Abstract
DNA methylation is one of the most stable but dynamically regulated epigenetic marks that act as determinants of cell fates during embryonic development through regulation of various forms of gene expression. DNA methylation patterns must be faithfully propagated throughout successive cell divisions in order to maintain cell-specific function. We have recently demonstrated that Uhrf1-dependent ubiquitylation of histone H3 at lysine 23 is critical for Dnmt1 recruitment to DNA replication sites, which catalyzes the conversion of hemi-methylated DNA to fully methylated DNA. In this review, we provide an overview of recent progress in understanding the mechanism underlying maintenance DNA methylation.
Keywords: cell cycle; DNA methylation; DNA methyltransferase 1, histone; Ubiquitin
The methylation of CpG dinucleotide in vertebrates is implicated in several key biological processes, including developmental Programming of gene expression, X chromosome inactivation, genomic imprinting, and silencing of repetitive elements (1–5). These are mainly regulated by formation of the repressive chromatin structure (6). CpG dinucleotides tend to cluster within CpG islands, which preferentially localize to transcription start sites (TSSs) of the majority of human genes (7, 8). In embryonic cells, most of CpG dinucleotides are methylated (9–11). In contrast, in somatic cells, CpG sites in CpG islands usually remain unmethylated independently of gene expression, although most CpG sites in non-CpG islands including repetitive elements are methylated (12). However, regions of methylated DNA have been correlated with tissue- or cell-specific expression of several genes across the genome (13–16). In addition, DNA methylation has been shown to be dynamic, and capable of temporary change at promoter CpG sites (17, 18). Therefore, in individual differentiated cells, regulated propagation of the DNA methylation pattern during cell proliferation is pivotal to maintaining tissue- or cell-specific functions.
Global DNA methylation patterns appear to be established by at least three DNA methyltransferases (Dnmts): Dnmt1, Dnmt3A, and Dnmt3B. Dnmt3A and 3B in association with Dnmt3L which lacks a methyltransferase domain, are thought to function as de novo DNMTs. Both these enzymes are responsible for establishing the pattern of DNA methylation during early embryonic development (19–21). They are highly expressed in embryonic stem cells and markedly downregulated during differentiation processes (22). Dnmt1 was originally reported by Bestor et al. (23) and is believed to be the primary enzyme responsible for copying the methylation pattern after DNA replication. Dnmt1 has the replication foci targeting sequence (RFTS), which could ensure its localization to replication sites (24), and interacts with proliferating cell nuclear antigen (PCNA) (25). Dnmt1 also shows a preference for hemi-methylated DNA in which one strand is methylated, although it has de novo DNA methyltransferase activity (26). This set of features is the reason why DNMT1 is often referred to as the ‘maintenance methyltransferase’.
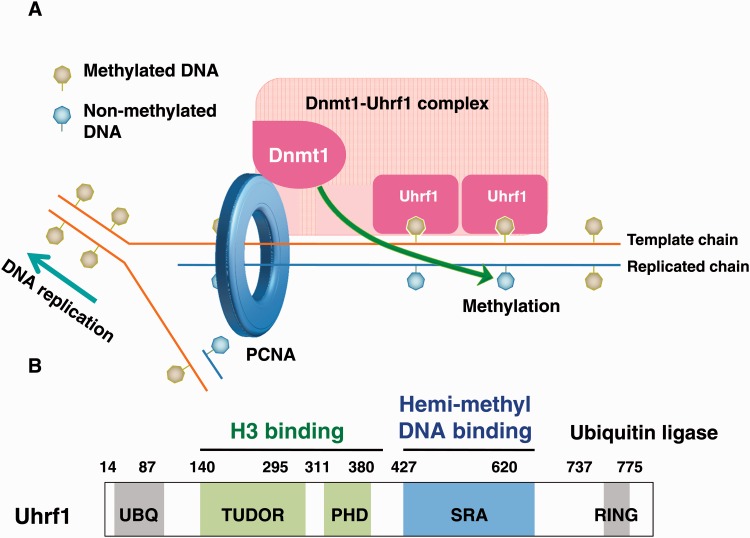
There is increasing evidence that Ubiquitin-like PHD and RING finger 1 (Uhrf1, also known as ICBP90 and Np95) also plays a fundamental role during maintenance DNA methylation (27, 28) (Fig. 2A). The Uhrf1 protein binds to hemi-methylated DNA via its SET- and RING-associated (SRA) domain (29–31). Recent studies revealed that Uhrf1 also recognizes H3K9 trimethylation (H3K9me3) and unmodified H3R2 through its tandem TUDOR and plant homeodomain (PHD) (32, 33). Furthermore, Uhrf1 possesses a RING (Really Interesting New Gene) finger domain at its carboxyl terminal region, through which Uhrf1 ubiquitylates histone H3 at K23 or K18 (34, 35). Dnmt1 preferentially binds to these ubiquitylations. Thus, the ability of Uhrf1 to ubiquitylate histon H3 facilitates and ensures Dnmt1 recruitment and maintenance of DNA methylation pattern (34). In this review, we summarize recent progress in the regulation of maintenance DNA methylation and discuss its possible implication in cellular differentiation.
Fig. 2.
Uhrf1 plays an essential role in the recruitment of Dnmt1 to hemi-methylated DNA regions during DNA replication. (A) Methylated cytosine cannot be replicated by DNA replication machinery, generating hemi-methylated DNA. Uhrf1 could specifically bind to hemi-methylated DNA. Uhrf1 then recruits Dnmt1 around the sites of hemi-methylated DNA although molecular mechanism underlying this recruitment remains to be elusive. The recruited Dnmt1 then catalyzes the conversion of himi-metylated DNA to full methylated DNA. (B) Uhrf1 contains various unique domains. TUDOR and PHD domains function as a single unit for readout of H3K9 methylation. SRA domain is a hemi-methylated DNA binding domain, flipping 5-methylcytosine out of the DNA helix. The carboxyl-terminal RING finger domain ubiquitylates H3K23 in a DNA replication-dependent manner. The amino-terminal ubiquitin like domain (UBQ) presents a highest homology with ubiquitin molecule, but its function remains elusive.
An In Vitro Cell-Free System Using Xenopus Egg Extracts is a Powerful Tool for Biochemical Analysis of Epigenetic Regulation During Cell Cycle Progression
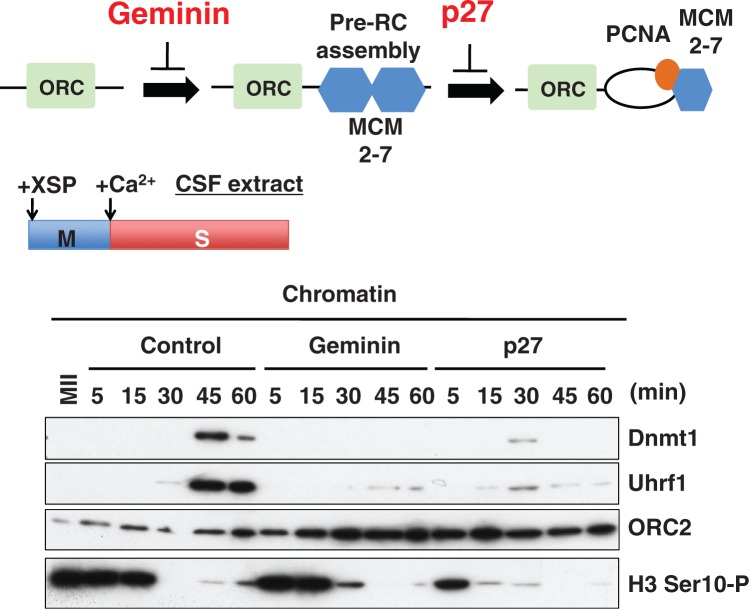
In vitro cell-free systems using Xenopus egg extracts are frequently applied to analyze stable genomic inheritance such as cell cycle progression (36, 37), DNA replication (38), and activation of checkpoints (39), and have provided marked insights into the molecular basis of these mechanisms. However, it has not been clear whether these in vitro systems are useful for the analysis of epigenetic inheritance. Recently, we established a cell-free system using Xenopus interphase egg extracts for analysis of maintenance DNA methylation (40). When demembranated sperm chromatin was incubated with egg extracts in the presence of [3H]-labeled S-adenosyl methionine as a donor of methyl group, a marked accumulation of radioactivity in sperm DNA was observed, suggesting that sperm DNA was actually methylated. Importantly, when DNA replication was blocked by the addition of DNA replication inhibitors, accumulation of radioactivity in sperm DNA was almost completely abrogated, suggesting that this type of DNA methylation is dependent on ongoing DNA replication. Under these conditions, Uhrf1 and Dnmt1 bound to replicating chromatin in a DNA replication-dependent manner (Fig. 1). Immunodepletion of Uhrf1 from egg extracts using specific antibodies against Uhrf1 resulted in a marked suppression of DNA methylation, likely due to impaired recruitment of Dnmt1 to the chromatin. Taken together, the results indicated that incorporation of radioactivity in sperm DNA corresponds to bona fide maintenance DNA methylation and that this maintenance is successfully reproduced in this cell-free system. Our results also highlight the possibility that cell-free system would be a powerful tool for the biochemical analysis of epigenetic regulation including DNA methylation and histone modification during cell cycle progression as predicted by Banaszynski et al. (41).
Fig. 1.
DNA replication and Uhrf1-dependent chromatin loading of Dnmt1 in Xenopus egg extracts. Schematic representation of Geminin and p27 functions in DNA replication. ORC: origin recognition complex, Pre-RC: pre-replication complex, MCM: minichromosome maintenance (upper panel). Meiotic metaphase II (MII) arrested cytostatic factor (CSF) extracts were incubated with demembraned Xenopus sperm (XSP) nuclei to assemble mitotic chromatin for 30 min. The extracts arrested in MII were released into interphase by addition of CaCl2, leading to degradation of mitotic cyclins by the APC/C, exit from mitosis and initiation of DNA replication (middle panel). Loading of Dnmt1 and Uhrf1 is dependent on DNA replication. DMSO (Control), Geminin or GST-p27 were added to the activated CSF extracts for inhibition of DNA replication. Sperm chromatin fractions at the indicated times and mitotic chromatin (-Ca2+) were analyzed by immunoblotting (IB) H3 Ser10-P: phosphorylation of H3 at Ser 10, a marker of mitosis (lower panels).
Role of Uhrf1 in the Coupling of DNA Replication with DNA Methylation
Uhrf1 was first identified as a nuclear phosphoprotein (a phosphorylated protein localized in nucleus) whose expression was regulated in a cell cycle-dependent manner (42). It was also reported to colocalize with PCNA during S phase (43) and its expression was markedly high in various cancer cells (44), suggesting its role in the regulation of cell proliferation and tumorigenesis. Later, Uhrf1 was found to be essential for the maintenance DNA methylation through the recruitment of Dnmt1 to DNA replication sites (27, 28) (Fig. 2A). Although Uhrf1 contains multiple functional domains (Fig. 2B), its unique ability is conferred by its SRA domain. Uhrf1 shows preferential affinity for hemi-methylated DNA via the SRA domain. With respect to Dnmt1 recruitment, Uhrf1 was reported to bind to Dnmt1, but this interaction is contestable because the Dnmt1-binding domain of Uhrf1 has not been incontrovertibly determined (28, 45). In addition, from the structures of SRA-hemi-methylated DNA and Dnmt1-hemi-methylated DNA complexes (29–31, 46, 47), when Uhrf1 binds to hemi-methylated DNA, it appears unlikely that Dnmt1 catalyzes the conversion of hemi-methylated DNA to fully methylated DNA because unmethylated target cytosine is preferentially recognized by the NRK finger of the SRA domain. Therefore, it is highly predictable that once Uhrf1 detects hemi-methylated DNA, an unidentified ‘message’ would be transmitted to Dnmt1, allowing the dissociation of Uhrf1 from hemi-methylated DNA. It should be noted that recent studies have proposed a role for Dnmt3A and 3B in maintenance DNA methylation patterns in somatic cells, especially in terms of repeat regions and imprinted genes (48, 49)
In addition to the SRA domain, tandem TUDOR and PHD domains of Uhrf1 also play an essential role in maintenance DNA methylation (50, 51). These domains function as a single functional unit and provide a defined readout of H3K9 methylation (52). Uhrf1 binding to methylated-H3K9 during mitosis targets it to chromatin and maintains the stability of Dnmt1, ensuring the maintenance DNA methylation. Intriguingly, either hemi-methylated DNA or methylated H3K9 binding of Uhrf1 can target Dnmt1 for maintenance DNA methylation and the collaboration of both forms of binding further ensures high fidelity maintenance DNA methylation (51). Thus, although the structural basis of collaboration of the SRA, and tandem TUDOR and PHD domains remains elusive, histone codes appear to play fundamental role in the regulation of maintenance DNA methylation.
The RING Finger Domain of Uhrf1 is Required for the Ubiquitylation of H3K23 in a DNA Replication-Dependent Manner, which is Prerequisite for the Recruitment of Dnmt1 to DNA Sites
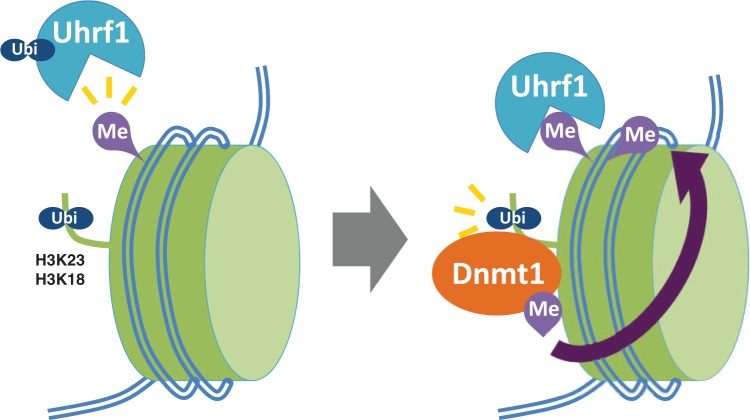
As described above, in order to identify the ‘message’ from hemi-methylated DNA-bound Uhrf1 to Dnmt1, we further analyzed maintenance DNA methylation using a cell-free system. Intriguingly, Uhrf1 ubiquitylated H3K23 and this ubiquitylation was dramatically enhanced by Dnmt1 depletion (34). Dnmt1 is directly capable of binding to ubiquitylated H3 through an RFTS domain previously identified as a region required for the colocalization of Dnmt1 to DNA replication sites (24), suggesting its role in the recruitment of Dnmt1 to hemi-methylated DNA regions. Similar Uhrf1-mediated ubiquitylation of H3 was also observed in mammalian cells (34). Consistent with results from the in vitro cell-free system, the RING finger domain of Uhrf1 is indispensable for H3K23 ubiquitylation and the recruitment of Dnmt1 to DNA replication sites and for the maintenance of global DNA methylation as well as that at retrotransposons in mammals (34, 35). Very recent study has identified an ubiquitin-interacting-motif (UIM) in RFTS of Dnmt1 (35). The ubiquitin binding ability of UIM is required for Dnmt1 recruitment to DNA replication foci, consistent with idea that Dnmt1 is tethered to DNA methylation sites through regulatory ubiquitylation of H3 by Uhrf1. In this study, Uhrf1 was reported to ubiquitylate H3 at K18 as well as K23. Taken together, our results suggest an attractive model for the regulation of maintenance DNA methylation as follows (Fig. 3). Fully methylated DNA cannot be precisely replicated during S phase and generates hemi-methylated DNA after DNA replication. Uhrf1 specifically binds to hemi-methylated DNA regions generated after DNA replication and ubiquitylates H3K23 or H3K18. Dnmt1 then directly binds to ubiquitylated H3K23 or H3K18, on recruitment to DNA replication sites. The recruited Dnmt1 catalyzes the conversion of hemi-methylated DNA to fully methylated DNA. This model suggests the existence of regulatory mechanisms of passive DNA demethylation by blocking methylation of newly synthesized DNA through acetylation or methylation at H3K23 or H3K18 thereby antagonizing ubiquitylation at the same sites and thus suppressing maintenance DNA methylation.
Fig. 3.
A proposed model of the regulation of maintenance DNA methylation through Uhrf1-mediated H3K23 and/or H3K18 ubiquitylations. After DNA replication, existing fully methylated DNA cannot be replicated by a canonical DNA replication system, generating hemi-methylated DNA. Uhrf1 specifically binds to hemi-methylated DNA and ubiquitylates H3K23 and/or H3K18. Ubiquitylations of H3K23 and/or H3K18 specifically recruit Dnmt1 at the sites of DNA replication. The recruited Dnmt1 then catalyzes the conversion of hemi-methylated DNA to fully methylated DNA.
Possible Implications of Regulation of Maintenance DNA Methylation in Cellular Differentiation
One possible explanation for the acquisition of site-specific hypomethylation patterns during cellular differentiation would be that DNA demethylation occurs preferentially at defined regions, while ignoring the remaining portions of the genome. This site-specific DNA demethylation could be mediated by DNA-binding transcription factors and chromatin modifiers that reduce local accesses of Dnmt1 or Uhrf1 during DNA replication through antagonized modification of H3K23 or H3K18 against ubiquitylation. Intriguingly, in plants, histone acetyltransferase IDM1/ROS4 targets H3K23 and H3K18 to promote active DNA demethylation (52, 53), suggesting that acetylated H3K23 and H3K18 marks have a critical role in the prevention of maintenance of DNA methylation at a subset of genomic regions. It has been reported that K23 is a major acetylated site of H3 in mammalian (54) and Drosophila cells (55) whose acetylation was predominantly detected in actively transcribed regions that are poor in the active marks acetyl-H3K9, acetyl-H3K16, and tri-methyl H3K4 in Drosophila (55). Interestingly, acetylation of K23 is reported to be catalyzed by p300/CBP and GCN5, both of which are sequence-specific transcription factor-associated co-activators (56, 57), suggesting that extra-cellular or intrinsic differentiation signals may lead to site-specific DNA demethylation. In mammalian cells, K23 is also a site subjected to propionylation (58). In addition, it is tempting to surmise that acetylated H3K23 or H3K18 binding protein(s) might also play a counteracting role together with Uhrf1-dependent H3 ubiquitylation in disrupting the mechanism of maintenance DNA methylation. Taken together, elucidation of the regulatory mechanisms underlying post-translational modifications at H3K23 or H3K18, including ubiquitylation, acetylation, propionylation, and possibly methylation may provide key clues to understanding the regulation of cellular differentiation.
Another unsolved question is a physiological importance of deubiquitin enzymes (DUBs) involved in the removal of ubiquitin from H3 in maintenance DNA methylation. Candidates for such DUBs include Usp7 (Ubiquitin specific processing protease 7) that forms a complex with Dnmt1 as well as Uhrf1 and stabilizes these proteins by suppressing polyubiquitylation and following proteasomal degradation (59, 60). One could expect that Usp7 also deubiquitylates H3 to regulate Dnmt1 binding to chromatin at DNA methylation sites. Thus, it should be cue to identify DUBs responsible for deubiquitylation of H3 for better understanding the mechanisms underlying dynamics of proper recruitment of Dnmt1 to sites of maintenance DNA methylation during DNA replication.
Funding
M.N. was supported by a Grant-in-Aid for Scientific Research on Innovative Area ‘Cell fate control’, Scientific Research (A), and Challenging Exploratory Research from MEXT Japan.
Conflict of Interest
None declared.
References
- 1.Holliday R., Pugh J.E. (1975) DNA modification mechanisms and gene activity during development. Science 187, 226–232 [PubMed] [Google Scholar]
- 2.Nafee T.M., Farrell W.E., Carroll W.D., Fryer A.A., Ismail K.M. (2008) Epigenetic control of fetal gene expression. BJOG 115, 158–168 [DOI] [PubMed] [Google Scholar]
- 3.Goll M.G., Bestor T.H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 [DOI] [PubMed] [Google Scholar]
- 4.Chang S.C., Tucker T., Thorogood N.P., Brown C.J. (2006) Mechanisms of X chromosome inactivation. Front. Biosci. 11, 852–866 [DOI] [PubMed] [Google Scholar]
- 5.Suzuki M.M., Bird A. (2008) DNA methylation landscape: Provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476 [DOI] [PubMed] [Google Scholar]
- 6.Ng H.H., Bird A. DNA methylation and chromatin modification (1999) Curr . Opin. Genet. Dev. 9, 158–163 [DOI] [PubMed] [Google Scholar]
- 7.Bird A.P. (1986) CpG-rich islands and the function of DNA methylation. Nature 321, 209–213 [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Leung F.C. (2004) An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics 20, 1170–1177 [DOI] [PubMed] [Google Scholar]
- 9.Smith Z.D., Meissner A. (2013) DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 [DOI] [PubMed] [Google Scholar]
- 10.Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., Yan J., Ren X., Lin S., Li J., Jin X., Shi X., Liu P., Wang X., Wang W., We Y., Li X., Guo F., Wu X., Fan X., Yong J., Wen L., Xie S.X., Tang F., Qiao J. (2014) The DNA methylation landscape of human early embryos. Nature 511, 606–610 [DOI] [PubMed] [Google Scholar]
- 11.Smith Z.D., Chan M.M., Humm K.C., Karnik R., Mekhoubad S., Regev A., Eggan K., Meissner A. (2014) DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takai D., Jones P.A. (2002) Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 99, 3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar V., Biswas D.K. (1988) Dynamic state of site-specific DNA methylation concurrent to altered prolactin and growth hormone gene expression in the pituitary gland of pregnant and lactating rats. J. Biol. Chem. 263, 12645–12652 [PubMed] [Google Scholar]
- 14.Shen L., Kondo Y., Guo Y., Zhang J., Zhang L., Ahmed S., Shu J., Chen X., Waterland R.A., Issa J-P. J. (2007) Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 3, 2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitamura E., Igarashi J., Morohashi A., Hida N., Oinuma T., Nemoto N., Song F., Ghosh S., Held W.A., Yoshida-Noro C., Nagase H. (2007) Analysis of tissue-specific differentially methylated regions (TDMs) in humans. Genomics 89, 326–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yagi S., Hirabayashi K., Sato S., Li W., Takahashi Y., Hirakawa T., Wu G., Hattori N., Hattori N., Ohgane J., Tanaka S., Liu X.S., Shiota K. (2008) DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 18, 1969–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doi A., Park I.H., Wen B., Murakami P., Aryee M.J., Irizarry R., Herb B., Ladd-Acosta C., Rho J., Loewer S., Miller J., Schlaeger T., Daley G.Q., Feinberg A.P. (2009) Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41, 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., Gnirke A., Jaenisch R., Lander E.S. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okano M., Bell D.W., Haber D.A., Li E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 20.Bourc’his D., Xu G.L., Lin C.S., Bollman B., Bestor T.H. (2001) Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 [DOI] [PubMed] [Google Scholar]
- 21.Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X. (2007) Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen T., Li E. (2004) Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 60, 55–89 [DOI] [PubMed] [Google Scholar]
- 23.Bestor T.H., Laudano A., Mattaliano R., Ingram V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzyme is related to bacterial restriction methyltransferases. J. Mol. Biol. 203, 971–983 [DOI] [PubMed] [Google Scholar]
- 24.Leonhardt H., Page A.W., Weier H.U., Bestor T.H. (1992) A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71, 865–873 [DOI] [PubMed] [Google Scholar]
- 25.Araujo F.D., Croteau S., Slack A.D., Milutinovic S., Bigey P., Price G.B., Zannis-Hadjopoulos M., Szyf M. (2001) The DNMT1 target recognition domain resides in the N terminus. J. Biol. Chem. 276, 6930–6936 [DOI] [PubMed] [Google Scholar]
- 26.Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet. 9, 2395–2402 [DOI] [PubMed] [Google Scholar]
- 27.Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Mitsuya K., Okano M., Koseki H. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 [DOI] [PubMed] [Google Scholar]
- 28.Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 [DOI] [PubMed] [Google Scholar]
- 29.Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M. (2008) Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821 [DOI] [PubMed] [Google Scholar]
- 30.Avvakumov G.V., Walker J.R., Xue S., Li Y., Duan S., Bronner C., Arrowsmith C.H., Dhe-Paganon S. (2008) Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455, 822–825 [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto H., Horton J.R., Zhang X., Bostick M., Jacobsen S.E., Cheng X. (2008) The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455, 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothbart S.B., Dickson B.M., Ong M.S., Krajewski K., Houliston S., Kireev D.B., Arrowsmith C.H., Strahl B.D. (2013) Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 27, 1288–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu L., Li Z., Wang P., Lin Y., Xu Y. (2011) Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res. 21, 1379–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., Shimamura S., Arita K., Kodama T., Ishikawa F., Koseki H., Nakanishi M. (2013) Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502, 249–253 [DOI] [PubMed] [Google Scholar]
- 35.Qin W., Wolf P., Lin N., Link S., Smets M., Mastra F.L., Forme I., Pichler G., Horl D., Fellinger K., Spada F., Bonapace I.M., Imhof A., Harz H., Leonhardt H. (2015) DNA methylateon requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 25, 911–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunphy W.G., Newport J.W. (1988) Unraveling of mitotic control mechanisms. Cell 55, 925–928 [DOI] [PubMed] [Google Scholar]
- 37.Coleman T.R., Dunphy W.G. (1994) Cdc2 regulatory factors. Curr. Opin. Cell Biol. 6, 877–882 [DOI] [PubMed] [Google Scholar]
- 38.Tutter A.V., Walter J.C. (1996) Chromosomal DNA replication in a soluble cell-free system derived from Xenopus eggs. Methods Mol. Biol. 322, 121–137 [DOI] [PubMed] [Google Scholar]
- 39.Kumagai A., Dunphy W.G. (2000) Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell 6, 839–849 [DOI] [PubMed] [Google Scholar]
- 40.Banaszynski L.A., Allis C.D., Shechter D. (2010) Analysis of histones and chromatin in Xenopus laevis egg and oocyte extracts. Methods. 51, 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujimori A., Matsuda Y., Takemoto Y., Hashimoto Y., Kubo E., Araki R., Fukukmura R., Mita K., Tatsumi K., Muto M. (1998) Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm. Genome 9 1032–1035 [DOI] [PubMed] [Google Scholar]
- 42.Uemura T., Kubo E., Kanari Y., Ikemura T., Tatsumi K., Muto M. (2000) Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell Struct. Funct. 25, 149–159 [DOI] [PubMed] [Google Scholar]
- 43.Mousli M., Hopfner R., Abbady A.Q., Monte D., Jeanblanc M., Oudet P., Louis B., Bronner C. (2003) ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br. J. Cancer. 89, 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Achour M., Jacq X., Ronde P., Alhosin M., Charlot C., Chataigneau T., Jeanblanc M., Macaluso M., Giordano A., Hughes A.D., Schini-Kerth V.B., Bronner C. (2008) The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene 27, 2187–2197 [DOI] [PubMed] [Google Scholar]
- 45.Takeshita K., Suetake I., Yamashita E., Suga M., Narita H., Nakagawa A., Tajima S. (2011) Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc. Natl Acad. Sci. USA. 108, 9055–9059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song J., Teplova M., Ishibe-Murakami S., Patel D.J. (2012) Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 335, 709–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones P.A., Liang G. (2009) Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 10, 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeong S., Liang G., Sharma S., Lin J.C., Choi S.H., Han H., Yoo C.B., Egger G., Yang A.S., Jones P.A. (2009) Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol. Cell Biol. 29, 5366–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothbart S.B., Krajewski K., Nady N., Tempel W., Xue S., Badeaux A.I., Barsyte-Lovejoy D., Martinez J.Y., Bedford M.T., Fuch S.M., Arrowsmith C.H., Strahl B.D. (2012) Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 19, 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X., Gao Q., Li P., Zhao Q., Zhang J., Li J., Koseki H., Wong J. (2013) UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 4, 1563. [DOI] [PubMed] [Google Scholar]
- 51.Rothbart S.B., Dickson B.M., Ong M.S., Krajewski K., Houliston S., Kireev D.B., Arrowsmith C.H., Strahl B.D. (2013) Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is regulated for the epigenetic inheritance of DNA methylation. Genes Dev. 27, 1288–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qian W., Miki D., Zhang H., Liu Y., Zhang X., Tang K., Kan Y., La H., Li X., Li S., Zhu X., Shi X., Zhang K., Pontes O., Chen X., Liu R., Gong Z., Zhu J.K. (2012) A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336, 1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X., Qian W., Zhao Y., Wang C., Shen J., Zhu J.K., Gong Z. (2012) Antisilencing role of the RNA-directed DNA methylateon pathway and a histone acetyltransferase in Arabidopsis. Proc. Natl Acad. Sci. USA 109, 11425–11430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue K., Song J., Wei H., et al. Synchronous behaviors of CBP and actylations of lysine 18 and lysine 23 on histone H3 during porcine oocyte first meiotic division. Mol. Reprod. Dev. 2010; 77: 605–614 [DOI] [PubMed] [Google Scholar]
- 55.Keller WA., Ramos E., Van Bortle K., Takenaka N., Corces VG. Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters. Genome Res. 2012; 22: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kikuchi H., Takami Y., Nakayama T. GCN5: a supervisor in all-inclusive control of vertebrate cell cycle progression through transcription regulation of various cell cycle-related genes. Gene. 2005; 347: 83–97 [DOI] [PubMed] [Google Scholar]
- 57.Bodai L., Zsindely N., Gaspar R., Kristo I., Komonyi O., Boros I.M. (2012) Ecdysone induced gene expression is associated with acetylation of histone H3 lysine 23 in Drosophila melanogaster. PLoS One 7, e40565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu B., Lin Y., Darwanto A., Song X., Xu G., Zhang K. (2009) Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem. 284, 32288–32295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Felle M., Joppien S., Nemeth A., Diermeier S., Thalhammer V., Dobner T., Kremmer E., Kappler R., Langst G. (2011) The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucl. Acid Res. 39, 8355–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma H., Chen H., Guo X., Wang Z., Sowa M.E., Zheng L., Hu S., Zeng P., Guo R., Diao J., Lan F., Harper J.W., Shi Y.G., Xu Y., Shi Y. (2012) M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc. Natl Acad. Sci. USA. 109, 4828–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]