Abstract
Methylobacterium extorquens AM1 is an aerobic facultative methylotroph known to secrete pyrroloquinoline quinone (PQQ), a cofactor of a number of bacterial dehydrogenases, into the culture medium. To elucidate the molecular mechanism of PQQ biosynthesis, we are focusing on PqqE which is believed to be the enzyme catalysing the first reaction of the pathway. PqqE belongs to the radical S-adenosyl-l-methionine (SAM) superfamily, in which most, if not all, enzymes are very sensitive to dissolved oxygen and rapidly inactivated under aerobic conditions. We here report that PqqE from M. extorquens AM1 is markedly oxygen-tolerant; it was efficiently expressed in Escherichia coli cells grown aerobically and affinity-purified to near homogeneity. The purified and reconstituted PqqE contained multiple (likely three) iron-sulphur clusters and showed the reductive SAM cleavage activity that was ascribed to the consensus [4Fe-4S]2+ cluster bound at the N-terminus region. Mössbauer spectrometric analyses of the as-purified and reconstituted enzymes revealed the presence of [4Fe-4S]2+ and [2Fe-2S]2+ clusters as the major forms with the former being predominant in the reconstituted enzyme. PqqE from M.extorquens AM1 may serve as a convenient tool for studying the molecular mechanism of PQQ biosynthesis, avoiding the necessity of establishing strictly anaerobic conditions.
Keywords: Methylobacterium extorquens AM1, PQQ, PqqE, radical SAM enzyme
Pyrroloquinoline quinone (PQQ) is an aromatic, tricyclic o-quinone that serves as a cofactor for a number of prokaryotic dehydrogenases, e.g. alcohol or glucose dehydrogenase (1, 2). The biosynthesis of PQQ is achieved in a series of reactions catalysed by enzymes encoded by genes located on pqq operon(s). The PQQ biosynthetic genes from several bacteria such as Acinetobacter calcoaceticus (3), Methylobacterium organophilum DSM 760 (4), Klebsiella pneumoniae (5), Pseudomonas fluorescens CHA0 (6) [reclassified as P.protegens CHA0 (7)], Methylobacterium extorquens AM1 (8), Enterobacter intermedium 60-2G (9) [reclassified as Kluyvera intermedia (10)] and Gluconobacter oxydans 621H (11) were reported and more than 125 bacterial species with PQQ biosynthetic capability were identified by bioinformatics analysis (12). Four to seven genes organized in operon(s) are responsible for PQQ biosynthesis in different bacteria (13). In K.pneumoniae, operon pqqABCDEF is involved in PQQ production. Among six genes, only pqqA, pqqC, pqqD and pqqE are exclusively required for PQQ production (14). Despite the fact that PQQ has been found decades ago, only little is known about its biosynthetic pathway. Although the putative function of each of the genes of pqq operon(s) has been proposed based on sequence analyses and homology modelling (15), the only biochemically confirmed function is that of pqqC, which encodes a protein catalysing oxidation of the PQQ biosynthetic intermediate, 3a-(2-amino-2-carboxyethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic acid to PQQ as the final step of PQQ biosynthesis (14, 16, 17). It was also shown that conserved tyrosine and glutamic acid residues that are part of a peptide PqqA serve as precursors for synthesis of PQQ backbone (18). It has been originally reported that PqqA is not required for PQQ production in M.extorquens AM1 as the mutant of PqqA showed PQQ production (19). However, it was later found that there are additional two pqqA genes located in other loci far from the PQQ biosynthesis operon that possibly compensated the loss of pqqA in the operon in this bacterium (20). PqqE belongs to the radical S-adenosyl-l-methionine (SAM) enzyme superfamily and presumably catalyses the first reaction of PQQ biosynthesis, i.e. formation of the carbon-carbon bond between the tyrosine and glutamic acid residues of PqqA through the radical formation. The ability of PqqE to cross-link the two amino acid residues was however not demonstrated yet and prior modification of PqqA may be required (21). The PqqD domain of PqqC/D from M.extorquens AM1 was just recently found to function as a chaperone for PqqA and interact with PqqE (22).
Similar to other radical SAM enzymes characterized to date, PqqE from K.pneumoniae is susceptible to rapid deactivation by oxygen due to the oxidation of [4Fe-4S] clusters that are present in the enzyme (21). PqqE expressed heterologously in Escherichia coli cells under aerobic conditions was exclusively found in inclusion bodies, whereas the soluble and functional form of PqqE was successfully overexpressed and isolated only under strictly anaerobic conditions. The anaerobically prepared PqqE rapidly degraded upon exposure to oxygen. In this article, we describe efficient expression and biochemical characterization of PqqE from an aerobic facultative methylotroph M.extorquens AM1, which in contrast to the K.pneumoniae PqqE, is remarkably tolerant to oxygen.
Experimental Procedures
Materials
SAM, DTT, FeCl3 and LB broth were purchased from Sigma Aldrich (Munich, Germany). Li2S, ammonium iron (III) citrate and 57Fe2O3 (96.64% isotopic enrichment) were purchased from Alfa Aesar (Karlsruhe, Germany), Lach-Ner (Neratovice, Czech Republic) and ISOFLEX (San Francisco, CA), respectively.
Bacterial strains and growth conditions
Methylobacterium extorquens AM1 was maintained on Hypho agar plate (23). Escherichia coli strains used in cloning experiments were maintained on LB plate supplemented with kanamycin and chloramphenicol at 50 and 35 µg/ml, respectively. The stock solution of 1 M 57FeCl3 was prepared by completely dissolving 57Fe2O3 in 6 M HCl using a hot plate. The solution was evaporated to near dryness and then an appropriate volume of 1M HCl was added to obtain 1 M 57FeCl3. The concentration of Fe3+ in solution was measured using the method of Fish (24).
DNA techniques
Genomic DNA was isolated from cells grown to the early stationary phase by a modified method of Marmur (25). Restriction enzyme digestion, DNA ligation and other DNA modifications were performed according to the manufacturer’s recommendations. Preparation of plasmid DNA from E.coli strains and other general molecular biology procedures were performed as described by Sambrook et al. (26). PCR for cloning purpose was performed using a ‘Phusion High-Fidelity PCR Kit’ (NEB, Ipswich, MA), whereas a GoTaq PCR Master Mix (Promega, Madison, WI) was used for analytical purpose. DNA fragments separated in agarose gel were purified using a Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI) or a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). All restriction enzymes were purchased from NEB (Ipswich, MA).
Construction of PqqE-expressing vectors
The gene encoding PqqE was amplified by PCR from genomic DNA of M.extorquens AM1 with each of the two specific primer sets containing restriction sites (underlined): NcoI-pqqE-F1 (5′-ATACCATGGATGCACCGACACCCGC-3′) and BamHI-pqqE-R1 (5′-ATAGGATCCCAAAGGGGTGCCTTTTCGCTC-3′) for His6-tag fusion at the C-terminus or NdeI-pqqE-F1 (5′-ATACATATGAATGCACCGACACCCGC-3′) and BamHI-pqqE-R2 (5′-ATAGGATCCTCAAAGGGGTGCCTTTTCGC-3′) for His6-tag fusion at the N-terminus. The PCR products were then digested with NcoI and BamHI or NdeI and BamHI and ligated to pET28b(+) plasmid digested with the same sets of restriction enzymes. The resulting plasmid, pET-pqqE-C and pET-pqqE-N, were then transformed into E.coli TOP10 (Invitrogen, Waltham, MA) as the cloning host. For construction of the C32S mutant of PqqE, plasmid pET-pqqE-N was used as a template for PCR. PCR was performed with Tks Gflex DNA polymerase (Takara, Tokyo, Japan) using phosphorylated primers containing a mismatched nucleotide (shown in bold face): PqqE_C32-S32_F1 (5′-CTGCCCGCTGCGCAGCCCATACTGCTCG-3′) and PqqE_C32-S32_R1 (5′-CGGTGCGTCAGCTCGGCGAGCAGAC-3′). The linearized PCR products were then self-ligated with T4 DNA Ligase (NEB, Ipswich, MA). The ligation mixtures were transformed into E.coli DH5α and the DNA sequences of the mutant plasmids were confirmed by DNA sequencing.
Expression of PqqE
Expression of PqqE was carried out with E.coli BL21 (DE3) Star (Invitrogen, Waltham, MA) and E.coli Rosetta 2 (DE3) (Novagen, Cambridge, MA). The cells harboring pET-pqqE-N were grown in LB medium with kanamycin when using E.coli BL21 (DE3) Star as the expression host or with kanamycin and chloramphenicol when using E.coli Rosetta 2 (DE3) as the expression host. The cells were grown in 500 ml of LB medium in a 1-l flask at 37°C and 150 rpm until reaching the OD600 of 0.9. To induce expression, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the final concentration of 0.1 mM, and the medium was supplemented with 0.1 mM of Fe3+. Expression was carried out at 18°C and 120 rpm for 16–18 h. Cells were harvested by centrifugation at 5700 g for 5 min. The cells were washed with distilled water and stored at −20°C until use.
Purification of PqqE
All purification steps were carried out at room temperature (∼22°C) and either under aerobic or anaerobic conditions. The steps of aerobic and anaerobic purification were identical. Aerobic purification was performed under laboratory conditions using buffers and solutions in which dissolved O2 was not removed, whereas all steps of anaerobic purification were carried out in a glove box (SICCO, Grünsfeld, Germany) with buffer solutions that were bubbled with N2 gas (O2 content 1.5 ppm) for at least 30 min inside the glove box. The inlet pressure of N2 gas flown into the glove box was set to about 0.3 bars throughout the process. The gaseous O2 level inside the box was monitored with a GMH 3691 Digital Oxymeter (GHM Messtechnik GmbH, Regenstauf, Germany). The cells were suspended in the deoxygenated equilibration buffer (50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 10 mM imidazole and 10% (v/v) glycerol) and disrupted inside the glove box with BugBuster supplemented with Benzonase (Novagen, Cambridge, MA) according to the manufacturer recommendation. The disrupted cell suspension was transferred to an air-tight sealed centrifugation tube and the cell debris was removed by centrifugation at 16,700 g for 15 min at 4°C. The tube was then transferred to the anaerobic glove box and supernatant was applied onto a Protino Ni-NTA Agarose affinity column (Macherey-Nagel, Düren, Germany) with a column volume of 3 ml, which had been pre-equilibrated with the deoxygenated equilibration buffer. PqqE protein appeared as dark brownish band in the column that facilitated PqqE protein purification. Non-specific proteins bound to the resin were removed with at least 2 volumes of the washing buffer (50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 50 mM imidazole and 10% (v/v) glycerol). PqqE was then eluted with the elution buffer (50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 135 mM imidazole and 10% (v/v) glycerol). The buffer in purified PqqE solution was then exchanged with buffer A (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT and 10% (v/v) glycerol) using an Amicon Ultra-15 Centrifugal Filter Unit with 10 kDa cut-off (Millipore, Darmstadt, Germany). The purified PqqE was immediately used for the subsequent experiments in most cases, but occasionally stored at −80°C until use.
Preparation of sample for Mössbauer spectroscopy
Cells of E.coli Rosetta 2 (DE3) harboring pET-pqqE-N were cultivated as a seed culture in LB medium for 6–8 h. Then 50 ml of the seed culture were transferred into 500 ml of fresh LB medium in 1-l flask and cultivated until the OD600 reached 0.9. IPTG and 57Fe3+ were added both to the final concentration of 0.1 mM and the culture was cultivated at 18°C for 16–18 h. PqqE was then purified as described in ‘Purification of PqqE’ under anaerobic conditions. The purified PqqE was concentrated to gain the final PqqE concentration of 1.4 mM and subjected to Mössbauer spectral measurements after freezing in liquid nitrogen.
Reconstitution of Fe-S clusters
Reconstitution of Fe-S clusters was carried out under anaerobic conditions inside a glove bag (Sigma-Aldrich, Munich, Germany) at 4°C, basically according to a published method (27). The purified PqqE in buffer A was incubated with 100 equivalents of DTT with gentle stirring for 1 h. Ten equivalents of ammonium iron (III) citrate were carefully added to the enzyme solution and then incubated with gentle stirring for 5 min. Finally, 10 equivalents of Li2S were added and the mixture was then incubated with gentle stirring overnight. Precipitates formed during the reconstitution reaction were removed by centrifugation at 17,500 g. The resulting supernatant was applied onto a Sephadex G-25 column (10-ml column volume), which was then eluted with buffer A. The eluted fraction with dark brownish colour was pooled as the reconstituted PqqE. This reconstituted PqqE was immediately used for the subsequent experiments in most cases, but occasionally stored at −80°C until use. Reconstitution with 57Fe3+ for Mössbauer spectral measurements was also carried out in a similar manner using an 57FeCl3 solution (in 1 M HCl) after adjusting the pH to about 5 with 1 M NaOH and 1 M ammonium citrate, instead of using ammonium iron (III) citrate (natural iron contains 91.72% (w/w) 56Fe that is Mössbauer silent).
SAM cleavage assay
Assays of PqqE for the reductive homolytic cleavage of SAM were carried out at a room temperature (∼22°C) in the anaerobic glove box. All buffers were bubbled with N2 gas and placed inside the box at least 30 min before use. Sodium dithionite solution (1 M) in buffer A was prepared freshly immediately before the reaction. The reaction mixtures contained 2.5 mg/ml (57.2 µM) purified PqqE in buffer A. The Fe-S clusters of PqqE were reduced by addition of sodium dithionite to the final concentration of 25 mM for 10 min. The enzyme reaction was started by addition of SAM to the final concentration of 200 µM. The reaction was stopped after 30 min (unless otherwise described) by heating at 80°C for 10 min. The protein precipitates were removed by centrifugation at 17,500 g for 5 min and the supernatant was analysed for the formation of 5′-deoxyadenosine (5′dA) using HPLC with UV detection at 259 nm. Reaction product, 5′dA, was determined on a Symmetry C18 column (2.1 × 150 mm, 5 µm; Waters, Milford) connected to an Alliance 2695 high performance liquid chromatograph (Waters) and a Waters 2996 photodiode array detector. The column was eluted by a linear gradient of 15 mM ammonium formate, pH 4.0 (A) and methanol (B) using the following solvent mixture: 0–5 min, 5–30% B, 5–6 min, 30–100% B, 6–7 min 100% B. The flow rate was 0.25 ml/min and the column temperature was 30°C. The concentration of product was determined by a calibration curve method using authentic standard compound. Identification of product (5′dA) formed by SAM cleavage was performed by Q-TOF mass spectrometer (Waters) connected to HPLC.
Protein, iron and sulphur analyses
Protein concentration was measured by the Bradford method (28) with a Bradford Protein Assay Kit (BioRad, Hercules, CA) and bovine serum albumin as the standard. For determination of the Bradford correction factor (29), the purified PqqE (80 µl, 5.49 ± 0.27 mg/ml) was precipitated with 10% (v/v) TCA at −20°C for 30 min and washed twice with cold acetone to remove the bound [Fe-S] clusters. The precipitates dried at 40°C were then dissolved at 80°C in 80 µl of 8 M urea containing 1 mM DTT (5.24 ± 0.17 mg/ml; recovery, 95%). As the urea solution exhibited a spectrum of simple protein without absorption above 350 nm, the protein concentration was calculated to be 5.37 mg/ml from the absorbance at 278 nm using the molar extinction coefficient of 57,870 M−1cm−1 obtained by the web-based tool, ProtParam (30), assuming that all Cys residues are reduced. The Bradford correction factor (5.37/5.24 = 1.02) thus obtained was used for all the subsequent enzyme characterizations. SDS–PAGE was carried out according to the method of Laemmli (31). Iron and acid labile sulphide contents were measured as described by Fish (24) and Beinert (32), respectively.
UV-Vis absorption and Mössbauer spectroscopies
UV-Vis absorption spectra were measured using a Shimadzu UV-2401 PC spectrophotometer (Shimadzu, Kyoto, Japan). The 57Fe Mössbauer spectra were measured employing a homebuilt Mössbauer spectrometer (33), operating at a constant acceleration mode and equipped with a 57Co(Rh) source. Low-temperature Mössbauer spectra of the samples were recorded at 5 K without applying an external magnetic field (zero-field) using a Spectromag (Oxford Instruments) cryomagnetic system with the Mössbauer spectrometer attached to the system. The acquired Mössbauer spectra were recorded with 512 channels and processed (i.e. noise filtering and fitting) using the MossWinn software program (34). The isomer shift values were referred to α-Fe foil sample at room temperature. During data analysis the recoilless factor was assumed to be identical for all iron sites within the experimental error of Mössbauer technique.
Bioinformatics analysis
Initially a total of 141 amino acid sequences was manually retrieved from PqqE homologs currently registered in the structure-function linkage database (35), by selecting one representative species from all bacterial genera having a pqqE gene. Then 26 PqqE sequences used for multiple sequence alignment were selected on the basis of the phylogenetic tree generated for the initial 141 PqqE homologs (Supplementary Fig. S1) to cover all representative sequences in each major branch of the tree and five sequences that have unique 3-amino acid insertion located near the SPASM domain (vide infra) including PqqE of K.pneumoniae. Multiple sequence alignment was carried out with a MEGA6 software (36) using the MUSCLE algorithm (37), and the resultant alignment figure was generated using ESPript 3.0 (38).
Results
Overexpression of PqqE in E. coli
To achieve overexpression of recombinant PqqE from M.extorquens AM1, the PCR-amplified 1155-bp gene encoding PqqE was cloned into an expression vector, pET28b(+), with a His6-tag fused at the N- or C-terminus of the protein. The expression of PqqE was first attempted in E.coli BL21 (DE3) Star cells grown aerobically followed by IPTG-induction at 37°C. However, we could observe only insignificant production of the recombinant protein corresponding to the His6-tagged PqqE with an expected molecular mass of ∼43.7 kDa (deduced from amino acid sequence) by SDS–PAGE, even after examining various expression conditions [e.g. induction temperatures at 15–37°C and varying cultivation time after induction (data not shown)]. Notifying that the pqqE gene sequence of M.extorquens AM1 contains the codons that are rarely used by typical E.coli strains, we next switched to use E.coli Rosetta 2 (DE3) strain carrying seven additional tRNA genes for the rare codons (39) as the PqqE expression host. With the E.coli Rosetta 2 (DE3) strain, expression level of PqqE was markedly increased (Fig. 1A), as compared with the control cells with the pET28b(+) vector alone (not shown). Although a considerable amount of the overexpressed PqqE protein was also detected in the insoluble fraction (Fig. 1A), the amount of soluble PqqE was much higher than that expressed in E.coli BL21 (DE3) strain. We then surveyed culture conditions for efficient expression of PqqE using E.coli Rosetta 2 (DE3) as the host by changing induction temperature (15–37°C) and extent of aeration (shaking speed 90–150 rpm) and found that overnight expression at 18°C with rotary shaking at 120 rpm was optimum for production of PqqE exhibiting SAM cleavage activity (as described later), probably because the E.coli Fe-S cluster-inserting system such as the Isc (iron-sulphur cluster) machinery (40) functioned efficiently under these conditions. Furthermore, the C-terminally His6-tagged PqqE expressed at 18°C in the soluble fraction showed no SAM cleavage activity. Therefore, the N-terminally His6-tagged PqqE (designated PqqE-N) was used in the subsequent experiments. Transformed E.coli cells were grown in the medium supplemented with 0.1 mM ferric ions for loading iron into PqqE-N. Approximately 1.5–2.2 g of wet cells were obtained from 500 ml of LB medium by cultivation at 18°C for 16–18 h after induction with 0.1 mM IPTG.
Fig. 1.
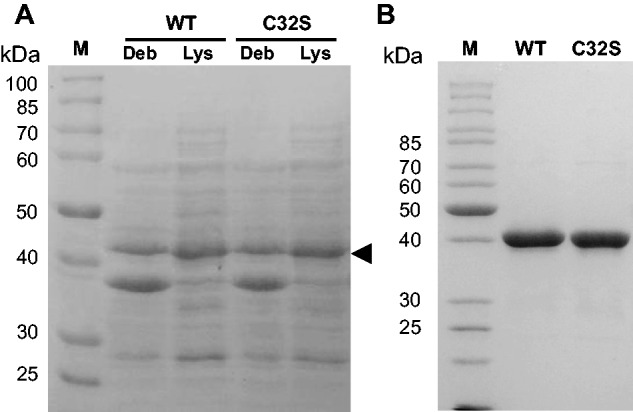
Expression and purification of recombinant PqqE. (A) SDS–PAGE analysis of cell debris (Deb) and lysate (Lys) of E. coli Rosetta 2 (DE3) cells expressing wild-type (WT) and C32S mutant (C32S) of PqqE from M.extorquens AM1. Fifteen microliters of cell lysate and debris (dissolved in 5 µl of sample buffer) were applied in each well of a 10% (w/v) gel. A triangle indicates the protein band corresponding to the expressed PqqE-N. (B) SDS–PAGE analysis of the purified wild-type (WT) and C32S mutant (C32S) of PqqE-N (5 µg protein/well). M: PageRuler marker (Thermo Scientific, Waltham, MA).
Purification and reconstitution of PqqE-N
In an early stage of investigation, we disrupted the cultured E.coli cells by ultrasonic oscillation, purified PqqE-N under anaerobic conditions, and obtained the enzyme that showed considerable SAM cleavage activities when assayed as described above (0.227 ± 0.074 nmol/min/mg). However, to avoid possible deactivation of the radical SAM enzyme PqqE that might occur during sonication under atmospheric conditions, we subsequently employed anaerobic disruption by using a protein extraction reagent (BugBuster) in combination with the addition of nuclease (Benzonase) to degrade nucleic acids that stack the column packed with an affinity resin. When conducted in an anaerobic chamber, this disruption method was proven to give reproducible results of purification of PqqE-N with nearly constant SAM cleavage activities (see below). Although affinity purification of PqqE-N using a single column of nickel-chelating resin at room temperature (∼22°C) either under aerobic or anaerobic conditions gave very similar results in terms of SAM cleavage activity and iron/sulphur contents as described later, we routinely purified PqqE-N in an anaerobic chamber filled with N2-gas. The dark brownish colour derived from the Fe-S cluster was helpful for monitoring the purification of PqqE-N by affinity chromatography. By employing appropriate imidazole concentrations for washing-out unbound or weakly bound proteins and for eluting the enzyme efficiently, PqqE-N was purified to near homogeneity (purity, >95% on SDS–PAGE; Fig. 1B) by the single step of affinity chromatography. The final yield of the as-purified PqqE-N was 2.4–4.3 mg/g of wet cells. We noted that freezing/thawing of the as-purified PqqE-N in the buffer without glycerol resulted in the precipitation of most proteins; addition of 10% (v/v) glycerol was essential to prevent protein precipitation. Furthermore, chemical reconstitution of the as-purified enzyme was performed immediately after purification to avoid unpredictable damage of the Fe-S clusters and associated loss of the SAM cleavage activity during storage. Reconstitution with 10-fold molar excess of ferric and sulphide ions was accompanied with the slight formation of protein precipitates but a sufficient amount of the soluble reconstituted PqqE-N was obtained by passing through a small gel filtration column of Sephadex G-25 (recovery ∼70% of the as-purified enzyme subjected to reconstitution). The as-purified and reconstituted PqqE-N preparations thus obtained were used freshly for the subsequent experiments. Otherwise, they were stored at −80°C until use.
UV-Vis absorption spectra and Fe/S contents
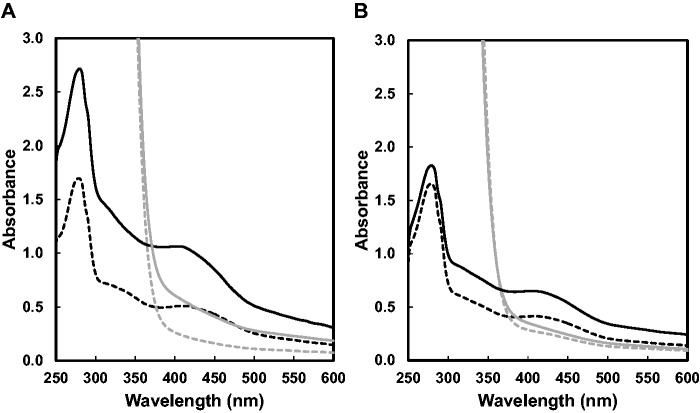
The as-purified PqqE-N exhibited a UV-Vis absorption spectrum typical for a protein that contains the Fe-S cluster with an absorption peak at 400–420 nm and a shoulder at 300–350 nm, as shown in Fig. 2A. A low and broad absorption band was also observed around 600 nm. Reconstitution resulted in a further increase of not only the broad absorption bands above 300 nm but also the absorption at 280 nm, which overlaps with the protein absorption band. Using a molar extinction coefficient at 410 nm reported for the typical [4Fe-4S] clusters (15–17 mM−1cm−1 on a per cluster basis) (41, 42) and protein concentrations determined by the Bradford method and corrected with the Bradford correction factor (29), the contents of the [4Fe-4S] cluster were calculated to be ∼1.4 and 2.9 moles per mole of the as-purified and reconstituted PqqE-N, respectively. Colorimetric iron/sulphur analyses also indicated the presence of multiple (likely 3) [4Fe-4S] clusters in the reconstituted PqqE-N (Table I). These results are consistent with the presence of three Fe-S cluster-binding sites conserved in the PqqE homologs (as described later) and also indicate that the [4Fe-4S] clusters are inserted, though partially (about a half), into these sites by the aid of the Fe-S inserting system(s) inherently contained in E.coli cells. The absorption band above 400 nm disappeared after incubation of PqqE-N with an excess amount of sodium dithionite (Fig. 2A), suggesting reduction of the Fe-S clusters.
Fig. 2.
UV-Vis absorption spectra of the as-purified and reconstituted wild-type and C32S mutant of PqqE-N. (A) UV-Vis absorption spectra of the as-purified (black broken line) and reconstituted (black solid line) wild-type PqqE-N are shown with those 20-min after reduction with excess sodium dithionite (grey solid and broken lines). (B) UV-Vis absorption spectra of the as-purified (black broken line) and reconstituted (black solid line) C32S mutant of PqqE-N are shown with those 20-min after reduction with excess sodium dithionite (grey solid and broken lines). Enzyme concentrations were all adjusted to 1 mg/ml in buffer A.
Table I.
SAM cleavage activities and Fe/S contents
| PqqE-N | As-purified |
Reconstituted |
||||
|---|---|---|---|---|---|---|
| SAM cleavage activity (nmol/min/mg) | Fe/PqqE-N (mol/mol) | S/PqqE-N (mol/mol) | SAM cleavage activity (nmol/min/mg) | Fe/PqqE-N (mol/mol) | S/PqqE-N (mol/mol) | |
| Wild-type | 0.418 ± 0.175 (n = 30) | 5.36 ± 0.11 (n = 7) | 8.75 ± 0.21 (n = 7) | 0.609 ± 0.156 (n = 29) | 12.7 ± 1.0 (n = 7) | 16.4 ± 1.3 (n = 7) |
| C32S mutant | 0 | 3.84 ± 0.65 (n = 7) | 5.30 ± 0.33 (n = 7) | 0 | 10.5 ± 1.3 (n = 7) | 12.1 ± 1.5 (n = 7) |
SAM cleavage activity
Most radical SAM enzymes catalyse the reductive cleavage of SAM into methionine and the 5′-deoxyadenosyl radical that abstracts a hydrogen atom either from the substrate in their respective coupled reactions or from protein or solvent (a proton plus an electron) in the uncoupled reaction, forming the final stable product, 5′dA (43–47). The reductive SAM cleavage activity of the as-purified and reconstituted PqqE-N was determined by measuring 5′dA in the reaction mixture by liquid chromatography. Control experiments indicated that the reaction proceeded only when the dithionite-reduced PqqE-N was incubated with SAM under strictly anaerobic conditions. The product obtained after the reaction was identified as 5′dA by comparison with the standard, showing an absorption spectrum and a mass-to-charge ratio for the singly charged positive ion as well as the retention time in the HPLC analysis, all identical with those of the reaction product. The SAM cleavage reaction by the dithionite-reduced reconstituted PqqE-N proceeded by following pseudo-first-order kinetics when measured for 4 h (data not shown), giving an observed rate constant (kobs) of 0.0130 ± 0.0011 min−1. The specific SAM cleavage activities of the as-purified and reconstituted enzymes (Table I) are considerably higher than the anaerobically as-purified K.pneumoniae PqqE [0.24–0.28 nmol min−1 mg−1, estimated from the reported reaction rates at given enzyme concentrations (21)]. By measuring the initial rates of 5′ dA production at different SAM concentrations (reaction for 10 min), the apparent Km for SAM was determined to be 60.1 ± 13.1 μM. Remarkably, the M.extorquens AM1 enzyme purified under the fully aerobic conditions showed a very high SAM cleavage activity (corresponding to ∼70% of the activity of the anaerobically purified enzyme) with comparable iron/sulphur contents, indicating that PqqE from M.extorquens AM1 is considerably stable under aerobic conditions. Nevertheless, the as-purified PqqE-N stored at 4°C in an unsealed vessel gradually lost its initial dark brownish colour and most of the SAM cleavage activity after about a week, as investigated in more detail in a later section.
Site-specific mutagenesis of the canonical [4Fe-4S] cluster-binding cysteine residue
Multiple sequence alignment of PqqE homologs has suggested the possible binding residues for [4Fe-4S] clusters, as described later. The CX3CX2C motif located near the N-terminus of PqqE is believed to be the consensus motif involved in binding of the [4Fe-4S] cluster that is responsible for the reductive SAM cleavage activity (hence often called ‘radical SAM cluster’) (43–47). To confirm this for PqqE, we mutated the central Cys residue (Cys32) in this motif to Ser by site-directed mutagenesis. The C32S mutant of PqqE-N was expressed and purified in the same way as the wild-type enzyme under anaerobic conditions (Fig. 1B). Although UV-Vis absorption spectra (Fig. 2B) of the as-purified and reconstituted C32S mutant indicated the presence of Fe-S clusters (estimated to be ∼1.1 and 1.8 moles of [4Fe-4S] cluster per mol of as-purified and reconstituted enzymes, respectively), no SAM cleavage activity was detected at all in both the as-purified and reconstituted C32S mutant of PqqE-N (Table I), clearly indicating that one [4Fe-4S] cluster that is involved in the SAM cleavage activity was lost by the Cys32-to-Ser substitution, although the other Cys-rich regions conserved in the C-terminal SPASM domain are still able to bind auxiliary Fe-S clusters, as described later. The decrease in the Fe/S contents as compared with those in the wild-type enzyme (Table I) also supports the absence of one [4Fe-4S] cluster and the presence of two [4Fe-4S] clusters in the reconstituted C32S mutant enzyme.
Mössbauer spectral analysis
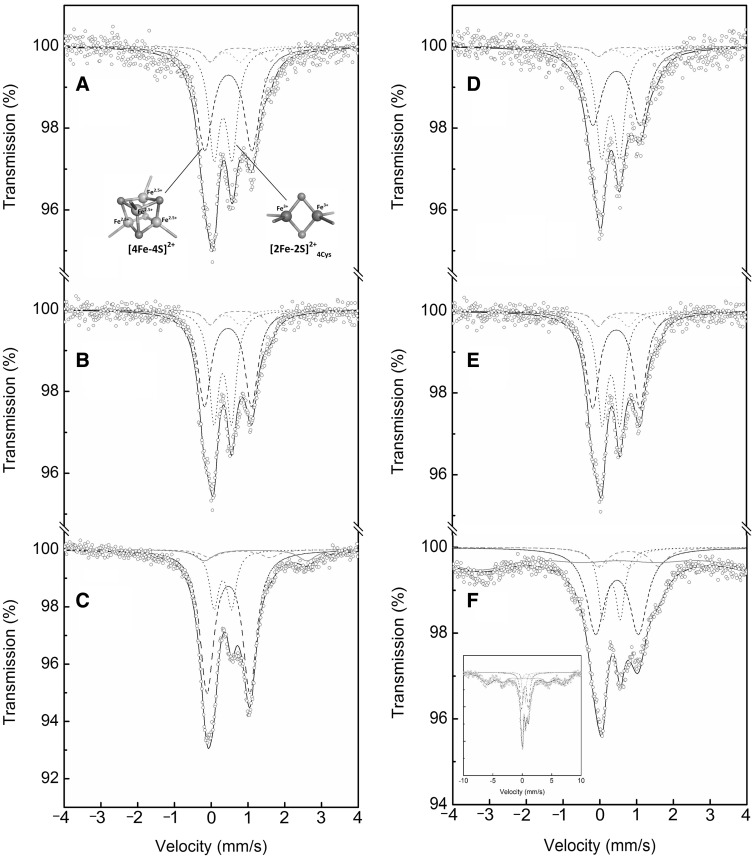
In order to analyse the type(s) of Fe-S clusters bound to the enzyme, Mössbauer spectroscopy was first conducted with the as-purified wild-type and C32S mutant enzymes of PqqE-N. Mössbauer spectroscopy is an element selective and local method when metal nucleus acts as a probe monitoring physical and chemical features of its surroundings through hyperfine interactions of electric and magnetic nature and therefore is very suitable for studying the structure and physicochemical properties of Fe-S clusters (48). Both the as-purified wild-type and C32S mutant enzymes of PqqE-N were prepared from the cultures enriched with 57Fe3+ and subjected to the low-temperature Mössbauer spectroscopy. The zero-field 57Fe Mössbauer spectra of the enzyme samples (concentrated to 1.4 mM) recorded at 5 K are shown in Fig. 3 and the values of the Mössbauer hyperfine parameters such as isomer shift (δ) and quadrupole splitting (ΔEQ), derived from the spectral fitting, are summarized in Table II. Although UV-Vis absorption spectra suggested that the as-purified wild-type and C32S mutant enzymes of PqqE-N contained 1.4 and 1.1 mole of [4Fe-4S] cluster per mole of enzyme, respectively, as described above (Fig. 2), both enzymes exhibited almost identical Mössbauer spectra, indicating a complex superimposed spectrum of different species of 57Fe. The spectra were fitted well with two major quadrupole doublets assignable to [4Fe-4S]2+ (Stotal = 0) and [2Fe-2S]2+ (Stotal = 0) clusters and two minor unassigned doublets having hyperfine parameters identical with those reported for the [4Fe-4S]2+-pyruvate complex (δ = 0.78 mm/s, ΔEQ = 1.62 mm/s for the ferrous site of a localized pair, and δ = 0.38 mm/s, ΔEQ = 0.81 mm/s for its ferric counterpart) (49) (Fig. 3A and B). The [4Fe-4S]2+ quadrupole doublet consists of two sets of {Fe2.5+-Fe2.5+} interacting species and accounts for ∼50% of the total signal in both the wild-type and C32S mutant of PqqE-N (Table II). Another prominent signal is assigned to the [2Fe-2S]2+ cluster containing two Fe3+ species ligated by four Cys residues ([2Fe-2S]2+4Cys), which is assumed to be derived either from incomplete cluster building at the [4Fe-4S]2+ cluster-binding sites by E.coli or from decomposition of the [4Fe-4S]2+ cluster during enzyme purification, or both (50). Alternatively, the [2Fe-2S]2+4Cys cluster may be a normal form bound in the auxiliary [4Fe-4S] cluster-binding sites conserved in the C-terminal half of the PqqE sequence (see below). Based on the relative areas (RA, Table II) of Mössbauer signals, which reflect the number of 57Fe atoms detected, the [4Fe-4S]2+/[2Fe-2S]2+ molar ratio is calculated to be about 0.7 [= (52/4)/(36/2)] and 0.6 [= (47/4)/(39/2)] in the as-purified wild-type and C32S mutant enzymes, respectively, indicating that the [2Fe-2S]2+ cluster is the more abundant form in both the wild-type and C32S mutant enzymes and that the [4Fe-4S]2+ cluster content is slightly higher in the wild-type enzyme than in the C32S mutant. The two minor unassigned doublets have hyperfine parameters that do not coincide with the values reported for ordinary protein-bound Fe-S clusters (48); thus the identity of the cluster(s) remains to be settled.
Fig. 3.
Mössbauer spectra of the wild-type and C32S mutant of PqqE-N. 57Fe Mössbauer spectra of (A) the as-purified wild-type PqqE-N, (B) the as-purified C32S mutant of PqqE-N, (C) the reconstituted wild-type PqqE-N, (D) the as-purified wild-type PqqE-N exposed to O2 for 1 day, (E) the as-purified C32S mutant of PqqE-N exposed to O2 for 1 day and (F) the reconstituted wild-type PqqE-N exposed to O2 for 1 day, all recorded at 5 K. In (A–F), a wide doublet (dashed line) is the simulated spectrum for [4Fe-4S]2+ cluster, a narrow doublet (dotted line) is the simulated spectrum for [2Fe-2S]2+4Cys cluster, small doublets (grey dashed and dotted lines) are those for the two minor unassigned doublets having hyperfine parameters identical with those reported for the [4Fe-4S]2+-pyruvate complex (48), a broad doublet (grey solid line) and the distributed sexted (grey solid line) are those for the free (non-cluster) high-spin Fe2+ and ferrihydrite (hydrous ferric oxyhydroxide), respectively, and black solid line is the sum of these simulated spectra. In (A), structure models for [4Fe-4S]2+ and [2Fe-2S]2+4Cys clusters drawn by PyMOL (Schrödinger, LLC) are shown. In (F), the spectrum covering the whole sexted of ferrihydrite is shown in a wider velocity range (inset).
Table II.
Summary of the Mössbauer hyperfine parametersa
| Sample | Doublet componentb | δ ± 0.01 (mm/s) | ΔEQ ± 0.01 (mm/s) | Hhf ± 0.3 (T) | RA ± 1 (%) | Cluster assignment, spin S | |
|---|---|---|---|---|---|---|---|
| Wild-type | as-purified | (I) | 0.47 | 1.29 | — | 52 | [4Fe-4S]2+, S = 0 |
| (II) | 0.32 | 0.49 | — | 36 | [2Fe-2S]2+, S = 0 | ||
| (III) | 0.78 | 1.62 | — | 6 | UAc | ||
| (IV) | 0.38 | 0.81 | — | 6 | UAc | ||
| 1 day exposure to O2 | (I) | 0.46 | 1.28 | — | 49 | [4Fe-4S]2+, S = 0 | |
| (II) | 0.30 | 0.49 | — | 41 | [2Fe-2S]2+, S = 0 | ||
| (III) | 0.78 | 1.62 | — | 5 | UAc | ||
| (IV) | 0.38 | 0.81 | — | 5 | UAc | ||
| C32S mutant | as-purified | (I) | 0.46 | 1.28 | — | 47 | [4Fe-4S]2+, S = 0 |
| (II) | 0.31 | 0.48 | — | 39 | [2Fe-2S]2+, S = 0 | ||
| (III) | 0.78 | 1.62 | — | 7 | UAc | ||
| (IV) | 0.38 | 0.81 | — | 7 | UAc | ||
| 1 day exposure to O2 | (I) | 0.47 | 1.29 | — | 43 | [4Fe-4S]2+, S = 0 | |
| (II) | 0.32 | 0.52 | — | 43 | [2Fe-2S]2+, S = 0 | ||
| (III) | 0.78 | 1.62 | — | 7 | UAc | ||
| (IV) | 0.38 | 0.81 | — | 7 | UAc | ||
| Wild-type | reconstituted | (I) | 0.46 | 1.17 | — | 66 | [4Fe-4S]2+, S = 0 |
| (II) | 0.31 | 0.48 | — | 17 | [2Fe-2S]2+, S = 0 | ||
| (III) | 0.78 | 1.62 | — | 5 | UAc | ||
| (IV) | 0.38 | 0.81 | — | 5 | UAc | ||
| (V) | 1.20 | 2.78 | — | 7 | Free Fe2+, S = 2 | ||
| 1 day exposure to O2 | (I) | 0.47 | 1.17 | — | 25 | [4Fe-4S]2+, S = 0 | |
| (II) | 0.31 | 0.48 | — | 10 | [2Fe-2S]2+, S = 0 | ||
| (III) | 0.78 | 1.62 | — | 4 | UAc | ||
| (IV) | 0.38 | 0.81 | — | 4 | UAc | ||
| (VI) | 0.50 | −0.05 | 40.0 | 57 | Ferrihydrite | ||
aδ is the isomer shift, ΔEQ is the quadrupole splitting, Hhf is the hyperfine magnetic field, and RA is the relative spectral area of individual components identified during spectral fitting, assuming an identical recoilless factor for all iron sites within the experimental error of Mössbauer technique. bComponents (I–VI) represent wide doublet (dashed line), narrow doublet (dotted line), minor unassigned doublets (grey dashed and dotted lines), free (non-cluster) high-spin Fe2+ (grey solid line), and ferrihydrite (hydrous ferric oxyhydroxide) (grey solid line) in Fig. 3, respectively. cUnassigned.
Next, the 57Fe-containing as-purified wild-type enzyme was further reconstituted chemically with 57Fe3+ and its zero-field Mössbauer spectrum was measured at 5 K (Fig. 3C). The reconstituted enzyme exhibited a more complex spectrum than the as-purified enzyme with an additional signal derived from the non-cluster iron with the hyperfine parameters characteristic of free high-spin (S = 2) Fe2+ ions (δ = 1.20 mm/s, ΔEQ = 2.78 mm/s) (51), which are known to be often bound onto the protein surface after chemical reconstitution (29). The most notable difference from the as-purified enzyme was that the reconstituted enzyme contained predominantly the [4Fe-4S]2+ clusters, accounting for ∼66% of the total signal (Table II). The relative content of the [2Fe-2S]2+ cluster decreased to ∼17% of the total signal, yielding the [4Fe-4S]2+/[2Fe-2S]2+ molar ratio of ∼1.9. This suggests that the reconstituted enzyme contains one [2Fe-2S]2+ and two [4Fe-4S]2+ clusters. The signals of the two minor unassigned doublets observed in the as-purified enzyme were also detected in the reconstituted enzyme. These results show that the chemical reconstitution resulted in an increase of the [4Fe-4S]2+ cluster content, which is fully consistent with the significant increases in the SAM cleavage activity (Table I) and the intensity of 410-nm absorption band (Fig. 2A) after reconstitution. The results also suggest that the [2Fe-2S]2+ cluster initially observed in the as-purified enzyme was not converted into the [4Fe-4S]2+ cluster.
Sensitivity to O2
The sensitivity of PqqE-N to O2 was examined by measuring the SAM cleavage activity of the enzyme continuously exposed to air. The as-purified and reconstituted enzyme solutions (10 mg/ml) were placed in 5-ml glass vials without a cap for 5 min to equilibrate enzyme solution with atmospheric oxygen and then kept at 4°C with a loosely placed cap to allow the air freely enter the vial. A 26-µl aliquot of each sample was taken from the vial at appropriate time intervals (1–7 days), blown with N2 gas for 1 min, diluted 5-folds with deoxygenated buffer A and assayed for the remaining SAM cleavage activity in the anaerobic box; control experiments using the samples kept at 4°C under anaerobic conditions were run for both the as-purified and reconstituted PqqE-N. UV-Vis absorption spectra of PqqE-N were also recorded at the same time intervals during exposure to O2.
As shown in Fig. 4A and B, the SAM cleavage activity of the as-purified and reconstituted PqqE-N decreased to ∼48% and 27%, respectively, of their original activity after 1 day of the O2-exposure and most of the activity was lost after 7 day exposure in both enzyme samples. The as-purified and reconstituted PqqE-N control samples (without exposure to O2) were relatively stable when kept under anaerobic conditions, retaining ∼70% and 30%, respectively, of their original activities after 7 days. Thus the continued mild exposure to O2 facilitated the inactivation of PqqE-N, though gradually (in a day scale). It was also found that the absorbance at 400–420 nm decreased significantly after 1 day of the O2-exposure for both the as-purified and reconstituted enzymes, followed by a gradual decrease until day 7 (Fig. 4C and D, insets). Since the absorption in the 400–420 nm region is mainly ascribed to the [4Fe-4S] clusters (41, 42), these results indicate the facilitated degradation of the [4Fe-4S] clusters in PqqE-N by exposure to O2, particularly that of the radical SAM cluster directly participating in the SAM cleavage reaction. Combined with the finding that the enzyme purified under fully aerobic conditions had a significantly high SAM cleavage activity as described above, these results clearly show that PqqE from M.extorquens AM1 has a very high tolerance to O2. This property is quite unusual for the radical SAM superfamily of enzymes that are generally very sensitive to O2. It seems likely that the more oxygen sensitive [4Fe-4S] cluster predominantly present in the reconstituted enzyme is first degraded rapidly and then the semistable [2Fe-2S] cluster formed from the [4Fe-4S] cluster is further degraded slowly by exposure to oxygen. Although the [2Fe-2S] cluster is inactive for the reductive SAM cleavage, the activity detected in the later phase of O2-exposure may be due to the reversion of the remaining [2Fe-2S] cluster to [4Fe-4S] by reduction with dithionite added in the SAM cleavage assay, as reported previously for MOCS1A, an oxygen-sensitive iron-sulphur protein involved in human molybdenum cofactor biosynthesis (42). Furthermore, the multiple (likely 3) Fe-S clusters contained in PqqE-N may have different tolerances to O2 with the radical SAM [4Fe-4S] cluster having the least one.
Fig. 4.
Effect of exposure to oxygen on SAM cleavage activity and absorption spectra of PqqE-N. SAM cleavage activities of the as-purified (A) and reconstituted (B) PqqE-N were measured at 1–7 days of continuous mild exposure to O2 (n = 6) and are shown with gray bars. Control samples (n = 3) kept under anaerobic conditions for the same period were also assayed and are shown with white bars. Activities at day 0 refer to the original activities before exposure to O2. Absorption spectra of the as-purified (C) and reconstituted (D) PqqE-N (0.5 mg/ml) recorded at 1–7 days of the continuous exposure to O2 (black to light gray dashed lines for the spectra at 1–7 days) are shown with those measured before the O2-exposure (solid lines). Arrows indicate the direction of the spectral change. Insets: The absorbance value at 410 nm are plotted against time (in days).
To examine if the Fe-S clusters bound in the enzyme change their forms by exposure to O2, the as-purified wild-type and C32S mutant enzymes of PqqE-N were exposed to atmospheric O2 for 1 day at 4°C and subjected to the Mössbauer spectral analysis (Fig. 3D and E). The [4Fe-4S]2+ signal was found to decrease slightly by 1-day exposure to O2, concomitant with the slight increase of the [2Fe-2S]2+ signal (Table II). In marked contrast, the [4Fe-4S]2+ and [2Fe-2S]2+ signals significantly decreased and increased, respectively, in the reconstituted wild-type enzyme after 1-day exposure to O2 (Fig. 3F and Table II), yielding the [4Fe-4S]2+/[2Fe-2S]2+ molar ratio of ∼1.3. This indicates that the [4Fe-4S]2+ cluster is more preferentially degraded by O2-exposure than the [2Fe-2S]2+ cluster, supporting the conclusion derived from the UV-Vis absorption spectral change and the decrease of SAM cleavage activity as described above. The Mössbauer spectrum also showed that the 57Fe2+ ions adventitiously bound in the reconstituted enzyme as well as those contained in the Fe-S clusters were considerably oxidized to non-crystalline iron oxide/hydroxide (known as ferrihydrite), exhibiting a characteristic signal of a distributed sexted (52) (Fig. 3F, inset). These results show that the [4Fe-4S]2+ cluster in PqqE-N is degraded to free irons via the semistable [2Fe-2S]2+ cluster by exposure to O2, like most radical SAM enzymes investigated so far (42, 53–56). Signal magnitudes of the two minor unassigned doublets observed in the Mössbauer spectra were little affected by exposure to O2.
Multiple sequence alignment of PqqE homologs
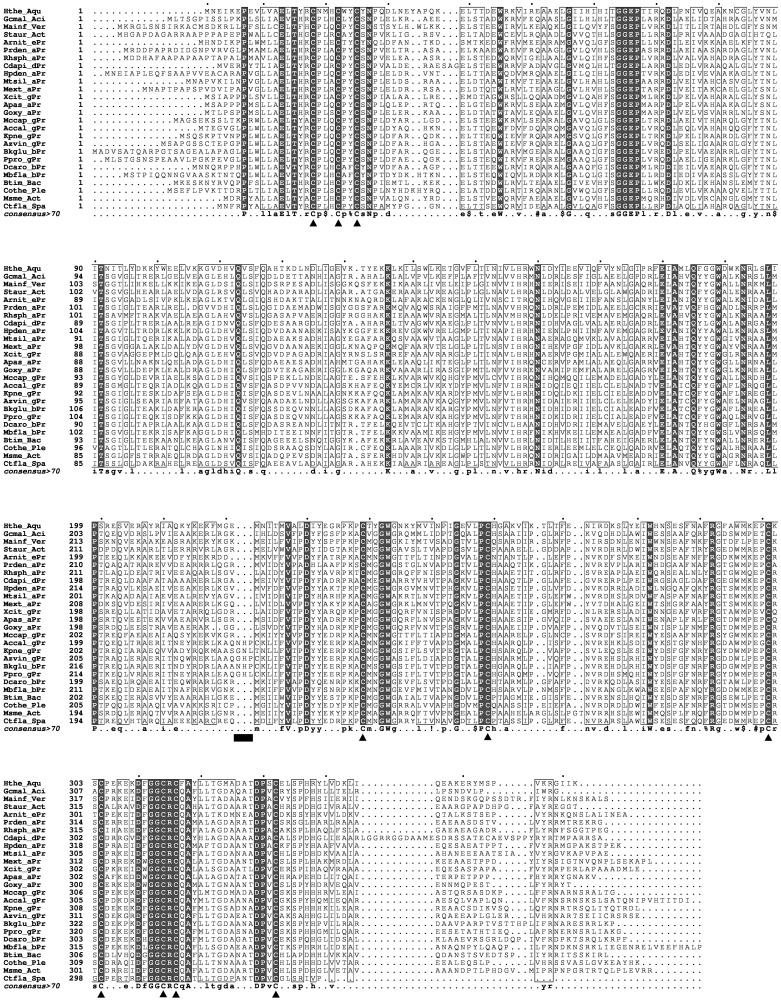
PqqE is a founding member of the subgroup designated ‘SPASM/twitch domain containing’ of radical SAM superfamily, together with subtilosin A synthesizing enzyme, AlbA; anaerobic sulfatase maturating enzyme, anSME; and mycofactocin synthesizing enzyme, MtfC (57, 58). About 12,270 members currently classified into this subgroup (35) are characterized by sharing the seven-cysteine motif (CX9–15GX4CXnCX2CX5CX3CXnC, where n is an unspecified number of amino acid residues) in the C-terminal half (59), which is likely involved in binding of auxiliary [4Fe-4S] clusters (60, 61). Indeed, multiple sequence alignment of 26 PqqE homologs from various bacterial species selected from the list of PqqE sequences (1055 sequences now registered) in the structure-function linkage database (35) reveals complete conservation of the seven-cysteine motif including an invariant glycine residue (Fig. 5). Thus, it is very likely that PqqE could contain auxiliary [4Fe-4S] clusters in addition to the radical SAM [4Fe-4S] cluster bound at the N-terminal signature CX3CX2C motif (43–47). The Fe/S contents measured in the reconstituted PqqE-N (Table I) support the presence of likely three [4Fe-4S] clusters. However, it is premature to conclude that only [4Fe-4S] clusters are bound to all the predicted Fe-S cluster-binding sites, as the Mössbauer spectral analysis indicated that not only [4Fe-4S]2+ but also [2Fe-2S]2+4Cys clusters are bound in the as-purified and reconstituted enzymes (Table II), although it is possible that the latter cluster is derived either from incomplete cluster building or from decomposition by oxygen. Besides these cysteine residues, the GGE motif, predicted to be involved in binding of SAM, is also totally conserved in all PqqE homologs (Fig. 5). Moreover, there is a unique 3-amino acid insertion located 17 amino acid residues upstream from the first cysteine residue of the seven-cysteine motif of PqqE sequences from K.pneumoniae and other closely related species (total 19 sequences out of the 141 sequences analysed), but not in other PqqE sequences. Although the function of these extra amino acid residues is unknown at the moment, it is tempting to speculate that the enzymes having this 3-amino acid insertion and showing high sequence identities to K. pneumoniae PqqE may be particularly sensitive to oxygen; however, further studies are needed to validate this assumption.
Fig. 5.
Multiple sequence alignment of PqqE homologs. Multiple sequence alignment was conducted for PqqE homologs from the following 26 bacterial species selected from the list of PqqE homologs registered in the structure-function linkage database (35) (from top to bottom): Hydrogenobacter thermophilus (Hthe_Aqu, GI: 502728863), Granulicella mallensis (Gcmal_Aci, GI: 504031789), Methylacidiphilum infernorum V4 (Mainf_Ver, GI: 189186106), Streptomyces aurantiacus JA 4570 (Staur_Act, GI: 514326545), Arcobacter nitrofigilis (Arnit_ePr, GI: 502901597), Paracoccus denitrificans PD1222 (Prden_aPr, GI: 119374856), Rhodobacter sphaeroides 2.4.1 (Rhsph_aPr, GI: 77388782), Chondromyces apiculatus DSM 436 (Cdapi_dPr, GI: 599567665), Hyphomicrobium denitrificans (Hpden_aPr, GI: 505410849), Methylocella silvestris (Mtsil_aPr, 501585892), M.extorquens AM1 (Mext_aPr, GI: 259016309), Xanthomonas citri subsp. citri UI6 (Xcit_gPr, GI: 780544983), Acetobacter pasteurianus (Apas_aPr, GI: 504270198), G.oxydans (Goxy_aPr, GI: 499571767), Methylococcus capsulatus (Mccap_gPr, GI: 499263186), Acinetobacter calcoaceticus (Accal_gPr, GI: 488042189), K.pneumoniae (Kpne_gPr, GI: 490288534), Azotobacter vinelandii (Azvin_gPr, GI: 502036033), Burkholderia glumae ATCC 33617 (Bkglu_bPr, GI: 779720809), P.protegens Pf-5 (Ppro_gPr, GI: 68347261), Dechloromonas aromatica (Dcaro_bPr, GI: 499606721), Methylobacillus flagellatus KT (Mbfla_bPr, GI: 91710020), Bacillus timonensis (Btim_Bac, GI: 497969539), Chroococcidiopsis thermalis (Cothe_Ple, GI: 504967445), Mycobacterium smegmatis (Msme_Act, GI: 500048568) and Chthoniobacter flavus (Ctfla_Spa, GI: 494037346). Bacterial classes are abbreviated by 3 characters and shown following underbars: Bac, Bacilli; Act, Actinobacteria; Ple, Pleurocapsales; aPr, Alphaproteobacteria; gPr, Gammaproteobacteria; Aci, Acidobacteria; dPr, Deltaproteobacteria; Spa, Spartobacteria; bPr, Betaproteobacteria; ePr, Epsilonproteobacteria; Ver, Verrucomicrobia (phylum); and Aqu, Aquificae. In the alignment, fully conserved residues are highlited by black background and the residues whose global similarity scores calculated from all groups are >0.7 (threshold) are boxed. Three Cys residues conserved in the N-terminal consensus motif (CX3CX2C) and seven Cys residues conserved in the C-terminal SPASM domain are indicated by triangles. The 3-amino acid insertion is marked with a black bar.
Discussion
It is widely accepted that almost all radical SAM enzymes utilize the 5′-deoxyadenosyl radical produced by the homolytic cleavage of SAM to catalyse a variety of subsequent coupled reactions (43–47). As described above, the [4Fe-4S]2+ cluster bound in the N-terminal signature CX3CX2C motif conserved in all radical SAM enzymes plays an essential role in this SAM cleavage reaction. It is also well-known that the [4Fe-4S]2+ clusters are easily oxidized by O2 and gradually degraded to different forms, often leading to the loss of enzymatic activity, particularly when the canonical [4Fe-4S]2+ cluster in radical SAM proteins is damaged. Hence, the sensitivity of [4Fe-4S]2+ clusters to O2 has been studied for several radical SAM enzymes (42, 62–65). For these O2-sensitive radical SAM enzymes, exposure to O2 generally causes degradation of the [4Fe-4S]2+ clusters within a short period (minutes to hours). The common intermediates of the O2-damaged [4Fe-4S]2+ cluster are [2Fe-2S]2+ or [3Fe-4S]0/1+, both of which are the semistable forms of Fe-S clusters in the presence of O2. For example, the [4Fe-4S]2+ cluster in the fumarate and nitrate reduction regulatory protein FNR of E.coli is degraded into [2Fe-2S]2+ after exposure to O2 (62). Of the two [4Fe-4S]2+ clusters contained in MOCS1A, the N-terminal one is rapidly degraded to [2Fe-2S]2+, whereas the C-terminal one is degraded to [3Fe-4S]0 (42). Rapid conversion of [4Fe-4S]2+ to [3Fe-4S]1+ in a ferrous ion transporter FeoC of K.pneumoniae has been observed within 1 h of O2-exposure, followed by complete degradation after 25 h (65). Moreover, the protein was found in an apo-form without any Fe-S clusters when it was purified in an atmospheric environment. Although the [4Fe-4S]3+ form, one-electron oxidized state of [4Fe-4S]2+ cluster, is generally vary labile and further easily degraded to [3Fe-4S]0, the high-potential iron-sulphur proteins (HiPIPs) contain an O2-tolerant [4Fe-4S]3+ cluster, when purified under aerobic conditions (66). It has been assumed that the [4Fe-4S]3+ cluster in HiPIPs is resistant to oxidative damage because of its location in the protein interior shielded from the solvent (66, 67). In marked contrast to the large majority of radical SAM proteins studied so far, PqqE from M.extorquens AM1 is unusually tolerant to oxygen; it can be purified even under fully aerobic conditions in a soluble form retaining significant SAM cleavage activity, as described in this article. To elucidate the mechanism of O2-tolerance of M.extorquens AM1 PqqE, it may be intriguing to compare the structural difference between the AM1 enzyme and PqqE from K.pneumoniae that is extremely sensitive to oxygen (21). The differences in oxygen sensitivity may depend on charge and spin states of the Fe-S clusters as well as on their spatial fixation by the enzyme’s scaffold (68).
In conclusion, we have shown here that the as-purified PqqE of M.extorquens AM1 is markedly tolerant to O2. The O2-tolerance is displayed unless the particularly O2-sensitive canonical [4Fe-4S]2+ cluster is fully installed by chemical reconstitution. In this respect, the described enzyme may serve as a convenient tool for studying the molecular mechanism of PQQ biosynthesis, avoiding the necessity of establishing strictly anaerobic conditions.
Supplementary Data
Supplementary Data are available at JB Online.
Funding
This research was supported by grants LO1204 and LO1305 from the National Program of Sustainability I from the Ministry of Education, Youth and Sports, Czech Republic and by the project POST-UP II, reg. No. CZ.1.07/2.3.00/30.0041. J.F., K.T. and T.Y. were supported by the Operational Program Education for Competitiveness - European Social Fund (project CZ.1.07 / 2.3.00 / 20.0165).
Conflict of Interest
None declared.
Supplementary Material
Glossary
Abbreviations
- 5′dA
5′-deoxyadenosine
- HiPIPs
high-potential iron-sulphur proteins
- IPTG
isopropyl-β-d-thiogalactopyranoside
- PQQ
pyrroloquinoline quinone
- SAM
S-adenosyl-l-methionine
References
- 1.Duine J.A. (1991) Quinoproteins: enzymes containing the quinonoid cofactor pyrroloquinoline quinone, topaquinone or tryptophan-tryptophan quinone. Eur. J. Biochem. 200, 271–284 [DOI] [PubMed] [Google Scholar]
- 2.Adachi O., Ano Y., Toyama H., Matsushita K. (2007) Biooxidation with PQQ- and FAD-dependent dehydrogenases. in Modern Biooxidation (Schmid R.D., Urlacher V.B. eds.) pp. 1–41. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 3.Goosen N., Horsman H.P., Huinen R.G., de Groot A., van de Putte P. (1989) Genes involved in the biosynthesis of PQQ from Acinetobacter calcoaceticus. Antonie Van Leeuwenhoek 56, 85–91 [DOI] [PubMed] [Google Scholar]
- 4.Biville F., Turlin E., Gasser F. (1989) Cloning and genetic analysis of six pyrroloquinoline quinone biosynthesis genes in Methylobacterium organophilum DSM 760. Microbiology 135, 2917–2929 [DOI] [PubMed] [Google Scholar]
- 5.Meulenberg J.J., Sellink E., Riegman N.H., Postma P.W. (1992) Nucleotide sequence and structure of the Klebsiella pneumoniae pqq operon. Mol. Gen. Genet. 232, 284–294 [DOI] [PubMed] [Google Scholar]
- 6.Schnider U., Keel C., Voisard C., Défago G., Haas D. (1995) Tn5-directed cloning of pqq genes from Pseudomonas fluorescens CHA0: mutational inactivation of the genes results in overproduction of the antibiotic pyoluteorin. Appl. Environ. Microbiol. 61, 3856–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramette A., Frapolli M., Saux M.F.-L., Gruffaz C., Meyer J.-M., Défago G., Sutra L., Moënne-Loccoz Y. (2011) Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 34, 180–188 [DOI] [PubMed] [Google Scholar]
- 8.Toyama H., Chistoserdova L., Lidstrom M.E. (1997) Sequence analysis of pqq genes required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AM1 and the purification of a biosynthetic intermediate. Microbiology 143, 595–602 [DOI] [PubMed] [Google Scholar]
- 9.Kim C.H., Han S.H., Kim K.Y., Cho B.H., Kim Y.H., Koo B.S., Kim Y.C. (2003) Cloning and expression of pyrroloquinoline quinone (PQQ) genes from a phosphate–solubilizing bacterium Enterobacter intermedium. Curr. Microbiol. 47, 457–461 [DOI] [PubMed] [Google Scholar]
- 10.Pavan M.E., Franco R.J., Rodriguez J.M., Gadaleta P., Abbott S.L., Janda J.M., Zorzópulos J. (2005) Phylogenetic relationships of the genus Kluyvera: transfer of Enterobacter intermedius Izard et al. 1980 to the genus Kluyvera as Kluyvera intermedia comb. nov. and reclassification of Kluyvera cochleae as a later synonym of K. intermedia. Int. J. Syst. Evol. Microbiol. 55, 437–442 [DOI] [PubMed] [Google Scholar]
- 11.Hölscher T., Görisch H. (2006) Knockout and overexpression of pyrroloquinoline quinone biosynthetic genes in Gluconobacter oxydans 621H. J. Bacteriol. 188, 7668–7676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Y.-Q., Bonnot F., Imsand E.M., RoseFigura J.M., Sjölander K., Klinman J.P. (2012) Distribution and properties of the genes encoding the biosynthesis of the bacterial cofactor, pyrroloquinoline quinone. Biochemistry 51, 2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klinman J.P., Bonnot F. (2014) Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ. Chem. Rev. 114, 4343–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velterop J.S., Sellink E., Meulenberg J.J., David S., Bulder I., Postma P.W. (1995) Synthesis of pyrroloquinoline quinone in vivo and in vitro and detection of an intermediate in the biosynthetic pathway. J. Bacteriol. 177, 5088–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puehringer S., Metlitzky M., Schwarzenbacher R. (2008) The pyrroloquinoline quinone biosynthesis pathway revisited: a structural approach. BMC Biochem. 9, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magnusson O.T., Toyama H., Saeki M., Rojas A., Reed J.C., Liddington R.C., Klinman J.P., Schwarzenbacher R. (2004) Quinone biogenesis: structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone. Proc. Natl. Acad. Sci. U.S.A. 101, 7913–7918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnusson O.T., Toyama H., Saeki M., Schwarzenbacher R., Klinman J.P. (2004) The structure of a biosynthetic intermediate of pyrroloquinoline quinone (PQQ) and elucidation of the final step of PQQ biosynthesis. J. Am. Chem. Soc. 126, 5342–5343 [DOI] [PubMed] [Google Scholar]
- 18.Houck D.R., Hanners J.L., Unkefer C.J. (1991) Biosynthesis of pyrroloquinoline quinone. 2. Biosynthetic assembly from glutamate and tyrosine. J. Am. Chem. Soc. 113, 3162–3166 [Google Scholar]
- 19.Toyama H., Lidstrom M. E. (1998) pqqA is not required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AM1. Microbiology 144, 183–191 [DOI] [PubMed] [Google Scholar]
- 20.Vuilleumier S., Chistoserdova L., Lee M.-C., Bringel F., Lajus A., Zhou Y., Gourion B., Barbe V., Chang J., Cruveiller S., Dossat C., Gillett W., Gruffaz C., Haugen E., Hourcade E., Levy R., Mangenot S., Muller E., Nadalig T., Pagni M., Penny C., Peyraud R., Robinson D.G., Roche D., Rouy Z., Saenampechek C., Salvignol G., Vallenet D., Wu Z., Marx C.J., Vorholt J.A., Olson M.V., Kaul R., Weissenbach J., Médigue C., Lidstrom M.E. (2009) Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS One 4, e5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wecksler S.R., Stoll S., Tran H., Magnusson O.T., Wu S.-P., King D., Britt R.D., Klinman J.P. (2009) Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry 48, 10151–10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latham J.A., Iavarone A.T., Barr I., Juthani P.V., Klinman J.P. (2015) PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway. J. Biol. Chem., 290, 12908–12918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okubo Y., Skovran E., Guo X., Sivam D., Lidstrom M.E. (2007) Implementation of microarrays for Methylobacterium extorquens AM1. Omics 11, 325–340 [DOI] [PubMed] [Google Scholar]
- 24.Fish W.W. (1988) Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 158, 357–364 [DOI] [PubMed] [Google Scholar]
- 25.Marmur J. (1961) A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3, 208–218 [Google Scholar]
- 26.Sambrook J., Russell D.W. (2001) Molecular Cloning: a Laboratory Manual. 3rd edn Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Flühe L., Knappe T.A., Gattner M.J., Schäfer A., Burghaus O., Linne U., Marahiel M.A. (2012) The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 8, 350–357 [DOI] [PubMed] [Google Scholar]
- 28.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 29.Lanz N.D., Grove T.L., Gogonea C.B., Lee K.-H., Krebs C., Booker S.J. (2012) RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins. Methods Enzymol. 516, 125–152 [DOI] [PubMed] [Google Scholar]
- 30.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. (2005) Protein identification and analysis tools on the ExPASy server. in The Proteomics Protocols Handbook, Humana Press . (Walker J.M. ed.) pp. 571–607. Humana Press, New York [Google Scholar]
- 31.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 32.Beinert H. (1983) Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 33.Pechoušek J., Jančík D., Frydrych J., Navařík J., Novák P. (2012) Setup of Mössbauer spectrometers at RCPTM. in Mössbauer Spectroscopy in Materials Science. (Tucek J., Machala L. eds.). AIP Conference Proceedings , Vol. 1489 pp. 186–193. Czech Republic, Olomouc [Google Scholar]
- 34.Klencsár Z., Kuzmann E., Vértes A. (1996) User-friendly software for Mössbauer spectrum analysis. J. Radioanal. Nucl. Chem. 210, 105–118 [Google Scholar]
- 35.Akiva E., Brown S., Almonacid D.E., Barber A.E., Custer A.F., Hicks M.A., Huang C.C., Lauck F., Mashiyama S.T., Meng E.C., Mischel D., Morris J.H., Ojha S., Schnoes A.M., Stryke D., Yunes J.M., Ferrin T.E., Holliday G.L., Babbitt P.C. (2014) The structure-function linkage database. Nucleic Acids Res. 42, D521–D530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robert X., Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tegel H., Tourle S., Ottosson J., Persson A. (2010) Increased levels of recombinant human proteins with the Escherichia coli strain Rosetta (DE3). Protein Expr. Purif. 69, 159–167 [DOI] [PubMed] [Google Scholar]
- 40.Zheng L., Cash V.L., Flint D.H., Dean D.R. (1998) Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273, 13264–13272 [DOI] [PubMed] [Google Scholar]
- 41.Agar J.N., Krebs C., Frazzon J., Huynh B.H., Dean D.R., Johnson M.K. (2000) IscU as a scaffold for iron-sulfur cluster biosynthesis: sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry 39, 7856–7862 [DOI] [PubMed] [Google Scholar]
- 42.Hänzelmann P., Hernández H.L., Menzel C., García-Serres R., Huynh B.H., Johnson M.K., Mendel R.R., Schindelin H. (2004) Characterization of MOCS1A, an oxygen-sensitive iron-sulfur protein involved in human molybdenum cofactor biosynthesis. J. Biol. Chem. 279, 34721–34732 [DOI] [PubMed] [Google Scholar]
- 43.Frey P.A., Hegeman A.D., Ruzicka F.J. (2008) The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 [DOI] [PubMed] [Google Scholar]
- 44.Roach P. L. (2011) Radicals from S-adenosylmethionine and their application to biosynthesis. Curr. Opin. Chem. Biol. 15, 267–275 [DOI] [PubMed] [Google Scholar]
- 45.Booker S.J. (2012) Radical SAM enzymes and radical enzymology. Biochim. Biophys. Acta 1824, 1151–1153 [DOI] [PubMed] [Google Scholar]
- 46.Broderick J.B., Duffus B.R., Duschene K.S., Shepard E.M. (2014) Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J., Woldring R.P., Román-Meléndez G.D., McClain A.M., Alzua B.R., Marsh E.N.G. (2014) Recent advances in radical SAM enzymology: new structures and mechanisms. ACS Chem. Biol. 9, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandelia M.-E., Lanz N.D., Booker S.J., Krebs C. (2015) Mössbauer spectroscopy of Fe/S proteins. Biochim. Biophys. Acta 1853, 1395–1405 [DOI] [PubMed] [Google Scholar]
- 49.Perche-Letuvée P., Kathirvelu V., Berggren G., Clemancey M., Latour J.-M., Maurel V., Douki T., Armengaud J., Mulliez E., Fontecave M., Garcia-Serres R., Gambarelli S., Atta M. (2012) 4-Demethylwyosine synthase from Pyrococcus abyssi is a radical-S-adenosyl-L-methionine enzyme with an additional [4Fe-4S]+2 cluster that interacts with the pyruvate co-substrate. J. Biol. Chem. 287, 41174–41185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandramouli K., Unciuleac M.-C., Naik S., Dean D.R., Huynh B.H., Johnson M.K. (2007) Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry 46, 6804–6811 [DOI] [PubMed] [Google Scholar]
- 51.Mulder D.W., Ortillo D.O., Gardenghi D.J., Naumov A.V., Ruebush S.S., Szilagyi R.K., Huynh B.-H., Broderick J.B., Peters J.W. (2009) Activation of HydAΔEFG requires a preformed [4Fe-4S] cluster. Biochemistry 48, 6240–6248 [DOI] [PubMed] [Google Scholar]
- 52.Murad E., Schwertmann U. (1980) The Mössbauer spectrum of ferrihydrite and its relations to those of other iron oxide. Am. Mineral. 65, 1044–1049 [Google Scholar]
- 53.Duin E.C., Lafferty M.E., Crouse B.R., Allen R.M., Sanyal I., Flint D.H., Johnson M.K. (1997) [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry 36, 11811–11820 [DOI] [PubMed] [Google Scholar]
- 54.Broderick J.B., Duderstadt R.E., Fernandez D.C., Wojtuszewski K., Henshaw T.F., Johnson M.K. (1997) Pyruvate formate-lyase activating enzyme is an iron−sulfur protein. J. Am. Chem. Soc. 119, 7396–7397 [Google Scholar]
- 55.Ollagnier S., Meier C., Mulliez E., Gaillard J., Schuenemann V., Trautwein A.X., Mattioli T., Lutz M., Fontecave M. (1999) Assembly of 2Fe-2S and 4Fe-4S clusters in the anaerobic ribonucleotide reductase from Escherichia coli. J. Am. Chem. Soc. 121, 6344–6350 [Google Scholar]
- 56.Pierrel F., Hernandez H.L., Johnson M.K., Fontecave M., Atta M. (2003) MiaB protein from Thermotoga maritima. Characterization of an extremely thermophilic tRNA-methylthiotransferase. J. Biol. Chem. 278, 29515–29524 [DOI] [PubMed] [Google Scholar]
- 57.Haft D.H., Basu M.K. (2011) Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J. Bacteriol. 193, 2745–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haft D.H. (2011) Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners. BMC Genomics 12, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjdia A., Subramanian S., Leprince J., Vaudry H., Johnson M.K., Berteau O. (2010) Anaerobic sulfatase-maturating enzyme: a mechanistic link with glycyl radical-activating enzymes? FEBS J. 277, 1906–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grove T.L., Lee K.-H., St Clair J., Krebs C., Booker S.J. (2008) In vitro characterization of AtsB, a radical SAM formylglycine-generating enzyme that contains three [4Fe-4S] clusters. Biochemistry 47, 7523–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldman P.J., Grove T.L., Sites L.A., McLaughlin M.I., Booker S.J., Drennan C.L. (2013) X–ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc. Natl. Acad. Sci. U.S.A. 110, 8519–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khoroshilova N., Popescu C., Münck E., Beinert H., Kiley P.J. (1997) Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. U.S.A. 94, 6087–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crack J.C., den Hengst C.D., Jakimowicz P., Subramanian S., Johnson M.K., Buttner M.J., Thomson A.J., Le Brun N.E. (2009) Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD. Biochemistry 48, 12252–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tilley G.J., Camba R., Burgess B.K., Armstrong F.A. (2001) Influence of electrochemical properties in determining the sensitivity of [4Fe-4S] clusters in proteins to oxidative damage. Biochem. J. 360, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsueh K.-L., Yu L.-K., Chen Y.-H., Cheng Y.-H., Hsieh Y.-C., Ke S.-C., Hung K.-W., Chen C.-J., Huang T.-H. (2013) FeoC from Klebsiella pneumoniae contains a [4Fe-4S] cluster. J. Bacteriol. 195, 4726–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benning M.M., Meyer T.E., Rayment I., Holden H.M. (1994) Molecular structure of the oxidized high-potential iron-sulfur protein isolated from Ectothiorhodospira vacuolata. Biochemistry 33, 2476–2483 [DOI] [PubMed] [Google Scholar]
- 67.Agarwal A., Li D., Cowan J.A. (1995) Role of aromatic residues in stabilization of the [Fe4S4] cluster in high-potential iron proteins (HiPIPs): physical characterization and stability studies of Tyr-19 mutants of Chromatium vinosum HiPIP. Proc. Natl. Acad. Sci. U.S.A. 92, 9440–9444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruska M.K., Stiebritz M.T., Reiher M. (2013) Analysis of differences in oxygen sensitivity of Fe-S clusters. Dalton Trans. 42, 8729–8735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.