Abstract
Although well documented for mammalian G-protein-coupled receptors, alternate functionalities and associated alternate signalling remain to be unequivocally established for the Saccharomyces cerevisiae pheromone Ste2p receptor. Here, evidence supporting alternate functionalities for Ste2p is re-evaluated, extended and quantified. In particular, strong mating and constitutive signalling mutations, focusing on residues S254, P258 and S259 in TM6 of Ste2p, are stacked and investigated in terms of their effects on classical G-protein-mediated signal transduction associated with cell cycle arrest, and alternatively, their impact on downstream mating projection and zygote formation events. In relative dose response experiments, accounting for systemic and observational bias, mutational-derived functional differences were observed, validating the S254L-derived bias for downstream mating responses and highlighting complex relationships between TM6-mutation derived constitutive signalling and ligand-induced functionalities. Mechanistically, localization studies suggest that alterations to receptor trafficking may contribute to mutational bias, in addition to expected receptor conformational stabilization effects. Overall, these results extend previous observations and quantify the contributions of Ste2p variants to mediating cell cycle arrest versus downstream mating functionalities.
Keywords: alternate functionalities, G-protein-coupled receptor, pheromone mating, site-directed mutagenesis, yeast
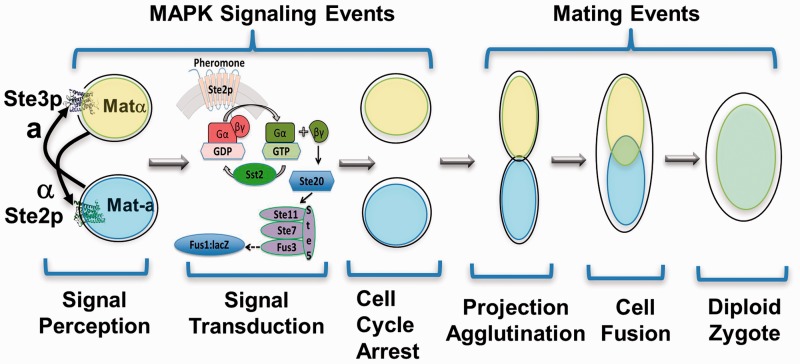
Saccharomyces cerevisiae pheromone G-protein-coupled receptors (GPCRs), Ste2p and Ste3p, mediate mating responses through binding of opposite mating type pheromones (1, 2). The Ste2p signal transduction pathway, which includes G-protein activation and stimulation of downstream mitogen-activated protein kinase (MAPK) cascades, culminates in cell cycle arrest. Subsequent to cell cycle arrest, the yeast undergoes mating, which includes projection formation and agglutination, followed by cell fusion (plasmogamy and karyogamy) to form the diploid zygote (Fig. 1). The Ste2p signalling pathway has served as a model par excellence for GPCR signal transduction, where the Gβγ protein subunits, the Ste5p scaffold and the RGS protein (Sst2p; regulators of G-protein signalling) were all first identified in yeast, and subsequently found in human GPCR pathways (3, 4). It has also served as a simplified system for analysis of human GPCRs and G-proteins which can be expressed in yeast and linked to the mating-associated pathway, enabling mechanistic analyses and characterization of orphan receptors in an isolated system (2).
Fig. 1.
Overview of the events that comprise the Saccharomyces cerevisiae pheromone mating response.
However, recent reports have highlighted the existence of important alternate functionalities and associated alternate signalling pathways for many human GPCRs. These alternate functionalities and pathways are linked to many of the known side effects associated with drugs today, and must be addressed in future drug development. For example, distinct functionalities (vasodilation and cardiac performance) have been linked to a biased-signalling mechanism of action for the angiotensin receptor (5). G-protein signalling mediates the vasodilation, while strong cardiac performance has been linked to signalling through β-arrestin. Similar biased functionalities and signalling mechanisms have been elucidated for a variety of mammalian GPCRs, by application of alternate ligands or select mutations of the receptors, that influence the GPCR conformation such that it preferentially interacts with either β-arrestin or G-proteins (6, 7). These effects are referred to as mutational- and ligand-derived bias, respectively.
A review of the literature does highlight some evidence supporting alternate Ste2p functionalities. In particular, alternate roles for Ste2p in mating events that occur downstream of cell cycle arrest have been proposed. Reports supporting this have included demonstration of roles for pheromone gradients in projection formation (8, 9), high concentrations of pheromone in prezygote cell fusion (10, 11) and observed localization of the receptor to the mating projection (12). Furthermore, inhibition of mating projection formation and subsequent mating events in Ste2p null yeast which otherwise maintain strong G-protein-mediated cell cycle arrest functionalities (4, 13) have been reported. A number of potential mechanisms by which Ste2p might be mediating these downstream events have been proposed, such as the existence of distinct high and low affinity pheromone-binding sites (14), transactivation (15) or direct mediation of membrane fusion (16). However, now, the possibility of biased signalling for the yeast pheromone receptors cannot be ignored. This is particularly true in light of the recent demonstration of a direct interaction between α-arrestins and Ste2p in internalization events (17).
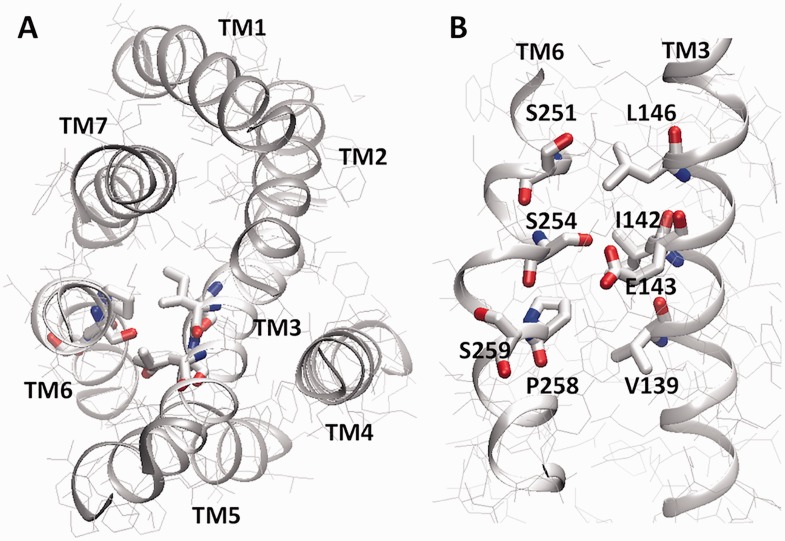
In addition to these genetic studies, a number of site-specific mutations in Ste2p have been shown to yield differential effects on G-protein-mediated MAPK signalling and diploid zygote formation. Some examples of these include mutations I24C and I29C in the N-terminus, yielding a phenotype that has strong G-protein-dependent MAPK signalling and cell cycle arrest activity, but no actual mating functionality (18). Similarly, mutation S251L, in the central region of transmembrane (TM) 6, also yielded normal signalling, but a weak-mating phenotype (19) (Fig. 2). In contrast, other TM6 mutations such as S254L produced normal signalling, but exceptionally strong mating, while the double mutation P258L/S259L yielded high levels of basal signalling or constitutive signalling activity (CSA), with normal inducible signalling, but weak mating (19, 20). Although these mutations highlight the possibility of alternate functionalities for Ste2p, their significance remains to be addressed comparatively to account for observational and systemic bias (6).
Fig. 2.
Model of Ste2p. Coordinates are from Eilers et al. (28). (A) Three-dimensional arrangement predicted for the Ste2p TM bundle, as viewed from the extracellular surface. The close proximity of TM6 and TM3 is highlighted. (B) Residues comprising the interactions between TM6 and TM3, including those targeted for mutagenesis in this report.
In this report, the possibility of alternate Ste2p-mediated functionalities is investigated by mutation-derived functional bias experiments. Results validate previously proposed alternate functionalities for Ste2p in post-cell cycle arrest mating events, while accounting for systemic and functional bias, and suggest that alterations in trafficking and localization may in part contribute to these effects.
Materials and Methods
Chemicals
All chemical were obtained from Sigma-Aldrich unless otherwise noted below. CSM-His-Ura, CSM-Leu-Ura and CSM-His-Leu-Ura were obtained from MP Biomedicals, USA. Bacto-Agar, Bacto-Tryptone and Bacto-Yeast Extract were from BD, Canada. CSM-Lys-Met was obtained from Sunrise Science products, USA. Mutagenic primers were obtained from Integrated DNA Technologies Inc. (USA).
Strains and plasmids
Escherichia coli strain DH5α and Top10 cells were used as hosts to propagate and amplify plasmids for different experiments. All yeast strains used in this study are described in Table I with their relevant genotype and source. For site-directed mutagenesis, STE2 was introduced into the BAMH1 site of pUC19 (New England Biolabs, USA), which was used as the template for all mutagenic reactions. The STE2 gene was mutated using the QuickChange site-directed mutagenesis (Agilent technologies, Canada) kit according to the manufacturer’s instructions. The wild type (WT) and mutated STE2 genes were first cloned into the pENTR/D-TOPO vector by blunt-end PCR amplification and cloning according to the manufacturer’s specifications (Life Technologies, USA). Primers included Ste2-Forward 5′-CACCATGTCTGATGCGGCTC TTC-3′ and Ste2-Reverse 5′-TAAATTATTATTATCTTCAGT CCAGAACTTTCTGGCTTCCTC-3′, which places a 5′-CACC-3′ sequence at the 5′ end of the forward primer. Target genes were subsequently recombined into the pDEST52 (Life Technologies, USA) or pAG426Gal-GFP (Addgene, Cambridge, MA) vectors according to Gateway specifications. Recombination in pAG426Gal-GFP yielded a Ste2p–GFP (Green Fluorescent Protein) fusion with the GFP added at the 3′end of the receptor. The WT and mutant STE2 containing expression plasmids were transformed into select receptor null yeast strains by the LiAc/PEG-mediated transformation method (21). Positive transformants were selected by growth on –Ura media.
Table I.
Description and source of yeast strains used in different experiments
| Strain | Genotype | Source |
|---|---|---|
| SCYO60 | MATα his3-11,15 leu2-3,112 trp 1-1 ura3-1 kan1-100 ade2-1 are1::Leu-2 | S. Sturley (Columbia University, NY) |
| SCYO61 | MATa his3-11,15 leu2-3,112 trp 1-1 ura3-1 kan1-100 ade2-1 are1::his-3 | S. Sturley (Columbia University, NY) |
| SY1793 | Mat α STE3 mfa1delta mfa2delta FUS1P::HIS3 ura3-52 leu2-3,112 ade1 | N. Davis (Wayne State University, MI) |
| JKY78 | MATa far1 bar1::hisG ste2::LEU2 lys2::FUS1-lacZ arg4 his3 leu2 lys2o trp1 ura3 | (19) |
| JKY79 | MATa bar1::hisG ste2::LEU2 ste5-3tslys2::FUS1-lacZ ade2-1o ade3 cry1 his4-580a leu2lys2o trp1a tyr1o ura3 SUP4-3ts | (19) |
| JKY-127-36-1 | MATa bar1::hisG far1 sst2-1 ste2D mfa1::LEU2mfa2::his51 ade2 his3 leu2 ura | (19) |
| LM102 | MATa bar1 his4 leu2 trp1 met1 ura3 FUS1-lacZ:URA3 ste2-dl | (27) |
FUS1-lacZ gene induction assay
Ste2p mutants were quantitatively assessed for MAPK signalling activity using the FUS1-lacZ gene induction assay as described previously (22). JKY78, JKY79 and JKY127-36-1 strains have the FUS1-lacZ reporter cassette integrated into their chromosomal DNA (19), while the WT SCY060 strain was made to express the FUS1-lacZ reporter gene from the uracil selectable vector pSB234 (23). Yeast strains were grown (5 ml cultures) overnight, in CSM (complete synthetic media) minimal media, lacking uracil, with 2% glucose. STE2 gene expression was induced the following morning by transfer of the cells into an equal volume of rich induction media [YPD (Yeast extract (1%) Peptone (2%) Dextrose (2%)] media with 2% galactose + 1% raffinose). After 4–5 hr growth in the rich induction media, yeast cultures were either treated with the indicated concentration of α-factor (Zymo Research), including a second dose 45 min later, or not treated. After 2 hr of incubation with or without pheromone, yeast cells were centrifuged and re-suspended in 300 μl Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCL, 1 m Mg2SO4) and the optical density (OD) measured at 600 nm. All samples were then frozen in liquid nitrogen and stored at −20°C. For analysis, samples were thawed, incubated at 30°C for 30 min and diluted with the addition of 700 μl Z-buffer containing β mercaptoethanol + 0.2% sarcosyl to permeabilize the samples. O-nitrophenyl-β-d-galactopyranoside (4 mg/ml) was added and the mixture was incubated at 30°C until a yellow colour developed (∼2–3 hr). A total of 400 μl of 1.5 M Na2CO3 was added to stop the reaction. The samples were then centrifuged (10 min at 18,000 × g) and the OD at 420 nm measured. The relative β-galactosidase activity was calculated assuming β-galactosidase units = OD420 × 1,000/t × v × OD600, where t is the time and v is the volume. All experiments were conducted in triplicate.
Quantitative mating efficiency assay
Strains were cultured overnight in minimal CSM media, lacking uracil or leucine for selection, but containing 2% glucose. These were transferred the following morning to rich induction media for 4–5 h. WT MATα SCY060 (-Leu) cells (2 × 107 cells) were mixed with pDEST52-Ste2p (WT and mutants) transformed MATa JKY78, JKY79 or JKY-127-36-1 (-Ura) cells (8 × 107 cells) to initiate mating, and then filtered using 0.22 μm filter paper. Equal quantities of each strain were independently filtered to serve as negative controls. The filter papers were placed on rich induction media plates and incubated for 5 hr at 30°C. Collected cells were washed and sonicated in a water bath for 30 sec in phosphate buffered saline. A total of 5 × 104 cells as well as 3- and 9-fold dilutions of these were plated on CSM minimal media plates lacking both uracil and leucine for 48 hr at 30°C. The number of single zygote colonies was then quantified and the mating efficiencies calculated according to the method described previously (24).
Cell cycle arrest and mating projection formation assays
Yeast strain LM102 was transformed with WT and mutant Ste2p expression constructs and was grown overnight on CSM minimal media, lacking leucine. The cultures were inoculated into rich induction media for 4–5 hr. Logarithmic phase cultures were then treated with different concentrations (ranging from 10−10 to 10−5 M) of α-factor for 4 and 7.5 hr. The cultures were sonicated in a water bath for 3 min and cells fixed in 3.7% formaldehyde solution (F-79; Fisher Scientific) for 1 hr at room temperature. Cells were washed thrice with 1× PBS (Phosphate Buffered Saline) solutions and stored at 4°C. The total number of cells was determined using a Z1 threshold Coulter counter (Coulter, FL, USA) according to the manufacturer’s instructions. Yeast cells treated with α-factor for 4 hr were evaluated for percent-unbudded cells (representative of cell cycle arrest) under a 60X LEICA DMR contrast phase microscope. Yeast cells treated with ligand for 7.5 hr were examined for the proportion of cells with projection formation (representative of mating projection formation). The yeast cultures were diluted 20 times and 10 μl of sample was applied under the microscope. Approximately 200 yeast cells were observed using C-chip disposable hemo-cytometer chambers (Incyto, Korea) for counting the unbudded cells and projections.
Localization of Ste2p by confocal microscopy
Yeast strains expressing GFP-tagged receptors were cultured overnight in minimal media to maintain the vector. Cells were washed with TE buffer (10 mM Tris, 1 mM ethylenediaminetetraacetic acid, pH 7.4) and sterile water and re-suspended in an equal volume of rich induction media. After 4–5 hr of growth, cells were treated (or not) with 1.5 μM α-factor ligand for different time periods. The cells were mounted on microscope slides with 1.5 μl of cell suspension mixed with 25 μl of hard mounting media and covered with a coverslip. Cells were fixed in 3.7% final concentration of formaldehyde solution (F-79; Fisher Scientific) for 1 h at room temperature before mounting in hard-set media. The slides were left undisturbed overnight at 4°C and examined by confocal microscopy (LEICA SP5, 63×, oil immersion lens), with excitation at 488 nm wavelength and emission monitored at 493–582 nm wavelength. For each mutant, at least three cells were imaged, looking at the top and bottom surfaces of each cell in focus to determine z-thickness. Subsequently, two-dimensional images (16 bit, 512 × 512, 24.6 × 24.6 micron, both brightfield and epifluorescent) were taken at the centre of the yeast cell in focus with the same zoom factor (10), power and gain settings (750) across all images. Fluorescence was assessed using ImageJ software (25). The peripheral region was defined as 0.5 micron from the yeast plasma membrane towards the inside of the cell.
Bias plot and statistics
Data for the bias plot was taken from experiments depicted in Fig. 5. The average functional responses (n = 3; signal transduction or mating projection formation) detected in the presence of 0 pM, 100 pM, 1 nM, 10 nM, 100 nM and 1 μM pheromone were normalized on a scale of 0–1 to obtain relative functional response values for each mutant. The obtained relative functional responses were then plotted against each other (relative signalling versus relative mating) for each mutant, at each concentration of pheromone tested. All statistical analyses through this report were performed using GraphPad Prism 6 (GraphPad, CA, USA) software. All data are reported as mean ± SEM. Treatment groups were compared using either t-test or analysis of variance after assumptions of normality and equal variance were met for all data analysed. In case of proportionate data, arc-sine transformation was done to normalize the data before applying statistical tests. Differences between multiple groups for different endpoints were tested using Fisher’s least significant difference and probabilities ≤0.05 were considered significant.
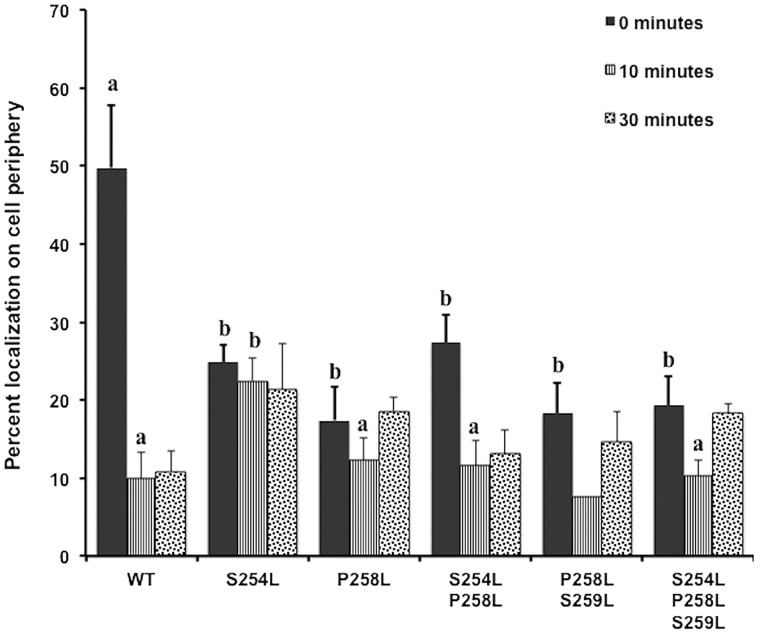
Fig. 5.
Localization of Ste2p WT and mutant receptors. The JKY78 strain was transformed with GFP-tagged WT or mutant and evaluated by confocal microscopy before and after addition of α-factor ligand. The amount of GFP signal in the peripheral cell membrane region (within 0.5 micron of the membrane) was assessed. The results represent the average 3–13 images for each sample. P < 0.001 was used as the cutoff value to define significant difference between groups. Values with common superscripts are not significantly different.
Results
Functionalities of Ste2p variants with TM6 substitutions
To investigate mutational-derived functional bias in Ste2p, basal (CSA) and ligand-induced MAPK signalling, as well as mating abilities of a selection of TM6 mutants, targeting residues Ser 254, Pro 258 and Ser 259 were initially characterized (Fig. 2). In addition to the previously reported S254L, P258L and double P258L/S259L mutants (19, 20), the double S254L/P258L mutant and the triple S254L/P258L/S259L mutants were also evaluated. This allowed consideration of the effect of incorporating a ‘strong mating’ mutation into weak-mating CSA mutants.
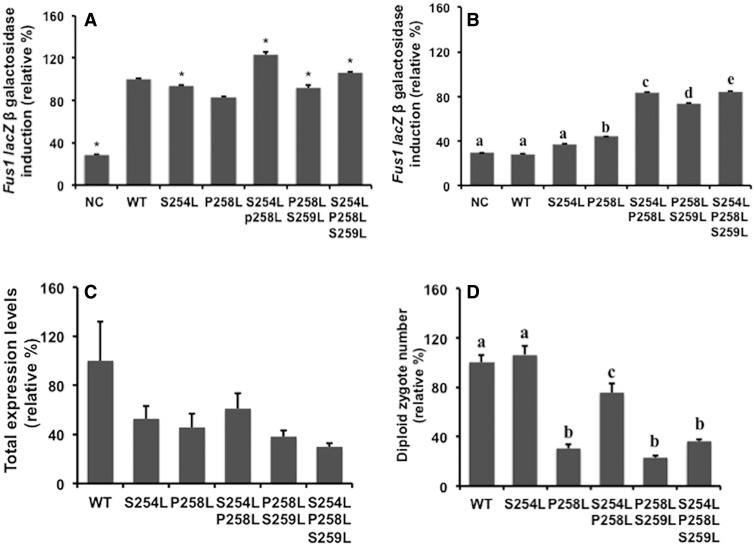
As determined by the FUS1-lacZ reporter gene induction assay carried out in MATa JKY78 cells, ligand-induced MAPK signalling was strong in all mutants, similar to that observed for WT (Fig. 3A). In contrast, basal signalling activity of the mutants varied dramatically (Fig. 3B). As expected, the WT receptor showed no basal activity above the empty vector control sample, while P258L and the double mutant P258L/S259L showed ∼15% and 50% increases in relative basal activity respectively, consistent with a previous report (20). Interestingly, although S254L yielded less than a 10% increase in basal signalling activity on its own, stacking this mutation with P258L in particular yielded a very significant increase of almost 40% relative basal activity above P258L alone. Incorporation of S254L into the double P258L/S259L mutant yielded basal activity similar to that of the double S254L/P258L mutant. Finally, the enhanced constitutive activity of the mutants was shown to not be a consequence of enhanced receptor surface expression, as the expression of mutants, determined by confocal microscopy, looking at C-terminal GFP-tagged receptors, was at least 50% decreased compared with WT GFP-tagged receptor (Fig. 3C). Western analysis (Supplementary Fig. S1) shows that all the mutants investigated are expressed at approximately comparable levels with the WT receptor. These results were validated in additional cell lines, JKY79 and JKY127-36-1 (Supplementary Figs S2 and S3, respectively). JKY79 does not include the far1 deletion and carries a temperature-sensitive mutation in a post-receptor component of the pheromone signalling pathway, ste5-3ts, allowing accumulation of receptor following galactose induction, while maintaining inhibition of the pheromone pathway, demonstrating that galactose and receptor alone are not yielding the basal MAPK signalling activity. JKY127-36-1 cells contain both sst2 and mfα1 deletions, controlling for autocrine signalling and demonstrating that the CSA is not a consequence of a defect in Sst2p-mediated adaptation.
Fig. 3.
Characterization of Ste2p mutants overexpressed in the JKY78 receptor null strain. (A) Ligand-stimulated Ste2p-induced MAPK signalling. Cultures expressing WT or mutant versions of STE2, or empty vector (NC), were treated with 1.5 µM α-factor and induction of the FUS1-lacZ reporter gene assayed. Comparisons between mutant values with those of WT were made using one-way analysis of variance test. *P < 0.001 was used as the cutoff value to define significant difference between mutant values compared with the WT. (B) Basal Ste2p-induced MAPK signalling. As for (A) but FUS1-lacZ reporter gene activity was measured in the absence of added α-factor ligand. P < 0.001 was set as cutoff value to indicate significant difference between groups. Values with no common superscript are different. (C) Ste2p expression levels. WT STE2 and mutant genes were tagged with GFP expressed in JKY78 cells and total cellular fluorescence measured by confocal microscopy. (D) Quantitative mating assay. WT MATα SCYO6O was mated with cultures expressing WT or mutant Ste2p. The number of diploid zygote nuclei was quantified. All results represent three independent assays, each done in triplicate. P < 0.001 was used as the cutoff value to define significant difference between groups. Values with no common superscript are different (P < 0.001).
Evaluation of the mating ability of these same MATa mutant Ste2p (-ura) expressing strains was subsequently carried out by assessing numbers of diploid zygotes formed following mating with the SCY060 WT MATα (-leu) strain, by selection on media lacking both uracil and leucine (Fig. 3D). WT and the previously characterized S254L, strong mating mutant (19), both showed strong mating as expected, with the double mutant S254L/P258L showed moderately reduced mating compared with these. P258L alone as well as the double mutant P258L/S259L and the triple mutant all showed significantly reduced mating, although some low level of mating was observed in all cases. Again, similar results were obtained using cell line JKY79 (Supplementary Fig. S2D). Interestingly, when tested for basal mating activity, which might be expected for mutants with high basal MAPK signalling, by mating against a MATα strain (SY1793) that is mfα1-mfα2 null (i.e. no pheromone ligand produced), no evidence of any basal mating was detected (data not shown). Interestingly, other weak mating mutants, I24C and S251L, were not rescued by stacking of the strong mating S254L mutation with these (Supplementary Fig. S4).
Dose response analyses demonstrate TM6 mutation-derived functional bias
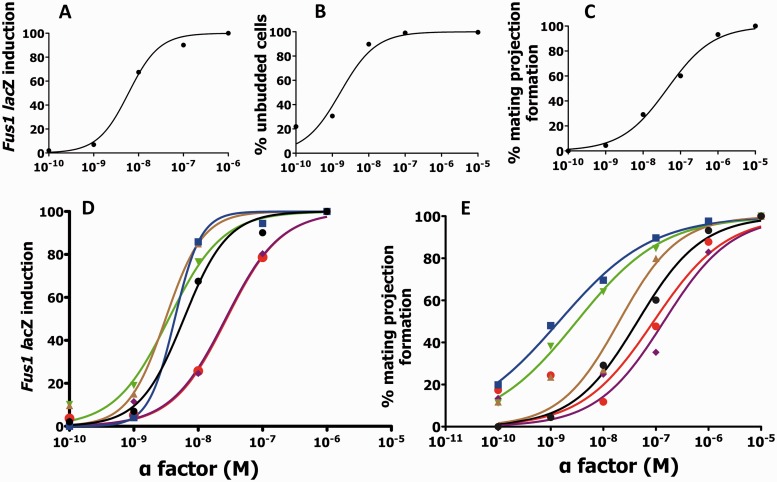
The putative alternate functionality of Ste2p in downstream mating events was tested according to current standards (6), by comparing relative α-factor dose responses for classical G-protein-mediated signal transduction to downstream alternate mating events. Initial dose response experiments carried out on Ste2p null yeast overexpressing WT Ste2p yielded EC50 values of 1.7 nM for elicitation of cell cycle arrest and 45 nM for elicitation of mating projection formation (Fig. 4B and C; Table II). Mating projection formation is the furthest downstream trait that can be detected in the presence of exogenous α-factor and absence of an actual mating partner, and thus was used instead of the direct quantitation of diploid zygotes as carried out in our preliminary analyses. These EC50 values, while not identical, are both in the same range and relative affinity as those reported previously for the same experiments (∼0.25 and ∼14 nM, respectively) (14). Differences between the original values and those reported here are likely attributable to variations in cell lines and other experimental conditions used (observational bias). Interestingly, based on this earlier work, it was proposed that Ste2p might play an alternate role in mating events, mediated by two distinct (high affinity versus lower affinity) pheromone binding sites (14). To further calibrate and investigate this possibility, the dose response for WT Ste2p in the FUS1-lacZ β-galactosidase reporter assay was also determined. With an EC50 of 6 nM (Fig. 4A; Table II), while again in the same range as that obtained for cell cycle arrest above, this value is somewhat higher. These results emphasize that direct comparison of WT Ste2p dose responses between assays cannot be used to evaluate receptor signalling or functional bias due to the interference of observational and systemic bias.
Fig. 4.
α-Factor-induced signal transduction and mating dose responses. Dose response curves for the yeast strain LM102 expressing WT Ste2p for FUS1-lacZ β galactosidase activity (A), cell cycle arrest (B) and mating projection formation (C) yielding EC50’s of 6, 1.7 and 45 nM, respectively. (D) Ligand-stimulated Ste2p-induced MAPK signalling dose responses for WT and mutant Ste2p’s. Yeast strains expressing WT or mutant Ste2p were incubated with the indicated concentration of α-factor and assayed for FUS1-lacZ induction. (E) Mating projection formation dose responses for WT and mutants Ste2p’s. Mating projection formation was assessed for the yeast strain LM102 expressing WT or mutant Ste2p following incubation with α-factor. Project formation was quantified microscopically. WT (filled circle) S254L (filled square) P258L (filled triangle), S254L/P258L (filled inverted triangle), P258L/S259L (filled rhombus), S254L/P258L/S259L (◯).
Table II.
Functional analysis of Ste2p WT and mutants
| Mutation | Constitutive activity (CA)a |
MAPK signalling activity: FUS1-lacZ induction |
Mating activity: mating projection formation |
Relative activity ratio | |||
|---|---|---|---|---|---|---|---|
| CA | Fold change | log EC50 ± SE (n = 3) | Fold EC50 | log EC50 ± SE (n = 3) | Fold EC50 | Mat:Sig | |
| WT | 31 ± 2 | 1 | −8.2 ± 0.3 | 1 | −7.3 ± 0.1 | 1 | 1 |
| S254L | 33 ± 1 | 1.1 | −8.4 ± 0.4 | 1.4 | −8.8 ± 0.2 | 32 | 23.2 |
| P258L | 47 ± 1 | 1.5 | −8.4 ± 0.5 | 1.6 | −7.6 ± 0.3 | 1.8 | 1.1 |
| S254L/P258L | 67 ± 0.3 | 2.2 | −8.4 ± 0.7 | 1.7 | −8.5 ± 0.2 | 14.5 | 8.5 |
| P258L/S259L | 62 ± 1 | 2.0 | −7.5 ± 0.8 | 0.2 | −6.8 ± 0.3 | 0.35 | 1.7 |
| S254L/P258L/S259L | 69 ± 2 | 2.2 | −7.5 ± 0.8 | 0.2 | −7.0 ± 0.2 | 0.5 | 2.1 |
aCA = total basal activity/Emax × 100.
However, precedent for alternate functionalities for GPCRs has now been set in the literature, and standards for demonstrating alternate functionality based on ligand and mutation-mediated bias have been reported (6, 7). Essentially, evaluation of dose responses of an array of ligands or receptor mutants relative to a single standard enables comparison of two assays by accounting for systemic and observational biases that might otherwise mislead comparative interpretations. Thus, dose response data and EC50 values for all the mutants described herein against both the FUS1-lacZ β-galactosidase reporter assay and the mating projection formation assays were obtained. The FUS1-lacZ β-galactosidase reporter assay was selected over cell cycle arrest as the representative assay for classical signal transduction, based on it being more reliable and more efficient for ease of reproducibility. The mutants showed MAPK signalling dose responses approximately comparable with WT (EC50’s in the range of 3.5–4.3 nM) with the exception of those containing the S259L mutations, which showed a 5-fold reduction in response (EC50’s of 27 and 28 nM, respectively; Fig. 4D; Table II). In contrast, mating projection responses were more variable, with the strong mating mutant, S254L, yielding the most potent response representing a 32-fold increase in EC50 (EC50 1.4 nM; Fig. 4E; Table II) consistent with the observed trend in diploid zygote formation (Fig. 3D). Overall, these results provide strong evidence in support of α-factor-dependent alternate functionalities for Ste2p in downstream mating functionalities.
TM6 substitutions modified ligand-stimulated receptor internalization
Although increased mating and constitutive activity of the mutants was not associated with increased receptor expression (Fig. 3C), it is well established that ligand binding and activation of the receptor stimulates receptor internalization (26, 27). Indeed, in other GPCR systems, internalization has been shown to be critical for mediation of alternate signalling through β-arrestin (7). Thus, the effect of TM6 mutations on ligand-stimulated Ste2p internalization was assessed by confocal microscopic evaluation of Ste2p null yeast expressing WT and mutant C-terminal GFP-tagged Ste2p (Fig. 5 and Supplementary Fig. S5). WT Ste2p demonstrated the expected internalization in as little as 10 min after stimulation with exogenous α-factor, with levels remaining low for at least 30 min. Levels of peripheral localized mutant receptors at time 0 were lower than WT levels, consistent with the overall lower expression levels. All mutations containing the P258L variation demonstrated similar initial responses to ligand stimulation as WT, with peripheral levels dropping to about 10% of the total. However, unlike WT, by 30 min most of these P258L containing mutants were beginning to see receptor content back in the periphery. In contrast, 10 min after application of ligand, the single S254L mutant showed only a very slight decrease in localization to the periphery, and these levels were maintained for at least 30 min.
Discussion
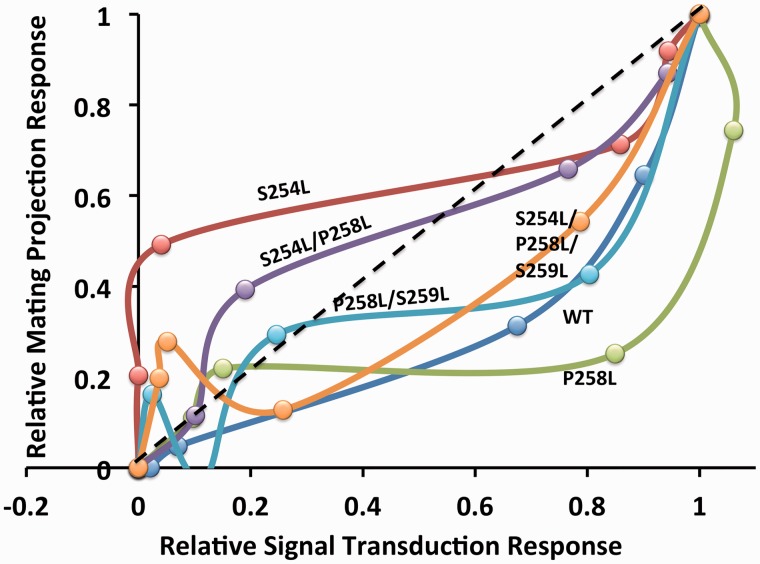
Towards validating reported alternate functionalities of Ste2p (10, 12, 14, 16, 18) according to current standards (6), a relative mutational-derived bias method was applied. This method accounts for systemic and observational bias that makes direct comparison of data between assays otherwise impossible. Indeed, examination of a bias plot of the relative responses of WT and the Ste2p variants in MAPK signalling versus mating projection formation assays highlights the presence of significant observational and systemic bias (Fig. 6). In particular, the degree of curvature of the WT data to the right of the diagonal is representative of the bias that exists between the assays used in this study. Although there are likely a number of sources of observational bias, including differences in cell lines and sensitivity issues in detection methodologies, the degree of curvature towards the signalling response suggests the presence of systemic bias potentially associated with proposed differences in ligand concentrations needed to stimulate MAPK signalling versus mating functionalities of Ste2p (10, 12). Indeed at 50% relative response for MAPK signal transduction, the WT receptor shows only half as much (∼25%) response in the mating assay. This confirms that data arising from each of the two assays for a given Ste2p variant cannot be directly compared in a meaningful manner. It is only when the data for a given variant are compared relative to some standard that real functional bias can be detected. Here, comparative data in the form of relative activity ratios (Table II) and a bias plot (Fig. 6) are reported, which provide un-equivocal quantitative evidence of functional bias in Ste2p variants.
Fig. 6.
Bias plot of relative functional responses of Ste2p WT and mutant receptors. Averaged projection formation dose response data for Ste2p WT and mutants was normalized and plotted as a function of the averaged, normalized data for the same receptor’s MAPK signal transduction dose response. Points comprising each curve represent relative functional responses detected at 0 pM, 100 pM, 1 nM, 10 nM, 100 nM and 1 μM pheromone. The dashed black line represents a theoretical state of non-bias, where responses in the two assays are equivalent. Curvature away from this diagonal represents bias towards one or the other response.
The dramatic shift in the S254L bias curve, relative to WT, above the diagonal towards the mating response axis provides a clear illustration of the S254L variant receptor’s functional bias for mating (Fig. 6). Mirroring the relative activity ratio of 23 (Table II), the full extent of the effect of this mutation is more evident here in the bias plot, where half maximal signalling yields 60% of the mating response. Further to this, the lack of effect of S254L alone on MAPK signal transduction relative to WT (EC50 values in Table II) emphasizes that Leu at residue 254 contributes to a mechanism or ligand-binding site that selectively mediates one function (mating), and not the others tested herein.
The extent of impairment to mating (bias for signalling) imposed by the P258L variation (with CSA; Fig. 3D; Table II) is illustrated by the overall shift in the P258L bias curve even more towards the signal transduction response compared with WT (Fig. 6). This is further emphasized by a similar shift in the S254L/P258L double mutant curve back towards the diagonal, relative to S254L alone. That the S254L/P258L curve only shifts as far as the diagonal, and not as far to the right as WT or P258L alone, further highlights the potency of the S254L bias for mating. Further comparison of the P258L data, however, suggests the possibility of differential effects on diploid zygote formation compared with mating projection formation, highlighting the possibility of two distinct, receptor-mediated downstream events in mating. Although both functionalities are downstream of the classical G-protein-mediated cell cycle arrest and might be expected to respond similarly to mutations, comparison of the data obtained here suggests otherwise. For mutation P258L, diploid zygote numbers show a significant decrease compared with WT (>60% decrease; Fig. 3D), while mating projection formation numbers at given concentrations of ligand are approximately comparable with WT (Fig. 4E). Whether this difference is simply related to observational bias (a distinct possibility not accounted for here), a role for some aspect of the mating partner cell (e.g. localization of a ligand concentration gradient) or represents mechanistic differences associated with roles for Ste2p early in mating projection and later in diploid zygote formation remains to be determined. Finally, it should be noted that slight increases in MAPK signalling dose response observed for these three mutants (S254L, P258L, and double S254L/P258L mutant; Table II) might account for the WT-like ligand-induced signalling activity of these mutants (Fig. 3A), despite reduced overall expression levels (Supplementary Fig. S1) and the relatively low level of peripheral localization of the mutants (Fig. 5).
Indeed, comparison of receptor trafficking and internalization for WT and the mutants highlight a number of interesting differences that may in part contribute to functional bias. P258L containing mutants were found to recover content back into the cell periphery significantly faster than WT (Fig. 5). Whether this increased rate of recovery is linked to higher CSA levels remains to be determined. As well, whether the constant levels of peripherally localized S254L are a product of an increased rate of trafficking/turnover or decreased rate of internalization remains to be determined. At the same time, whether the increased localization to the periphery following ligand stimulation contributes to the mating functionality bias of the S254L mutant also remains to be investigated.
For obvious reasons dose response analyses of CSA is not possible to evaluate, limiting what can be interpreted from the results presented here. Non-the-less an overview of potential mechanisms of action related to CSA and functional bias are presented below.
From a structure–function perspective, the P258L variation is proposed to stimulate CSA activity through release of the P258-induced kink in TM6 (Fig. 2B) (20, 28), where the straight helix stabilizes a conformation in the cytosolic domain similar to that achieved by ligand binding to WT. It is, in this context, interesting then to consider the loss of ligand-induced mating ability (according to diploid zygote formation levels) of P258L containing mutants with the stabilization/increase of basal signalling activity (Fig. 3B and D; Table II). Notably, beyond the data reported here, an S254F variation, while having a strong CSA effect, was also reported to have very weak ligand-induced mating, like P258L (19). Interestingly, although the double mutant S258L/S259L does show heightened CSA compared with P258L alone (Table II), consistent with previous reports (19, 20), both of the S259L-containing mutants have significantly reduced ligand dose responses for all functionalities, including signal transduction (Table II) as well as diploid zygote formation (Fig. 3D) and mating projection formation (Table II). The outcome of this general loss of ligand responsiveness is represented in the bias plot, with both S259L containing mutants yielding bias curves shifted back towards WT (Fig. 6). Overall, although a unique effect of these CSA mutations is to stabilize the cytosolic domain in an orientation that stimulates basal MAPK signalling, they also inhibit ligand-induced mating (and in the case of S259L ligand-induced MAPK signalling as well), suggesting that (i) the conformation of the cytosolic domain of the receptor is critical to mediating its alternate mating functionality and (ii) the MAPK signalling conformation is distinct (and in a sense competing) with the mating conformation. Furthermore, the S259L mutation highlights that potent CSA conformations can even compete with ligand-induced signalling conformations.
Not too surprisingly, stacking the strong mating mutation, S254L, with the CSA mutations (P258L) at least partially rescued the very weak mating phenotype of the CSA mutants. However, unexpectedly, stacking S254L also stimulated (doubled to almost WT-like ligand stimulated levels) the relative CSA activity induced by P258L, despite that fact that S254L on its own did not induce any significant change in CSA activity previously (19), or in the experiments reported here (Fig. 3B; Table II). The effect of S254L on CSA (in the presence of P258L) is similar to the effect of the alternate large space filling variation at S254F, as well as a variation at the adjacent residue Q253L, which showed 2.4- and 4.6-fold increases in basal activity respectively on their own (19). That strong basal signalling can co-exist with at least moderately strong ligand-induced mating suggests a more complex mechanism than non-redundant cytosolic domain conformations mediating alternate signalling. Looking again at the structural model (Fig. 2B) (28), clearly S254F introduces sufficient bulk to cause a steric clash with residues L146, I142 and E143 in TM3, likely leading to a significant shift in the orientation of TM6 and/or TM3 and like the P258L mutation stabilizing a cytosolic domain conformation similar to the WT ligand-induced conformation. Thus while the S254L mutation does not introduce sufficient bulk in itself to elicit a CSA effect, the structural impact of the P258L mutation may shift the 254 side chain closer to TM3 such that the added bulk in the S254L variation is in this instance sufficient to mediate additional conformational impact on the cytosolic domain conformation like S254F. That the effect of the two residues appear to be additive for CSA suggests that their effects may be mediated through different conformational effects, perhaps with P258L primarily modulating TM6 and the S254L mutation modulating the orientation of the neighbouring TM3.
But what of the mechanism of strong mating? On the one hand, it may be possible that S254L on its own introduces sufficient bulk to clash with residues L146, I142 and D143 in TM3, eliciting a small but significant conformational change in the cytosolic domain, stabilizing the mating conformation without impeding the MAPK signalling conformation. One might also speculate that the S254L mutation enables an inducible (more redundant) cytosolic conformation that can be induced to interact with alternate signalling partners in their presence. As well, although the possibility of S254L having an impact on the extracellular surface seems unlikely (no effect on extracellular I24C, Supplementary Fig. S4), it also cannot be strictly eliminated. And indeed the impact of S254L on the extracellular domain would fit with the possibility of there being two different α-factor ligand binding sites, a high-affinity site associated with MAPK signalling, and a lower affinity site associated with mating functionalities described here (14). Although also consistent with a previous report indicating a 5-fold increase for S254L α-factor affinity (19), modulation of the extracellular domain by S254L remains to be demonstrated.
Conclusion
Together these analyses of the effects of variations in the TM6 of Ste2p confirm mutational-derived bias for different functionalities of Ste2p modulating different events in yeast mating. Although the classical MAPK signal transduction pathway is very well defined, elucidation of mechanisms and signalling pathways associated with Ste2p functionalities in downstream mating events such as projection and diploid zygote formations remain enigmatic. Will yeast arrestins be linked to alternate Ste2p receptor functionalites, as is the case in mammalian systems? Although this remains to be tested, a recent report has highlighted roles for arrestin-like proteins in Ste2p desensitization (17), setting the stage for possible roles for arrestins in mediating Ste2p-linked signalling pathways downstream of classical MAPK signalling. Overall, possible continued relevance of the yeast pheromone-mating pathway as a simplified model for GPCR research, in the broader context of alternate functionalities for GPCRs, is supported based on validation of alternate functionalities for Ste2p in modulation of mating events.
Supplementary Data
Supplementary Data are available at JB Online.
Acknowledgements
The WT strains, SCYO6O and SCYO61 were kindly donated by Prof. S. Sturely (Columbia University, New York) and the T24D strain was from Dr Nicholas G. Davis (Wayne State University, Detroit). The LM102 cell line was kindly provided by Dr Jeffery Becker (University of Tennessee, Knoxville). Addgene yeast expression plasmid (Addgene#14203) was prepared in the laboratory of Susan Lindquist (Howard Hughes Medical Institute) and deposited in the non-profit plasmid repository.
Glossary
Abbreviations
- CSA
constitutive signalling activity
- CSM
complete synthetic media
- GPCR
G-protein-coupled receptor
- MAPK
mitogen-activated protein kinase
- OD
optical density
- Ste2p
Saccharomyces cerevisiae Mata pheromone receptor
- Ste3p
Saccharomyces cerevisiae Matα pheromone receptor
- TM
transmembrane
Funding
This work was funded by a Natural Science and Engineering Research Council - Discovery Grant (261683-06/2012) to M.C.L., and the National Research Council of Canada.
Conflict of Interest
None declared.
References
- 1.Xue C., Hsueh Y.-P., Heitman J. (2008) Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol. Rev. 32, 1010–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ladds G., Goddard A., Davey J. (2005) Functional analysis of heterologous GPCR signalling pathways in yeast. Trends Biotechnol. 23, 367–373 [DOI] [PubMed] [Google Scholar]
- 3.Dietzel C., Kurjan J. (1987) Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: a model for desensitization to pheromone. Mol. Cell. Biol. 7, 4169–4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whiteway M., Hougan L., Thomas D.Y. (1990) Overexpression of the STE4 gene leads to mating response in haploid Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Violin J.D., DeWire S.M., Yamashita D., Rominger D.H., Nguyen L., Schiller K., Whalen E.J., Gowen M., Lark M.W. (2010) Selectively engaging β-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J. Pharmacol. Exp. Ther. 335, 572–579 [DOI] [PubMed] [Google Scholar]
- 6.Kenakin T., Christopoulos A. (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat. Rev. Drug Discov. 12, 205–216 [DOI] [PubMed] [Google Scholar]
- 7.Reiter E., Ahn S., Shukla A.K., Lefkowitz R.J. (2012) Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu. Rev. Pharmacol. Toxicol. 52, 179–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segall J.E. (1993) Polarization of yeast cells in spatial gradients of alpha mating factor. Proc. Natl. Acad. Sci. U S A. 90, 8332–8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barkai N., Rose M.D., Wingreen N.S. (1998) Protease helps yeast find mating partners. Nature 396, 422–423 [DOI] [PubMed] [Google Scholar]
- 10.Brizzio V., Gammie A.E., Nijbroek G., Michaelis S., Rose M.D. (1996) Cell fusion during yeast mating requires high levels of a-factor mating pheromone. J. Cell. Biol. 135, 1727–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elia L., Marsh L. (1996) Role of the ABC transporter Ste6 in cell fusion during yeast conjugation. J. Cell. Biol. 135, 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson C.L., Konopka J.B., Hartwell L.H. (1991) S. cerevisiae alpha pheromone receptors activate a novel signal transduction pathway for mating partner discrimination. Cell 67, 389–402 [DOI] [PubMed] [Google Scholar]
- 13.Schrick K., Garvik B., Hartwell L.H. (1997) Mating in Saccharomyces cerevisiae: the role of the pheromone signal transduction pathway in the chemotropic response to pheromone. Genetics 147, 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore S.A. (1983) Comparison of dose-response curves for alpha factor-induced cell division arrest, agglutination, and projection formation of yeast cells. Implication for the mechanism of alpha factor action. J. Biol. Chem. 258, 13849–13856 [PubMed] [Google Scholar]
- 15.White J.M., Rose M.D. (2001) Yeast mating: Getting close to membrane merger. Curr. Biol. 11, R16–R20 [DOI] [PubMed] [Google Scholar]
- 16.Shi C., Kaminskyj S., Caldwell S., Loewen M.C. (2007) A role for a complex between activated G protein-coupled receptors in yeast cellular mating. Proc. Natl. Acad. Sci. U S A. 104, 5395–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvaro C.G., O’Donnell A.F., Prosser D.C., Augustine A.A., Goldman A., Brodsky J.L., Cyert M.S., Wendland B., Thorner J. (2014) Specific α-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein coupled receptor Ste2. Mol. Cell. Biol. 34, 2660–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi C., Kendall S.C., Grote E., Kaminskyj S., Loewen M.C. (2009) N-terminal residues of the yeast pheromone receptor, Ste2p, mediate mating events independently of G1-arrest signaling. J. Cell. Biochem. 107, 630–638 [DOI] [PubMed] [Google Scholar]
- 19.Dube P., Konopka J.B. (1998) Identification of a polar region in transmembrane domain 6 that regulates the function of the G protein-coupled alpha-factor receptor. Mol. Cell. Biol. 18, 7205–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konopka J.B., Margarit S.M., Dube P. (1996) Mutation of Pro-258 in transmembrane domain 6 constitutively activates the G protein-coupled alpha-factor receptor. Proc. Natl. Acad. Sci. U S A. 93, 6764–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gietz D., St Jean A., Woods R.A., Schiestl R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acid Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kippert F. (1995) A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol. Lett. 128, 201–206 [DOI] [PubMed] [Google Scholar]
- 23.Jin H., McCaffery J.M., Grote E. (2008) Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. J. Cell. Biol. 180, 813–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sprague G.F., Jr. (1991) Assay of yeast mating reaction. Methods Enzymol. 194, 77–93 [DOI] [PubMed] [Google Scholar]
- 25.Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hicke L., Zanolari B., Riezman H. (1998) Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J. Cell. Biol. 141, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim K.M., Lee Y.H., Akal-Strader A., Uddin M.S., Hauser M., Naider F., Becker J.M. (2012) Multiple regulatory roles of the carboxy terminus of Ste2p a yeast GPCR. Pharmacol. Res. 65, 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eilers M., Hornak V., Smith S.O., Konopka J.B. (2005) Comparison of class A and D G protein-coupled receptors: common features in structure and activation. Biochemistry 44, 8959–8975 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.