Abstract
Tibetan sheep have lived on the Tibetan Plateau for thousands of years; however, the process and consequences of adaptation to this extreme environment have not been elucidated for important livestock such as sheep. Here, seven sheep breeds, representing both highland and lowland breeds from different areas of China, were genotyped for a genome-wide collection of single-nucleotide polymorphisms (SNPs). The FST and XP-EHH approaches were used to identify regions harbouring local positive selection between these highland and lowland breeds, and 236 genes were identified. We detected selection events spanning genes involved in angiogenesis, energy production and erythropoiesis. In particular, several candidate genes were associated with high-altitude hypoxia, including EPAS1, CRYAA, LONP1, NF1, DPP4, SOD1, PPARG and SOCS2. EPAS1 plays a crucial role in hypoxia adaption; therefore, we investigated the exon sequences of EPAS1 and identified 12 mutations. Analysis of the relationship between blood-related phenotypes and EPAS1 genotypes in additional highland sheep revealed that a homozygous mutation at a relatively conserved site in the EPAS1 3′ untranslated region was associated with increased mean corpuscular haemoglobin concentration and mean corpuscular volume. Taken together, our results provide evidence of the genetic diversity of highland sheep and indicate potential high-altitude hypoxia adaptation mechanisms, including the role of EPAS1 in adaptation.
The Tibetan Plateau represents 25% of the landmass of China. It is the largest high-altitude area on earth, with an average altitude exceeding 4,500 m1. Such a high-altitude environment has produced many unique highland species. Compared with neighbouring and lowland populations, the native people and animals have adapted to this habitat, which has a 43% lower partial pressure of oxygen2 and 40% higher ultraviolet radiation3. Several animals, such as Tibetan antelopes4 and Tibetan Mastiffs5,6, have recently been shown to exhibit adaptation to living in this challenging environment. China has many sheep breeds, such as Tibetan and Mongolian sheep, that are widely distributed from the highlands to lowlands, many of which have evolved over centuries or even millennia7,8. Therefore, they also represent ideal organisms to study plateau adaptability.
To date, the genome sequences of Tibetan antelopes4, Tibetan wild boars9, yaks10 and ground tits11 have been generated, and several important pathways and functional categories have been identified, including energy metabolism and oxygen transmission, response to hypoxia, DNA repair and ATPase production. In addition, the mechanisms underlying plateau adaptability have been explored using population surveys of SNP data, successfully identifying candidate genes for genetic adaptation to the Tibetan Plateau. Recent studies have demonstrated that the positively selected haplotypes of EGLN1 and PPARA were significantly associated with the low haemoglobin content of Tibetan people, which is a unique characteristic of this extreme high-altitude population12. One single-nucleotide polymorphism (SNP) in EPAS1, encoding a transcription factor involved in response to hypoxia, was identified; for this SNP, there was a 78% frequency difference between the Tibetan and Han populations, representing the largest allele frequency difference observed for any human gene to date13. Recently, variations in EPAS1 were identified as high-altitude adaptations in the Tibetan Mastiff 5,6. Additionally, other hypoxia-related genes were identified in yaks (ADAM17, ARG2 and MMP3), Tibetan antelopes (ADORA2A, CCL2, ENG, PIK3C2A, PKLR, ATP12A and NOS3) and Tibetan wild boars (ALB, ECE1, GNG2 and PIK3C2G)4,9,10. In addition, Qu found 11 candidate positively selected genes (HIFAN, MTOR, SRF, TXNRD2, WNT7B, ANGP4, ADAM9, PSMD2, LRRC7, MDH1B and LRRK1) associated with a hypoxia response in ground tits11. Almost all of the identified genes mentioned above belong to the list of 247 hypoxia genes that are priority candidates for adaptation to high-altitude hypoxia12; however, each species may have different candidate genes. This phenomenon might indicate differences between species in the specific genes selected, even in the same environment. Thus, Tibetan sheep might differ from other species with respect to the molecular mechanisms of high-altitude adaptation. However, there has been no research on the adaptation of Tibetan sheep to the high-altitude plateau.
Data from several amphibians and mammals have indicated that modifications of haemoglobin (Hb) function often play a key role in regulating an adaptive response to high-altitude hypoxia14,15,16. For example, the haemoglobin concentration of Andean individuals who are native to different elevations is higher than those of Tibetan and sea-level residents17,18. In addition, improved oxygen (O2) loading and unloading are physiological marks of high-altitude adaptation19. The O2-loading capacity is determined by the Hb-O2 affinity. An increased Hb-O2 affinity helps maintain pulmonary O2 loading at a sufficient to maximal level of tissue oxygenation under conditions of extreme hypoxia20. Many highland animals have haemoglobin with high O2 affinity, such as alpacas21, Andean geese22, bar-headed geese23,24 and deer mice25,26.
Although many studies have focused on highland humans and animals, the genetic mechanisms underlying the adaptation of domesticated animals to the Tibetan Plateau have been rarely studied. In this study, a genome-wide investigation of the adaptation of Tibetan sheep to the high-altitude plateau was conducted using the Illumina Ovine SNP50K Bead Chip assay. We performed whole-genome SNP sweeps to study the adaptive evolution of high-altitude sheep by analyzing seven breeds. We identified candidate genes using selective sweep mapping and revealed potential genetic mechanisms of high-altitude adaptation in sheep compared with other species. We also more deeply analyzed gene evolution by investigating genes and networks as well as the genetic diversity of EPAS1.
Results
Population structure
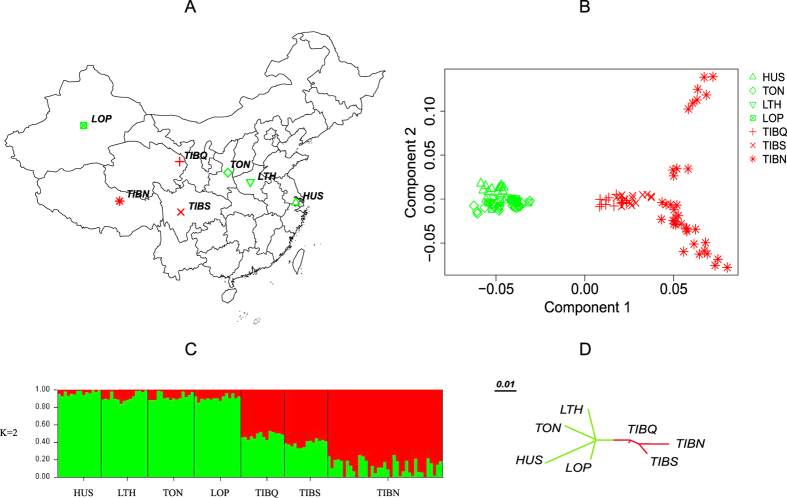
To assess the relationships between the animals and breeds under investigation, we applied multidimensional scaling (MDS) to analyze 122 individuals genotyped at 20,632 autosomal SNPs. The individuals were representative of seven native Chinese sheep breeds: Hu sheep (HUS), Tong sheep (TON), Large-tailed Han sheep (LTH), Lop sheep (LOP), Tibetan sheep of the Qinghai (TIBQ), Sichuan (TIBS) and Nagqu (TIBN). (Figure 1A, Supplemental Table S1). The first dimension (Component 1) separated domestic individuals into two broad non-overlapping clusters (Fig. 1B). The clusters corresponded well with the geographic origin of each breed within the cluster. Our cluster results showed that the TIBQ, TIBS and TIBN breeds were grouped together as the highland group (G1) and that the HUS, TON, LTH and LOP breeds belonged to the lowland group (G2). To explore further the relationships obtained using the MDS approach, we performed a population structure analysis using the program STRUCTURE 2.3.427 to obtain a model-based unsupervised hierarchical clustering of the individuals arising from a user-defined number of ancestral populations (K). The number of populations was varied from K = 2 to 4, and the largest change in the log of the likelihood function (ΔK) was found when K = 2 (Fig. S1), which suggests that there is likely a small number of ancestral groups. Additionally, the individuals from low-altitude breeds appeared distinct from those sampled from high-altitude breeds (Fig. 1C, Fig. S2), which was consistent with the MDS results. To further confirm the phylogenetic relationships among the sheep breeds, a neighbour-joining tree28 based on the pairwise genetic distances was constructed. The tree also indicated that high-altitude (TIBQ, TIBS and TIBN) and low-altitude breeds (TON, HUS, LOP and TLH) were split into two distinct branches (Fig. 1D).
Figure 1. Description of the location and genetic relationships of samples.
(A) Sampling and habitats of the seven Tibetan sheep breeds. The map was created using the R package ‘maptools’, version: 0.8–37, URL: http://r-forge.r-project.org/projects/maptools/. (B) Plots for the first (Component 1) and second (Component 2) dimensions revealed the clustering of all individuals. (C) Genome-wide admixtures inferred by STRUCTURE 2.3.4. The results from K = 2 are shown; (D) Neighbour-joining (NJ) phylogenetic tree for the seven breeds based on pairwise FST. Each population is represented by a different symbol and colour label: high-altitude breeds are indicated in red, and low-altitude breeds are indicated in blue. The abbreviations for the seven breeds are shown in Supplemental Table S1.
Selection signal is correlated with high altitude
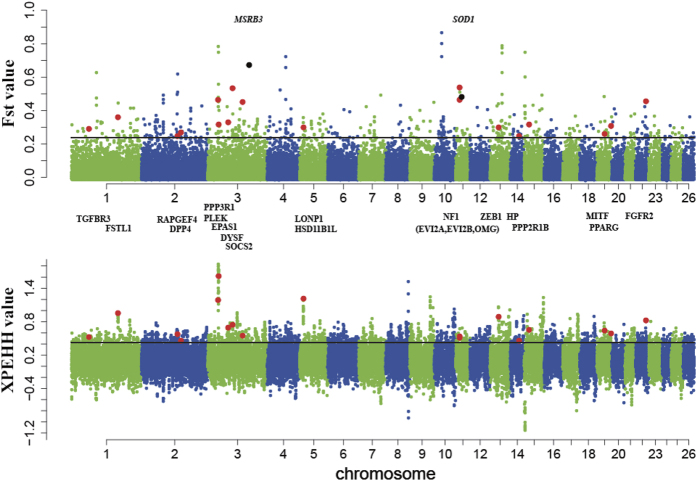
FST, which can reveal a high coefficient of genetic differentiation, has been widely used to identify selective signals among whole-genome SNPs29. We evaluated the population differentiation and calculated the FST for each SNP between G1 and G2, ultimately identifying 464 SNPs with extremely high values (FST > 0.242, top 1%) from the total tested (46,355 SNPs, Supplemental Table S2). Moreover, we used FST outlier tests by LOSITAN30 to identify loci with levels of differentiation between the G1 and G2 pairs that were higher or lower than expected under a neutral model, and approximately 96% of the outlier loci were the same as the top 1% FST value SNPs obtained using the Genepop software31 (Supplemental Table S2). Moreover, 40,652 pairwise XP-EHH (Cross Population Extend Haplotype Homozygosity Test) values, which are haplotype-based parameters, were calculated for each SNP32. In total, 2000 unique SNPs were within the top 5% of XP-EHH values (XP-EHH > 0.415) (Supplemental Table S3). The SNPs that were identified by high FST and XP-EHH values produced a subset of 171 SNPs, which represented 152 candidate genes with strong signatures of selection (Supplemental Table S4). In addition, 70 SNPs with high FST but null XP-EHH values were included, representing 84 candidate genes (Supplemental Table S5). Finally, genome-wide polymorphisms revealed chromosomal regions that contained 236 genes with evidence of positive selection.
We performed Gene Ontology (GO) enrichment analyses on the highlighted candidate genes in Tibetan sheep populations (Table 1). These genes are involved in response to hypoxia (GO:0001666) and several other biological processes, such as cell morphogenesis involved in neuron differentiation (GO:0048667), axonogenesis (GO:0007409), tissue homeostasis (GO:0001894) and cell maturation (GO:0048469), all of which likely play roles in the local adaptation of Tibetan sheep. To further understand the functions of the candidate genes, we compared them with several genes that are considered likely to be associated with high-altitude adaptation, which led us to prioritize a set of 317 functional candidate genes (Supplemental Table S6). In our Tibetan sheep breeds, we found that the set of candidate loci contained EPAS1, NF1, LONP1, SOD1 and DPP4 as well as the homologous genes PPARG and TGFBR3 of the priority candidate genes.
Table 1. GO terms enriched with candidate genes for high-altitude adaptation in Tibetan sheep.
| ID | Term | P value | P Bonferroni value | Associated genes |
|---|---|---|---|---|
| GO:0048667 | cell morphogenesis involved in neuron differentiation | 6.60E-05 | 0.0017 | CHN1, CNP, EPHA8, FGFR2, NUMBL, OMG, SOD1, SPTBN4, TBR1 |
| GO:0022829 | wide pore channel activity | 7.23E-05 | 0.0017 | GJA3, GJB2, VDAC2 |
| GO:0007409 | axonogenesis | 1.26E-04 | 0.0029 | CHN1, CNP, EPHA8, FGFR2, NUMBL, OMG, SPTBN4, TBR1 |
| GO:0016877 | ligase activity, forming carbon-sulfur bonds | 5.51E-04 | 0.0121 | ACLY, ACSL6, SLC27A2 |
| GO:0005681 | spliceosomal complex | 6.95E-04 | 0.0146 | DHX38, HNRNPK, RALY, SMNDC1, TXNL4B, U2AF1 |
| GO:0001666 | response to hypoxia | 2.71E-02 | 0.0271 | EPAS1,CRYAA, LONP1 |
| GO:0001894 | tissue homeostasis | 2.71E-02 | 0.0271 | EPAS1, PTHLH, SOD1 |
| GO:0048469 | cell maturation | 2.71E-02 | 0.0271 | EPAS1, EPHA8, PPARG |
Hypoxia-inducible factors (HIFs) are transcription factors that respond to changes in the available oxygen in the cellular environment under high-altitude conditions. EPAS1, also known as HIF2a, is a member of the HIF family that responds to changes in available oxygen in the cellular environment under high-altitude conditions33. Three candidate genes, FGFR2, NF1 and RasGef4, function in the Ras/ERK signalling pathway, which commonly promotes angiogenesis with the HIF pathway under hypoxia34 and may have been selected for this function. Ras is indirectly activated by FGFR235, and RasGef436,37 activates Rap1, which regulates the proliferation and migration of human umbilical vein endothelial cells via the ERK and Akt pathways. NF1 is a negative regulator of the Ras signal transduction pathway38, and the loss of NF1 results in the activation of the Ras signalling pathway, leading to the aberrant growth of haematopoietic cells39. DYSF and ZEB1 are also target genes of HIFs. The genetic loss of dysferlin (encoded by DYSF) caused an abrogated angiogenic response to vascular endothelial growth factor40. A study found that ZEB1 is critical in the regulation of the macrovascular angiogenic response but not that of microvascular angiogenesis41. Moreover, under hypoxic conditions, HIF-1α up-regulates the expression of proteins that induce TWIST1 and ZEB142,43. However, in different animals, different genes affecting angiogenesis have been detected, such as DAM1710, NOS34 and PLXNA46 in yaks, Tibetan antelope and Tibetan Mastiffs, respectively.
HIFs were discovered because of their ability to stimulate transcription of the erythropoietin (EPO) gene during hypoxia at high altitudes43,44,45. Here, SOCS2 and LONP1 were also identified as candidate selected genes associated with the primary role of HIFs in promoting the hypoxic EPO response. SOCS2 regulates EPO-enhanced neuronal differentiation46 and, together with SOCS3, can regulate EPO signalling in response to hypoxia47. Lon peptidase 1 (LONP1) is a multifunctional ATP-dependent protease that mainly participates in mitochondrial proteolysis, is involved in heme biosynthesis48,49 and can regulate the EPO gene50. Under conditions of reduced O2 availability, HIF-1α reciprocally regulates COX4 subunit expression by activating the transcription of the gene encoding LONP151.
Candidate genes participating in energy metabolism have been identified regularly in animals living in high-altitude areas4,10,11,52. In Tibetan sheep, we identified a few genes associated with energy metabolism that were under positive selection. DPP4 is a key enzyme involved in glycolysis, where it degrades GLP-1, which is powered by the hydrolysis of ATP, and plays crucial roles in maintaining glucose stability53,54,55. PPARG, another selected gene, reactivates adipogenesis and transcriptionally activates LPP1 expression, indicating a potential role in the metabolism of phospholipids56,57. Substantial evidence has shown that mitochondrial function is altered in high-altitude adaptations58. A change is initiated by reduced oxygen delivery to cells during metabolism, and reactive oxygen species (ROS) are produced by hypoxic mitochondria, which stimulate the activation of HIF-1 and HIF-259,60. SOD1, the anti-oxidant superoxide dismutase 1, which activates ATP production61 and reduces mitochondrial ROS production62, was identified as being under positive selective pressure in Tibetan sheep.
Hypoxia causes an influx of Ca2+ and an increase in intracellular Ca2+ concentrations by opening store-operated Ca2+ (SOC) channels, which results in increased cell contraction63. In our study, RYR3 was found to encode the ryanodine receptor 3, a calcium channel, and showed selective signatures in a study in Tibetan Mastiffs6. The calcineurin genes (PPP3R1 and PPP2R1B), which encode protein phosphatases, play crucial roles in regulating Ca2+/calmodulin. Calcineurin can maintain vascular structure and function, and vascular smooth muscle cell proliferation. Calcineurin is a unique calcium/calmodulin-regulated protein phosphatase that functions as a key mediator of the hypertrophic response of the heart. PPP3R1 (protein phosphatase 3) and PPP2R1B (protein phosphatase 2) are regulatory subunits of calcineurin that are important for phosphatase activity64.
Tibetan sheep develop low partial oxygen pressure and are exposed to high levels of ultraviolet radiation. Another important candidate selected gene is microphthalmia-associated transcription factor (MITF). Hypoxia up-regulates cyclooxygenase-2, leading to the prostaglandin E(2)-mediated loss of MITF in cervical stromal cells65. In our study, MITF, which is associated with melanogenesis, was detected as positively selected in Tibetan sheep. MITF was also identified as being subject to high selective pressure in the world’s sheep breeds66.
HSD11B1L, HP, TGFBR3 and MSRB3 were also identified as candidate selected genes in Tibetan sheep. HSD11B1, which encodes an isoform of the enzyme 11beta-hydroxysteroid dehydrogenase, acts exclusively as an NAD-dependent dehydrogenase in activating cortisol to cortisone67, whose activity is down-regulated under high-altitude hypoxia conditions68. In Tibetan sheep, another isoform product of HSD11B1L, which is believed to act predominantly as an oxo-reductase using NADP(H) as a cofactor to generate cortisone69, was identified as a candidate selected gene; this result indicates that 11beta-HSD1a is also involved in the adaptive response to high altitudes. TGFBR3 is associated with TGF-β, which is an HIF-responsive product that is suspected of playing a role in cancer70,71. TGFBR3 is involved in regulating cell growth and differentiation and in inflammatory responses72,73,74. Haptoglobin (Hp; encoded by HP) is a plasma glycoprotein, the main biological function of which is to bind free Hb and prevent the loss of iron and subsequent kidney damage following intravascular haemolysis75. The gene encoding methionine sulfoxide reductase B3 (MSRB3) has been identified as a gene associated with altitude in dogs5, although its association with ear types has also been previously reported76,77. In the world’s sheep breeds, MSRB3 on sheep chromosome 3 was noted to develop high selection pressure66.
Some particularly extreme values for either FST or XP-EHH were also noticed (Fig. 2). For FST, three genes that had undergone strongly selection were reported in previous studies66,78. PPP1CC (FST = 0.78, chromosome 13) is a positional candidate locus for skeletal muscle strength phenotypes79. RXFP2 (FST = 0.72, chromosome 10) is a candidate gene for sheep horns80. BMP2 (FST = 0.63, chromosome 13) is associated with body size traits81. Additionally, for XP-EHH, four genes with strong signatures of selection were detected in the highland group. WDR92 (XP-EHH = 1.83, chromosome 3) promotes apoptosis induced by tumour necrosis factor-α (TNF-α)82. PNO1 (XP-EHH = 1.62, chromosome 3) is vital to normal cell function83. PARK2 (XP-EHH = 1.30, chromosome 8) is expressed primarily in the nervous system and is part of the multi-protein E3 ubiquitin ligase complex84. OSR2 (XP-EHH = 1.25, chromosome 9) is a key intrinsic regulator of palatal growth and patterning85.
Figure 2. Genome-wide distribution of FST and XP-EHH values.
Red dots represent sites showing significant signals in both the FST and XP-EHH approaches; black dots represent sites showing significant signal in the FST approach only. The symbols for candidate genes for adaptation to high-altitude hypoxia in the map are shown in bold and italics.
EPAS1 mutations and physiological associations
To further study the functions of the core factor EPAS1 in high-altitude adaption, we measured six haematological parameters in a high-altitude breed Tibetan sheep breed (TIBQ) and a low-altitude sheep breed (LTH). There were no significant differences between males and females within the breeds (Supplemental Table S7). In the high-altitude breeds, the red blood cell (RBC) count, haemoglobin (HGB) concentration, haematocrit (HCT) concentration, mean corpuscular volume (MCV) and mean corpuscular haemoglobin (MCH), but not the mean corpuscular haemoglobin concentration (MCHC), were higher or significantly higher than those of the low-altitude breeds (Supplemental Table S7). This result was similar to that found in dogs5. EPAS1 polymorphisms in native Tibetan people are associated with lower haemoglobin concentrations13.
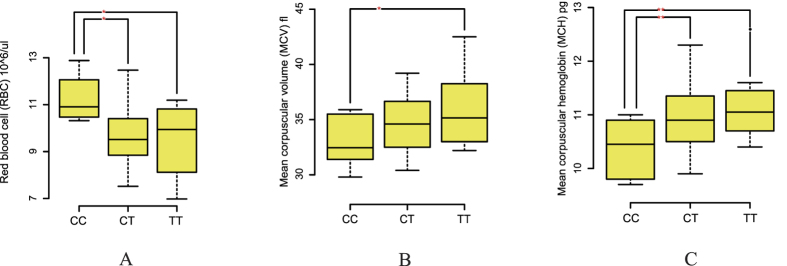
To understand the relationship between EPAS1 and haematological parameters, we designed 10 pairs of primers and detected 12 mutations in the exons of EPAS1 using Sanger sequencing. Ten mutations were in the coding region, including three non-synonymous mutations, six synonymous mutations, and an AGC insert; two SNPs were in the 3′ untranslated regions (Supplemental Table S8). Chi-squared tests showed that the frequencies of the 1st, 2nd, 5th and 11th mutations were significantly different (P < 0.05) between the populations (Supplemental Table S9). In the Tibetan breeds, alleles of these four SNPs were tested for association with haematological parameters, and only the 11th SNP showed a significant difference. The CC genotype was associated with a significantly higher RBC count and significantly lower MCV and MCH than the TT genotype (Fig. 3).
Figure 3.
Genotype–phenotype association with the (A) RBC, (B) MCV and (C) MCH parameters for the 11th SNP in EPAS1 in the Tibetan breeds. *Indicates a significant difference between the genotypes; **Indicates an extremely significant difference between the genotypes.
Discussion
Tibetan sheep are a hypoxia-tolerant species that live in an extremely inhospitable high-altitude environment, which has high ultraviolet radiation and a low partial pressure of oxygen compared with low-altitude areas86. In our study, we genotyped 122 sheep from seven breeds from high and low altitudes using a 50K SNP chip. SNP diversity was examined within the samples of native sheep that were the first to be domesticated by humans. This was the first study to characterize the genetic polymorphisms and evolution of Tibetan sheep.
The examination of a greater number of SNPs allowed STRUCTURE and MDS to robustly detect a distinct geographic pattern within the breeds genotyped. In our study, the seven Chinese indigenous breeds could be divided into two groups (G1 and G2) based on their genetic structure. G1 included Tibetan sheep (TIBQ, TIBS and TIBN) that live in the Qinghai Tibet plateau mountainous area, with an altitude over 3000 m, whereas Mongolian sheep (HUS, TON, LTH and LOP), which live at an altitude under 1000 m on the plain and basin, were clustered in G2 (Fig. 1A).
To detect the potential mechanisms of adaptation to high altitudes and ultraviolet radiation, we searched for SNPs that showed evidence of specific selection between the Tibetan sheep breeds that live at high altitude and the breeds that live at low altitude by using the FST and XP-EHH methods, which both have good statistical power for detecting selection signals among genome-wide SNP genotypes, although the sample sizes are limited87,88. Genome-wide polymorphisms revealed chromosomal regions that contained 236 genes with evidence of positive selection. Their GO annotation showed that the three genes examined (CRYAA, EPAS1 and LONP1) are involved the hypoxia response (GO:0001666, P Bonferroni < 0.05). Additionally, neuron development (GO:0048667, GO:0007409, P Bonferroni < 0.05) was affected by the highland environment. Moreover, we found 13 genes among the candidate genes (NF1, PPP3R1, HSD11B1L, ZEB1, SOCS2, DYSF, PPP2R1B, MITF, RAPGEF4, MSRB3, TGFBR3, HP and DPP4) that have other plausible biological functions associated with high-altitude adaptation (Fig. 2).
Several of the selected genes in Tibetan sheep have also been identified in other species. For example, EPAS1 was selected in both Tibetan humans12 and Tibetan Mastiffs5,6. MSRB3 was selected in Tibetan Mastiffs and modern sheep5,66. Moreover, MITF showed the strongest balancing selection signal among the world’s sheep breeds66. The major function of MITF, as the target of novel melanoma amplification89, is melanocyte differentiation, which might play a critical role in melanogenesis90. These results indicate that MITF might be associated with adaptation to high ultraviolet levels at high altitudes.
Other candidate genes demonstrating signals of selection did not overlap among different high-altitude species, although they were inferred to have similar functions. In this study, DPP453,54,55, PPARG58,59 and SOD161 were associated with energy metabolism. FGFR235, NF138 and RasGef437,38 were found in the Ras/ERK signalling pathway, which plays a role in hypoxia, along with the HIF pathway, by promoting angiogenesis. Additionally, DYSF and ZEB1 play important roles in angiogenesis. SOCS246,47 and LONP51 were also identified as candidate selected genes associated with the primary role of HIFs in promoting the EPO response under hypoxia. PPP3R1 and PPP2R1B64 play crucial roles in regulating Ca2+/calmodulin. HP75 is a plasma glycoprotein whose main biological function is to bind free haemoglobin and prevent the loss of iron. HSD11B1L67 is believed to predominantly act in vivo as an oxo-reductase using NADP(H) as a cofactor to generate cortisone.
Furthermore, we constructed a complex network of plausible pathways of positively selected genes for high-altitude adaptation in Tibetan sheep based on the functions of these genes, which are involved in hypoxia, energy metabolism, angiogenesis, Ca2+ metabolism, cortisone generation, erythropoietin and iron homeostasis under high-altitude conditions (Fig. 4). Network analysis demonstrated that most of the scanned candidate hypoxia-response genes are regulated by the HIF signalling pathway, thus indicating the vital role that EPAS1 plays in high-altitude adaptation in Tibetan sheep. This result is similar to that observed in dogs5,6. Processes mediated by these HIFs have been detected in both humans and Tibetan Mastiffs in response to high-altitude conditions and include iron homeostasis91, erythropoiesis44, vascular permeability92,93, glycolytic protein angiogenesis and metabolism94,95,96,97. This result indicates that different high-altitude animals have similar signatures of adaptive evolution in response to the environment.
Figure 4. Complexity of plausible pathways of positively selected genes in high-altitude adaptation in Tibetan sheep.
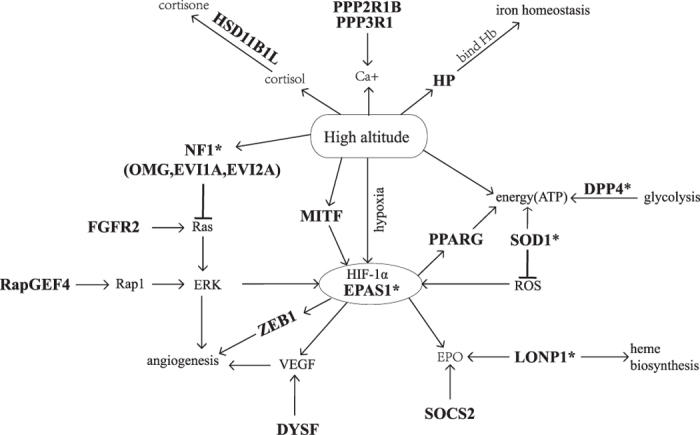
The symbols for candidate genes for adaptation to high-altitude in the map are shown in bold. Candidate loci marked with an asterisk (*) represent the hypoxia priority candidate genes. Candidate selected genes are associated with hypoxia, energy metabolism, angiogenesis, Ca2+ metabolism, generating cortisone, erythropoietin and iron homeostasis under high-altitude conditions.
Interestingly, EPAS1 was selected in Tibetans, Tibetan Mastiffs and Tibetan sheep. This indicates that parallel evolution at the molecular level may exist among these species. With the assistance of their human companions, dogs dispersed into new environments as human civilization expanded during the Paleolithic period98, a finding that is consistent with the time at which dogs were domesticated. Humans settled on highlands approximately 25,000 years ago99. The domestication of sheep occurred during the Neolithic period, approximately 9000 years ago100, in nearby regions in Iran and in a region of northern Iraq101.
EPAS1 plays an important role in high-altitude adaption in humans and dogs5,13. Tibetans and Tibetan Mastiffs display higher blood flow for oxygen delivery5,102, and this gene is related to the haemoglobin concentration in Andeans103 and Tibetans13,104,105,106. Therefore, we propose that similar mechanisms may have facilitated sheep adaptation to high-altitude hypoxia. In this study, we also found that EPAS1 had a high positive selection signal in Tibetan sheep breed. One limitation of this study is its relatively small sample size; consequently, the conclusions cannot be extrapolated to all sheep populations. In this study, we examined 12 mutations in the exons of EPAS1 in detail. However, only four SNPs with different allele frequencies between high- and low-altitude breeds were identified. Mutations in the EPAS1 coding region and phenotypes are not associated with haematologic parameters; however, in the 3′ untranslated regions, one SNP did show an association: the CC genotype was associated with a significantly higher RBC count and significantly lower MCV and MCH compared to the TT genotype in Tibetan sheep. MCV was found to differ among humans living at high altitudes and at sea level, with significantly higher values associated with humans living at higher altitudes107. These results indicate that increased MCV and MCH are associated with the enhanced ability to carry oxygen under conditions of high-altitude hypoxia. Consistent with this observation, reports of EPAS1 mutations in human patients have been exclusively associated with haematological phenotypes, especially alterations in MCH104. Our results indicate that the allele targeted by selection likely confers a functionally relevant adaptation to the hypoxic environment at high altitudes, although the precise physiological mechanism remains to be discovered.
Methods
Ethics Statement
All animals were handled according to the Guidelines for the Biological Studies Animal Care and Use Committee, People’s Republic of China. Animal experiments were approved by the Animal Ethics Committee of the Institute of Animal Sciences of Chinese Academy of Agricultural Sciences.
Genotyping and quality control
For the six Chinese sheep breed data set (Supplemental Table S1), blood samples were collected from 12 HUS, 15 TON, 15 LTH, 15 LOP, 14 TIBQ and 14 TIBS sheep. These animals were recently used in the registration and recording system of the National Center for Preservation and Utilization of Genetic Resources of Domestic Animals, National Animal Husbandry Service, Beijing, China. All experimental and surgical procedures were approved by the Biological Studies Animal Care and Use Committee, People’s Republic of China. In total, 87 DNA samples were extracted by the IQS DNA extraction method and were genotyped using the Illumina Ovine SNP 50K Bead Chip assay system at Capital Bio Corporation (Beijing, China). The second data set used in this study came from the Ovine HapMap project of the International Sheep Genomics Consortium and is publicly available at http://www.sheephapmap.org. The Chinese TIBN breed including 37 individuals was selected in the current study (Supplemental Table S1). We combined the two data sets and obtained 122 individuals and 49,034 SNPs. SNPs that could not pass the following three criteria were removed: (1) SNPs with a minor allele frequency >0.01; (2) maximum per-SNP missing rate < 0.05; and (3) SNPs on chromosome X. After quality control, there were 122 subjects and 46,355 SNPs in the analyzed dataset. Nucleotide diversity π values (Supplemental Table S1) were calculated for each breed using the Bio::PopGen::Statistics package in BioPerl (v1.6.1)108. Among the 7 breeds, LOP showed the highest diversity (π = 7.84 × 10−6), and TIBN had the lowest diversity (π = 7.32 × 10−6).
Population analysis
Before analysis, all the SNPs were pruned using the indep-pairwise option, with a non-overlapped window size of 25 SNPs, a step of five SNPs, and a pairwise r2 threshold of 0.05, resulting in 20,632 independent SNP markers. MDS was performed using PLINK 1.07109. Pairwise identity-by-state distances were calculated among all the individuals using the 20,632 independent SNP markers, and MDS components were obtained using the mds-plot option, based on the identity-by-state matrix. Population structure was evaluated using STRUCTURE 2.3.427. All animals were analyzed in triplicate for K = 2–4. All analyses were performed with a burn-in length of 20,000, followed by 30,000 MCMC (Markov Chain Monte Carlo) replications for each K-value. The solutions for each K were visualized using DISTRUCT 1.1110.
Priority candidates for adaptation to high-altitude hypoxia
Except for the strongest, clearest selective signals, it is difficult to confidently distinguish true signals from false positives using population genetic data alone111. Thus, we generated a set of priority functional candidate loci; 247 genes were obtained from Simonson et al.12, and we searched for 128 genes using his approach, from GO categories, which were associated with hypoxia using the Bos taurus background (http://amigo.geneontology.org/amigo). The resulting 317 functional candidate loci, including those from Homo sapiens and B. taurus, are listed in Supplemental Table S6.
Statistical analysis
Before statistical analysis, TIBQ, TIBS and TIBN were clustered into G1, whereas TON, HUS, LOP and TLH were clustered into G2. FST values per-SNP were calculated using Genepop 4.2.231. The formulae proposed by Weir and Cockerham29 were used to analyze a single locus. The FST value can theoretically range from 0 to 1. Next, the outlier tests were implemented in LOSITAN, which was run using 50,000 simulations, a ‘neutral’ mean FST, confidence intervals of 95% and a false discovery rate of 0.1, using the infinite alleles model30.
To determine if there was selection in G1, we computed the XP-EHH values using haplotype information in the xpehh program from http://hgdp.uchicago.edu/Software/. The XP-EHH derives from the idea of extended haplotype homozygosity (EHH), which is defined as the probability that two randomly chosen extended haplotypes carrying a given core haplotype are homozygous112. XP-EHH is used to test whether the site is homozygous in one population but polymorphic in another population through comparison of EHH scores for one core SNP between two populations112. A negative XP-EHH score indicates that selection occurred in the reference population, whereas a positive XP-EHH score indicates that selection occurred in the observed population.
Haplotypes were estimated with fastphase 1.4113. We used population label information to estimate the phased haplotype background and the following options for each chromosome: -Ku40 -Kl10 -Ki10. Candidate region annotations were obtained from Ovis 3.1 of the sheep genome from NCBI (ftp://ftp.ncbi.nih.gov/genomes/Ovis_aries/protein/). To perform functional enrichment of the candidate genes, which was required by the ClueGO plugin of Cytoscape 3.2.1114 using Symbol ID as input parameters, the background organism selected B. taurus. P values less than 0.05 after Bonferroni correction for multiple testing were considered statistically significant.
Physiological measurement and association analysis
To verify the accuracy of the experiments, we also sampled Tibetan and Mongolian sheep from different altitudes. Jugular venous blood samples from 30 one-year-old TIBQ sheep (16 males and 14 females) that lived at least 3000 m high in Gansu Province and 31 one-year-old LTH sheep (16 males and 15 females) that lived no higher than 100 m in altitude in Beijing were collected to measure the six haematological parameters: RBC, HGB, HCT, MCV, MCH, and MCHC using a BC-2800Vet Auto Hematology Analyzer (Mindray Co., Ltd, Shenzhen, China). For the association analysis, 10 primer pairs were designed (Supplemental Table S10), and 19 EPAS1 exons were amplified. Twelve mutations were detected and genotyped by traditional Sanger sequencing. We next used the general linear model with sex as a covariate in SPSS 20.0 to determine the association between haematological parameters and each genotype in the Tibetan breeds. The least square difference test was used for post-hoc analyses.
Additional Information
How to cite this article: Wei, C. et al. Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Sci. Rep. 6, 26770; doi: 10.1038/srep26770 (2016).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the International Sheep Genomics Consortium for permitting using Ovine HapMap genotype data. We are also grateful to Prof. Dongmin Liu (CAAS) for reviewing the manuscript. This research was supported by the National Key Technology R&D Program of China (2011BAD28B05-2) and Production and storage of livestock and poultry genetic material and measurement of genetic distance (Agricultural Finance Development (2013), No. 36).
Footnotes
Author Contributions C.W., H.W. and G.L. drafted the manuscript. Y.M., G.L., J.L., Z.L., R.L. and S.Z. performed the experiments and participated in phenotype data collection. H.W., F.Z., G.W. and J.K. carried out the population genetic studies. L.Z., M.W. and J.C. contributed reagents and materials. H.W. provided the map. C.W., L.D. and C.L. conceived the study, participated in its design and coordination and helped draft the manuscript. All of the authors read and approved the final manuscript.
References
- Thompson L. G. et al. A high-resolution millennial record of the south asian monsoon from himalayan ice cores. Science 289, 1916–1920 (2000). [DOI] [PubMed] [Google Scholar]
- Peacock A. J. Oxygen at high altitude. Brit Med J (Bmj) 317, 1063–1066 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumthaler M., Ambach W. & Ellinger R. Increase in solar UV radiation with altitude. J Photoch Photobio B 39, 130–134 (1997). [Google Scholar]
- Ge R. L. et al. Draft genome sequence of the Tibetan antelope. Nat Commun 4, 1858, doi: 10.1038/ncomms2860 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X. et al. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 24, 1308–1315, doi: 10.1101/gr.171876.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Population variation revealed high-altitude adaptation of tibetan mastiffs. Mol Biol. Evol. 31, 1200–1205, doi: 10.1093/molbev/msu070 (2014). [DOI] [PubMed] [Google Scholar]
- Linxin D. Animal Genetic Resources In China (China Agriculture Press, 2011). [Google Scholar]
- Cai D.-W., Han L., Zhang X.-L., Zhou H. & Zhu H. DNA analysis of archaeological sheep remains from China. J Archaeol Sci. 34, 1347–1355 (2007). [Google Scholar]
- Li M. et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet 45, 1431–1438, doi: 10.1038/ng.2811 (2013). [DOI] [PubMed] [Google Scholar]
- Qiu Q. et al. The yak genome and adaptation to life at high altitude. Nat Genet 44, 946–949, doi: 10.1038/ng.2343 (2012). [DOI] [PubMed] [Google Scholar]
- Qu Y. et al. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun 4, 2071, doi: 10.1038/ncomms3071 (2013). [DOI] [PubMed] [Google Scholar]
- Simonson T. S. et al. Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75, doi: 10.1126/science.1189406 (2010). [DOI] [PubMed] [Google Scholar]
- Yi X. et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78, doi: 10.1126/science.1190371 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. F. Hemoglobin function and physiological adaptation to hypoxia in high-altitude mammals. J Mammal 88, 24–31 (2007). [Google Scholar]
- Weber R. E. High-altitude adaptations in vertebrate hemoglobins. Resp Physiol Neurobi 158, 132–142, doi: 10.1016/j.resp.2007.05.001 (2007). [DOI] [PubMed] [Google Scholar]
- Weber R. E. & Fago A. Functional adaptation and its molecular basis in vertebrate hemoglobins, neuroglobins and cytoglobins. Resp Physiol Neurobi 144, 141–159, doi: 10.1016/j.resp.2004.04.018 (2004). [DOI] [PubMed] [Google Scholar]
- Beall C. M. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. P Natl Acad Sci. USA 104, Suppl 1, 8655–8660, doi: 10.1073/pnas.0701985104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall C. M. et al. An Ethiopian pattern of human adaptation to high-altitude hypoxia. P Natl Acad Sci. USA 99, 17215–17218, doi: 10.1073/pnas.252649199 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. F., Scott G. R. & Cheviron Z. A. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J Exp Biol. 213, 4125–4136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. F. & Moriyama H. Mechanisms of hemoglobin adaptation to high altitude hypoxia. High Alt Med Biol. 9, 148–157, doi: 10.1089/ham.2007.1079 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinini M., Kleinschmidt T., Jurgens K. D. & Braunitzer G. Primary structure and oxygen-binding properties of the hemoglobin from guanaco (Lama guanacoe, Tylopoda). Biol. Chem Hoppe Seyler 371, 641–648 (1990). [DOI] [PubMed] [Google Scholar]
- Jessen T. H., Weber R. E., Fermi G., Tame J. & Braunitzer G. Adaptation of bird hemoglobins to high altitudes: demonstration of molecular mechanism by protein engineering. P Natl Acad Sci. USA 88, 6519–6522 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Hua Z., Liang X., Xu Q. & Lu G. The crystal structure of bar-headed goose hemoglobin in deoxy form: the allosteric mechanism of a hemoglobin species with high oxygen affinity. J Mol Biol. 313, 123–137, doi: 10.1006/jmbi.2001.5028 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. The crystal structure of a high oxygen affinity species of haemoglobin (bar-headed goose haemoglobin in the oxy form). J Mol Biol. 255, 484–493, doi: 10.1006/jmbi.1996.0040 (1996). [DOI] [PubMed] [Google Scholar]
- Storz J. F. et al. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. P Natl Acad Sci. USA 106, 14450–14455 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. F. et al. The molecular basis of high-altitude adaptation in deer mice. Plos Genet 3, e45, doi: 10.1371/journal.pgen.0030045 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M. & Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno W. J., Socci N. D. & Halpern A. L. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol Biol. Evol. 17, 189–197 (2000). [DOI] [PubMed] [Google Scholar]
- Weir B. S. & Cockerham C. C. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370, doi: 10.2307/2408641 (1984). [DOI] [PubMed] [Google Scholar]
- Antao T., Lopes A., Lopes R. J., Beja-Pereira A. & Luikart G. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. Bmc Bioinformatics 9, 323, doi: 10.1186/1471-2105-9-323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8, 103–106, doi: 10.1111/j.1471-8286.2007.01931.x (2008). [DOI] [PubMed] [Google Scholar]
- Sabeti P. C. et al. Positive natural selection in the human lineage. Science 312, 1614–1620, doi: 10.1126/science.1124309 (2006). [DOI] [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T. & Maglott D. R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, D501–504, doi: 10.1093/nar/gki025 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet E. et al. ERK activation upon hypoxia: involvement in HIF-1 activation. Febs Lett 468, 53–58 (2000). [DOI] [PubMed] [Google Scholar]
- Wilkie A. O. Cancer drugs to treat birth defects. Nat Genet 39, 1057–1059, doi: 10.1038/ng0907-1057 (2007). [DOI] [PubMed] [Google Scholar]
- Coughlin J. J., Stang S. L., Dower N. A. & Stone J. C. RasGRP1 and RasGRP3 regulate B cell proliferation by facilitating B cell receptor-Ras signaling. J Immunol 175, 7179–7184 (2005). [DOI] [PubMed] [Google Scholar]
- Yamashita S. et al. CalDAG-GEFIII activation of Ras, R-ras, and Rap1. J Biol. Chem. 275, 25488–25493, doi: 10.1074/jbc.M003414200 (2000). [DOI] [PubMed] [Google Scholar]
- McCormick F. Ras signaling and NF1. Curr Opin Genet Dev 5, 51–55 (1995). [DOI] [PubMed] [Google Scholar]
- Bollag G. et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet 12, 144–148, doi: 10.1038/ng0296-144 (1996). [DOI] [PubMed] [Google Scholar]
- Sharma A. et al. A new role for the muscle repair protein dysferlin in endothelial cell adhesion and angiogenesis. Arterioscl Throm Vas 30, 2196–2204, doi: 10.1161/ATVBAHA.110.208108 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. et al. MicroRNA-200C and -150 play an important role in endothelial cell differentiation and vasculogenesis by targeting transcription repressor ZEB1. Stem Cells 31, 1749–1762, doi: 10.1002/stem.1448 (2013). [DOI] [PubMed] [Google Scholar]
- Martin A. & Cano A. Tumorigenesis: Twist1 links EMT to self-renewal. Nat Cell Biol. 12, 924–925, doi: 10.1038/ncb1010-924 (2010). [DOI] [PubMed] [Google Scholar]
- Semenza G. L., Nejfelt M. K., Chi S. M. & Antonarakis S. E. Hypoxia-inducible nuclear factors bind to an enhancer element located 3’ to the human erythropoietin gene. P Natl Acad Sci. USA 88, 5680–5684 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitsinou P. P. et al. Hepatic HIF-2 regulates erythropoietic responses to hypoxia in renal anemia. Blood 116, 3039–3048, doi: 10.1182/blood-2010-02-270322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. L. & Wang G. L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 12, 5447–5454 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Erythropoietin up-regulates SOCS2 in neuronal progenitor cells derived from SVZ of adult rat. Neuroreport 15, 1225–1229 (2004). [DOI] [PubMed] [Google Scholar]
- Linossi E. M., Babon J. J., Hilton D. J. & Nicholson S. E. Suppression of cytokine signaling: the SOCS perspective. Cytokine Growth F R 24, 241–248, doi: 10.1016/j.cytogfr.2013.03.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H. Molecular mechanism of heme biosynthesis. Tohoku J Exp. Med. 183, 83–99 (1997). [DOI] [PubMed] [Google Scholar]
- Tian Q. et al. Lon peptidase 1 (LONP1)-dependent breakdown of mitochondrial 5-aminolevulinic acid synthase protein by heme in human liver cells. J Biol. Chem. 286, 26424–26430, doi: 10.1074/jbc.M110.215772 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. A., Dunning S. P. & Bunn H. F. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science 242, 1412–1415 (1988). [DOI] [PubMed] [Google Scholar]
- Fukuda R. et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122, doi: 10.1016/j.cell.2007.01.047 (2007). [DOI] [PubMed] [Google Scholar]
- Cai Q. et al. Genome sequence of ground tit Pseudopodoces humilis and its adaptation to high altitude. Genome Biol. 14, R29, doi: 10.1186/gb-2013-14-3-r29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S. S. et al. Regulation of dipeptidyl peptidase 4 production in adipocytes by glucose. Diabetes Metab Syndr Obes 7, 185–194, doi: 10.2147/DMSO.S62610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman R. R., Paul S. K., Bethel M. A., Matthews D. R. & Neil H. A. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med. 359, 1577–1589, doi: 10.1056/NEJMoa0806470 (2008). [DOI] [PubMed] [Google Scholar]
- Zheng T. P. et al. Increased plasma DPP4 activities predict new-onset atherosclerosis in association with its proinflammatory effects in Chinese over a four year period: A prospective study. Atherosclerosis 235, 619–624, doi: 10.1016/j.atherosclerosis.2014.05.956 (2014). [DOI] [PubMed] [Google Scholar]
- Ren D., Collingwood T. N., Rebar E. J., Wolffe A. P. & Camp H. S. PPARgamma knockdown by engineered transcription factors: exogenous PPARgamma2 but not PPARgamma1 reactivates adipogenesis. Gene Dev 16, 27–32, doi: 10.1101/gad.953802 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz-Aydogan H. et al. Different effects of PPARA, PPARG and ApoE SNPs on serum lipids in patients with coronary heart disease based on the presence of diabetes. Gene 523, 20–26, doi: 10.1016/j.gene.2013.03.136 (2013). [DOI] [PubMed] [Google Scholar]
- Hoppeler H., Vogt M., Weibel E. R. & Fluck M. Response of skeletal muscle mitochondria to hypoxia. Exp. Physiol. 88, 109–119 (2003). [DOI] [PubMed] [Google Scholar]
- LaRochelle W. J. et al. PDGF-D, a new protease-activated growth factor. Nat Cell Biol. 3, 517–521, doi: 10.1038/35074593 (2001). [DOI] [PubMed] [Google Scholar]
- Scortegagna M. et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1−/− mice. Nat Genet 35, 331–340, doi: 10.1038/ng1266 (2003). [DOI] [PubMed] [Google Scholar]
- Coussee E. et al. G37R SOD1 mutant alters mitochondrial complex I activity, Ca(2+) uptake and ATP production. Cell Calcium 49, 217–225, doi: 10.1016/j.ceca.2011.02.004 (2011). [DOI] [PubMed] [Google Scholar]
- van Patot M. C. & Gassmann M. Hypoxia: adapting to high altitude by mutating EPAS-1, the gene encoding HIF-2alpha. High Alt Med. Biol. 12, 157–167, doi: 10.1089/ham.2010.1099 (2011). [DOI] [PubMed] [Google Scholar]
- Wang Y. X. & Zheng Y. M. ROS-dependent signaling mechanisms for hypoxic Ca(2+) responses in pulmonary artery myocytes. Antioxid Redox Sign 12, 611–623, doi: 10.1089/ars.2009.2877 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. et al. Identification of a novel 5-base pair deletion in calcineurin B (PPP3R1) promoter region and its association with left ventricular hypertrophy. Am Heart J 150, 845–851, doi: 10.1016/j.ahj.2004.12.004 (2005). [DOI] [PubMed] [Google Scholar]
- Hari Kishore A., Li X. H. & Word R. A. Hypoxia and PGE(2) regulate MiTF-CX during cervical ripening. Mol Endocrinol 26, 2031–2045, doi: 10.1210/me.2012-1100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijas J. W. et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. Plos Biol. 10, e1001258, doi: 10.1371/journal.pbio.1001258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson J. W. & Stewart P. M. Cortisol metabolism and the role of 11beta-hydroxysteroid dehydrogenase. Best Pract Res. Cl En 15, 61–78, doi: 10.1053/beem.2000.0119 (2001). [DOI] [PubMed] [Google Scholar]
- Heiniger C. D. et al. Hypoxia causes down-regulation of 11 beta-hydroxysteroid dehydrogenase type 2 by induction of Egr-1. Faseb J 17, 917–919, doi: 10.1096/fj.02-0582fje (2003). [DOI] [PubMed] [Google Scholar]
- Atanasov A. G., Nashev L. G., Schweizer R. A., Frick C. & Odermatt A. Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type 1 as an oxoreductase. Febs Lett 571, 129–133, doi: 10.1016/j.febslet.2004.06.065 (2004). [DOI] [PubMed] [Google Scholar]
- Kaelin W. G. The von Hippel-Lindau tumor suppressor protein: roles in cancer and oxygen sensing. Cold Spring Harb Sym 70, 159–166, doi: 10.1101/sqb.2005.70.001 (2005). [DOI] [PubMed] [Google Scholar]
- Kaelin W. G. Jr. & Ratcliffe P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30, 393–402, doi: 10.1016/j.molcel.2008.04.009 (2008). [DOI] [PubMed] [Google Scholar]
- Bonewald L. F. Regulation and regulatory activities of transforming growth factor beta. Crit Rev Eukar Gene 9, 33–44 (1999). [PubMed] [Google Scholar]
- Flanders K. C. & Wakefield L. M. Transforming growth factor-(beta)s and mammary gland involution; functional roles and implications for cancer progression. J Mammary Gland Biol. 14, 131–144, doi: 10.1007/s10911-009-9122-z (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letterio J. J. & Roberts A. B. Regulation of immune responses by TGF-beta. Annu Rev Immunol 16, 137–161, doi: 10.1146/annurev.immunol.16.1.137 (1998). [DOI] [PubMed] [Google Scholar]
- Beutler E., Gelbart T. & Lee P. Haptoglobin polymorphism and iron homeostasis. Clin Chem 48, 2232–2235 (2002). [PubMed] [Google Scholar]
- Boyko A. R. et al. A simple genetic architecture underlies morphological variation in dogs. Plos Biol. 8, e1000451, doi: 10.1371/journal.pbio.1000451 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaysse A. et al. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. Plos Genet 7, e1002316, doi: 10.1371/journal.pgen.1002316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. et al. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. Bmc Genomics 16, 194, doi: 10.1186/s12864-015-1384-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windelinckx A. et al. Identification and prioritization of NUAK1 and PPP1CC as positional candidate loci for skeletal muscle strength phenotypes. Physiol Genomics 43, 981–992, doi: 10.1152/physiolgenomics.00200.2010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. E. et al. Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol Ecol 20, 2555–2566, doi: 10.1111/j.1365-294X.2011.05076.x (2011). [DOI] [PubMed] [Google Scholar]
- Fan B. et al. Genome-wide association study identifies loci for body composition and structural soundness traits in pigs. Plos One 6, e14726 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki M. et al. Monad, a WD40 repeat protein, promotes apoptosis induced by TNF-alpha. Biochemical And Biophysical Research Communications 342, 568–572, doi: 10.1016/j.bbrc.2006.02.009 (2006). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Pno1 Tissue-Specific Expression and Its Functions Related to the Immune Responses and Proteasome Activities. Plos One 7, e46093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrina M. I. et al. Effective quantitative real-time polymerase chain reaction analysis of the parkin gene (PARK2) exon 1–12 dosage. Bmc Med Genet 8, 1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y. et al. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development 131, 3207–3216 (2004). [DOI] [PubMed] [Google Scholar]
- Petropoulos E. & Timiras P. Biological effects of high altitude as related to increased solar radiation, temperature fluctuations and reduced partial pressure of oxygen. Progress in biometeorology. Division A: Progress In Human Biometeorology 1, 295 (1974). [PubMed] [Google Scholar]
- Huff C. D., Harpending H. C. & Rogers A. R. Detecting positive selection from genome scans of linkage disequilibrium. Bmc Genomics 11, 8, doi: 10.1186/1471-2164-11-8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K. et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19, 826–837, doi: 10.1101/gr.087577.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway L. A. et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436, 117–122, doi: 10.1038/nature03664 (2005). [DOI] [PubMed] [Google Scholar]
- Takeda K. et al. Lipocalin-type prostaglandin D synthase as a melanocyte marker regulated by MITF. Biochem Bioph Res. Co 339, 1098–1106, doi: 10.1016/j.bbrc.2005.11.125 (2006). [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C., Nizet V. & Johnson R. S. Role of the hypoxia inducible factors HIF in iron metabolism. Cell Cycle 7, 28–32 (2008). [DOI] [PubMed] [Google Scholar]
- Chen L., Endler A. & Shibasaki F. Hypoxia and angiogenesis: regulation of hypoxia-inducible factors via novel binding factors. Exp Mol Med 41, 849–857, doi: 10.3858/emm.2009.41.12.103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. et al. Cobalt promotes angiogenesis via hypoxia-inducible factor and protects tubulointerstitium in the remnant kidney model. Lab Invest 85, 1292–1307, doi: 10.1038/labinvest.3700328 (2005). [DOI] [PubMed] [Google Scholar]
- Biswas S. et al. Effects of HIF-1alpha and HIF2alpha on Growth and Metabolism of Clear-Cell Renal Cell Carcinoma 786-0 Xenografts. J Oncol 2010, 757908, doi: 10.1155/2010/757908 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin E. B. et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol. 29, 4527–4538, doi: 10.1128/MCB.00200-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohet R. V. & Garcia J. A. Keeping the engine primed: HIF factors as key regulators of cardiac metabolism and angiogenesis during ischemia. J Mol Med (Berl) 85, 1309–1315, doi: 10.1007/s00109-007-0279-x (2007). [DOI] [PubMed] [Google Scholar]
- Tormos K. V. & Chandel N. S. Inter-connection between mitochondria and HIFs. J Cell Mol Med 14, 795–804, doi: 10.1111/j.1582-4934.2010.01031.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germonpré M. et al. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. J Archaeol Sci. 36, 473–490 (2009). [Google Scholar]
- Zhao M. et al. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. P Natl Acad Sci. USA 106, 21230–21235, doi: 10.1073/pnas.0907844106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Handley L. J. et al. Genetic structure of European sheep breeds. Heredity 99, 620–631, doi: 10.1038/sj.hdy.6801039 (2007). [DOI] [PubMed] [Google Scholar]
- Zeder M. A. Animal domestication in the Zagros: a review of past and current research. Paleorient 25, 11–25 (1999). [Google Scholar]
- Erzurum S. C. et al. Higher blood flow and circulating NO products offset high-altitude hypoxia among Tibetans. P Natl Acad Sci. USA 104, 17593–17598, doi: 10.1073/pnas.0707462104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A. W. et al. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum Genomics 4, 79–90 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall C. M. et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. P Natl Acad Sci. USA 107, 11459–11464, doi: 10.1073/pnas.1002443107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol. Evol. 28, 1075–1081, doi: 10.1093/molbev/msq290 (2011). [DOI] [PubMed] [Google Scholar]
- Xu S. et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol. Evol. 28, 1003–1011, doi: 10.1093/molbev/msq277 (2011). [DOI] [PubMed] [Google Scholar]
- Al-Sweedan S. A. & Alhaj M. The effect of low altitude on blood count parameters. Hematol Oncol Stem Cell Ther 5, 158–161, doi: 10.5144/1658-3876.2012.158 (2012). [DOI] [PubMed] [Google Scholar]
- Stajich J. E. et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12, 1611–1618 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, doi: 10.1086/519795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4, 137–138 (2004). [Google Scholar]
- Teshima K. M., Coop G. & Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Res. 16, 702–712, doi: 10.1101/gr.5105206 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C. et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P. & Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78, 629–644, doi: 10.1086/502802 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093, doi: 10.1093/bioinformatics/btp101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.