector (GE Healthcare Biosciences) to generate the GST-PEX9 DNA. To generate the PEX9 mutants, we performed similar protocols but used the primers indicated in Table 1. For GST-B1, GST-B2, GST-B3, and GST-B4, we engineered an internal EcoRI site in the forward primers (GAATTC) and a XhoI site (CTCGAG) in the reverse primers. For GST-ΔB1, GST-ΔB2, GST-ΔB3, and GST-ΔB4, we designed internal primers with the initial and final sequences of each removed blade, and we amplified the complete construction from the GST-PEX9 plasmid. All obtained sequences were confirmed by DNA sequencing.
TABLE 1.
Primers designed to generate the various GST-PEX9 recombinant proteins
F, forward; R, reverse.
| Protein | Primers (5′ → 3′) |
|---|---|
| PEX9 F | GAATTCCCCTTTGAGTCCGGTGGACG |
| PEX9 R | CTCGAGCTAGTCCTCAGGGCACTGCA |
| B1B2 F | GAATTCCCCTTTGAGTCCGGTGGACG |
| B1B2 R | CTCGAGTCACCTGGGCCACGTC |
| B3B4 F | GAATTCAGCCGACGTGGCCCAG |
| B3B4 R | CTCGAGCTAGTCCTCAGGGCACTGCA |
| B1 F | GAATTCCCCTTTGAGTCCGGTGGACG |
| B1 R | CTCGAGTCCAGCTTGCGGGGCA |
| B2 F | GAATTCACCGCTGGACTCGGTCTTTG |
| B2 R | CTCGAGTCACCTGGGCCACGTC |
| B3 F | GAATTCAGCCGACGTGGCCCAG |
| B3 R | CTCGAGTCCAAAGGCACCCCGG |
| B4 F | GAATTCTTTGGACACGCACGACG |
| B4 R | CTCGAGCTAGTCCTCAGGGCACTGCA |
| ΔB1 F | TCTAGAGCTGGACTCGGTCTTTGA |
| ΔB1 R | TCTAGAGATCCACGCGGAACCAGAT |
| ΔB2 F | TCTAGAAGTGGCAGGGGGAAGATG |
| ΔB2 R | TCTAGAGCCACTTGTCGGCGATA |
| ΔB3 F | TCTAGAGACGTCTTCCAGTACCGAGAG |
| ΔB3 R | TCTAGAGCCCAGCTTGTCCAGA |
| ΔB4 F | TCTAGAGACTGACTGACGATCTGCCTC |
| ΔB4 R | TCTAGATCCGGGGATCCACCAT |
Expression and Purification of GST and GST Fusion Proteins
Recombinant proteins were prepared as previously described for GST and GST-PEX9 (14). Briefly, protein constructs were expressed in DH5α Escherichia coli competent cells by induction with isopropyl-1-thio-β-d-galactopyranoside. Recombinant proteins were specifically induced in this system and produced in sufficient amount for the desired experimentation. Bacteria cultures were lysed by sonication in 1.5 m NaCl, 0.5 m Tris, 50 mm Na2 EDTA, 10% Triton, and centrifuged. GST, GST-B1, GST-B2, and GST-B3 were soluble in this buffer and were purified using a glutathione-agarose matrix (Sigma-Aldrich). All other fusion proteins appeared in inclusion bodies and were solubilized in PBS, supplemented with 1% sarkosyl. These fusion proteins did not bind to glutathione-agarose and were purified by SDS-PAGE and electroelution. Purity and identity of the proteins were confirmed by SDS-PAGE and Western blotting. Purified fusion proteins were renatured by extensive dialysis against PBS.
Cell Adhesion and Soluble Binding Assays
Adhesion assays were performed in 96-well plates coated with 0.5% BSA or various concentrations of appropriate proteins. 1 × 105 MEC-1 cells were incubated with 1.4 ng/ml 2′,7′-bis(carboxyethyl)-5(6′)-carboxyfluorescein-acetoxymethyl ester (BCECF-AM; Molecular Probes) for 20 min; suspended in RPMI 1640, 0.5% BSA (adhesion medium); and added to the coated wells. After 60 min at 37 °C, attached cells were lysed with PBS, 0.1% SDS and quantified using a fluorescence analyzer (BMG Labtech, Offenburg, Germany). For binding assays in solution, 1 × 105 cells were incubated in 100 μl of adhesion medium containing the proteins under investigation and incubated for 30 min at 4 °C. After washing with ice-cold medium, the cells were incubated with anti-GST polyclonal antibodies (30 min, 4 °C), washed with cold PBS, and incubated (30 min, 4 °C) with Alexa 488-labeled secondary antibodies. Surface-bound proteins were analyzed by flow cytometry.
Incubation of PEX9 Proteins with Active MMP-9
MMP-9 in 50 mm Tris, 150 mm NaCl, 5 mm CaCl2, 0.01% Tween 20 buffer was activated with the catalytic domain of MMP-3 (Millipore) at a MMP-3:MMP-9 ratio of 1:100. GST-PEX9 (1 μg) was incubated with active MMP-9 for 100 min, and aliquots of the reaction were taken every 10 min. For the analysis of nonmuscular actin and αB-crystallin cleavage, 2 μg of the recombinant proteins were incubated with active MMP-9 (enzyme:substrate ratio of 1:20) at 37 °C for 2, 8, and 24 h in the presence or absence of GST-PEX9. The incubated fractions were analyzed on 12% SDS-PAGE gels and visualized by staining with Coomassie Brilliant Blue R-250 (Sigma).
Gelatin Zymography Analysis
Samples of 20 ng of recombinant purified MMP-9 were analyzed on 7.5% polyacrylamide gels containing 0.1% gelatin (Sigma). The amount of the MMP-9 added in the gel was within the linear range of the zymogram, and this was evaluated by testing a dilution series of MMP-9 by zymography analysis. After electrophoresis, the gels were rinsed three times for 30 min in 2.5% Triton X-100 and once for 30 min in distilled water, followed by overnight incubation in 50 mm Tris, pH 7.5, 200 mm NaCl, 10 mm CaCl2 at 37 °C. To analyze the effect of GST-PEX9 recombinant proteins, mutant proteins were added to the incubation buffers overnight at a final concentration of 0.4 μm. The gels were stained with 0.2% Coomassie Blue, and areas of gelatinolysis were visualized as transparent lysis zones. The bands were quantitated on a densitometer (Molecular Dynamics) using the Quantity-OneTM program (Bio-Rad). Control MMP-9 levels were normalized to 100%.
FITC-Gelatin Degradation Assay
60 nm activated MMP-9 in 50 mm Tris, 150 mm NaCl, 5 mm CaCl2, 0.01% Tween 20 was added to 96 FITC-gelatin-coated wells (100 μg/ml) in the absence or presence of 0.4 μm recombinant GST-PEX9 proteins. After 24 h at 37 °C, the wells were washed with PBS, and fluorescence was measured using a fluorescence analyzer (BMG Labtechnologies, Offenburg Germany). The background fluorescence of the conditioned medium added to unlabeled gelatin-coated wells was subtracted from all values. To analyze the capacity of the GST-PEX9 proteins on gelatin degradation by leukemic cells, 96-well plates were coated with 100 μg/ml of FITC-labeled gelatin and the various GST-PEX9 proteins and fixed with 0.5% glutaraldehyde. After washing with PBS, 70% ethanol/PBS and medium, 1 × 105 MMP-9–MEC-1 cells were added to the wells. After 24 h, the cells were removed with several washing steps, and the fluorescence was measured.
DQ-Gelatin or Fluorigenic Peptide Degradation Assay
A DQ-gelatin degradation assay was performed as previously described (21). Briefly, to measure the degradation in fluorescence units, 0.1 nm of active MMP-9 was added to a solution of 2.5 μg/ml of DQ-gelatin in a black 96-well plate. To analyze the maximal enzyme velocity in these assays, several concentrations of the DQ-gelatin substrate (20, 10, 5, 2.5, 0.75, 0.5, and 0.25 μg/ml) were added to the 96-well plate. To test the effect of the GST-PEX9 constructs, 0.4 μm of the recombinant proteins were added to the wells. The plates were immediately placed in the fluorescence reader (FL600 microplate fluorescence reader; Biotek, Highland Park, IL), and fluorescence was measured every 10 min for 2 h at 37 °C (excitation, 485 nm; emission, 530 nm). All of the data were corrected by subtraction of the negative controls. In all the experiments, the maximal velocity of the enzyme was the velocity observed during the first 10 min of the assay. Graphs and calculations were obtained with Prism 5 (GraphPad Software, Inc.).
To test the degradation of a small fluorogenic peptide we used DNPPro-Cha-Gly-Cys(Me)-His-Ala-Lys(N-Me-Abz)-NH2 (excitation, 365 nm; emission, 450 nm). This substrate is cleaved by MMP-9 into a single cleavage product: Dnm-Pro-Cha-Gly (22). Activated MMP-9 was used at a concentration of 1 nm, and the fluorigenic peptide was used at 10 μg/ml. Fluorescence was measured every 10 min for 2 h in the fluorescence reader.
ELISA
High binding 96-well plates were coated with 5 μg of gelatin overnight at 4 °C. After washing, recombinant proteins were added at a final concentration of 0.4 μm and incubated for 1 h at 37 °C; then the plates were washed, and anti-GST polyclonal antibody at 5 μg/ml was added to the wells. After incubation for 1 h at 37 °C, the plate was incubated with secondary anti-rabbit HRP antibody for 30 min, washed, and developed.
Results
The Human proMMP-9 Hemopexin Domain Inhibits Gelatin Degradation by MMP-9
Roeb et al. (17) previously showed that the murine PEX9 domain inhibits MMP-9 activity in gelatin zymography assays. To investigate the molecular basis of this observation, we prepared a fusion protein containing GST and human PEX9 (residues 508–707 of proMMP-9; Fig. 1A). After purification, the GST-PEX9 protein ran as a single 50-kDa band in SDS gels, and its identity was confirmed by Western blotting analysis with an anti-MMP-9 antibody (Fig. 1B). Recombinant GST alone was also prepared by bacterial expression (Fig. 1B) and was used as control. We previously showed that the resulting GST-PEX9 protein, but not GST alone, mediates CLL cell adhesion and soluble cell binding via specific cell surface receptors (14, 23). GST-PEX9 also specifically inhibits chemotaxis and transendothelial migration of leukemic B cells and induced survival signals upon binding to α4β1 integrin (14, 23). Therefore, the purified GST-PEX9 protein fully retained its biological activities and was suitable for further functional analyses.
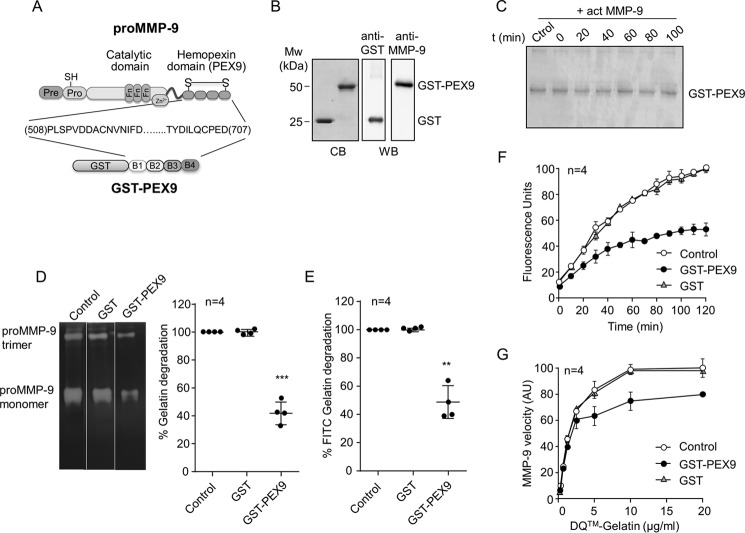
FIGURE 1.
Effect of human GST-PEX9 on the degradation of gelatin by MMP-9. A, schematic drawing of the structural domains of proMMP-9. The four structural blades of PEX9 (B1–B4) are indicated. The GST-PEX9 fusion protein prepared in this study contains amino acid residues 508–707 of human proMMP-9. B, SDS-PAGE (10% acrylamide) and Western blotting (WB) analyses of the purified GST and GST-PEX9 proteins. C, 1 μg of GST-PEX9 was incubated with 1 nm active MMP-9 for the indicated times. The samples were analyzed by 10% SDS-PAGE and stained with Coomassie Blue (CB). D, gelatin zymography analysis of 20 ng of MMP-9 (within the linearity of the assay) in the presence or absence of 0.4 μm GST-PEX9. A representative zymography is shown. E, 60 nm of MMP-9 was added to FITC-gelatin-coated plates, in the absence or presence of 0.4 μm GST or GST-PEX9. After 24 h at 37 °C, fluorescence was determined, and control (Ctrol) values were normalized to 100. F, effect of GST and GST-PEX9 (0.4 μm) on the conversion of DQ-gelatin into fluorogenic gelatin. G, maximal enzyme velocity as a function of the amount of substrate. The maximal velocity of each point is the first read time point at 10 min, and these measurements were within the linearity of the assay. The bars represent standard deviation. AU, arbitrary units. **, p < 0.01; ***, p < 0.001.
In initial experiments, we determined whether MMP-9 was able to cleave GST-PEX9. The GST-PEX9 protein (1 μg) was incubated with active MMP-9 (1 nm), and samples were taken every 10 min for a time interval of 100 min. Analyses by SDS-PAGE followed by protein staining with Coomassie Blue showed that MMP-9 did not degrade the GST-PEX9 protein (Fig. 1C). With the knowledge that GST-PEX9 remained intact in the presence of active MMP-9, we analyzed the effect of GST-PEX9 on gelatin degradation by MMP-9, using three different techniques: gelatin zymography, degradation of FITC-gelatin in 96-well plate format, and DQ-gelatin degradation assay. In all these experiments, we also tested the effect of purified GST as negative controls.
Gelatin zymography analyses demonstrated that adding GST-PEX9 (0.4 μm) reduced gelatin degradation by MMP-9 by ∼50% (Fig. 1D), whereas GST had no effect. To study whether GST-PEX9 inhibited the degradation of FITC-gelatin, GST-PEX9 was incubated with active MMP-9 in microtiter plate wells coated with FITC-gelatin, and the fluorescence was measured after 24 h. GST-PEX9, but not GST, significantly decreased gelatin degradation by MMP-9 (average 50% decrease) (Fig. 1E). The inhibitory function of PEX9 was also demonstrated with a dye-quenched gelatin (DQ-gelatin) degradation assay (21). As shown in Fig. 1F, GST-PEX9 significantly reduced the level of fluorescence in a time-dependent manner, during the 120 min of the assay. GST-PEX9 also reduced the MMP-9 enzyme velocity in this assay, whereas no inhibition was seen after incubation with GST (Fig. 1G). Altogether, these results established that the human PEX9 domain inhibits the degradation of gelatin by MMP-9, in agreement with previous results with murine PEX9 (17).
The PEX9 Domain Does Not Act as an MMP-9 Antagonist
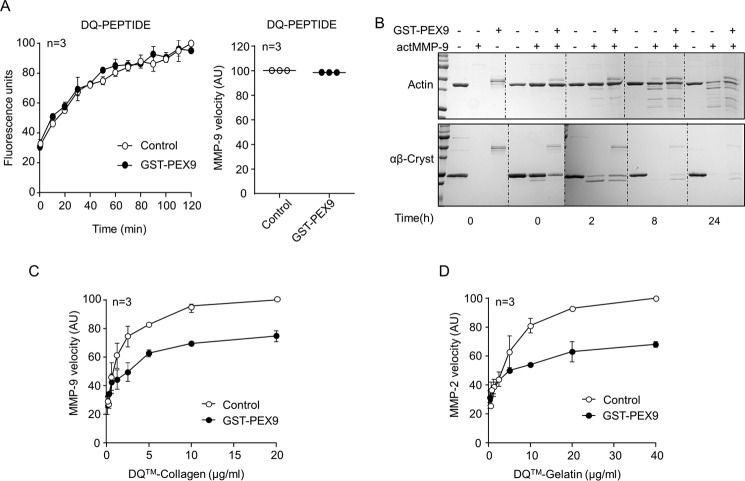
To determine whether PEX9 is an MMP-9 antagonist and blocks its catalytic activity or, alternatively, binds to gelatin and prevents its degradation by MMP-9, we tested different MMP-9 substrates and different gelatinases. In contrast to the inhibitory effect on the degradation of macromolecular gelatin (Fig. 1, D–G), GST-PEX9 was unable to block the proteolysis of a small DQ-fluorogenic peptide (FP) by MMP-9 (Fig. 2A). Moreover, the enzyme velocity from the same assays was similar in the presence or absence of GST-PEX9 (Fig. 2A).
FIGURE 2.
Role of GST-PEX9 on the degradation of MMP-9 and MMP-2 substrates. A, 1 nm MMP-9 was incubated with 2.5 μg/ml DQ-peptide in the absence or presence of 0.4 μm GST-PEX9. The conversion of DQ-peptides into fluorogenic peptides was monitored on a fluorescence reader. The fluorescence and the maximal velocity of the enzyme (initial velocity at the 10-min read point) from the same experiment are shown in the two graphs. B, preparations of 2 μg of actin or αB-crystallin (αB-Cryst) were incubated with active MMP-9 (actMMP-9) in the absence or presence of 0.4 μm GST-PEX9. After the indicated times at 37 °C, cleaved proteins were analyzed by SDS-PAGE and Coomassie Blue staining. C and D, velocity of the degradation of DQ-collagen by MMP-9 (C) and DQ-gelatin by MMP-2 (D). Maximum velocity (initial velocity time point at 10 min) was normalized to 100, and enzymes were used at 0.1 nm. AU, arbitrary units.
We next analyzed whether GST-PEX9 affected the proteolytic activity of MMP-9 on other MMP-9 substrates, such as αB-crystallin (24) and nonmuscular actin (6). To this end, we performed kinetic assays incubating αB-crystallin and actin with active MMP-9 in the presence or absence of GST-PEX9 and evaluated protein degradation at 2, 8, and 24 h. As shown in Fig. 2B, the presence of PEX9 did not influence the degradation of actin by MMP-9 and only minimally delayed the proteolysis of αB-crystallin by this enzyme. In contrast to these results, GST-PEX9 inhibited the degradation of DQ-collagen, another MMP-9 substrate (Fig. 2C). Because DQ-gelatin can also be degraded by MMP-2 (gelatinase A), we tested whether PEX9 also inhibited DQ-gelatin proteolysis by this enzyme. Fig. 2D shows that GST-PEX9 was able to inhibit DQ-gelatin degradation by MMP-2 at similar levels as observed for MMP-9. Altogether, these results corroborate the role of PEX9 as an inhibitor of gelatin/collagen degradation and establish that PEX9 is not a direct antagonist of the catalytic activity of MMP-9.
Blades 1 and 4 of the PEX9 Domain Are Responsible for the Gelatin Degradation Inhibitory Activity of PEX9
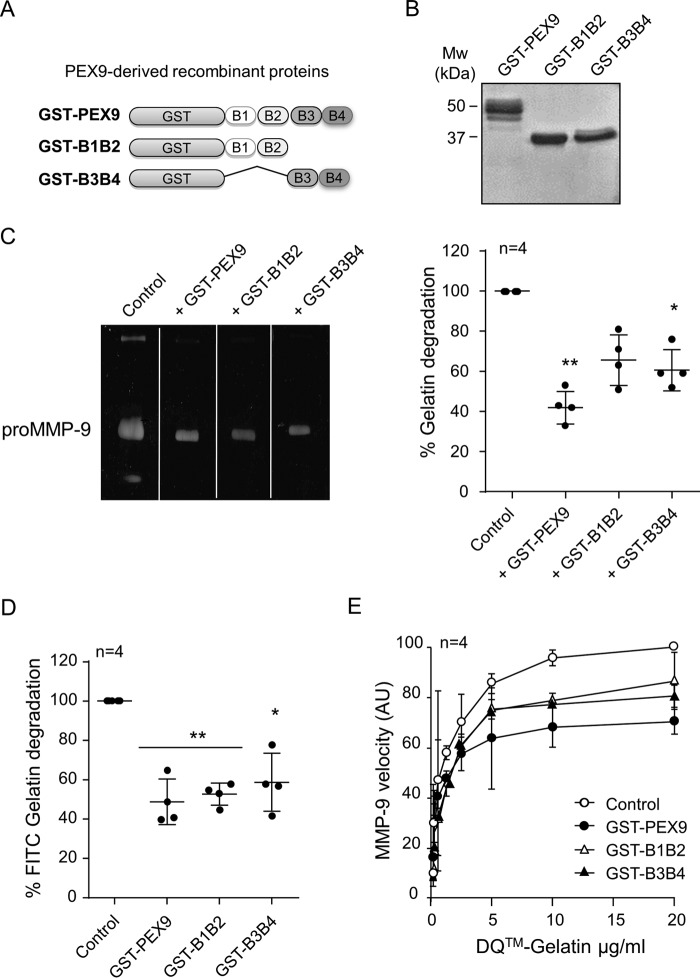
The PEX9 domain contains a four-blade β-propeller structure (11), schematically shown in Figs. 1A and 3A. To define the region involved in the inhibition of gelatin degradation, we prepared two GST fusion proteins containing deletions of blades 3 and 4 (GST-B1B2 protein, proMMP-9 residues 508–613) or blades 1 and 2 (GST-B3B4 protein, proMMP-9 residues 609–707) (Fig. 3A). The purity of these proteins was confirmed by SDS-PAGE analyses (Fig. 3B). We previously showed that purified GST-B1B2 and GST-B3B4 mediated CLL cell adhesion by binding to CD44 and α4β1 integrin, respectively (14, 23). Moreover, we also identified specific cell binding sequences in both proteins (14, 23). The soluble proteins also specifically bound to cells and inhibited CLL cell chemotaxis and transendothelial migration (14, 23), thus confirming the retention of their biological activity. GST-B1B2 and GST-B3B4 were therefore tested as inhibitors of gelatin degradation using different assays. In gelatin zymography analysis, GST-PEX9, GST-B1B2, and GST-B3B4 (all at 0.4 μm) inhibited gelatin degradation by 60, 35, and 40%, respectively (Fig. 3C). Similar results were obtained with the FITC-gelatin degradation assay. In this case, all three proteins, GST-PEX9, GST-B1B2, and GST-B3B4 reduced the gelatinolytic activity of MMP-9 to ∼50%, compared with the control (Fig. 3D). To further confirm these results, we analyzed the inhibitory role of these recombinant proteins in the DQ-gelatin degradation assay. Fig. 3E shows that GST-PEX9, GST-B1B2, and GST-B3B4 similarly inhibited the velocity of the DQ-gelatin degradation by MMP-9. These data indicated that both regions B1B2 and B3B4 appeared to contribute to the inhibitory function of PEX9.
FIGURE 3.
Effect of the B1B2 and B3B4 regions of PEX9 on the inhibition of gelatin degradation by MMP-9. A, schematic drawing of the truncated GST-B1B2 and GST-B3B4 fusion proteins prepared in this study. B, SDS-PAGE analysis of the purified GST fusion proteins, visualized by Coomassie Blue staining. C, representative gelatin zymography analysis using 20 ng of proMMP-9 and with or without the indicated proteins (0.4 μm each). D, 60 nm of MMP-9 was added to FITC-gelatin-coated plates in the absence or presence of 0.4 μm of the indicated proteins. After 24 h at 37 °C, fluorescence was determined, and control values were normalized to 100. E, effect of the indicated proteins on the conversion of DQ-gelatin into fluorogenic gelatin. The lines show the maximal enzyme velocity evolution as a function of the amount of substrate. The bars represent standard deviation. *, p < 0.05; **, p < 0.01. AU, arbitrary units.
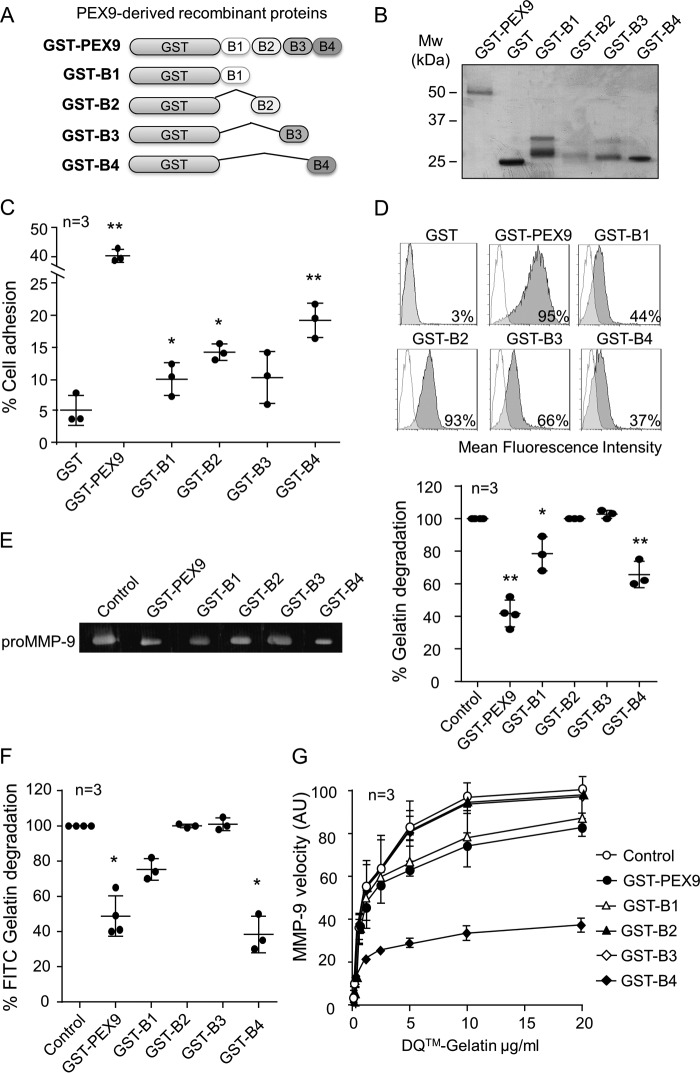
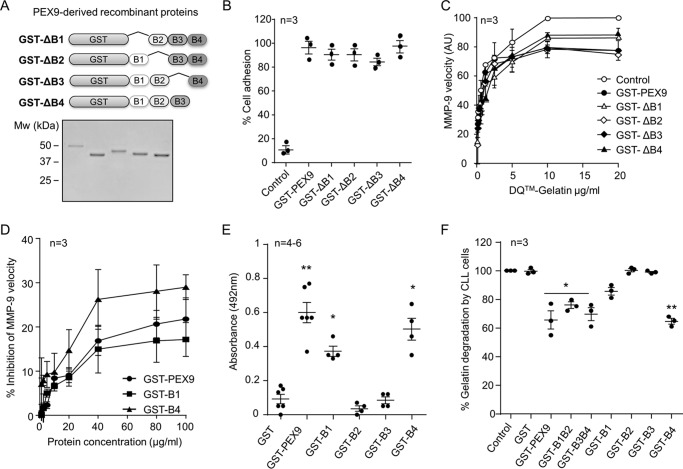
To identify the specific sequence within PEX9 that binds to gelatin, we prepared alternative recombinant proteins, each containing only one blade of the PEX9 domain (GST-B1, GST-B2, GST-B3, and GST-B4) (Fig. 4, A and B). To first confirm that these proteins retained their biological function, we analyzed their ability to mediate cell adhesion and soluble binding. Fig. 4C shows that all four PEX9 blades mediated adhesion of MEC-1 cells, whereas GST did not. As expected, the level of cell adhesion to the individual blades was lower than adhesion to the entire PEX9 domain (Fig. 4C). Additionally, these fusion proteins, but not GST, were able to bind to MEC-1 cells in suspension, as detected with an anti-GST antibody (Fig. 4D).
FIGURE 4.
Blades B1 and B4 of PEX9 inhibit gelatin degradation by MMP-9. A, schematic drawing of the truncated recombinant GST fusion proteins. B, SDS-PAGE analysis of the purified proteins shown in A. C, BCECF-AM-labeled MEC-1 cells were added to wells coated with the indicated proteins (0.4 μm), and adhesion was quantitated as explained. D, MEC-1 cells were incubated for 30 min with or without the indicated proteins (0.8 μm) and analyzed by flow cytometry using anti-GST antibodies. E, representative gelatin zymography analysis of 20 ng of purified recombinant proMMP-9 in the presence or absence of the indicated proteins. The values (arbitrary units) represent the averages of three different experiments. F, 60 nm of MMP-9 was added to FITC-gelatin-coated plates in the absence or presence of the indicated proteins. After 24 h, the fluorescence was determined. G, effect of the indicated proteins on the conversion of DQ-gelatin into fluorogenic gelatin. The graph represents the maximal enzyme velocity evolution as a function of the amount of substrate. *, p < 0.05; **, p < 0.01. AU, arbitrary units.
Having confirmed that the purified GST-B1, GST-B2, GST-B3, and GST-B4 proteins were functional, we tested their ability to block gelatin degradation. Fig. 4E shows for a representative experiment how GST-B1 and GST-B4, but not GST-B2 or GST-B3 (all at 0.4 μm), inhibited gelatin degradation in a gelatin zymography assay. By comparison of the four single blade proteins, GST-B4 was found to be more effective, blocking gelatin degradation by 42%, whereas incubation with GST-B1 inhibited by 25%. In parallel experiments, GST-PEX9 diminished the degradation of gelatin by 60% (average of three independent experiments) (Fig. 4C). We further compared the effect of the recombinant proteins in a FITC-labeled gelatin degradation assay. Again, GST-B4 was the most effective inhibitor, impairing FITC-gelatin degradation by MMP-9 by 75%, whereas GST-B1 diminished degradation by 25% (Fig. 4F). GST-B2 and GST-B3 had no effect. We next tested the ability of these proteins to inhibit DQ-gelatin degradation by MMP-9. In these assays, GST-B4 blocked DQ-gelatin degradation by 63%, whereas the effect of GST-B1 was lower and similar to that of the entire GST-PEX9 (Fig. 4G).
The preceding results indicated that blades B1 and B4 were the most effective inhibitory sequences in PEX9. To further confirm this, we generated GST fusion proteins containing deletions of each of the four blades in PEX9: GST-ΔB1, GST-ΔB2, GST-ΔB3, and GST-ΔB4 (Fig. 5A). The purity of these proteins was confirmed by SDS-PAGE analyses (Fig. 5A). These four fusion proteins supported adhesion of MEC-1 cells (Fig. 5B), confirming the retention of their biological activity. GST-ΔB1, GST-ΔB2, GST-ΔB3, and GST-ΔB4 proteins were then tested as inhibitors of DQ-gelatin degradation by MMP-9. Fig. 5C shows that deletion of the B1 or B4 blades reduced the inhibitory ability of PEX9 from 25% to 13 and 10%, respectively, whereas deletion of B2 or B3 had no effect.
FIGURE 5.
Further characterization of the effect of PEX9 blades B1 or B4 on gelatin degradation by MMP-9. A, schematic drawing of the truncated GST fusion proteins lacking specific blades of PEX9 and SDS-PAGE analysis of the purified proteins. B, cell adhesion of BCECF-AM-labeled MEC-1 cells to the indicated proteins (all at 0.4 μm). C, effect of the indicated mutant proteins on the conversion of DQ-gelatin into fluorogenic gelatin. The maximal enzyme velocity (initial velocity t = 10 min) evolution as a function of the amount of substrate is shown. D, dose-dependent inhibition of MMP-9-driven DQ-gelatin degradation by the indicated recombinant proteins. The graph shows the inhibition of the maximal velocity in each experiment, after normalizing control signals to 100. E, binding of the indicated proteins to 5 μg of immobilized gelatin determined by ELISA. F, effect of PEX9-derived proteins on the degradation of FITC-gelatin by MMP-9–MEC-1 cells. 1 × 105 MMP-9–MEC-1 cells were added to 96-well plates coated with FITC-gelatin, in the absence or presence of the indicated proteins. After 24 h at 37 °C, wells were washed, and the remaining fluorescence was determined. The values from control wells (FITC coated without cells) were normalized to 100. The bars represent standard deviation. AU, arbitrary units. *, p < 0.05; **, p < 0.01.
Further Characterization of the Inhibitory Activity of Blades B1 and B4 on Gelatin Degradation by MMP-9
Having identified blades 1 and 4 as the regions in PEX9 with inhibitory activity, we tested whether this activity was dose-dependent. The DQ-gelatin degradation assay was used for this purpose. Activated MMP-9 was incubated with constant concentrations of DQ-gelatin (20 μg/ml) in the absence or presence of increasing concentrations of GST-PEX9, GST-B1, or GST-B4. All three proteins inhibited gelatinolysis in a dose-dependent manner, and GST-B4 was the most effective at all concentrations tested (Fig. 5D). To confirm these results, we tested whether these proteins bound to gelatin using ELISAs. Fig. 5E shows that GST-PEX9, GST-B1, and GST-B4 bound to gelatin, whereas GST, GST-B2, and GST-B3 did not. These data therefore established the identity and specificity of blades 1 and 4 of PEX9 as inhibitors of gelatin degradation.
To place these biochemical findings in a biological context, we analyzed the inhibitory capacity of the PEX9 proteins on cell-induced degradation of FITC-gelatin. For these experiments, we used the leukemic cell line MEC-1, stably transfected with MMP-9 (MMP-9–MEC-1 cells) (19). We previously showed that these cells produce high amounts of MMP-9, most of which is in an active form (19). 1 × 105 MMP-9 transfected MEC-1 cells were incubated with FITC-gelatin in the presence or absence of the GST-PEX9 variants, and the resulting fluorescence was determined after 24 h. Fig. 5F shows that GST-PEX9 significantly inhibited FITC-gelatin degradation by 34%, compared with the control cells with no inhibitor. In agreement with the results obtained with purified MMP-9, recombinant proteins containing blades B1 and/or B4 inhibited the degradation of gelatin by MMP-9–MEC-1 cells, whereas GST-B2 and GST-B3 had no effect (Fig. 5F). Altogether, these results established that blade 4 and to a lesser extent blade 1 are the regions responsible for the inhibitory role of PEX9 in gelatin degradation.
Discussion
We have studied the effect of human PEX9 on MMP-9 proteolysis on several substrates. We report that PEX9 inhibits the degradation of gelatin but not of other MMP-9 substrates. This was due to the sequestration of gelatin by PEX9, which is therefore a binding competitor, but not an antagonist, of MMP-9.
The MMP family has been studied for many years, because of their role in many physiological and pathological processes. At first, the scientific community focused on the catalytic roles of these molecules, and most efforts were concentrated on developing inhibitors of the catalytic activity. The main problem with this was that the amino acid sequences of the catalytic domain are highly conserved among all MMPs and even within the ADAM and ADAMTS family (25, 26). Therefore, most of the inhibitors were not specific, inhibiting a range of MMPs and resulting in side effects. In recent years, it was demonstrated that most MMPs also have noncatalytic functions (27) and can act like ligands (28) or even like transcription factors (29, 30). Because of the failure of first generation MMP inhibitors, the search is now open toward inhibitors that target specific functions of one or more MMPs. More detailed information about the structure and function of single MMP domains (exosites) is therefore needed (25, 31).
The hemopexin domain of the MMPs is the main domain that drives the noncatalytic functions, because of its ability to interact with substrates, receptors, and inhibitors (11–13). For example, in chronic lymphocytic leukemia cells, PEX9 can drive intracellular signaling and survival independently of other MMP-9 domains (14, 28). Given the lower amino acid sequence homology in the hemopexin domains than in other domains among MMPs, we suggest that targeting this domain may be a useful and more specific approach to prevent MMP-9-mediated pathological functions. Indeed, strategies aimed at blocking the interaction of PEX with individual molecules are already in progress. For example, we recently identified the sequences in PEX9 responsible for the interaction of proMMP-9 with CD44 (FDAIAEIGNQLYLFKDGKYW, blade 1) and α4β1 integrin (FPGVPLDTHDVFQYREKAYFC, blade 4) in chronic lymphocytic leukemia cells (14, 23). Likewise, active sequences in blades 1 and 4 of MMP-14 were identified and shown to inhibit carcinoma cell migration, tumor metastasis, and angiogenesis (32).
In the present report, we have characterized a different function of PEX9, namely its ability to block gelatin and collagen degradation by MMP-9. Interestingly, this activity also involved structural blades 1 and 4, highlighting the importance of these regions in many PEX9 functions. Our data clearly show that the inhibitory function of PEX9 did not apply to the degradation of other MMP-9 substrates, such as a small fluorogenic peptide or αB-crystallin. This, together with the fact that PEX9 also abrogated gelatinolysis by MMP-2, indicates that PEX9 is not an antagonist of MMP-9 catalytic function, as previously proposed (17). Instead, PEX9 blocks gelatin degradation because it specifically binds to gelatin and probably not to other MMP-9 substrates. Indeed, murine PEX9 was previously shown to bind to gelatin, and the binding parameters have been reported (17, 18). Using recombinant proteins that lack or contain specific blades of human PEX9, in the present study we have extended the binding studies and have identified blade 4 and, to a lesser extent, blade 1 as the structural modules in PEX9 that bind to gelatin and inhibit MMP-9 gelatinolysis.
Degradation and remodeling of the extracellular matrix, mostly carried out by MMPs, play crucial roles in normal development but also in the migration and metastasis of cancer cells (33, 34). The extracellular matrix is therefore a dynamic structure whose components interact with specific integrin receptors and trigger intracellular signaling and cell responses. Consequently, depending on the composition of the extracellular matrix, these ligand-receptor interactions will induce different signaling, directly affecting cell behavior (35). To fully understand the behavior of different cell types, including metastatic cancer cells, new probes and also extracellular matrix molecules may lead to the development of new drugs against cancer. Until now, the research focus was to target the malignant cells and the MMPs, which degrade the extracellular matrix. It might be equally important to try to block specific components of this matrix, such as gelatin. Therefore the PEX9 blades, generated in this study, may become useful probes to study gelatinolysis and/or serve to generate specific inhibitors of gelatinolysis. Because this approach specifically targets gelatin catalysis, it might also serve to block the migration of metastatic cancer cells and of leukocytes in pathological inflammations. In summary, the present study helps to understand how MMP-9 and gelatin interact with each other and points toward new directions to develop new inhibitors of specific MMP functions.
Author Contributions
E. U-B. designed and performed most of the experiments, analyzed the data, and wrote the paper. J. V. provided valuable help and advice with some experiments. E. B. provided valuable help and advice with some experiments and the preparation of the figures. G. O. and A. G-P. designed and supervised the experiments, discussed the results, and critically reviewed the manuscript.
Acknowledgments
We thank Erik Martens for excellent technical assistance and Dr. Irene Amigo-Jiménez for useful comments and help with the protein purification procedures.
This work was supported by Grant SAF2012-31613 and Red Temática de Investigación Cooperativa en Cáncer Grant RD12/0036/0061 from the Ministry of Economy and Competitivity (Spain) (to A. G.-P.); by Grant S2010/BMD-2314 (to A. G.-P.) from the Comunidad de Madrid/European Union; and by the Concerted Research Actions Grant GOA 2013–2015 and the fund for Scientific Research of Flanders (to G. O.). The authors declare that they have no conflicts of interest with the contents of this article.
- MMP
- matrix metalloprotease
- CLL
- chronic lymphocytic leukemia
- BCECF-AM
- 2′,7′-bis(carboxyethyl)-5(6′)-carboxyfluorescein-acetoxymethyl ester.
References
- 1. Overall C. M., McQuibban G. A., and Clark-Lewis I. (2002) Discovery of chemokine substrates for matrix metalloproteinases by exosite scanning: a new tool for degradomics. Biol. Chem. 383, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 2. Overall C. M., Tam E. M., Kappelhoff R., Connor A., Ewart T., Morrison C. J., Puente X., López-Otín C., and Seth A. (2004) Protease degradomics: mass spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated DNA microarray of all human proteases and inhibitors. Biol. Chem. 385, 493–504 [DOI] [PubMed] [Google Scholar]
- 3. Butler G. S., and Overall C. M. (2009) Updated biological roles for matrix metalloproteinases and new “intracellular” substrates revealed by degradomics. Biochemistry 48, 10830–10845 [DOI] [PubMed] [Google Scholar]
- 4. Huesgen P. F., and Overall C. M. (2012) N- and C-terminal degradomics: new approaches to reveal biological roles for plant proteases from substrate identification. Physiol. Plant 145, 5–17 [DOI] [PubMed] [Google Scholar]
- 5. Cauwe B., Van den Steen P. E., and Opdenakker G. (2007) The biochemical, biological, and pathological kaleidoscope of cell surface substrates processed by matrix metalloproteinases. Crit. Rev. Biochem. Mol. Biol. 42, 113–185 [DOI] [PubMed] [Google Scholar]
- 6. Cauwe B., and Opdenakker G. (2010) Intracellular substrate cleavage: a novel dimension in the biochemistry, biology and pathology of matrix metalloproteinases. Crit. Rev. Biochem. Mol. Biol. 45, 351–423 [DOI] [PubMed] [Google Scholar]
- 7. Nabeshima K., Inoue T., Shimao Y., and Sameshima T. (2002) Matrix metalloproteinases in tumor invasion: role for cell migration. Pathol. Int. 52, 255–264 [DOI] [PubMed] [Google Scholar]
- 8. Löffek S., Schilling O., and Franzke C. W. (2011) Series “matrix metalloproteinases in lung health and disease”: biological role of matrix metalloproteinases: a critical balance. Eur. Respir. J. 38, 191–208 [DOI] [PubMed] [Google Scholar]
- 9. Coussens L. M., Fingleton B., and Matrisian L. M. (2002) Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 295, 2387–2392 [DOI] [PubMed] [Google Scholar]
- 10. Vandooren J., Van den Steen P. E., and Opdenakker G. (2013) Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit. Rev. Biochem. Mol. Biol. 48, 222–272 [DOI] [PubMed] [Google Scholar]
- 11. Cha H., Kopetzki E., Huber R., Lanzendörfer M., and Brandstetter H. (2002) Structural basis of the adaptive molecular recognition by MMP9. J. Mol. Biol. 320, 1065–1079 [DOI] [PubMed] [Google Scholar]
- 12. Piccard H., Van den Steen P. E., and Opdenakker G. (2007) Hemopexin domains as multifunctional liganding modules in matrix metalloproteinases and other proteins. J. Leukoc. Biol. 81, 870–892 [DOI] [PubMed] [Google Scholar]
- 13. Redondo-Muñoz J., Ugarte-Berzal E., García-Marco J. A., del Cerro M. H., Van den Steen P. E., Opdenakker G., Terol M. J., and García-Pardo A. (2008) α4β1 integrin and 190-kDa CD44v constitute a cell surface docking complex for gelatinase B/MMP-9 in chronic leukemic but not in normal B cells. Blood 112, 169–178 [DOI] [PubMed] [Google Scholar]
- 14. Ugarte-Berzal E., Bailón E., Amigo-Jiménez I., Vituri C. L., del Cerro M. H., Terol M. J., Albar J. P., Rivas G., García-Marco J. A., and García-Pardo A. (2012) A 17-residue sequence from the matrix metalloproteinase-9 (MMP-9) hemopexin domain binds α4β1 integrin and inhibits MMP-9-induced functions in chronic lymphocytic leukemia B cells. J. Biol. Chem. 287, 27601–27613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van den Steen P. E., Van Aelst I., Hvidberg V., Piccard H., Fiten P., Jacobsen C., Moestrup S. K., Fry S., Royle L., Wormald M. R., Wallis R., Rudd P. M., Dwek R. A., and Opdenakker G. (2006) The hemopexin and O-glycosylated domains tune gelatinase B/MMP-9 bioavailability via inhibition and binding to cargo receptors. J. Biol. Chem. 281, 18626–18637 [DOI] [PubMed] [Google Scholar]
- 16. Vandooren J., Born B., Solomonov I., Zajac E., Saldova R., Senske M., Ugarte-Berzal E., Martens E., Van den Steen P. E., Van Damme J., Garcia-Pardo A., Froeyen M., Deryugina E. I., Quigley J. P., Moestrup S. K., Rudd P. M., Sagi I., and Opdenakker G. (2015) Circular trimers of gelatinase B/matrix metalloproteinase-9 constitute a distinct population of functional enzyme molecules differentially regulated by tissue inhibitor of metalloproteinases-1. Biochem. J. 465, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roeb E., Schleinkofer K., Kernebeck T., Pötsch S., Jansen B., Behrmann I., Matern S., and Grötzinger J. (2002) The matrix metalloproteinase 9 (mmp-9) hemopexin domain is a novel gelatin binding domain and acts as an antagonist. J. Biol. Chem. 277, 50326–50332 [DOI] [PubMed] [Google Scholar]
- 18. Burg-Roderfeld M., Roderfeld M., Wagner S., Henkel C., Grötzinger J., and Roeb E. (2007) MMP-9-hemopexin domain hampers adhesion and migration of colorectal cancer cells. Int. J. Oncol. 30, 985–992 [DOI] [PubMed] [Google Scholar]
- 19. Bailón E., Ugarte-Berzal E., Amigo-Jiménez I., Van den Steen P., Opdenakker G., García-Marco J. A., and García-Pardo A. (2014) Overexpression of progelatinase B/proMMP-9 affects migration regulatory pathways and impairs chronic lymphocytic leukemia cell homing to bone marrow and spleen. J. Leukoc. Biol. 96, 185–199 [DOI] [PubMed] [Google Scholar]
- 20. Mira E., Lacalle R. A., Buesa J. M., de Buitrago G. G., Jiménez-Baranda S., Gómez-Moutón C., Martínez-A C., and Mañes S. (2004) Secreted MMP9 promotes angiogenesis more efficiently than constitutive active MMP9 bound to the tumor cell surface. J. Cell Sci. 117, 1847–1857 [DOI] [PubMed] [Google Scholar]
- 21. Vandooren J., Geurts N., Martens E., Van den Steen P. E., Jonghe S. D., Herdewijn P., and Opdenakker G. (2011) Gelatin degradation assay reveals MMP-9 inhibitors and function of O-glycosylated domain. World J. Biol. Chem. 2, 14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bickett D. M., Green M. D., Berman J., Dezube M., Howe A. S., Brown P. J., Roth J. T., and McGeehan G. M. (1993) A high throughput fluorogenic substrate for interstitial collagenase (MMP-1) and gelatinase (MMP-9). Anal. Biochem. 212, 58–64 [DOI] [PubMed] [Google Scholar]
- 23. Ugarte-Berzal E., Bailón E., Amigo-Jiménez I., Albar J. P., García-Marco J. A., and García-Pardo A. (2014) A novel CD44-binding peptide from the pro-matrix metalloproteinase-9 hemopexin domain impairs adhesion and migration of chronic lymphocytic leukemia (CLL) cells. J. Biol. Chem. 289, 15340–15349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Starckx S., Van den Steen P. E., Verbeek R., van Noort J. M., and Opdenakker G. (2003) A novel rationale for inhibition of gelatinase B in multiple sclerosis: MMP-9 destroys αB-crystallin and generates a promiscuous T cell epitope. J. Neuroimmunol. 141, 47–57 [DOI] [PubMed] [Google Scholar]
- 25. Hu J., Van den Steen P. E., Sang Q. X., and Opdenakker G. (2007) Matrix metalloproteinase inhibitors as therapy for inflammatory and vascular diseases. Nat. Rev. Drug. Discov. 6, 480–498 [DOI] [PubMed] [Google Scholar]
- 26. Van Wart H. E., and Birkedal-Hansen H. (1990) The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. U.S.A. 87, 5578–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia-Pardo A., and Opdenakker G. (2015) Nonproteolytic functions of matrix metalloproteinases in pathology and insights for the development of novel therapeutic inhibitors. Metalloproteinases Med. 2, 19–28 [Google Scholar]
- 28. Redondo-Muñoz J., Ugarte-Berzal E., Terol M. J., Van den Steen P. E., Hernández del Cerro M., Roderfeld M., Roeb E., Opdenakker G., García-Marco J. A., and García-Pardo A. (2010) Matrix metalloproteinase-9 promotes chronic lymphocytic leukemia b cell survival through its hemopexin domain. Cancer Cell 17, 160–172 [DOI] [PubMed] [Google Scholar]
- 29. Marchant D. J., Bellac C. L., Moraes T. J., Wadsworth S. J., Dufour A., Butler G. S., Bilawchuk L. M., Hendry R. G., Robertson A. G., Cheung C. T., Ng J., Ang L., Luo Z., Heilbron K., Norris M. J., Duan W., Bucyk T., Karpov A., Devel L., Georgiadis D., Hegele R. G., Luo H., Granville D. J., Dive V., McManus B. M., and Overall C. M. (2014) A new transcriptional role for matrix metalloproteinase-12 in antiviral immunity. Nat. Med. 20, 493–502 [DOI] [PubMed] [Google Scholar]
- 30. Eguchi T., Kubota S., Kawata K., Mukudai Y., Uehara J., Ohgawara T., Ibaragi S., Sasaki A., Kuboki T., and Takigawa M. (2008) Novel transcription-factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene. Mol. Cell Biol. 28, 2391–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sela-Passwell N., Rosenblum G., Shoham T., and Sagi I. (2010) Structural and functional bases for allosteric control of MMP activities: can it pave the path for selective inhibition? Biochim. Biophys. Acta 1803, 29–38 [DOI] [PubMed] [Google Scholar]
- 32. Zarrabi K., Dufour A., Li J., Kuscu C., Pulkoski-Gross A., Zhi J., Hu Y., Sampson N. S., Zucker S., and Cao J. (2011) Inhibition of matrix metalloproteinase 14 (MMP-14)-mediated cancer cell migration. J. Biol. Chem. 286, 33167–33177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Egeblad M., and Werb Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 34. Ethell I. M., and Ethell D. W. (2007) Matrix metalloproteinases in brain development and remodeling: synaptic functions and targets. J. Neurosci. Res. 85, 2813–2823 [DOI] [PubMed] [Google Scholar]
- 35. Hynes R. O. (2009) The extracellular matrix: not just pretty fibrils. Science 326, 1216–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]