Abstract
Amino-terminal enhancer of split (Aes) is a member of Groucho/Transducin-like enhancer (TLE) family. Aes is a recently found metastasis suppressor of colorectal cancer (CRC) that inhibits Notch signalling, and forms nuclear foci together with TLE1. Although some Notch-associated proteins are known to form subnuclear bodies, little is known regarding the dynamics or functions of these structures. Here, we show that Aes nuclear foci in CRC observed under an electron microscope are in a rather amorphous structure, lacking surrounding membrane. Investigation of their behaviour during the cell cycle by time-lapse cinematography showed that Aes nuclear foci dissolve during mitosis and reassemble after completion of cytokinesis. We have also found that heat shock cognate 70 (HSC70) is an essential component of Aes foci. Pharmacological inhibition of the HSC70 ATPase activity with VER155008 reduces Aes focus formation. These results provide insight into the understanding of Aes-mediated inhibition of Notch signalling.
Keywords: colorectal cancer, heat shock protein, Notch signalling, nuclear matrix, transcription
Colorectal cancer (CRC) is caused by genetic mutations in such genes as adenomatous polyposis coli (APC), KRAS, TP53, PIK3CA and FBXW7, and epigenetic silencing of tumour suppressor genes (1). Through these genetic and epigenetic alterations, normal colonic epithelial cells turn into cancer cells; ‘adenoma-carcinoma’ sequence (2). However, our understanding is still fragmentary when it comes to how cancer metastasizes. We have previously reported that Amino-terminal enhancer of split (Aes) as a novel metastasis suppressor of CRC (3). Aes is a truncated member of the Groucho/transducin-like enhancer of split (TLE) family, containing only the N-terminal Q (glutamine-rich) and glycine/proline-rich (GP) domains, and lacking the C-terminal three domains present in full-length Groucho/TLE proteins (4). Aes is an endogenous repressor of Notch signalling transcription (3); genetic loss of Aes in the intestinal epithelium of Apc+/Δ716 mice, a mouse model of familial adenomatous polyposis (5), causes invasion and intravasation of their intestinal tumours (3).
Notch signalling plays critical roles in development of both vertebrates and invertebrates (6, 7). Upon binding of a Notch ligand to the Notch receptor, the latter is cleaved by an ADAM protease and γ-secretase in succession, releasing the Notch intracellular domain (NICD). NICD then moves into the nucleus, and associates with the transcription factor Recombination signal binding protein for immunoglobulin kappa J region (RBPJ) together with Mastermind-like (MAML), thereby activating transcription of several genes (8, 9). Aes inhibits Notch signalling in CRC cells by converting active RBPJ transcription complex into an inactive form (3). Aes also forms nuclear matrix (NM)-associated foci together with TLE1, a full-length member of the Groucho/TLE family. In these foci, we can also find Notch transcription factor/cofactors RBPJ, MAML and NICD (3). There are some reports of Notch signalling-associated nuclear bodies in mammalian cells. For example, Notch-1 intracellular domain (N1ICD) and RBPJ are found in subnuclear bodies (10). Nuclear corepressor complex proteins SHARP, SKIP and SMRT also colocalize with RBPJ foci that are associated with NM and histone deacetylases (HDACs) (called matrix-associated deacetylase (MAD) body) (11–14). However, there is limited characterization of these Notch-associated nuclear foci.
Here, we have studied Aes nuclear foci by electron microscopy (EM) and time-lapse cinematography. Under an EM, Aes nuclear foci showed amorphous structure, lacking surrounding membrane. They dissolved during mitosis and reassembled after completion of cytokinesis. We have also found that phosphorylation of the serine residue 239 of TLE1 (TLE1 S239) is important for the focus formation. In addition, heat shock cognate 70 (HSC70) is one of the essential components of Aes nuclear foci. Namely, inhibition of the HSC70 ATPase activity reduces nuclear focus formation significantly.
Materials and Methods
Cell lines, animals and reagents
HCT116, RKO and HEK293T cells were obtained from American type culture collection. The identities of these cell lines were confirmed in July 2014 by short tandem repeat (STR) analysis (Takara Bio CDM Center, Mié, Japan). Apc+/Δ716 C57BL/6 mice have been described previously (5). Dithiobis [succinimidylpropionate] (DSP) and HSC70 inhibitor VER155008 were purchased from Pierce (Rockford, IL) and Tocris Bioscience (Bristol, UK), respectively. For transient transfection, we used Flag-tagged Aes and AU1-tagged TLE1 cDNAs placed in pCAG vector (3, 15), Aes cDNA placed in pAcGFP (Clontech, Palo Alto, CA) and myc-tagged Rbpj-associated molecule domain and intracellular domain of the Notch receptor (RAMIC) (a recombinant NICD) in pEFBosneo (16). cDNAs of Aes truncation mutants were constructed by PCR using wild-type Aes cDNA as a template, followed by sequencing. Expression construct for TLE1(S239A) was published previously (17).
Time-lapse cinematography
cDNAs encoding AcGFP-tagged Aes on its N-terminus and TLE1 were introduced into HCT116 cells using Lipofectamine LTX (Invitrogen, Carlsbad, CA) 24 h before observation. Fluorescent photomicrographs were taken every 3 min under a TCS SP5 confocal microscope (Leica Microsystems, Buffalo Grove, IL), and cinematographs were constructed in QuickTime software.
Immunofluorescence analysis
Adenomas from Apc+/Δ716 mice and cells in culture expressing Aes and/or TLE1 were processed and analysed as described (3). Antibodies sources: anti-Flag from Sigma (St Louis, MO), anti-AU1 from Covance (Princeton, NJ), anti-SC35 and anti-HSC70 from Abcam (Cambridge, UK), anti-C23 (nucleolin) from Santa Cruz (Santa Cruz, CA), anti-fibrillarin from Novus Biologicals (Littleton, CO), anti-Aes (3) and anti-HDAC3 from Cell Signalling Technology (Danvers, MA).
Cell fractionation
Cell fractionation was performed as reported previously (18, 19) with some modifications. Precise description can be seen in Supplementary Materials and Methods.
Mass spectrometry analysis
Mass spectrometry (MS) was performed as previously described (20). Briefly, after protein samples were separated by sodium dodecyl sulfate-polyacrylamde gel electrophoresis (SDS-PAGE), they were visualized by silver staining and excised as protein bands. These protein bands were digested with trypsin (Promega) in a buffer containing 50 mM ammonium bicarbonate (pH 8.0) and 2% acetonitrile overnight at 37°C and subjected to matrix-assisted laser desorption/ionization time-of-flight MS using an Ultraflex TOF/TOF (Bruker Daltonics, Billerica, MA).
Electron microscopy
Cells with nuclear foci were fixed with 4% paraformaldehyde and 2% glutaraldehyde at 4°C for 2 h. After incubation with 1% osmium tetroxide at 4°C for 1 h, samples were dehydrated and embedded in epoxy resin. Sections were cut and stained with uranyl acetate and lead citrate as reported (21, 22).
Luciferase reporter assay
Cells were transfected with a Notch signalling reporter construct pGa981-6 (16) and phRluc-CMV vector (Promega), using Lipofectamine LTX (Invitrogen). Luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega) with a MITHORAS LB 940 (Berthold Technologies, Bad Wildbad, Germany). The activities of firefly luciferase were normalized against those of Renilla luciferase.
Results and Discussion
Aes nuclear foci disappear during mitosis and lack surrounding membrane
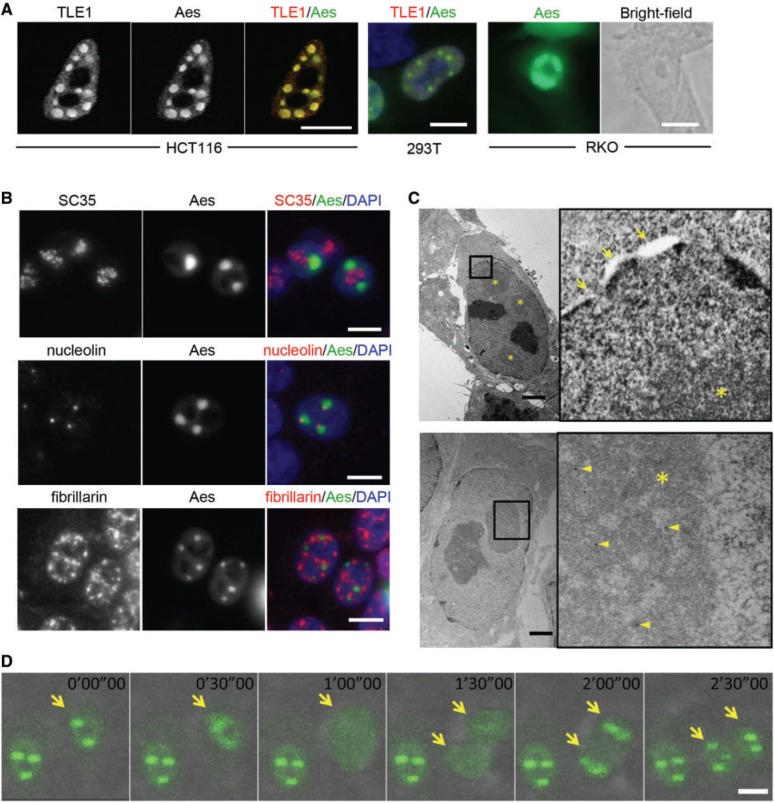
We have reported that Aes forms nuclear foci together with TLE1, one of the Drosophila Groucho homologues, in HCT116 and RKO human CRC cells and HEK293T cells in culture (Fig. 1A) as well as in adenoma cells of Apc+/Δ716 mouse in vivo (Supplementary Fig. S1) (3). Similar focal structures have been reported in the nucleus such as splicing bodies or nucleoli (23). To investigate the possible relationship between these nuclear structures and Aes nuclear foci, we performed immunofluorescence studies. As shown in Fig. 1B, Aes foci were distinct from splicing bodies (containing SC35) and nucleoli (containing nucleolin and fibrillarin). Interestingly, however, each nucleolin focus always colocalized with a part of an Aes focus (Fig. 1B, middle row). Because nucleolin is a matrix attached region-binding protein (24), it is possible that Aes nuclear foci bind to the NM through nucleolin. We also found that Aes nuclear foci contained HDAC3 (ref. 3 and Supplementary Fig. S1). Therefore, it is conceivable that Aes foci share some characteristics with MAD bodies. Further study is required to determine how much these two foci are similar structurally and functionally.
Fig. 1.
Characterization of Aes nuclear foci. (A) AU1-tagged TLE1 (AU1-TLE1) and Flag-tagged Aes (Flag-Aes) were expressed and detected by fluorescent antibodies for each tag in HCT116 (left), HEK293T (middle) and RKO (right) cells. Note that the nuclear shape and size are different among the cell lines shown. (B) SC35 (a marker for splicing bodies), nucleolin and fibrillarin (both nucleolar markers, but localized to different subregions) were immunostained in HCT116 cells where AcGFP-tagged Aes (AcGFP-Aes) and TLE1 were overexpressed and immunostained simultaneously. (C) Flag-Aes and TLE1 were expressed in HCT116 cells and photographed by EM (upper photos). For immuno-EM (lower photos), rabbit anti-Flag and 12-nm colloidal Gold-conjugated anti-rabbit antibodies were used to localize Flag-Aes (arrowheads). Boxed areas in left panels were enlarged (right panels). Arrows, nuclear membrane; asterisk, Aes nuclear foci. (D) Chronological changes of Aes nuclear foci in HCT116 cells where AcGFP-Aes and TLE1 were expressed. Arrows, a cancer cell with Aes nuclear foci that went through a cell division during the recording, and its daughter cells. Scale bars, 10 µm (A, B, D), 2 µm (C).
To further characterize the structure of Aes nuclear foci, we next examined them under an EM. As shown in Fig. 1C, the foci showed rather amorphous structures, lacking surrounding membranes (* in Fig. 1C, top photos). Immuno-EM confirmed that these foci contained Aes protein (Fig. 1C, arrowheads in bottom photos). Because nuclear structures such as nucleoli and heterochromatins show dynamic behaviours during mitosis (25), we next investigated Aes foci throughout the cell cycle using AcGFP-tagged Aes and time-lapse cinematography. Previously, we confirmed that AcGFP-tagged Aes as well as Flag-tagged Aes formed focal structures in the nucleus as wild-type Aes did (3). Notably, these Aes foci showed dynamic changes during the cell cycle: dissolving during mitosis and reassembling after cytokinesis (Fig. 1D and Supplementary Movie). These dynamic features of Aes foci suggest that their physicochemical nature and subnuclear localization are regulated during the cell cycle like other cell cycle-associated proteins (see later).
Aes Q domain and TLE1 are required for Aes nuclear focus formation
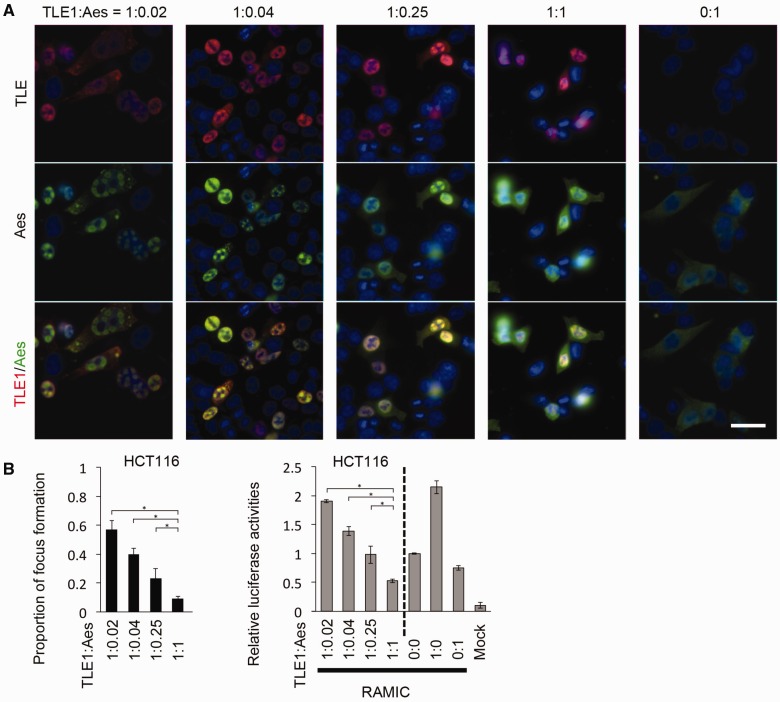
When TLE1 is not expressed exogenously, overexpressed Aes alone does not form nuclear foci but distributes diffusely in the cytoplasm of cultured cells (Fig. 2A; ratio of expression plasmids used in transient transfections, TLE1:Aes = 0:1). When overexpressed with TLE1 that is a transcription cofactor and exists mainly in the nucleoplasm, Aes moves into the nucleus and can form nuclear foci, although some remain diffusely in the nucleoplasm (Fig. 2A) (3). Accordingly, we investigated how TLE1 helps the formation of Aes foci. Unexpectedly, we found that the frequency of Aes focus formation decreased with increasing amounts of Aes relative to that of TLE1, with the Notch signalling transcription activity also decreasing concomitantly as determined with a luciferase reporter (Fig. 2A and B). Given our earlier observation that coexpression of Aes and TLE1 caused nuclear focus formation (3), it seemed counterintuitive that focus formation was decreased with relatively higher amounts of Aes. This may be explained that the soluble form Aes-TLE1 complex is responsible for the inhibitory effect on the Notch signalling-mediated transcription, with a fraction of Aes-TLE1 complex can be precipitated as nuclear foci. Such an interpretation is consistent with an example of toxicity caused by the soluble amyloid-beta (Aβ) oligomers rather than by its precipitates (26). We coin this interpretation as ‘wine tannin hypothesis’ because it is not the precipitated tannin, but rather the soluble form that causes an astringent taste in wine. This hypothesis may also be supported by our observation that Aes foci are completely dissolved during mitosis. It is interesting to speculate that Aes is involved in silencing Notch-dependent transcription in the mitotic phase during which mRNA transcription is reduced significantly (27).
Fig. 2.
Optimal focus formation in the presence of Aes and TLE1. (A) AU1-TLE1 and Flag-Aes were expressed in HCT116 cells at the ratios shown beneath the panels. When only Aes was expressed exogenously, focus formation could not be detected (0:1). (B) Upper graph shows the efficiency of Aes nuclear focus formation (focus-forming cells among the transfected cells) in HCT116 cells at the ratios of TLE1 and Aes shown in (A). Lower graph shows the analysis of Notch-mediated transcriptional activation using a luciferase reporter construct (pGa981-6; refs. 3 and 16) in HCT116 cells transfected with Aes and TLE1. (*P < 0.01)
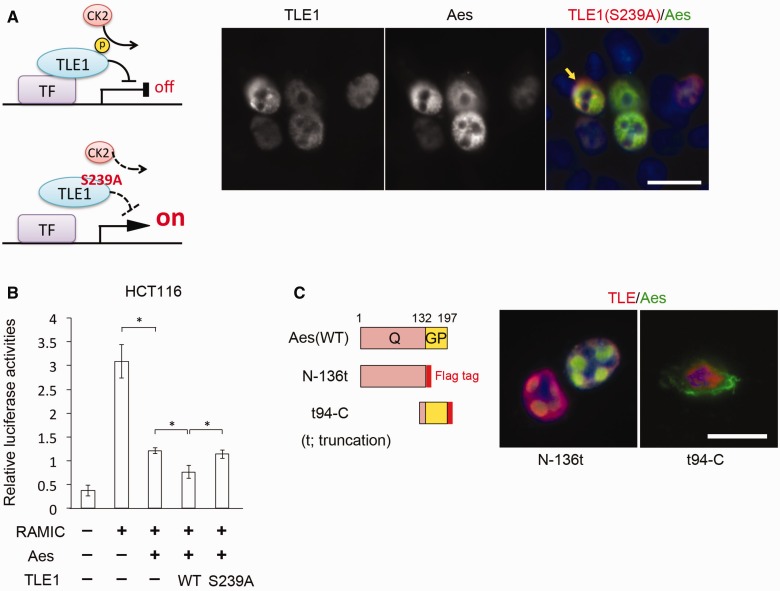
It is known that serine residue 239 (S239) of TLE1 is phosphorylated by casein kinase II (CK2), and its phosphorylation is important for TLE1 to repress transcription (Fig. 3A left) (17, 28). Therefore, we studied whether phosphorylation of TLE1(S239) was critical for the inhibition of Notch signalling-mediated transcription and/or focus formation with Aes. To this end, we overexpressed a mutant form of TLE1 of which S239 was converted to a structurally similar but unphosphorylatable amino acid alanine (S239A) (28) in HCT116 CRC cells. Compared with wild-type TLE1, S239A mutant did not show inhibition of Notch signalling transcription when co-expressed with Aes (Fig. 3B). At the same time, the cells expressing TLE1(S239A) showed significantly lower frequency of Aes focus formation than those expressing the wild type (2% versus 10% of focus forming cells among the transfected cells when the equal amounts of Aes and TLE1 cDNAs were transfected. A representative photograph is shown in Fig. 3A right). These results may appear contradictory to the ‘wine tannin hypothesis’ if the soluble form of Aes-TLE1 complex is responsible for transcriptional inhibition. We speculate that being soluble is a necessary but not the sufficient feature for Aes-TLE1 to inhibit transcription, and that TLE1(S239A) is unable to interact with the Rbpj transcription factor complex as with other transcription factors (17, 28).
Fig. 3.
Aes focus formation with mutated proteins. (A) Schematic representation of TLE1 transcriptional repression through phosphorylation of S239 of TLE1 (left), and an immunostaining photomicrograph when TLE1(S239A) was expressed together with AcGFP-Aes in HCT116 cells (right). TLE1(S239A) failed to form foci (∼2% of focus-forming cells among all transfected cells when the same amount of AcGFP-Aes and TLE1 cDNA were transfected). The arrow points to the cell expressing both TLE1(S239A) and Aes at high levels. TF, transcription factor; CK2, casein kinase II; p, phosphorylation of TLE1 S239. (B) Analysis of Notch-mediated transcriptional activation using a luciferase reporter assay (pGa981-6) (3) in HCT116 cells transfected with Aes and TLE1 (wild type or S239A) (*P < 0.01). WT, wild type. Values indicate the means ± SD (C) Schematic drawing of Aes deletion mutants (left) and immunostaining photomicrographs when Aes mutation constructs were expressed together with TLE1 in HCT116 cells (centre and right). t, truncation. WT, wild type. Values indicate the means ± SD Scale bars, 20 µm (A, C), 10 µm (D).
To study the interaction of Aes with TLE1 further, we next constructed a pair of Aes deletion mutants each of which lacked C-terminal (N-136t; t for truncation) or N-terminal (t94-C) residues (Fig. 3C left), and exogenously expressed them with TLE1 in HCT116 CRC cells. We found that Aes(N-136t) efficiently formed nuclear foci, whereas Aes(t94-C) did not (Fig. 3C right). This result is consistent with the observation that Aes and Groucho/TLE proteins oligomerize through their Q domains (29). Taken together, these results suggest that formation of Aes nuclear foci requires both the N-terminal Q domain of Aes and phosphorylation of S239 in TLE1 outside of its Q-GP domains.
Heat shock cognate 70 (HSC70) is an integral component of Aes nuclear foci
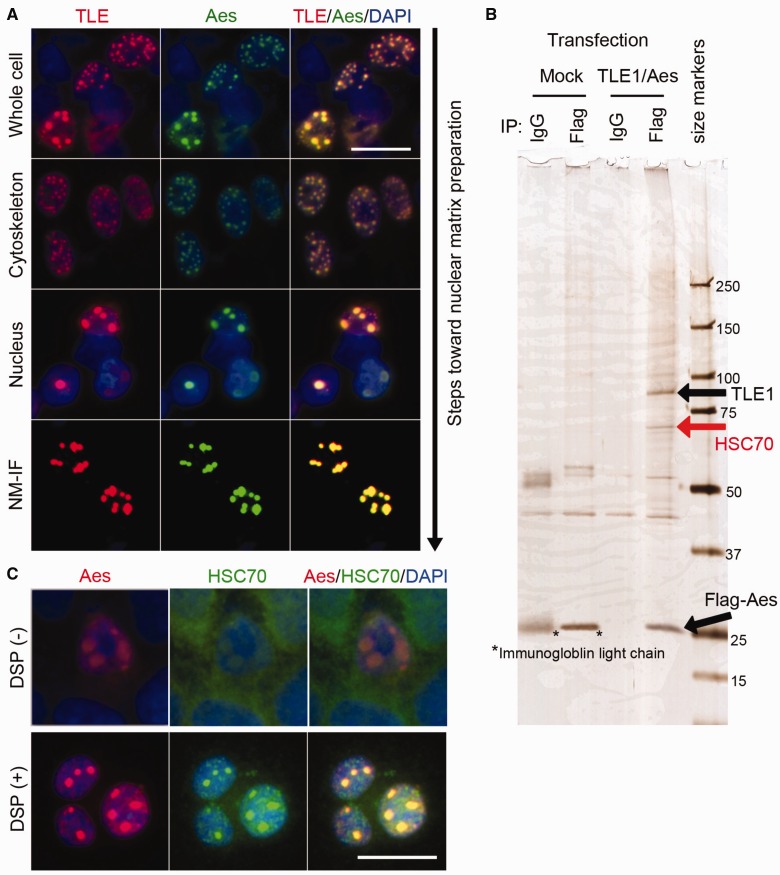
We have recently reported that Aes nuclear foci are associated with insoluble NM, and contain transcription factor/cofactors involved in Notch signalling including RBPJ, MAML1 and NICD, as well as TLE1, when all of them were exogenously expressed in cultured cells (3). To identify additional endogenous components of Aes nuclear foci, we overexpressed Flag-Aes and TLE1 in HCT116 cells, and isolated the NM fraction. To prevent loss of the focus components during the preparation process, we used chemical cross-linker DSP prior to subcellular fractionation. With DSP cross-linking, Aes nuclear foci remained associated with the insoluble NM while residual Aes in the nucleoplasm was washed away (Fig. 4A). We then subjected the NM fraction to immunoprecipitation using anti-Flag antibody, followed by SDS-PAGE (Fig. 4B). Silver staining showed an ∼70 kDa protein that was co-immunoprecipitated with Flag-Aes. A mass-spectrometry analysis identified it as HSC70, one of the heat shock proteins (HSPs). We confirmed colocalization of endogenous HSC70 with the Aes nuclear foci by immunofluorescence analysis of HCT116 cells (Fig. 4C).
Fig. 4.
HSC70 is a component of Aes foci. (A) Immunostaining photomicrographs of Aes nuclear foci associated with the NM. AU1-TLE1 and Flag-Aes were expressed in HCT116 cells and the cells were permeabilized and washed in cytoskeleton (CSK), EXT (extraction) and digestion (DIG) buffers as described in Supplementary Materials and Methods, followed by immunostaining. CSK, Nucleus and NM-intermediate filament (NM-IF) indicate the products obtained after washing with CSK, EXT and DIG buffers, respectively. (B) Silver-stained SDS-PAGE gel loaded with immunoprecipitates of NM fractions. After HCT116 cells were transfected with Flag-Aes and TLE1 or Mock empty control, and treated with 2 mM DSP, the NM fraction was prepared and immunoprecipitated with anti-Flag antibody or control IgG. Immunoprecipitates were separated by SDS-PAGE, followed by silver staining. Size markers, kDa. (C) HCT116 cells were transfected with Flag-Aes and TLE1 and treated with or without 2 mM DSP, followed by staining for endogenous HSC70 as well as for Flag-Aes and nuclei (DAPI). Scale bars, 20 µm.
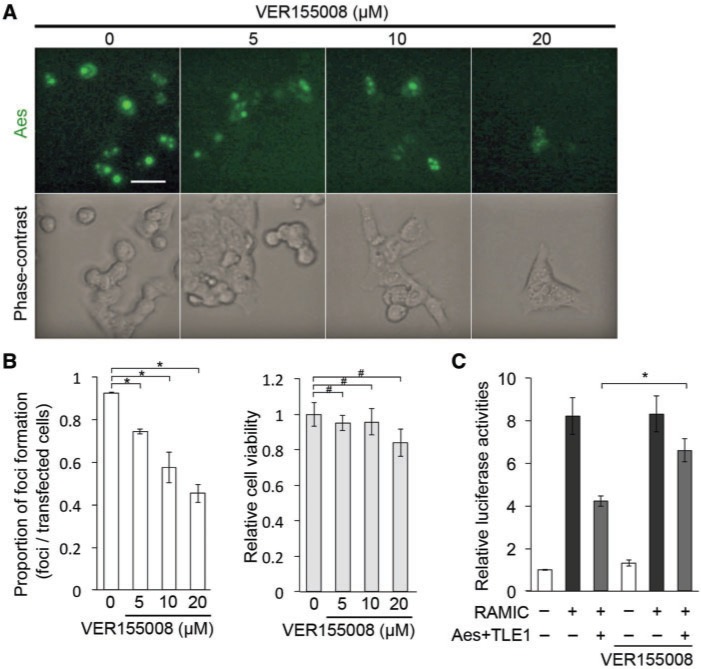
HSC70 ATPase activity is essential for Aes nuclear focus formation
HSC70 is a member of the HSP70 family, and is a molecular chaperone. Although most other HSP70 family members are induced when cells are under stress such as a high temperature, HSC70 is expressed constitutively, suggesting its essential role in normal cellular physiology and homeostasis (30). Therefore, we hypothesized that HSC70 is important for the Aes nuclear focus structure. To test this possibility, we treated HCT116 cells with VER155008, an HSC70 inhibitor that binds to the ATPase pocket of HSC70 to inhibit its function as a molecular chaperone (31). Notably, VER155008 inhibited nuclear focus formation in a dose-dependent manner without affecting cell viability (Fig. 5A and B). These results suggest that HSC70 ATPase activity is necessary for Aes-TLE focus formation and/or maintenance.
Fig. 5.
ATPase activity of HSC70 is important for Aes nuclear focus formation. (A) Effects of HSC70 inhibitor VER155008 on the formation of Aes nuclear foci. HCT116 cells were transfected with AcGFP-Aes and TLE1 followed by the treatment with VER155008 at the indicated concentrations for 24 h. Shown are immunofluorescence photomicrographs (top row) and phase-contrast photomicrographs (bottom row). (B) Proportion of Aes nuclear foci formed in the presence of VER155008 quantified from the data shown in (A) (left), and the cell viability assessed with the WST-1 assay when HCT116 cells were treated with VER155008 at the indicated concentrations for 24 h (right). *P < 0.01, #P > 0.05. (C) Analysis of Notch-mediated transcriptional activation using a luciferase reporter assay (pGa981-6) (3) in HCT116 cells transfected with Aes and TLE1, either in the absence or presence of VER155008 for 24 h (10 µM) (*P < 0.01). Values indicate the means ± SD Scale bar, 20 µm.
We also evaluated the effects of VER155008 on Notch signalling transcription. Together with TLE1, Aes inhibited Notch transcriptional activation by ∼50% in transfected cells mediated by RAMIC. Treatment with VER155008 suppressed this inhibitory effect by Aes and TLE1 slightly (Fig. 5C). Based on our wine tannin hypothesis that soluble form of Aes-TLE1 complex is responsible for inhibition of Rbpj-mediated transcription, reduction in the focus-associated fraction of Aes by VER155008 should increase the soluble Aes-TLE1 and therefore reduce the luciferase reporter activity; in fact it was ∼50% higher than that without VER15508 (Fig. 5C). Accordingly, the apparent suppression by VER155008 of the inhibition of Notch signalling transcription in the presence of Aes may be some indirect effects, and needs further investigation.
In conclusion, we interpret these results that HSC70 is an integral component of Aes nuclear foci, and its ATPase activity is important for the formation and/or maintenance of the Aes foci, although its role in Notch signalling-mediated transcription requires further study.
Supplementary Data
Supplementary Data are available at JB Online.
Acknowledgements
The authors thank T. Honjo for plasmids and suggestions, M. Okabe for plasmids, R. Sasaki for cinematography and K. Okamoto-Furuta for electron microscopy.
Glossary
Abbreviations
- Aes
Amino-terminal enhancer of split
- APC
Adenomatous polyposis coli
- CK2
casein kinase II
- CRC
colorectal cancer
- CSK
cytoskeleton
- DIG
digestion
- DSP
dithiobis [succinimidylpropionate]
- EM
electron microscope/microscopy
- EXT
extraction
- HDAC
Histone deacetylase
- HSC
Heat shock cognate
- MAD
matrix-associated deacetylase
- MAML
Mastermind-like
- NICD
Notch intracellular domain
- NM-IF
nuclear matrix-intermediate filament
- RBPJ
Recombination signal binding protein for immunoglobulin kappa J region
- RT
room temperature
- TLE
Transducin-like enhancer of split
- RAMIC
Rbpj-associated molecule domain and intracellular domain of the Notch receptor
Funding
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to M.S. and M.M.T.
Conflict of Interest
None declared.
References
- 1.Wood L.D., Parsons D.W., Jones S., Lin J., Sjöblom T., Leary R.J., Shen D., Boca S.M., Barber T., Ptak J., Siliman N., Szabo S., Dezso Z., Ustyanksky V., Nikolskaya T., Nikolsky Y., Karchin R., Wilson P.A., Kaminker J.S., Zhang Z., Croshaw R., Willis J., Dawson D., Shipitsin M., Wilson J.K., Sukumar S., Polyak K., Park B.H., Pethiyagoda C.L., Pant P.V., Ballinger D.G., Sparks A.B., Hartigan J., Smith D.R., Suh E., Papadopoulos N., Buckhaults P., Markowitz S.D., Parmigiani G., Kinzler K.W., Velculescu V.E., Vogelstein B. (2007) The genomic landscape of human breast and colorectal cancers. Science 318, 1108––1113. [DOI] [PubMed] [Google Scholar]
- 2.Taketo M.M. (2012) Roles of stromal microenvironment in colon cancer progression. J. Biochem. 151, 477–481 [DOI] [PubMed] [Google Scholar]
- 3.Sonoshita M., Aoki M., Fuwa H., Aoki K., Hosogi H., Sakai Y., Hashida H., Takabayashi A., Sasaki M., Robine S., Itoh K., Yoshida K., Kakizaki F., Kitamura T., Oshima M., Tateto M.M. (2011) Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell 19, 125–137 [DOI] [PubMed] [Google Scholar]
- 4.Miyasaka H., Choudhury B.K., Hou E.W., Li S.S. (1993) Molecular cloning and expression of mouse and human cDNA encoding AES and ESG proteins with strong similarity to Drosophila enhancer of split groucho protein. Eur. J. Biochem. 216, 343–352 [DOI] [PubMed] [Google Scholar]
- 5.Oshima M., Oshima H., Kitagawa K., Kobayashi M., Itakura C., Taketo M.M. (1995) Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyposis in mice carrying a truncated Apc gene. Proc. Natl Acad. Sci. U. S. A. 92, 4482–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurlbut G.D., Kankel M.W., Lake R.J., Artvanis-Tsakonas S. (2007) Crossing paths with Notch in the hyper-network. Curr. Opin. Cell Biol. 19, 166–175 [DOI] [PubMed] [Google Scholar]
- 7.Artavanis-Tsakonas S., Rand M.D., Lake R.J. (1999) Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 8.Osborne B.A., Minter L.M. (2007) Notch signaling during peripheral T-cell activation and differentiation. Nat. Rev. Immunol. 7, 64–75 [DOI] [PubMed] [Google Scholar]
- 9.Ilagan M.X., Kopan R. (2007) SnapShot: Notch signaling pathway. Cell 128, 1246. [DOI] [PubMed] [Google Scholar]
- 10.Hooper C., Chapple J.P., Lovestone S., Killick R. (2007) The Notch-1 intracellular domain is found in sub-nuclear bodies in SH-SY5Y neuroblastomas and in primary cortical neurons. Neurosci. Lett. 415, 135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao H.Y., Ordentlich P., Koyano-Nakagawa N., Tang Z., Downes M., Kintner C., Evans R.M., Kadesch T. (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12, 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downes M., Ordentlich P., Kao H.Y., Alvarez J.G.A., Evans R.M. (2000) Identification of a nuclear domain with deacetylase activity. Proc. Natl Acad. Sci. U. S. A. 97, 10330–10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oswald F., Kostezka U., Astrahantseff K., Bourteele S., Dillinger K., Zechner U., Ludwig L., Wilda M., Hameister H., Knöchel W., Liptay S., Schmid R.M. (2002) SHARP is a novel component of the Notch/RBP-Jκ signaling pathway. EMBO J. 21, 5417–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenzie G.J., Stevenson P., Ward G., Papadia S., Bading H., Chawla S., Privalsky M., Hardingham G.E. (2005) Nuclear Ca2+ and CaM kinase IV specify hormonal- and Notch-responsiveness. J. Neurochem. 93, 171–185 [DOI] [PubMed] [Google Scholar]
- 15.Okabe M., Ikawa M., Kominami K., Nakanishi T., Nishimune Y. (1997) ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 [DOI] [PubMed] [Google Scholar]
- 16.Kato H., Taniguchi Y., Kurooka H., Minoguchi S., Sakai T., Nomura-Okazaki S., Tamura K., Honjo T. (1997) Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124, 4133–4141 [DOI] [PubMed] [Google Scholar]
- 17.Nuthall H.N., Joachim K., Stifani S. (2004) Phosphorylation of serine 239 of GrouchoTLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol. Cell Biol. 24, 8395–8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fey E.G., Wan K.M., Penman S. (1984) Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: three-dimensional organization and protein composition. J. Cell Biol. 98, 1973–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fey E.G., Krochmalnic G., Penman S. (1986) The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J. Cell Biol. 102, 1654–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimoto R., Kataoka N., Okawa K., Ohmo M. (2009) Isolation and characterization of post-splicing lariat-intron complexs. Nucleic Acid Res. 37, 891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muraoka Y., Ohashi-Ikeda H., Nakano N., Hangai M., Toda Y., Okamoto-Furuta K., Kohda H., Kondo M., Terasaki H., Kakizuka A., Yoshimura N. (2012) Real-time imaging of rabbit retina with retinal degeneration by using spectral-domain optical coherence tomography. PLoS One 7, e36135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsubouchi A., Tsuyama T., Fujioka M., Kohda H., Okamoto-Furuta K., Aigaki T., Uemura T. (2009) Mitochondrial protein Preli-like is required for development of dendritic arbors and prevents their regression in the Drosophila sensory nervous system. Development 136, 3757–3766 [DOI] [PubMed] [Google Scholar]
- 23.Sleeman J.E., Trinkle-Mulcahy L. (2014) Nuclear bodies: new insight into assembly/dynamics and disease relevance. Curr. Opin. Biol. 28C, 76–83 [DOI] [PubMed] [Google Scholar]
- 24.Dickinson L.A., Kohwi-Shigematsu T. (1995) Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell Biol. 15, 456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolffe A.P., Hansen J.C. (2001) Nuclear visions: functional flexibility from structural instability. Cell 104, 631–634 [DOI] [PubMed] [Google Scholar]
- 26.Larson M.E., Lesné S.E. (2012) Soluble Aβ oligomer production and toxicity. J. Neurochem. 120, 125–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesfeld J.M., Forbes D.J. (1997) Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22, 197–202 [DOI] [PubMed] [Google Scholar]
- 28.Nuthall H.N., Joachim K., Palapariti A., Stifani S. (2002) A role for cell cycle-regulated phosphorylation in Groucho-mediated transcriptional repression. J. Biol. Chem. 277, 51049–51057 [DOI] [PubMed] [Google Scholar]
- 29.Jennings H., Ish-Horowicz D. (2008) The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 9, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stricher F., Macri C., Ruff M., Muller S. (2013) HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting. Autophagy 9, 1937–1954 [DOI] [PubMed] [Google Scholar]
- 31.Massey A.J., Williamson D.S., Browne H., Murray J.B., Dokumo P., Shaw T., Macias A.T., Daniels Z., Geoffroy S., Dopson M., Lavan P., Natassova N., Francis G.L., Graham C.J., Parsons R., Wang Y., Padfield A., Comer M., Drysdale M.J., Wood M. (2010) A novel small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother. Pharmacol. 66, 535–545 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.