Abstract
Although monoclonal antibodies have been used not only as analytical tools but also as biologic therapeutics, they cannot target intracellular proteins due to their large molecular size and low membrane permeability, which limit their applications. During previous attempts to delivery antibodies intracellularly, the low efficiency of escape from endosomes to the cytosol reduced the bioavailability of antibodies or antibody-conjugated effectors. Recently, we found that the fusogenic peptides (FPs) B18 and B55 from bindin, a sea urchin gamete recognition protein, facilitated the endosomal escape of FP-fused enhanced green fluorescent protein (eGFP) and/or of co-administered cargos such as dextrans [Niikura et al. A fusogenic peptide from a sea urchin fertilization protein promotes intracellular delivery of biomacromolecules by facilitating endosomal escape. J. Control. Release 2015;212:85-93]. In this study, we constructed FP-fused anti-epidermal growth factor receptor (EGFR) single-chain Fv (αEGFR[scFv]) proteins and evaluated their endosomal escape efficiency by utilizing a nuclear localization signal). When the FP-fused αEGFR[scFv] proteins were incubated with A431 cells, the estimated endosomal escape efficiency of αEGFR[scFv]-B18 was significantly higher than that of αEGFR[scFv] alone, suggesting that the B18 peptide facilitates endosomal escape of the conjugated scFv in cis. Moreover, αEGFR[scFv]-B55 promoted the intracellular uptake of co-administered eGFP and dextrans in trans. These results imply that B18- and B55-fused antibodies may be useful for the cell-specific intracellular delivery of biomacromolecules.
Keywords: antibody engineering, endosomal escape, fusogenic peptide, intracellular delivery, nuclear translocation
Antibodies have been used not only as analytical tools in molecular biology in vitro but also as biologic therapeutics. Because antigen-antibody interactions have considerably high affinity and specificity, they have been utilized to detect specific molecules in various immunoassays, such as immunoblotting (1), immunocytochemistry (2), enzyme-linked immunosorbent assay (ELISA) (3) and flow cytometry (4). In addition, antibodies with high in vivo stability and cytotoxic effector functions have been utilized to neutralize cytokines in autoimmune disease therapy and to remove abnormal cells in cancer therapy (5). However, the following drawbacks limit the application of antibodies for cellular analysis and therapeutics. First, because of their large and complex structure and extensive glycosylation, it is expensive to produce and purify antibodies during the manufacturing process. In addition, antibodies cannot target intracellular proteins due to their low membrane permeability. Although antibody fragments have been developed to overcome these drawbacks (6), their membrane permeability is still insufficient to target intracellular proteins.
Two approaches have thus far been adopted to target intracellular proteins using antibodies. ‘Intrabody’ is a method employing the intracellular expression of antibody fragments that then bind to intracellular proteins (7, 8). Because the intracellular antibody fragments are sufficiently expressed to bind its antigen, this approach meets the demands for molecular biological analysis in vitro. However, for therapeutic application, the vectors from which the antibody fragments are expressed must be delivered to the appropriate cells. ‘Transbody’ is another method that uses a cell-penetrating peptide (CPP) to deliver antibody fragments intracellularly (9, 10). Although CPPs have been employed for the intracellular delivery of various peptides and proteins, non-specific cell penetrations by CPPs causes instability in vivo due to their cationic charge of CPPs (11–13). Moreover, the low efficiency of endosomal escape is another limitation of intracellular delivery by CPPs (14, 15).
Additionally, antibody-drug conjugates (ADCs) have also used antibodies as a drug delivery system (DDS) to target-specific cells (16). With ADCs, antibodies are internalized after binding to receptors that are specifically expressed on the surface of target cells; they then release the conjugated drugs intracellularly. In this approach, although the antibodies can be internalized, few antibodies escape from endosomes to the cytosol, resulting in reduced drug efficacy, particularly in the case of immunotoxin (17) or immunoRNase (18), which are antibodies fused to a toxic protein or RNase, respectively.
As described above, the low endosomal escape efficiency of antibodies is a critical limitation that must be overcome to develop next-generation antibody therapeutics. Thus, we focused on using fusogenic peptides (FPs) to improve the endosomal escape efficiency of an antibody that recognizes a receptor. Because FPs exert membrane-disrupting activities through pH-dependent conformational changes (19, 20), they are expected to facilitate endosomal escape at acidic pH. We found recently that the FPs, B18 and B55, which are derived from bindin, a sea urchin gamete recognition protein, facilitated the endosomal escape of FP-fused enhanced green fluorescent protein (eGFP) and of co-administered cargos such as dextrans (21). In this study, we produced B18- and B55-fused αEGFR (epidermal growth factor receptor) single-chain Fvs (αEGFR[scFv]-B18 and -B55). We then investigated their specificity for the antigen and their endosomal escape efficiency by imaging analysis and quantitative analysis of immunoblotting utilizing a nuclear localization signal (NLS). Our results suggested that αEGFR[scFv]-B18 escaped from the endosomes and that αEGFR[scFv]-B55 facilitated the endosomal escape of co-administered macromolecules. Therefore, these FPs are expected to be valuable tools not only for improving the efficacy of ADCs with proteinaceous effectors but also for enabling antibodies to target the intracellular proteins of specific cells using bispecificity for cellular receptors and intracellular targets.
Materials and Methods
Construction of plasmids
All oligonucleotides were purchased from Eurofins Genomics (Tokyo, Japan). The primer sequences are shown in Supplementary Table SI. A DNA fragment containing the tac promoter derived from pGEX-6P-1 (GE Healthcare Bio-Sciences, Piscataway, NJ) was cloned along with a synthetic DNA sequence (Eurofins Genomics) that encoded an αEGFR[scFv] gene with an N-terminal bacterial pelB leader sequence whose VH and VL sequences were derived from monoclonal mouse anti-EGFR-528 IgG (22) and connected via a (G4S)3 linker into the BglII and XhoI sites of a pET20b(+) vector (Merck KGaA, Darmstadt, Germany), yielding pAnti-EGFR. The DNA sequence of pAnti-EGFR is shown in Supplementary Fig. S1.
The B18, B55 and TAT genes were each amplified from plasmids encoding eGFP fused to these peptides (21) (Supplementary Methods) using the primers B18-F and B18-R, B55-F and B55-R, and TAT-F and TAT-R, respectively. The NLS gene was amplified from a synthetic DNA NLS1 (Supplementary Table SI) derived from RNA-binding protein 10 (23), using the primers NLS-F and NLS-R. B18-NLS and TAT-NLS genes were amplified from the B18 and NLS1 genes, respectively, using the primers B18-F and B18-NLS-R, and TAT-NLS-F and TAT-R, respectively. The B55-NLS gene was amplified by overlap-extension PCR from the B55 gene and from the B55-NLS-adaptor (Supplementary Table SI) using primers B55-F and NLS-R. All of the PCR products described above were cloned into pAnti-EGFR at the BamHI and XhoI sites, resulting in pAnti-EGFR-B18, pAnti-EGFR-B55, pAnti-EGFR-TAT, pAnti-EGFR-NLS, pAnti-EGFR-B18-NLS, pAnti-EGFR-B55-NLS and pAnti-EGFR-TAT-NLS. The NLS gene was also cloned into pEGFP-B18 (Supplementary Methods) at the BamHI and XhoI sites, resulting in pEGFP-NLS.
The EGFR gene (NM_005228.3) was amplified from a cDNA library from A431 cells and was cloned into a pcDNA 3.1/Hygro(−) vector (Life Technologies, Carlsbad, CA) at the NheI and XhoI sites, resulting in pEGFR. The DNA sequences of all plasmids were confirmed using an Applied Biosystems 3130xl Genetic Analyzer (Life Technologies).
Protein expression and purification
All proteins except for eGFP and eGFP-NLS were expressed in Escherichia coli JM109 cells (Takara Bio, Shiga, Japan). eGFP and eGFP-NLS were expressed in BL21-CodonPlus-RIL (Agilent Technologies, Santa Clara, CA) and purified as previously described (Supplementary Methods).
A series of pAnti-EGFR-X expression vectors encoding the αEGFR[scFv] fusion proteins described above were transformed into bacterial cells, and one clone with each vector was cultivated in 5 ml of 2xYT medium containing 100 mg/ml ampicillin and 0.4% glucose at 37°C for 24 h. After cultivation of starter cultures, each starter culture was diluted in 400 ml of 2xYT medium containing 100 mg/ml ampicillin and 0.1% glucose was cultivated at 37°C until optical density at 600 nm reached 0.6. The cultures were then induced with IPTG (1 mM final) and grown for a further 5 h at 20°C. Next, the bacterial cells from each sample were collected in a 50-ml centrifuge tube and stored at −80°C until use. Tris-buffered saline (TBS) (50 mM Tris-HCl, pH 7.6, and 200 mM NaCl) supplemented with 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, St. Louis, MO) was added to each tube prior to sonication. Each sample was then centrifuged (8,500 × g, 10 min), and the supernatants were harvested.
Each αEGFR[scFv] fusion protein was purified using anti-FLAG M2-agarose (Sigma-Aldrich) or TALON metal affinity resin (Takara Bio). Phosphate-buffered saline (PBS) (Nacalai Tesque, Kyoto, Japan) supplemented with 400 mM NaCl and 1% Tween 20 was used to washing the beads, and PBS supplemented with 400 mM NaCl and 300 mM imidazole was used for elution from the beads. After purification, each buffer containing the purified samples was exchanged with PBS supplemented with 400 mM NaCl using Amicon Ultra-4 centrifugal filters (30 kDa; Merck Millipore, Billerica, MA). The expression of each protein was confirmed using 12.5% SDS–PAGE with Coomassie brilliant blue staining, and protein concentrations were measured using a BSA standard.
Cell line and cell culture
The human epidermoid carcinoma cell line A431 (RIKEN Cell Bank, Ibaraki, Japan), the human cervical cancer cell line HeLa (RIKEN Cell Bank) and the human embryonic kidney cell line HEK293 (RIKEN Cell Bank) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Nacalai Tesque) with 10% (v/v) foetal bovine serum (Nichirei Biosciences, Tokyo, Japan) and 1% (v/v) penicillin–streptomycin (Life Technologies) and were incubated at 37°C and 5% CO2 in static culture.
Enzyme-linked immunosorbent assay
The A431, HeLa and HEK293 cells (1.0 × 103) were seeded in 96-well plates (Thermo Fisher Scientific, Waltham, MA) and grown for 48 h prior to the experiments. Additionally, the HEK293 cells were transiently transfected with pEGFR using Lipofectamine 2000 (Life Technologies) for 24 h prior to the experiments. The medium in each well was replaced with DMEM containing 10% (v/v) foetal bovine serum, 1% (v/v) penicillin–streptomycin and 100 nM of each αEGFR[scFv] fusion protein. After a 1-h incubation, each well was washed three times with D-PBS (−) (Nacalai Tesque), and cells were fixed using a 4% paraformaldehyde phosphate buffer solution (Nacalai Tesque) for 30 min. After fixation, the cells were washed twice with D-PBS (−) and blocked with 3% (w/v) BSA in PBS for 30 min. Next, the cells were treated with horseradish peroxidase (HRP)-conjugated anti-FLAG IgG (Sigma-Aldrich) in 3% (w/v) BSA in PBS for 1 h at 4°C with gentle shaking. The cells were washed five times with D-PBS (−), and 100 µl of ELISA POD substrate TMB solution (Nacalai Tesque) was added to each well. After several minutes, the enzymatic reaction was stopped with 1 M H2SO4, and the absorbance of each sample was measured using a Safire microplate reader (Tecan, Männedorf, Switzerland).
Immunocytochemistry and fluorescent imaging
To investigate the binding specificity of αEGFR[scFv] fusion proteins to EGFR, HeLa cells were seeded in glass-bottomed dishes (AGC Techno Glass, Shizuoka, Japan) and incubated for 24 h. Additionally, the cells were transiently transfected with pEGFR using the GeneJuice transfection reagent (Merck Millipore) for 24 h prior to experiments. For colocalization analysis of the αEGFR[scFv] fusion proteins and EGFR, the A431 cells were seeded in glass-bottomed dishes 48 h prior to the experiments.
The medium for each sample was replaced with DMEM containing 10% (v/v) foetal bovine serum, 1% (v/v) penicillin–streptomycin and 500 nM of each αEGFR[scFv] fusion protein. After a 1-h incubation, the cells were washed three times with D-PBS (−) and fixed using 4% paraformaldehyde phosphate buffer solution for 30 min. After fixation, the cells were treated with 0.2% Triton X-100 in PBS for 5 min and washed twice with D-PBS (−). Next, the cells were blocked using 3% (w/v) BSA in PBS for 30 min before being treated with anti-FLAG M2 IgG (1/1,000) (Sigma-Aldrich) for αEGFR[scFv] fusion proteins and anti-EGFR-2232 IgG (1/200) (2232, Cell Signaling Technology, Danvers, MA) which recognizes the intracellular domain of EGFR in 3% (w/v) BSA in PBS at 4°C for 16 h. The cells were washed twice with D-PBS (–) and treated with CF488-conjugated anti-mouse IgG (1/1,000) (Biotium, Hayward, CA) and Alexa568-conjugated anti-rabbit IgG (1/1,000) (Life Technologies) to detect αEGFR[scFv] fusion proteins and EGFR, respectively. SlowFade Gold Antifade Mountant with DAPI (1/20) (Life Technologies) in 3% (w/v) BSA in PBS was also added, and cells were incubated at 4°C for 3 h. The cells were then washed twice with D-PBS (−) and observed by confocal microscopy (FV1000, Olympus, Tokyo, Japan) as quickly as possible. Quantitative analysis was performed using FV10-ASW software (Olympus), and relative fluorescence intensities were calculated using the ‘average’ parameters of the region of interest (ROI) defined in each cell.
Immunoblotting and quantitative analysis
The A431 cells were seeded in six-well dishes (AGC Techno Glass) 48 h prior to the experiments. The medium for each sample was replaced with DMEM containing 10% (v/v) foetal bovine serum and 1% (v/v) penicillin–streptomycin, along with 2 µg/ml of αEGFR-528 IgG (Santa Cruz Biotechnology, Dallas, TX) which recognizes the extracellular domain of EGFR, 20 or 100 nM of αEGFR[scFv], or 300 nM of each αEGFR[scFv] fusion protein. After a 4-h incubation, the cells were washed three times with D-PBS (−) and collected into microfuge tubes by scraping. Each sample contained 1.0 × 105 cells. The collected cells were solubilized and fractionated, using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific), into nuclear and non-nuclear (the remaining cell lysate, which contains cytoplasm) lysates. The proteins in each lysate were separated by 12.5% (for αEGFR[scFv] fusion proteins) or 7.5% (for EGFR) SDS–PAGE and were blotted on polyvinylidene fluoride membranes (Merck Millipore). After blotting, membranes were blocked with 5% skim milk in Tris-buffered saline with Tween 20 (TBS-T) for 1 h. For fractionation markers, membranes were treated (in 5% skim milk in TBS-T) with anti-Hsp90 IgG (1/400) (Abcam, Cambridge, MA) and anti-LaminB IgG (1/4,000) (Santa Cruz) at 4°C for 16 h with gentle shaking. For αEGFR[scFv] fusion proteins, membranes were treated with anti-FLAG M2 IgG (1/1,000). For EGFR, membranes were incubated with anti-EGFR-2232 IgG (1/1,000) in 5% BSA in TBS-T at 4°C for 16 h with gentle shaking. Next, membranes were washed five times with TBS-T before being incubated with appropriate HRP-conjugated secondary antibodies in 5% skim milk in TBS-T for 30 min. Membranes were again washed five times with TBS-T, and the antibodies were detected with Immunostar LD (Wako Pure Chemical Industries, Osaka, Japan) and analysed with a C-DiGit blot scanner (LI-COR Biosciences, Lincoln, NE). The amount of each αEGFR[scFv] was quantified using amino-terminal FLAG-BAP fusion protein (Sigma-Aldrich) standards.
The values for endosomal escape efficiency were calculated using the following equation:
In this equation, [E] represents the endosomal escape efficiency, AnNLS and AcNLS represent the amounts of an NLS-fused scFv protein in the nuclear (n) or non-nuclear (c) fractions, respectively, and An and Ac represent the amounts of an scFv fusion protein in the nuclear (n) or non-nuclear (c) fractions, respectively. [E] was calculated from simultaneous experiments using both FP- and FP-NLS-fused versions of a given αEGFR[scFv] protein. For example, the amounts of αEGFR[scFv]-B18 and αEGFR[scFv]-B18-NLS in the nuclear and the non-nuclear fractions were used to determine the [E] of the αEGFR[scFv]-B18.
Intracellular uptake of co-administered molecules
The A431 cells or A431-HeLa mixture cells were seeded in glass-bottomed dishes 48 h prior to the experiments. The medium for each sample was replaced with DMEM containing 10% (v/v) foetal bovine serum, 1% (v/v) penicillin–streptomycin, 300 nM of each αEGFR[scFv] fusion protein, 20 µM eGFP and Texas Red-conjugated dextran (M.W. 40,000) (Life Technologies) or 20 µM eGFP-NLS, and 1 µg/ml Hoechst 33342 (Life Technologies). For competitive inhibition, A431 cells were treated with 20 µM Texas Red-conjugated dextran (M.W. 40,000), 1.5 µM αEGFR[scFv] and/or 300 nM αEGFR[scFv]-B55. After a 24-h incubation, the cells were washed three times with D-PBS (−) and fixed using a 4% paraformaldehyde phosphate buffer solution for 30 min. After fixation, the cells were washed twice with D-PBS (−) and observed by confocal microscopy as soon as possible. Quantitative analysis was performed using FV10-ASW software, and the relative fluorescence intensities were calculated using the ‘average’ parameters of the ROI defined in each cell.
Results
Preparation of FP-fused scFv proteins and confirmation of the binding efficacy and specificity
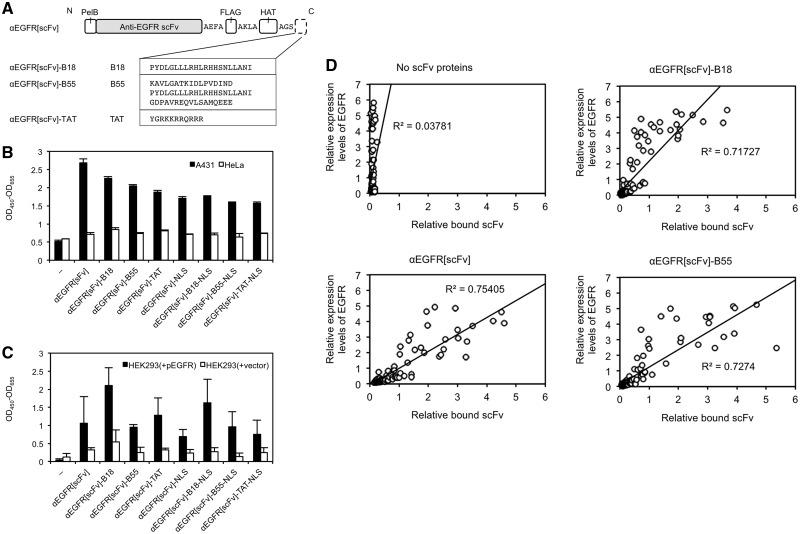
We designed and prepared four αEGFR[scFv] proteins fused to FP or CPP (Fig. 1A). The B18 and B55 peptides are FPs that facilitated the endosomal escape of cargo proteins in HeLa cells as shown in our previous study (21). The TAT, a CPP derived from the HIV-1 virus, were also used to compare the functions of the FPs with a CPP. All proteins with FLAG-tags and HAT-tags were expressed in E. coli and were purified using anti-FLAG antibody-conjugated agarose beads or Co2+ beads.
Fig. 1.
Preparation of FP-fused αEGFR scFv proteins with EGFR-binding activity. (A) Construction of four FP- or CPP-fused αEGFR[scFv] proteins with an N-terminal pelB signal peptide and a C-terminal FLAG tag and HAT tag, each connected with alanine-rich linkers. The amino acid sequences of B18, B55, TAT and HA2 are also shown. For FP-NLS-fused αEGFR[scFv] proteins, an NLS was also fused at the C-terminus of the FPs. (B and C) Binding assays of αEGFR[scFv] fusion proteins (100 nM each) to A431 or HeLa cells (B) or to HEK293 cells transfected with pEGFR or with a control vector (C) using ELISA with HRP-conjugated anti-FLAG IgG. N = 3 (‘−’: N = 2). Mean ± SD. The error bars in (C) were larger than in (B), probably because HEK293 cells were more easily detached from the ELISA plate during the washing procedure, in comparison with the A431 and HeLa cells. (D) Quantitative image analysis of immunocytochemistry to verify correlations between cell-bound αEGFR[scFv] fusion proteins and the expression levels of EGFR in individual HeLa cells. The relative fluorescence intensities of each fluorescent dye of the corresponding secondary antibodies are shown on each axis. The relative intensities were normalized with respect to the average of the sample treated with anti-EGFR[scFv] in each sample. Each plot indicates one HeLa cell. N = 93–113.
We then investigated the binding activity and EGFR specificity of the purified FP-fused αEGFR[scFv] proteins. First, we performed ELISAs using the purified αEGFR[scFv] fusion proteins and EGFR-expressing cells such as A431 cells [3 × 106 EGFR molecules per cell (24)] and HeLa cells [5 × 104 EGFR molecules per cell (24)]. The ELISA results showed that the binding efficacy of each αEGFR[scFv] fusion protein to the A431 cells was clearly greater than for the HeLa cells (Fig. 1B), probably because the EGFR expression level of A431 cells is 60 times higher than that of HeLa cells. To investigate whether each αEGFR[scFv] specifically binds to EGFR, we further performed ELISAs using HEK293 cells transfected with pEGFR or a control vector. The binding efficacy of each αEGFR[scFv] fusion protein to the EGFR-expressing HEK293 cells was higher than to HEK293 cells transfected with the control vector (Fig. 1C).
To further confirm the specific binding of the FP-fused αEGFR[scFv] proteins to EGFR, we used immunocytochemistry with the quantitative image analysis to verify the correlations between EGFR expression levels and the amounts of FP-fused αEGFR[scFv] proteins that bound to EGFR-expressing cells. We used HeLa cells transiently transfected with pEGFR, which should demonstrate different EGFR expression levels in the same dish. As expected, each αEGFR[scFv] fusion protein bound to HeLa cells according to their EGFR expression levels. The correlations for B18- and B55-fused αEGFR[scFv] proteins were similar to those for αEGFR[scFv] (Fig. 1D). These results indicate that the recombinant FP-fused αEGFR[scFv] proteins expressed in E. coli specifically bind to EGFR and that they can bind in an environment in which cells with different EGFR expression levels were cocultivated.
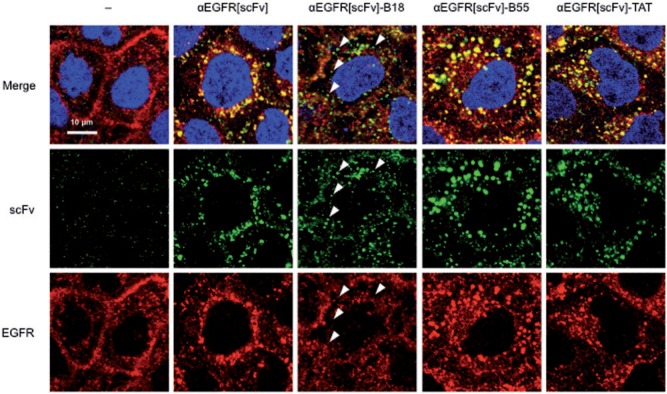
Localization of internalized FP-fused scFv proteins
Using purified FP-fused αEGFR[scFv] proteins that retained the EGFR-binding activity, we investigated the localization of internalized αEGFR[scFv] fusion proteins in EGFR-expressing cells. After incubation of each αEGFR[scFv] with A431 cells, the localization patterns of internalized scFv proteins and EGFR were detected by immunocytochemistry (Fig. 2). In contrast to αEGFR[scFv], which localized to endosome-like vesicles with EGFR (observed in yellow), αEGFR[scFv]-B18 localized to smaller particles and colocalized less frequently with EGFR (observed in green and indicated in Fig. 2 with arrowheads), suggesting that the αEGFR[scFv]-B18 partially escaped from the endosomes to the cytosol. αEGFR[scFv]-B55 localized with EGFR to endosome-like vesicles similar to αEGFR[scFv] but showed more accumulation on the vesicles (Fig. 2; αEGFR[scFv]-B55). For αEGFR[scFv]-TAT, there were almost no differences in the localization patterns of the scFvs and EGFR (Fig. 2; αEGFR[scFv]-TAT).
Fig. 2.
Confocal microscopic images of A431 cells treated with FP-fused αEGFR scFv proteins. The localization patterns of FP-fused αEGFR[scFv] proteins (300 nM each) and of EGFR in A431 cells were detected by immunocytochemistry using CF488-conjugated IgG (green) for scFv and Alexa 568-conjugated IgG (red) for EGFR. Cells were also stained with DAPI (blue). Arrowheads indicate small particles containing αEGFR[scFv]-B18 but not colocalized with EGFR.
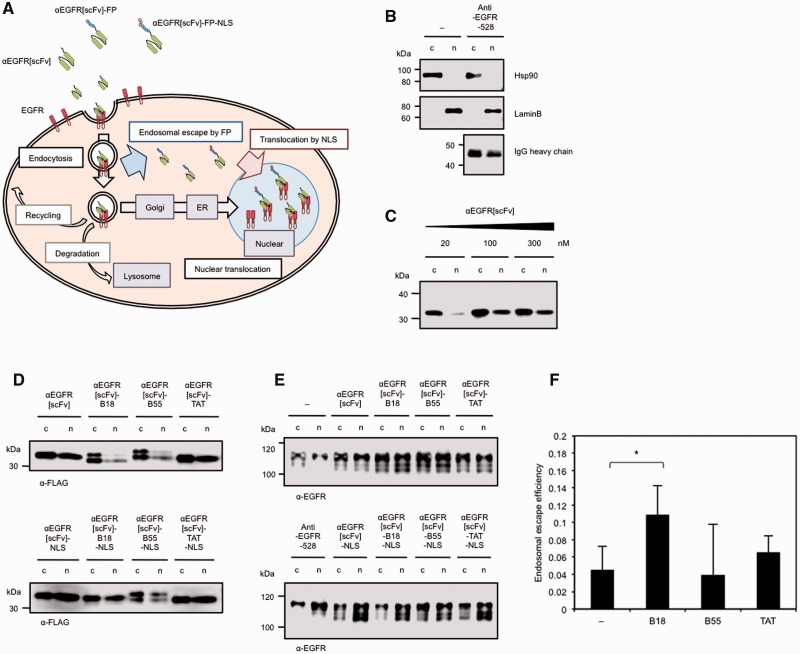
Estimation of the endosomal escape efficiency of FP-fused scFv proteins by utilizing NLS
Via quantitative analysis of the confocal microscopic images described above, we could not reveal distinct differences in either the fluorescence intensities of internalized scFv or in the colocalization of scFv and EGFR, as it was difficult to distinguish cytosolic signals from endosomal signals (data not shown). To more easily and quantitatively verify the endosomal escape efficiency of FP-fused αEGFR[scFv] proteins, we developed another approach using an NLS. If FP-fused scFv proteins are also fused to an NLS, scFvs that are internalized to the cytosol after endosomal escape are expected to then be transported into the nucleus by importin. The amounts of an scFv in the nuclear fraction can then be easily quantified. However, it has been reported that EGFR itself is also translocated to the nucleus with its ligands (25, 26). After internalization via ligand-induced endocytosis, the intracellular trafficking of EGFR is determined by endosomal sorting (26) to various pathways, such as recycling (27), degradation (28) and nuclear translocation (25). In the nuclear translocation pathway (Fig. 3A), EGFR is transported to the Golgi from the endosomes (29) and is subsequently transported to the endoplasmic reticulum (ER) (30). Thereafter, direct translocation from the ER to the nucleus was proposed as the ‘INTERNET’ model (26), in which EGFR is directly transported to the nucleus in a membrane-penetrating state by importin ß rather than through the cytosol by endosomal escape. In this model, Sec61 ß translocates EGFR from the inner nuclear membrane to the inside of the nucleus along with its ligands (31). Thus, it is expected that we could estimate the endosomal escape efficiency of FP-fused αEGFR[scFv] proteins by quantifying their nuclear localization and calculating the difference between the amounts of protein with and without an NLS. In principle, as shown in Fig. 3A, αEGFR[scFv] fusion proteins are endocytosed with EGFR after the internalization that is induced by binding to EGFR, and they are then partially translocated to the nucleus in the case of αEGFR[scFv]. On the other hand, FP-fused αEGFR[scFv] proteins are expected to escape from the endosomes to the cytosol during endosomal maturation, and the amount of scFv in the nucleus should consequently decrease (Fig. 3A). Moreover, FP-NLS-fused αEGFR[scFv] proteins enhance translocation from the cytosol to the nucleus after endosomal escape, and, consequently, the amount of scFv localized to the nucleus is expected to be restored to levels comparable to those of αEGFR[scFv]. Taking into account all of these effects, the endosomal escape efficiency of FP-fused αEGFR[scFv] proteins can be estimated as the ratio between the amounts of scFv protein with and without an NLS in the nucleus.
Fig. 3.
Estimating the endosomal escape efficiency of FP-fused αEGFR scFv proteins by utilizing an NLS. (A) Scheme for the intracellular trafficking of αEGFR[scFv], FP-fused αEGFR[scFv] (αEGFR[scFv]-FP) and FP-NLS-fused αEGFR[scFv] (αEGFR[scFv]-FP-NLS). (B) Confirmation of fractionated lysates and of the nuclear translocation of αEGFR-528 IgG as a ligand of EGFR. Immunoblotting was performed with anti-Hsp90, anti-LaminB and anti-mouse IgGs on fractionated lysates from A431 cells treated with 2 µg/ml of anti-EGFR-528 IgG for 4 h. The electrophoresis ratio between the non-nuclear fraction) (c) and the nuclear fraction (n) was 1:2. (C) Confirmation of the nuclear translocation of αEGFR[scFv] as a ligand of EGFR. Immunoblotting was performed with an anti-FLAG IgG on fractionated lysate from A431 cells treated with 20–300 nM of αEGFR[scFv] for 4 h. The electrophoresis ratio of (c) and (n) was 1:4. (D and E) Immunoblots with anti-FLAG IgG (D) or an anti-EGFR IgG recognizing the intracellular domain of EGFR (E) on fractionated lysates from A431 cells treated with 300 nM of each αEGFR[scFv] fusion protein or with 2 µg/ml of αEGFR-528 IgG for 4 h. The electrophoresis ratio between (c) and (n) was 1:4 (D) or 1:2 (E). (F) The endosomal escape efficiency of each αEGFR[scFv] fusion protein as estimated by the difference between the proportions of FP-fused αEGFR[scFv] proteins and the proportion of FP-NLS-fused αEGFR[scFv] proteins in the nuclear fraction. The ‘−’ sample indicates αEGFR[scFv] without a fused FP. N = 5. Mean ± SD. *P < 0.05.
According to this scheme, we first confirmed successful fractionation of cell lysates into nuclear and non-nuclear fractions. Indeed, the cytoplasmic marker Hsp90 was detected only in the non-nuclear fraction, whereas the nucleic marker LaminB was detected only in the nuclear fraction of lysates from A431 cells (Fig. 3B), indicating that the cell lysates were successfully separated into nuclear and non-nuclear fractions. In lysates from A431 cells treated with αEGFR-528, a parental IgG of the αEGFR[scFv] used in this study, IgG heavy chain was also detected in both the nuclear and non-nuclear fractions (Fig. 3B), supporting the idea that ligands bound to EGFR are also transported to the nucleus by the nuclear translocation pathway. We also investigated whether αEGFR[scFv] could be transported to the nucleus with EGFR in a similar manner to αEGFR-528. Indeed, αEGFR[scFv] was detected in both the nuclear and non-nuclear fractions of lysates from A431 cells treated with various concentrations of anti-EGFR[scFv] (Fig. 3C), indicating that αEGFR[scFv] could also be transported to the nucleus by the nuclear translocation pathway of EGFR. Because we observed that the amount of αEGFR[scFv] internalized to the A431 cells was saturated at 100–300 nM, we used each αEGFR[scFv] fusion protein at 300 nM in subsequent analyses.
To estimate of the endosomal escape efficiency of FP-fused scFv proteins according to the scheme described above, we constructed four additional FP-NLS-fused αEGFR[scFv] proteins and confirmed that these antibodies retained their EGFR-binding activity and specificity when expressed in E. coli (Fig. 1B and C). To quantitatively evaluate the amount of each αEGFR[scFv] fusion protein localized to the nucleus, each αEGFR[scFv] fusion protein was detected in fractionated lysates from A431 cells treated with each αEGFR[scFv] fusion protein by immunoblotting; the blots were then quantified. As expected, the proportions of αEGFR[scFv]-B18 and -B55 in the nuclear fraction decreased compared with αEGFR[scFv] (Fig. 3D and Table I). In lysates of A431 cells treated with αEGFR[scFv]-B18-NLS, decreased proportion of αEGFR[scFv]-B18 was restored, suggesting that αEGFR[scFv]-B18 partially escaped from endosomes to the cytosol. Conversely, the proportion of αEGFR[scFv]-TAT localized to the nucleus was comparable to that of αEGFR[scFv] (Fig. 3D and Table I), suggesting that the TAT peptide does not affect endosomal escape efficiency, although it does enhance binding to the cell surface by electrostatic interactions (11–13).
Table I.
The number of molecules of each αEGFR[scFv] fusion protein per cell in each fraction, as estimated by quantitative immunoblotting with anti-FLAG IgG.
| Mean ± SD (105 molecules/cell) |
||
|---|---|---|
| c | n | |
| αEGFR[scFv] | 25.0 ± 10.8 | 4.7 ± 2.5 |
| αEGFR[scFv]-NLS | 14.4 ± 6.1 | 3.5 ± 2.0 |
| αEGFR[scFv]-B18 | 14.8 ± 5.2 | 1.2 ± 0.6 |
| αEGFR[scFv]-B18-NLS | 8.3 ± 3.6 | 1.6 ± 0.6 |
| αEGFR[scFv]-B55 | 11.6 ± 3.2 | 1.2 ± 0.5 |
| αEGFR[scFv]-B55-NLS | 12.4 ± 5.0 | 1.8 ± 0.8 |
| αEGFR[scFv]-TAT | 18.6 ± 8.3 | 2.4 ± 1.0 |
| αEGFR[scFv]-TAT-NLS | 13.4 ± 5.4 | 2.6 ± 1.0 |
c, the cell lysate excluding nuclear proteins (non-nuclear fraction); n, nuclear fraction. N = 5.
We also detected the proportion of EGFR localized in the nucleus of A431 cells. When A431 cells were treated with αEGFR IgG or with scFv proteins, the proportion of EGFR localized in the nucleus increased (Fig. 3E), indicating that nuclear translocation of EGFR was promoted by ligand binding, as shown in previous studies (25, 29–31). In lysates of A431 cells treated with each FP-fused αEGFR[scFv] protein, an additional band was detected at a lower molecular weight than the bands from αEGFR-528-treated cells or from cells without antibody treatment. Although the reason for the appearance of this additional band remains unknown, it is suggested that there are some differences in the intracellular responses of A431 cells to treatment with αEGFR scFv and IgG.
Finally, we estimated the endosomal escape efficiency of each peptide fused to αEGFR[scFv] based on the difference between the proportions of scFv fusion proteins and NLS-fused scFv proteins localized to the nucleus of A431 cells. Our estimates showed that the endosomal escape efficiency of αEGFR[scFv]-B18 was significantly higher than that of αEGFR[scFv] (Fig. 3F and see equation in Materials and Methods), suggesting that the FP such as B18 peptide facilitates endosomal escape when fused to an αEGFR[scFv], although the efficiency is insufficient for the intracellular delivery of antibodies.
This result may be supported by the data in Fig. 1D in which the relative bound of αEGFR[scFv]-B18 estimated by immunocytochemistry and fluorescent imaging was somewhat smaller than those of other scFvs, while the binding activity of αEGFR[scFv]-B18 estimated by ELISA (Fig. 1B and C) was not low. This difference can be explained if the endosomal escape efficiency of αEGFR[scFv]-B18 is slightly higher than that of other scFvs, because diffuse fluorescent signals of αEGFR[scFv]-B18 into cytoplasm showed very low sensitivity (Fig. 2), so that the apparent total fluorescence of αEGFR[scFv]-B18 (Fig. 1D) may decrease in comparison with other scFvs.
Conversely, the endosomal escape efficiency of αEGFR[scFv]-B55 was similar to αEGFR[scFv] (Fig. 3F). Although the proportion of αEGFR[scFv]-B55 in the nucleus decreased (Fig. 3D and Table I), the proportion of αEGFR[scFv]-B55-NLS in the nucleus also remained at low levels, suggesting that the B55 peptide affected the nuclear translocation with EGFR (Fig. 3E) but did not facilitate the endosomal escape of the conjugated αEGFR[scFv].
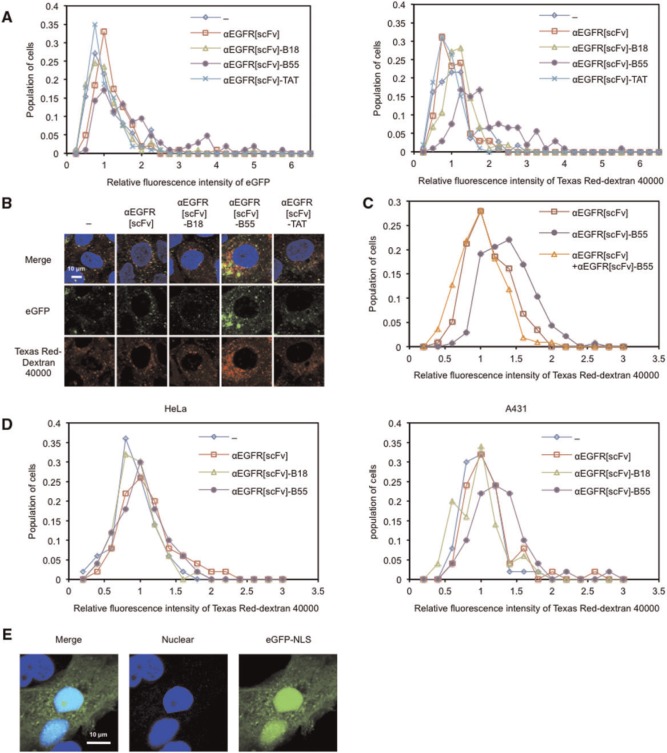
Intracellular uptake of co-administered molecules in the presence of FP-fused scFv proteins
There are two approaches for the delivery of cargoes into the cytosol using FPs. The first is a cis method that involves ‘direct (covalent) conjugation’ of FPs to cargoes as described above, and the second is a trans method that involves ‘simple co-incubation’ of FPs with cargoes or other membrane-disruptive reagents (32). We recently reported that an eGFP-fused B55 peptide facilitated the endosomal escape not only of the eGFP-fused protein itself (via the cis method) but also of various biomacromolecules co-administered with the eGFP-fused B55 (via the trans method) (21). Thus, we reasoned that αEGFR[scFv]-B55 might also facilitated the endosomal escape of co-administered molecules in trans. Indeed, the intracellular levels of eGFP and Texas Red-conjugated dextran increased in the presence of αEGFR[scFv]-B55 compared with αEGFR[scFv] (Fig. 4A and B), although the fluorescence of eGFP and dextrans were still low levels in most of cells possibly due to the insufficient efficiency of facilitating endosomal escape. To verify whether αEGFR[scFv]-B55-mediated endosomal escape of co-administered molecules is dependent on its antibody specificity, we performed a competitive inhibition assay. As a result, the fluorescence of Texas Red-dextran decreased in A431 cells treated with αEGFR[scFv] and αEGFR[scFv]-B55 compared with αEGFR[scFv]-B55 alone (Fig. 4C). In addition, αEGFR[scFv]-B55 promoted the intracellular uptake of co-administered dextrans only in A431 cells in the dish in which A431 and HeLa cells were cocultivated (Fig. 4D). Finally, when we used eGFP-NLS, cells displayed nuclear eGFP fluorescence in the presence of αEGFR[scFv]-B55 (Fig. 4E). These results suggest that αEGFR[scFv]-B55 facilitates the endosomal escape of co-administered molecules in EGFR-expressing cells specifically and that αEGFR[scFv]-B55 is suitable for the intracellular delivery of co-administered molecules via the trans method.
Fig. 4.
Intracellular uptake of eGFP in the presence of αEGFR[scFv]-B55. Cell-population histograms of eGFP (left panel) and Texas Red (right panel) fluorescence for each cell (A) and Confocal microscopic images (B) of A431 cells treated for 24 h with 20 µM eGFP (green) and Texas Red-conjugated dextran (M.W. 40,000) (red), 1 µg/ml Hoechst 33342 (blue) and 300 nM of each αEGFR[scFv] fusion protein. Relative fluorescence intensities were normalized with respect to the averages of the samples without scFv (‘−’) in each experiment. N = 101–111. (C) Cell-population histograms of Texas Red fluorescence for each cell of A431 cells treated for 24 h with 20 µM Texas Red-conjugated dextran (M.W. 40,000), 1.5 µM αEGFR[scFv] and/or 300 nM αEGFR[scFv]-B55. N = 110–136. (D) Cell-population histograms of Texas Red fluorescence for each cell of HeLa (left panel) and A431 (right panel) cells treated for 24 h with 20 µM Texas Red-conjugated dextran (M.W. 40,000) and 300 nM of each αEGFR[scFv] fusion protein. N = 50. (E) Confocal microscopic images of A431 cells treated for 24 h with 20 µM eGFP-NLS (green), 1 µg/ml Hoechst 33342 (blue) and 300 nM αEGFR[scFv]-B55.
Discussion
In this study, we suggested that fusogenic B18 and B55 peptides, when fused to αEGFR[scFv] proteins, can facilitate the endosomal escape of the scFv protein in cis or of co-administered molecules in trans, respectively. These results are similar to our previous results for FP-fused eGFP (21). Conversely, it was suggested that TAT peptide does not affect endosomal escape efficiency. Although cationic CPPs such as TAT have been used for intracellular delivery of various molecules, the efficiency of cytosolic macromolecular delivery mediated by TAT is quite low due to the poor endosomolytic activity of TAT (15). Therefore, it should be considered that, not only the intracellular uptake by endocytosis, the endosomal escape efficiency is also important in the cytosolic delivery of macromolecules.
To quantify the endosomal escape efficiency of FP-fused scFv proteins via the cis method, we compared the amounts of nuclear αEGFR[scFv] proteins with and without an NLS by immunoblotting. Because the nuclear translocation of proteins containing an NLS is induced only when the scFvs escape from endosomes, this method can also be applied to evaluate the endosomal escape efficiency of scFv proteins that recognize receptors other than EGFR and are internalized by endocytosis.
In future applications, the cis method may contribute to improved efficiency of ADCs, particularly for the immunotoxin (17) and immunoRNase (18), as these antibodies or their effectors are often required to reach the cytoplasm. Indeed, a previous report using immunoRNase indicated that the low endosomal escape efficiency of the active component of immunoRNase was the major limiting factor for its cytotoxic action (18). Quite recently, Kiesgen et al. reported that the cytotoxic activity of anti-EGFR immunoRNase against EGFR-expressing cells was enhanced by the additional fusion of a FP derived from dengue virus (33), but they did not reveal whether the FP-fused immunoRNase escaped from endosomes and did not consider nuclear translocation with EGFR. Because our study demonstrated that the endosomal escape efficiency of αEGFR[scFv]-B18 was significantly higher than that of αEGFR[scFv], fusion to the B18 peptide is also expected to promote the intracellular translocation of proteinaceous toxin- or RNase-conjugated antibodies.
Conversely, the trans method has the potential to be a novel DDS that is independent of individual medicines. In this method, it is expected that a B55-fused antibody can control the membrane permeability of specific cells that are recognized by the antibody, resulting that co-administered molecules are accumulated to the cells. This method has some advantages over conventional ADCs in that molecules with low permeability, such as peptides or proteins, can be specifically delivered to the cytosol of targeted cells without being limited by the amount or variety of molecules that can be conjugated to an antibody. Therefore, the B55 peptide may contribute to the assembly of such a novel antibody DDS.
Our present study suggested that αEGFR[scFv]-B18 facilitated its own endosomal escape in cis and suggested that αEGFR[scFv]-B55 could facilitate the endosomal escape of co-administered molecules in trans, which are similar to our previous results for eGFP-B18 and eGFP-B55 (21), respectively. The advantage of FP-fused antibodies over FP-fused eGFP is that allows target cell-specific intracellular delivery of various biomacromolecules. However, their activities of these fusion antibodies were somewhat low compared with that of FP-fused eGFP (21). One reason for this disadvantage may be that most of antibodies remain bound to antigens after internalization by endocytosis, likely reducing the endosomal escape efficiency. Thus, we anticipate that a pH-dependent antibody, which binds to an antigen at a neutral pH but dissociates from the antigen as the pH decreases during endosomal maturation (34, 35), will improve the facilitation of the endosomal escape for both the cis and trans methods. Such an FP-fused pH-dependent antibody will escape from endosomes more efficiently and will provide a novel antibody DDS to control the membrane permeability of target cells. Currently, we are studying the directed evolution of a pH-dependent αEGFR[scFv] by using mRNA display (36, 37), and the results will be published in the future.
Supplementary Data
Supplementary Data are available at JB Online.
Acknowledgements
We thank Dr Kotaro Oka for help with the study using the confocal microscope. We also thank Dr Kei Fujiwara for useful comments on the manuscript, and we thank the members of our laboratory for helpful discussions.
Funding
This work was supported in part by the Mochida Memorial Foundation for Medical and Pharmaceutical Research, by the Strategic Research Foundation Grant-aided Project for Private Universities (S1411003) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a Grant-in-Aid for Scientific Research (22360351, 25289298, 15K12545) from the Japan Society for the Promotion of Science. K.N. was supported by a KLL 2014 Ph.D. Program Research Grant (000038) from Keio University.
Conflict of Interest
None declared.
Glossary
Abbreviations
- ADC
antibody-drug conjugate
- CPP
cell-penetrating peptide
- DDS
drug delivery system
- eGFP
enhanced green fluorescent protein
- EGFR
epidermal growth factor receptor
- FP
fusogenic peptide
- NLS
nuclear localization signal
- scFv
single-chain Fv
References
- 1.Kurien B.T., Scofield R.H. (2003) Protein blotting: a review. J. Immunol. Methods 274, 1–15 [DOI] [PubMed] [Google Scholar]
- 2.Burry R.W. (2011) Controls for immunocytochemistry: an update. J. Histochem. Cytochem. 59, 6–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gan S.D., Patel K.R. (2013) Enzyme immunoassay and enzyme-linked immunosorbent assay. J. Invest. Dermatol. 133, e12. [DOI] [PubMed] [Google Scholar]
- 4.Baumgarth N., Roederer M. (2000) A practical approach to multicolor flow cytometry for immunophenotyping. J. Immunol. Methods 243, 77–97 [DOI] [PubMed] [Google Scholar]
- 5.Martin-Mateos M.A. (2007) Monoclonal antibodies in pediatrics: use in prevention and treatment. Allergol. Immunopathol. (Madr) 35, 145–150 [DOI] [PubMed] [Google Scholar]
- 6.Nelson A.L. (2010) Antibody fragments: hope and hype. MAbs 2, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J., Rabbitts T.H. (2014) Intracellular antibody capture: a molecular biology approach to inhibitors of protein-protein interactions. Biochim. Biophys. Acta 1844, 1970–1976 [DOI] [PubMed] [Google Scholar]
- 8.Kaiser P.D., Maier J., Traenkle B., Emele F., Rothbauer U. (2014) Recent progress in generating intracellular functional antibody fragments to target and trace cellular components in living cells. Biochim. Biophys. Acta 1844, 1933–1942 [DOI] [PubMed] [Google Scholar]
- 9.Poungpair O., Pootong A., Maneewatch S., Srimanote P., Tongtawe P., Songserm T., Tapchaisri P., Chaicumpa W. (2010) A human single chain transbody specific to matrix protein (M1) interferes with the replication of influenza A virus. Bioconjug. Chem. 21, 1134–1141 [DOI] [PubMed] [Google Scholar]
- 10.Phalaphol A., Thueng-In K., Thanongsaksrikul J., Poungpair O., Bangphoomi K., Sookrung N., Srimanote P., Chaicumpa W. (2013) Humanized-VH/VHH that inhibit HCV replication by interfering with the virus helicase activity. J. Virol. Methods 194, 289–299 [DOI] [PubMed] [Google Scholar]
- 11.Wang F., Wang Y., Zhang X., Zhang W., Guo S., Jin F. (2014) Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J. Control. Release 174, 126–136 [DOI] [PubMed] [Google Scholar]
- 12.Vivès E., Schmidt J., Pèlegrin A. (2008) Cell-penetrating and cell-targeting peptides in drug delivery. Biochim. Biophys. Acta 1786, 126–138 [DOI] [PubMed] [Google Scholar]
- 13.Jain M., Chauhan S.C., Singh A.P., Venkatraman G., Colcher D., Batra S.K. (2005) Penetratin improves tumor retention of single-chain antibodies: a novel step toward optimization of radioimmunotherapy of solid tumors. Cancer Res. 65, 7840–7846 [DOI] [PubMed] [Google Scholar]
- 14.Reissmann S. (2014) Cell penetration: scope and limitations by the application of cell-penetrating peptides, J . Pept. Sci. 20, 760–784 [DOI] [PubMed] [Google Scholar]
- 15.Erazo-Oliveras A., Muthukrishnan N., Baker R., Wang T.Y., Pellois J.P. (2012) Improving the endosomal escape of cell-penetrating peptides and their cargos: strategies and challenges. Pharmaceuticals 5, 1177–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panowksi S., Bhakta S., Raab H., Polakis P., Junutula J.R. (2014) Site-specific antibody drug conjugates for cancer therapy. MAbs 6, 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wayne A.S., Fitzgerald D.J., Kreitman R.J., Pastan I. (2014) Immunotoxins for leukemia. Blood 123, 2470–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schirrmann T., Frenzel A., Linden L., Stelte-Ludwig B., Willuda J., Harrenga A., Dübel S., Müller-Tiemann B., Trautwein M. (2014) Evaluation of human pancreatic RNase as effector molecule in a therapeutic antibody platform. MAbs 6, 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorieau J.L., Louis J.M., Schwieters C.D., Bax A. (2012) pH-triggered, activated-state conformations of the influenza hemagglutinin fusion peptide revealed by NMR. Proc. Natl. Acad. Sci. USA 109, 19994–19999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nouri F.S., Wang X., Dorrani M., Karjoo Z., Hatefi A. (2013) A recombinant biopolymeric platform for reliable evaluation of the activity of pH-responsive amphiphile fusogenic peptides. Biomacromolecules 14, 2033–2040 [DOI] [PubMed] [Google Scholar]
- 21.Niikura K., Horisawa K., Doi N. (2015) A fusogenic peptide from a sea urchin fertilization protein promotes intracellular delivery of biomacromolecules by facilitating endosomal escape. J. Control. Release 212, 85–93 [DOI] [PubMed] [Google Scholar]
- 22.Madej M.P., Coia G., Williams C.C., Caine J.M., Pearce L.A., Attwood R., Bartone N.A., Dolezal O., Nisbet R.M., Nuttall S.D., Adams T.E. (2012) Engineering of an anti-epidermal growth factor receptor antibody to single chain format and labeling by Sortase A-mediated protein ligation. Biotechnol. Bioeng. 109, 1461–1470 [DOI] [PubMed] [Google Scholar]
- 23.Xiao S.J., Wang L.Y., Kimura M., Kojima H., Kunimoto H., Nishiumi F., Yamamoto N., Nishio K., Fujimoto S., Kato T., Kitagawa S., Yamane H., Nakajima K., Inoue A. (2013) S1-1/RBM10: multiplicity and cooperativity of nuclear localisation domains. Biol. Cell 105, 162–174 [DOI] [PubMed] [Google Scholar]
- 24.Ichinose J., Murata M., Yanagida T., Sako Y. (2004) EGF signalling amplification induced by dynamic clustering of EGFR. Biochem. Biophys. Res. Commun. 324, 1143–1149 [DOI] [PubMed] [Google Scholar]
- 25.Li C., Iida M., Dunn E.F., Ghia A.J., Wheeler D.L. (2009) Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene 28, 3801–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y.N., Yamaguchi H., Hsu J.M., Hung M.C. (2010) Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene 29, 3997–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceresa B.P. (2006) Regulation of EGFR endocytic trafficking by rab proteins. Histol. Histopathol. 21, 987–993 [DOI] [PubMed] [Google Scholar]
- 28.Ceresa B.P., Bahr S.J. (2006) Rab7 activity affects epidermal growth factor:epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J. Biol. Chem. 281, 1099–1106 [DOI] [PubMed] [Google Scholar]
- 29.Du Y., Shen J., Hsu J.L., Han Z., Hsu M.C., Yang C.C., Kuo H.P., Wang Y.N., Yamaguchi H., Miller S.A., Hung M.C. (2014) Syntaxin 6-mediated Golgi translocation plays an important role in nuclear functions of EGFR through microtubule-dependent trafficking. Oncogene 33, 756–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y.N., Wang H., Yamaguchi H., Lee H.J., Lee H.H., Hung M.C. (2010) COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem. Biophys. Res. Commun. 399, 498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y.N., Yamaguchi H., Huo L., Du Y., Lee H.J., Lee H.H., Wang H., Hsu J.M., Hung M.C. (2010) The translocon Sec61beta localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J. Biol. Chem. 285, 38720–38729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angeles-Boza A.M., Erazo-Oliveras A., Lee Y.J., Pellois J.P. (2010) Generation of endosomolytic reagents by branching of cell-penetrating peptides: tools for the delivery of bioactive compounds to live cells in cis or trans. Bioconjug. Chem. 21, 2164–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiesgen S., Liebers N., Cremer M., Arnold U., Weber T., Keller A., Herold-Mende C., Dyckhoff G., Jäger D., Kontermann R.E., Arndt M.A., Krauss J. (2014) A fusogenic dengue virus-derived peptide enhances antitumor efficacy of an antibody-ribonuclease fusion protein targeting the EGF receptor. Protein Eng. Des. Sel. 27, 331–337 [DOI] [PubMed] [Google Scholar]
- 34.Igawa T., Ishii S., Tachibana T., Maeda A., Higuchi Y., Shimaoka S., Moriyama C., Watanabe T., Takubo R., Doi Y., Wakabayashi T., Hayasaka A., Kadono S., Miyazaki T., Haraya K., Sekimori Y., Kojima T., Nabuchi Y., Aso Y., Kawabe Y., Hattori K. (2010) Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat. Biotechnol. 28, 1203–1207 [DOI] [PubMed] [Google Scholar]
- 35.Chaparro-Riggers J., Liang H., DeVay R.M., Bai L., Sutton J.E., Chen W., Geng T., Lindquist K., Casas M.G., Boustany L.M., Brown C.L., Chabot J., Gomes B., Garzone P., Rossi A., Strop P., Shelton D., Pons J., Rajpal A. (2012) Increasing serum half-life and extending cholesterol lowering in vivo by engineering antibody with pH-sensitive binding to PCSK9. J. Biol. Chem. 287, 11090–11097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuda I., Kojoh K., Tabata N., Doi N., Takashima H., Miyamoto-Sato E., Yanagawa H. (2006) In vitro evolution of single-chain antibodies using mRNA display. Nucleic Acids Res. 34, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabata N., Sakuma Y., Honda Y., Doi N., Takashima H., Miyamoto-Sato E., Yanagawa H. (2009) Rapid antibody selection by mRNA display on a microfluidic chip. Nucleic Acids Res. 37, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.