Abstract
Super resolution imaging is becoming an increasingly important tool in the arsenal of methods available to cell biologists. In recognition of its potential, the Nobel Prize for chemistry was awarded to three investigators involved in the development of super resolution imaging methods in 2014. The availability of commercial instruments for super resolution imaging has further spurred the development of new methods and reagents designed to take advantage of super resolution techniques. Super resolution offers the advantages traditionally associated with light microscopy, including the use of gentle fixation and specimen preparation methods, the ability to visualize multiple elements within a single specimen, and the potential to visualize dynamic changes in living specimens over time. However, imaging of living cells over time is difficult and super resolution imaging is computationally demanding. In this review, we discuss the advantages/disadvantages of different super resolution systems for imaging fixed live specimens, with particular regard to cytoskeleton structures.
Keywords: microscopy, live cell imaging, cytoskeleton
Introduction
Visualizing proteins indirectly in cells and tissues at the light microscopic level using antibodies conjugated to fluorochromes revolutionized the field of cell biology 40 years ago. In the late 1980s and early 1990s, the use of commercial, user-friendly, confocal microscopes in combination with digital image acquisition systems dramatically improved the image quality of fluorescently labeled specimens and made image capture easier by avoiding the vagaries of the dark room. The technical benefits of confocal over “conventional” microscopy include the removal of the out-of-focus glare that interferes with what the imager really wishes to observe and an increase in specimen contrast. More recently, the ability to follow tagged molecules in live cells using both conventional and confocal fluorescence microscopy and the development of molecular biosensors has allowed cell biologists to study the dynamics of individual proteins and protein complexes with high precision. Conventional techniques used for light microscopy can achieve resolutions of up to ~100 nm in the image plane, as first detailed by Abbe 1, and about twice this value along the focal axis 2. In practice, most authors consider the resolution of commercially available microscopes to be ~200 nm in the image plane and ~500 nm in the axial dimension. So called “super resolution” is achieved with techniques that allow resolution beyond the diffraction limit of conventional optics 3. The term super resolution as applied to microscopy has been in use since at least as early as the 1960s 4. Resonance energy transfer 5, near field scanning optical microscopy 6, dual objective (4Pi) microscopy 7, total internal reflection microscopy 8, single molecule fluorescence localization 9, expansion microscopy 10, and several other strategies represent successful efforts to obtain structural or positional information from biological specimens at resolutions better than those afforded by conventional microscopy. However, many of these approaches are technically demanding or present significant limits to the type of specimens that can be examined. The last decade has seen the development of methods that closely resemble more familiar far-field and laser scanning confocal microscopy but allow direct visualization of subdiffraction size structures in fixed and living specimens. Current super resolution techniques provide resolutions of less than 10 nm in the image plane 11 and ~20 nm in the z-axis. The improved resolution offered by these methods has produced breathtaking images of the nuclear pore complex 12, 13, the tubular walls of microtubules 14, and many other structures.
Although the potential and allure of super resolution methods are indisputable, they also present new challenges to image acquisition, storage, and interpretation. For example, changing the resolution of an image from 200 nanometers to 20 nanometers in the image (xy) plane, while maintaining a fixed field of view, results in a 100-fold increase in image size. Extending these calculations to the third dimension, multispectral imaging and time (for live cells) reveals the potential extent of demands that super resolution microscopy can place on specimens and fluorophores as well as imaging and data processing hardware. Moreover, although computer and imaging technologies have advanced to the point where accumulating these data is feasible, human involvement is currently still needed to identify regions of interest for examination and analysis. Each of the methods used to achieve super resolution imaging also offer unique technical strengths and weaknesses to cell biologists. We direct the reader to several excellent recent reviews that provide an overview of the capabilities, advantages, and disadvantages of a variety of super resolution procedures in tabular form 15– 18.
Despite the inherent challenges, the use of super resolution microscopy is beginning to make an impact on a wide variety of biological topics. Among the subjects most likely to benefit from the application of super resolution imaging is the study of the cytoskeleton and its associated structures. Unlike many other cellular components, cytoskeletal filaments form anastomosing networks of fibers smaller than the resolution of conventional imaging methods. Techniques allowing enhanced resolution imaging of cytoskeletal structures, especially in live cells, are already advancing our knowledge of their formation and function. For example, super resolution has been a boon to investigators studying cytoskeletal rearrangements in bacteria 19, 20 and yeast cells 21, which have generally been too small to approach with conventional diffraction-limited imaging methods. Below, we briefly discuss the major methods for achieving super resolution as well as their strengths and weaknesses with emphasis on recent work in which these methods have been applied to address biological questions involving the cytoskeleton.
Localization microscopy
Stochastic optical reconstruction microscopy (STORM 22), photoactivated localization microscopy (PALM 23), fluorescence photoactivation localization microscopy (FPALM 24), and a growing number of related methods are techniques where fluorescent specimens are examined by activating a limited set of fluorophores at a time which must be separated by distances greater than the resolution limit of the microscope. A diffraction-limited image of the fluorophores is captured and the position and intensity of each fluorophore calculated at subdiffraction precision. Activated fluorophores are deactivated, and a new set of fluorophores is activated and imaged. After many images are collected, a completed super resolution image is calculated. The techniques differ in the type of fluorophore used. For example, PALM and FPALM are used to image expressed photoactivatable fluorescent proteins and fusion proteins 23, 24, while STORM is used to create images of fluorescent dyes and tags that can be switched between fluorescing and non-fluorescing states 22. In practice, these methods can produce the highest resolution of the available diffraction-unlimited techniques when applied to biological specimens, with two-dimensional resolutions of less than 20 nm frequently reported 12, 25– 27. However, overall image resolution and quality increase with the number of photons captured and fluorescent molecules examined. Therefore, these methods are limited by the time required to obtain a sufficient number of images – often tens of thousands – needed to create a final super resolution image. In addition, many conventional fluorophores and fluorescent proteins are not suitable for localization microscopy 14, 28, making some techniques such as multicolor staining methods challenging. Localization methods also require that fluorescent molecules or proteins within the specimen be detected individually, complicating the application of these techniques to densely labeled three-dimensional cytoskeletal arrays or arrays contained within thick samples exhibiting autofluorescence. To address this, some investigators have incorporated total internal reflection fluorescence illumination (TIRF), two photon illumination, or light sheet illumination to limit the volume of a specimen under inspection 23, 29– 31. However, these implementations increase the complexity of the instrumentation required. Despite these limitations, a large number of studies encountered in our review of current literature employ localization techniques. This may be because of both the superior resolution of the methods and the relatively simple hardware requirements which have allowed many investigators to build their own STORM or PALM/FPALM imaging systems. In addition, fluorophores, fluorescent proteins, buffering agents, and illumination strategies are being actively developed to extend localization to both multicolor labeling and other fluorescent imaging tasks (see 28, 32– 35 for representative reviews).
Among the most novel and dramatic discoveries made using localization methods is the periodic distribution of actin filaments and associated cytoskeletal proteins in axons first visualized using STORM imaging 36. The periodicity of this structural feature of neurons is below the resolution limit of conventional microscopy and was overlooked in thin section electron micrographs 37, 38. Other studies have further exploited single molecule/protein imaging to analyze the development and regulation of these arrays 39, 40. STORM and PALM methods are also being applied to the study of the structure of adhesion complexes such as hemidesmosomes 41, intercellular adherens junctions 42, 43, and the less well-defined adhesions formed by leucocytes 44, as well as actin cytoskeletal rearrangements that occur during endocytosis 45 and bacterial host cell invasion 46. In the case of adherens junctions, STORM analyses indicate that E-cadherin exists in clusters at sites of cell-cell interaction rather than as the “solid” belt typically observed by non-super resolution methods 42, 43. Similarly, dual-color PALM studies suggest that paxillin and vinculin form functionally distinct non-overlapping nanoaggregates in focal adhesions that are not detectable using conventional imaging methods 47.
Microtubules, 24 nm diameter tubes composed of protofilaments, are sparsely distributed at the edge of cultured cells and are often used as proof-of-concept targets by developers of super resolution imaging methods 48– 51. STORM and PALM imaging are beginning to provide new details regarding the function and organization of the microtubule cytoskeleton. For example, these techniques have been used to study the organization of centrosomal proteins in intact cells, the architecture of microtubules underlying the movement of organelles within living cells 52, and the interaction of kinesin motor proteins with microtubules in neuronal processes 53. A variant of PALM imaging has also been used to study the structure of EB1 at the distal tip of growing microtubules 54, and PALM has made possible the visualization of FtsZ, the bacterial homolog of eukaryotic tubulin, in distinct polymeric arrays in prokaryotes 20, 55.
Although relatively few studies have examined intermediate filament arrays using super resolution imaging, STORM has been used to investigate keratin, plectin, and integrins in hemidesmosomes formed by cultured keratinocytes 41. Additionally, desmin, a cytoskeletal protein mutated in clinically important cardiomyopathies, has been visualized using dual color PALM microscopy in cultured cardiomyocytes 56. In the latter study, the authors report a 10-fold increase in the resolution of desmin protein aggregates and filaments over non-super resolution light microscopic methods. More importantly, their super resolution images reveal that both mutant and wild-type desmin proteins are incorporated into the same filament, suggesting the possibility that changes in the mechanical properties of a “mixed” filament might be the cause of disease 56.
Structured illumination microscopy
Structured illumination microscopy (SIM) improves the resolution of light microscopy by illuminating a specimen with a defined regular pattern of diffraction-limited light and dark bands which create Moiré patterns when combined with the structure of a specimen 57. An image of the resulting interference pattern is created and recorded. The illuminating pattern is then rotated and further images captured. Finally, an image with improved resolution is calculated from the combined rotation series. Because the illuminating pattern is generated using wide-field optics, the technique exposes a specimen to illumination levels that are comparable to other wide-field microscopy methods (although multiple images must be captured for each view of the specimen). While many implementations of localization microscopy use total internal reflection illumination and are therefore limited to observation of structures within ~100 nm of an optical surface, SIM can resolve structures many microns deep within a specimen. SIM can also be used with any fluorescent probe and, unlike localization methods or laser scanning methods such as stimulated emission depletion microscopy (STED), complete images of a specimen are obtained at speeds determined by camera sensitivities. SIM is therefore one of the least phototoxic and most rapid methods for obtaining enhanced resolution images and has been used extensively in studies of living cells. A three-dimensional version of structured illumination allows for improved resolution in the z direction and has been used to visualize cytoskeletal structures in three dimensions. Both of these applications are discussed below.
The resolution achieved with SIM is generally only twofold greater than that offered by conventional microscopy 57, although some implementations allow SIM to achieve lateral resolutions as high as 50 nm 58, 59. However, even a twofold increase in resolution, combined with the power of multispectral fluorescent labeling methods, has allowed investigators to observe previously unresolved features of a wide variety of cytoskeletal structures. For example, SIM has been used to characterize the distribution of microtubules and associated structures in a variety of specimens that conventional diffraction-limited imaging methods have been unable to resolve well, including the neuromuscular junction 60, platelets 61, centrosomes 62, 63, and protists 64. SIM has also facilitated studies investigating the structure of striated muscle 65, 66 as well as actin and myosin filament organization in non-muscle cells 67– 70. Intermediate filament architecture and associated junctions have also been examined using this method 71, 72. Interestingly, although the localization-based imaging methods described above achieve higher resolution than SIM, the pointillized appearance of images produced by localization microscopy can obscure fine structural detail that may be visible using SIM. For example, SIM has revealed that focal adhesions comprise linear subarrays 73, a feature not readily visible in images published by investigators using interferometric (i)PALM to analyze the axial distribution of focal adhesion components 74– 76.
Finally, because SIM can be applied to microscopy of any fluorescent probe, it can be readily used for multispectral studies using conventional fluorophores. SIM has been used to examine cytoskeletal structures in triple-labeling studies of adherens junctions 77, 78, neuronal spines 79, centrioles 80, kinetochores 81, and endosomal vesicles 82. For example, SIM analyses of VASP, zyxin, testin, and other proteins surprisingly indicate that these tension-regulating proteins are likely recruited to adhesion junctions independent of the core adhesion complex 78.
Stimulated emission depletion microscopy
STED, reversible saturable optical fluorescence transitions microscopy (RESOLFT), and related techniques excite fluorophores in a diffraction-limited spot by a focused laser. Outlying fluorophores are converted to a non-fluorescent state by illumination with a second (depletion) laser in a manner that leaves a central, subdiffraction limited area of fluorophores unconverted 83– 85. The remaining still-fluorescent fluorophores are detected to create an image with resolutions far greater than those provided by conventional imaging methods. Some implementations of these techniques have yielded resolutions in biological specimens of <50 nm in the image plane and 150 nm in the axial dimension 86. Common implementations of STED are technically demanding and the depletion light energies are substantially greater than the illumination intensities required by other super resolution imaging methods. However, STED and similar systems closely resemble laser scanning confocal microscopes already employed by many investigators, and these methods can be applied to most commonly available fluorophores. Unlike localization methods and SIM, STED does not require calculations to generate enhanced resolution images, and image resolution can be readily varied by changing the raster scanning patterns used to visualize a specimen. Scanning rates achieved by STED are suitable for imaging of live specimens 87, 88, but the required depletion energies currently make extended live cell imaging a challenge. However, super resolution imaging can also be interchanged with conventional confocal scanning approaches simply by turning the depletion laser on or off.
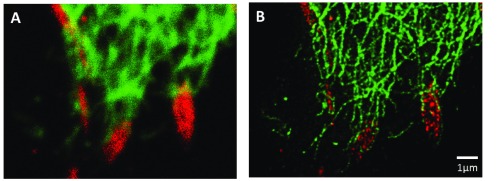
STED has been used to visualize the periodic ring structure of actin in neurons 89 and actin dynamics in dendritic spines of neurons in living brain tissue 90. Others have used STED to examine the actin-like MreB protein in bacteria 91 and the reorganization of actin filament arrays during activation of natural killer cells and T cells 92, 93. It has allowed the visualization of myosin mini-filament formation in mammalian non-muscle cells 94 and insight into the regulation of the actin cytoskeleton by intracellular signaling proteins 95, 96. Microtubules have been examined using STED in primary cilia 97, and the interphase microtubule array has been examined in muscle and non-muscle cells 98, 99. Finally, several groups have utilized STED microscopy to visualize elements of the intermediate filament network, including vimentin 100, 101, keratin 102, and nuclear lamins 103. In our laboratory, we have used STED to assay the relative localization of vimentin and focal adhesions in cultured epithelial cells. Whether vimentin interacts with focal adhesions has been controversial for years 104, 105. However, STED not only provides images with more detail than can be obtained using conventional confocal microscopy but also reveals a distinct pattern of organization of paxillin within a focal adhesion ( Figure 1). Moreover, in the STED image, vimentin filaments clearly wrap around each focal adhesion, an interaction that is not apparent in the confocal image.
Figure 1. Association of paxillin (Alexafluor 555, red) and vimentin (Oregon green 488, green) at focal adhesions located at the leading edge of a migrating A549 lung cell.
Conventional confocal imaging is shown in A while STED imaging of the same area is shown in B.
Live cells
The ability to study proteins in cells in the living state is one of the most significant advantages of light microscopy over other methods of analysis. However, as has been noted by many others (see 106– 108 for representative reviews), obtaining images of living cells that are doing something other than dying on the microscope is difficult and involves balancing the competing requirement of image quality and cell health. In addition, the study of cytoskeletal dynamics often requires rapid image acquisition, increasing the light exposure of live cells over time. Achieving resolutions greater than the diffraction limit of conventional microscopy ultimately requires capturing larger numbers of photons from smaller areas of live cells than is necessary for conventional resolution images, an effort that can clearly compromise the integrity of cells under examination 108. Fluorescently labeled cytoskeletal filaments are also notoriously photolabile 109. To date, many investigators of the cytoskeleton in live cells have combined super resolution microscopy of fixed cells with wide-field or confocal imaging of live cells 39, 44, 45, 52, 53, 64, 68, 69, 77, 79, 98, 110– 113 or have obtained single images of live cells using super resolution methods 56, 89. Although the latter avoid potential artifacts induced by fixation and immunostaining, this approach does not address dynamic changes in cell architecture.
The majority of publications in our survey that examined dynamic changes in cytoskeletal architecture employed SIM. Observations of the cytoskeleton in live cells have been made over time scales ranging from seconds to tens of minutes using this approach. The twofold resolution enhancement offered by SIM has allowed investigators to examine in previously impossible detail the growth of microtubules in living plant cells 114, actin retrograde flow in cultured insect cells 115, and the reorganization of cytoskeletal arrays in dividing yeast and bacterial cells 21, 91. Two-color applications of SIM in living cells have been used to demonstrate heterotypic assembly of myosin II isoforms 67, the spatial relationship between myosin IIA and alpha actinin 116, the clustering of receptors in adherens junctions 117, and that vimentin intermediate filaments move bi-directionally along microtubules 118.
Although capturing clear images of closely apposed structures using localization methods requires accumulation and processing of thousands of images over periods of many seconds to minutes, localization methods have also been applied to the study of rapidly changing cytoskeletal structures in live cells by sacrificing some image clarity to improve temporal resolution. For example, PALM has been used to quantify the addition and loss of individual paxillin proteins at focal adhesion sites with a spatial resolution of 60 nm and temporal resolution of 25 seconds 119. Proof-of-principle studies have also demonstrated that PALM can be used to visualize dynamic changes in fluorescent actin filaments over brief intervals 120 and the movement of fluorescently labeled endosomes along microtubules in living axons 121. In addition, PALM imaging platforms have been exploited by several groups that have followed changes in localization of single fluorescent cytoskeletal proteins over time, either alone or in combination with PALM, STORM, or STED techniques 122– 125. This appears to be a powerful, multimodal approach that can place the movement of individual proteins in the context of cytoskeletal architecture.
STED employs depletion lasers at energies that are largely incompatible with extended viewing of live cells, and, not surprisingly, we encountered few publications where this method has been used to image cytoskeletal arrays in vivo. Nonetheless, STED has great potential for imaging structures within complex tissues and has been used in a seminal study of actin dynamics in living neurons within brain tissue 90. Single images of microtubules within living cells have also been obtained at 60 nm resolution using STED 86. STED has also been applied to imaging using multiphoton excitation 126– 128 and total internal reflection microscopy 129. These methods limit light exposure to a sample and offer further potential for the application of STED illumination to live cells. We anticipate that as these complex instruments become more widely available, we will see an increase in the number of investigators employing them for studies of living cells.
The third dimension
Many initial implementations of super resolution methods did not provide an increase in resolution in the third, or axial, dimension of the microscope (see 18 for review). However, recent modifications to these methods have achieved impressive resolution enhancements in the third dimension. Three-dimensional SIM doubles the resolving ability of the light microscope in all dimensions while retaining its ability to obtain images rapidly with low light exposures 130. Although many studies have captured Z-stacks of images using super resolution methods and generated extended focus images from them, relatively few studies have analyzed the cytoskeleton in three dimensions using super resolution microscopy. However, three-dimensional SIM has been used to resolve actin filament arrays and microtubules in three dimensions using both fixed and live cultured cells 113, 116, 130, 131 as well as to study the structure of centrosomes 63, 80 and kinetochores 81. Additionally, this method has also been used to visualize the three-dimensional organization of FtsZ in dividing bacteria 132. Modification of STORM and PALM imaging platforms can achieve axial resolution of up to 20 nm by introducing axial astigmatism into the optical path 133 and by generating interference patterns from images obtained with paired, opposed objective lenses 134. Three-dimensional STORM and PALM have been used to study the movement of organelles along microtubules in live cells and the formation of FtsZ ring structures in live dividing bacteria 20, 52. A STORM imaging method employing axial astigmatism and dual objectives has been used to show that sheet-like cellular extensions in cultured cells contain two distinctly separate actin filament arrays each with unique patterns of actin filament organization 134. The development and regulation of these previously undetected cytoskeletal arrays have been further analyzed using similar approaches in normal cell movement 112, 116. iPALM can resolve structures less than 20 nm in diameter in three dimensions 27. The technique has recently been used to dissect the organization of cytoskeletal proteins associated with matrix adhesion devices termed focal adhesions at an unprecedented level of detail 74– 76. Impressively, the technique was able to resolve not only vertical stratification enriched for specific cytoskeletal components but also the polarized orientation of the N- and C-terminal ends of talin within focal adhesions of intact cells. Finally, STED microscopy has also been modified by the addition of dual depletion patterns, one oriented in the image plane and the other in the axial dimension, allowing for increases in resolution in the third dimension of up to 125 nm 135. STED and RESOLFT have also been extended to the third dimension using a dual objective imaging strategy 136.
Presently, most work using super resolution imaging has been conducted using single cells cultured on optical surfaces. However, previous work has shown that cell morphology and behavior can be dramatically altered in three-dimensional environments 137, 138. To date, the literature examining the cytoskeleton in cells in situ is largely limited to proof-of-principle studies. However, these efforts demonstrate the growing potential of super resolution methods. For example, STED has been used to view the dynamics of actin filament arrays within live neurons in 350 μM thick brain slices 90, and three-dimensional SIM has been used to visualize actin arrays in developing Drosophila 115. In other studies, labeled nuclear histones have been visualized in cells within 150 μM cell spheroids by combining single molecule localization methods with planar illumination 139. Both planar and multi-photon illumination methods have been coupled with structured illumination to view green fluorescent protein (GFP) expressed in living nematodes 140, 141. These methods improve visualization by restricting effective illumination to a single plane of interest within thick specimens. While these latter studies examined structures other than the cytoskeleton, these methods show promise for the analysis of the cytoskeleton at subdiffraction resolutions in situ.
Summary
Many of us have been fortunate to work as cell biologists during two major revolutions in imaging technology: the development of fluorescent proteins as tools for biologists and the development of confocal microscopy, which extracts clear, in-focus images in which the contaminating blur of out-of-focus structures has been removed. The astonishment and wonder with which we now view images created by super resolution microscopy suggest that we are experiencing yet a third revolution in the technology available to cell biologists. It is likely that for dual and triple labeling studies of cultured cells in two and three dimensions, as well as studies of bright, relatively slow-moving structures in live cells, super resolution imaging will soon replace confocal microscopy as the state of the art in much the same way that confocal microscopy replaced conventional wide-field imaging in the 1980s and 1990s. However, each of the approaches used to achieve images at better than diffraction-limited resolution have strengths and weaknesses. Studies of rapid cytoskeletal dynamics in live cells and three-dimensional studies are likely to present technical and biological challenges to practitioners of super resolution microscopy for some time. In addition, while the best super resolution light microscopic methods achieve resolutions of <10 nm, this is still 50–100-fold greater than the resolution afforded by electron microscopy 142, 143. Conventional fluorescence microscopes can also take advantage of a myriad of probes for physiological conditions and molecular interactions that have yet to be adapted to super resolution imaging methods. For the time being, no method addresses all possible experimental needs, and investigators will likely have to address the limitations of their super resolution instruments with complementary approaches involving conventional methods. Moreover, the dream of seeing individual protein complexes and their partners functioning in live cells within a complex three-dimensional organism remains unrealized. Nonetheless, we are seeing the development of instrumentation, computational methods, fluorescent probes, and novel methods at an amazing pace. We can only imagine what the next advances will be.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Omar Skalli, University of Memphis, Memphis, TN, USA
Jeffrey Segall, Albert Einstein College of Medicine , Bronx, NY, 10461, USA
Patricia Wadsworth, University of Massachusetts Amherst, Amherst, MA, USA
Funding Statement
This work was supported by grants from the National Science Foundation (IOS 1457368) to EAS and National Institutes of Health (AR054184) to Jonathan C.R. Jones. Zachery T. Colburn was supported, in part, by a Poncin Scholarship.
[version 1; referees: 3 approved]
References
- 1. Abbe E: Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Archiv f mikrosk Anatomie. 1873;9(1):413–8. 10.1007/BF02956173 [DOI] [Google Scholar]
- 2. Francon M: Phase-contrast microscopy. Progress in Microscopy.New York: Row, Peterson & Co,1961. [Google Scholar]
- 3. Kolobov MI: Quantum Imaging.New York, NY: Springer New York;2007. 10.1007/0-387-33988-4 [DOI] [Google Scholar]
- 4. Lohmann AW, Paris DP: Superresolution for Nonbirefringent Objects. Appl Opt. 1964;3(9):1037 10.1364/AO.3.001037 [DOI] [Google Scholar]
- 5. Stryer L: Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem. 1978;47:819–46. 10.1146/annurev.bi.47.070178.004131 [DOI] [PubMed] [Google Scholar]
- 6. Pohl DW, Courjon D: Near field optics. Springer Science & Business Media.2012;242. [Google Scholar]
- 7. Hell S: Double-scanning confocal microscope. European patent. 1990;491289. [Google Scholar]
- 8. Axelrod D: Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89(1):141–5. 10.1083/jcb.89.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yildiz A, Forkey JN, McKinney SA, et al. : Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300(5628):2061–5. 10.1126/science.1084398 [DOI] [PubMed] [Google Scholar]
- 10. Chen F, Tillberg PW, Boyden ES: Optical imaging. Expansion microscopy. Science. 2015;347(6221):543–8. 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rittweger E, Han KY, Irvine SE, et al. : STED microscopy reveals crystal colour centres with nanometric resolution. Nature Photon. 2009;3:144–7. 10.1038/nphoton.2009.2 [DOI] [Google Scholar]
- 12. Löschberger A, van de Linde S, Dabauvalle MC, et al. : Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J Cell Sci. 2012;125(Pt 3):570–5. 10.1242/jcs.098822 [DOI] [PubMed] [Google Scholar]
- 13. Schermelleh L, Carlton PM, Haase S, et al. : Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320(5881):1332–6. 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dempsey GT, Vaughan JC, Chen KH, et al. : Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8(12):1027–36. 10.1038/nmeth.1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fornasiero EF, Opazo F: Super-resolution imaging for cell biologists: concepts, applications, current challenges and developments. Bioessays. 2015;37(4):436–51. 10.1002/bies.201400170 [DOI] [PubMed] [Google Scholar]
- 16. Galbraith CG, Galbraith JA: Super-resolution microscopy at a glance. J Cell Sci. 2011;124(Pt 10):1607–11. 10.1242/jcs.080085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Godin AG, Lounis B, Cognet L: Super-resolution microscopy approaches for live cell imaging. Biophys J. 2014;107(8):1777–84. 10.1016/j.bpj.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schermelleh L, Heintzmann R, Leonhardt H: A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190(2):165–75. 10.1083/jcb.201002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biteen JS, Goley ED, Shapiro L, et al. : Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astigmatism. Chemphyschem. 2012;13(4):1007–12. 10.1002/cphc.201100686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holden SJ, Pengo T, Meibom KL, et al. : High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci U S A. 2014;111(12):4566–71. 10.1073/pnas.1313368111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dudin O, Bendezú FO, Groux R, et al. : A formin-nucleated actin aster concentrates cell wall hydrolases for cell fusion in fission yeast. J Cell Biol. 2015;208(7):897–911. 10.1083/jcb.201411124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rust MJ, Bates M, Zhuang X: Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods. 2006;3(10):793–5. 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Betzig E, Patterson GH, Sougrat R, et al. : Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–5. 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- 24. Hess ST, Girirajan TP, Mason MD: Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91(11):4258–72. 10.1529/biophysj.106.091116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Greenfield D, McEvoy AL, Shroff H, et al. : Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7(6):e1000137. 10.1371/journal.pbio.1000137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones SA, Shim SH, He J, et al. : Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8(6):499–508. 10.1038/nmeth.1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shtengel G, Galbraith JA, Galbraith CG, et al. : Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A. 2009;106(9):3125–30. 10.1073/pnas.0813131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chozinski TJ, Gagnon LA, Vaughan JC: Twinkle, twinkle little star: photoswitchable fluorophores for super-resolution imaging. FEBS Lett. 2014;588(19):3603–12. 10.1016/j.febslet.2014.06.043 [DOI] [PubMed] [Google Scholar]
- 29. Baddeley D, Crossman D, Rossberger S, et al. : 4D super-resolution microscopy with conventional fluorophores and single wavelength excitation in optically thick cells and tissues. PLoS One. 2011;6(5):e20645. 10.1371/journal.pone.0020645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen BC, Legant WR, Wang K, et al. : Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346(6208):1257998. 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cella Zanacchi F, Lavagnino Z, Faretta M, et al. : Light-sheet confined super-resolution using two-photon photoactivation. PLoS One. 2013;8(7):e67667. 10.1371/journal.pone.0067667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lippincott-Schwartz J, Patterson GH: Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends Cell Biol. 2009;19(11):555–65. 10.1016/j.tcb.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shcherbakova DM, Sengupta P, Lippincott-Schwartz J, et al. : Photocontrollable fluorescent proteins for superresolution imaging. Annu Rev Biophys. 2014;43:303–29. 10.1146/annurev-biophys-051013-022836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wiedenmann J, Gayda S, Adam V, et al. : From EosFP to mIrisFP: structure-based development of advanced photoactivatable marker proteins of the GFP-family. J Biophotonics. 2011;4(6):377–90. 10.1002/jbio.201000122 [DOI] [PubMed] [Google Scholar]
- 35. Wu B, Piatkevich KD, Lionnet T, et al. : Modern fluorescent proteins and imaging technologies to study gene expression, nuclear localization, and dynamics. Curr Opin Cell Biol. 2011;23(3):310–7. 10.1016/j.ceb.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu K, Zhong G, Zhuang X: Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339(6118):452–6. 10.1126/science.1232251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones SL, Korobova F, Svitkina T: Axon initial segment cytoskeleton comprises a multiprotein submembranous coat containing sparse actin filaments. J Cell Biol. 2014;205(1):67–81. 10.1083/jcb.201401045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watanabe K, Al-Bassam S, Miyazaki Y, et al. : Networks of polarized actin filaments in the axon initial segment provide a mechanism for sorting axonal and dendritic proteins. Cell Rep. 2012;2(6):1546–53. 10.1016/j.celrep.2012.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ganguly A, Tang Y, Wang L, et al. : A dynamic formin-dependent deep F-actin network in axons. J Cell Biol. 2015;210(3):401–17. 10.1083/jcb.201506110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhong G, He J, Zhou R, et al. : Developmental mechanism of the periodic membrane skeleton in axons. eLife. 2014;3:e04581. 10.7554/eLife.04581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nahidiazar L, Kreft M, van den Broek B, et al. : The molecular architecture of hemidesmosomes, as revealed with super-resolution microscopy. J Cell Sci. 2015;128(20):3714–9. 10.1242/jcs.171892 [DOI] [PubMed] [Google Scholar]
- 42. Erami Z, Timpson P, Yao W, et al. : There are four dynamically and functionally distinct populations of E-cadherin in cell junctions. Biol Open. 2015;4(11):1481–9. 10.1242/bio.014159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu Y, Kanchanawong P, Zaidel-Bar R: Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions. Dev Cell. 2015;32(2):139–54. 10.1016/j.devcel.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 44. Wee JL, Schulze KE, Jones EL, et al. : Tetraspanin CD37 Regulates β 2 Integrin-Mediated Adhesion and Migration in Neutrophils. J Immunol. 2015;195(12):5770–9. 10.4049/jimmunol.1402414 [DOI] [PubMed] [Google Scholar]
- 45. Picco A, Mund M, Ries J, et al. : Visualizing the functional architecture of the endocytic machinery. eLife. 2015;4:e04535. 10.7554/eLife.04535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Han JJ, Kunde YA, Hong-Geller E, et al. : Actin restructuring during Salmonella typhimurium infection investigated by confocal and super-resolution microscopy. J Biomed Opt. 2014;19(1):16011. 10.1117/1.JBO.19.1.016011 [DOI] [PubMed] [Google Scholar]
- 47. Shroff H, Galbraith CG, Galbraith JA, et al. : Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci U S A. 2007;104(51):20308–13. 10.1073/pnas.0710517105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kner P, Chhun BB, Griffis ER, et al. : Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6(5):339–42. 10.1038/nmeth.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vicidomini G, Hernández IC, d'Amora M, et al. : Gated CW-STED microscopy: a versatile tool for biological nanometer scale investigation. Methods. 2014;66(2):124–30. 10.1016/j.ymeth.2013.06.029 [DOI] [PubMed] [Google Scholar]
- 50. York AG, Parekh SH, Dalle Nogare D, et al. : Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nat Methods. 2012;9(7):749–54. 10.1038/nmeth.2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang X, Chen X, Zeng Z, et al. : Development of a reversibly switchable fluorescent protein for super-resolution optical fluctuation imaging (SOFI). ACS Nano. 2015;9(3):2659–67. 10.1021/nn5064387 [DOI] [PubMed] [Google Scholar]
- 52. Bálint Š, Verdeny Vilanova I, Sandoval Álvarez Á, et al. : Correlative live-cell and superresolution microscopy reveals cargo transport dynamics at microtubule intersections. Proc Natl Acad Sci U S A. 2013;110(9):3375–80. 10.1073/pnas.1219206110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakata T, Niwa S, Okada Y, et al. : Preferential binding of a kinesin-1 motor to GTP-tubulin-rich microtubules underlies polarized vesicle transport. J Cell Biol. 2011;194(2):245–55. 10.1083/jcb.201104034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xia P, Liu X, Wu B, et al. : Superresolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol Biol Cell. 2014;25(25):4166–73. 10.1091/mbc.E14-06-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buss J, Coltharp C, Huang T, et al. : In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol. 2013;89(6):1099–120. 10.1111/mmi.12331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brodehl A, Hedde PN, Dieding M, et al. : Dual color photoactivation localization microscopy of cardiomyopathy-associated desmin mutants. J Biol Chem. 2012;287(19):16047–57. 10.1074/jbc.M111.313841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gustafsson MG: Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198(Pt 2):82–7. 10.1046/j.1365-2818.2000.00710.x [DOI] [PubMed] [Google Scholar]
- 58. Gustafsson MG: Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U S A. 2005;102(37):13081–6. 10.1073/pnas.0406877102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rego EH, Shao L, Macklin JJ, et al. : Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proc Natl Acad Sci U S A. 2012;109(3):E135–43. 10.1073/pnas.1107547108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pielage J, Cheng L, Fetter RD, et al. : A presynaptic giant ankyrin stabilizes the NMJ through regulation of presynaptic microtubules and transsynaptic cell adhesion. Neuron. 2008;58(2):195–209. 10.1016/j.neuron.2008.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Aslan JE, Phillips KG, Healy LD, et al. : Histone deacetylase 6-mediated deacetylation of α-tubulin coordinates cytoskeletal and signaling events during platelet activation. Am J Physiol Cell Physiol. 2013;305(12):C1230–9. 10.1152/ajpcell.00053.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fu J, Glover DM: Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2(8):120104. 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. He R, Huang N, Bao Y, et al. : LRRC45 is a centrosome linker component required for centrosome cohesion. Cell Rep. 2013;4(6):1100–7. 10.1016/j.celrep.2013.08.005 [DOI] [PubMed] [Google Scholar]
- 64. Chen CT, Kelly M, Leon JD, et al. : Compartmentalized Toxoplasma EB1 bundles spindle microtubules to secure accurate chromosome segregation. Mol Biol Cell. 2015;26(25):4562–76. 10.1091/mbc.E15-06-0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fernandes I, Schöck F: The nebulin repeat protein Lasp regulates I-band architecture and filament spacing in myofibrils. J Cell Biol. 2014;206(4):559–72. 10.1083/jcb.201401094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Granzier HL, Hutchinson KR, Tonino P, et al. : Deleting titin's I-band/A-band junction reveals critical roles for titin in biomechanical sensing and cardiac function. Proc Natl Acad Sci U S A. 2014;111(40):14589–94. 10.1073/pnas.1411493111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Beach JR, Shao L, Remmert K, et al. : Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24(10):1160–6. 10.1016/j.cub.2014.03.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brown AC, Oddos S, Dobbie IM, et al. : Remodelling of cortical actin where lytic granules dock at natural killer cell immune synapses revealed by super-resolution microscopy. PLoS Biol. 2011;9(9):e1001152. 10.1371/journal.pbio.1001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Santiago-Medina M, Gregus KA, Nichol RH, et al. : Regulation of ECM degradation and axon guidance by growth cone invadosomes. Development. 2015;142(3):486–96. 10.1242/dev.108266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Whalen K, Reitzel AM, Hamdoun A: Actin polymerization controls the activation of multidrug efflux at fertilization by translocation and fine-scale positioning of ABCB1 on microvilli. Mol Biol Cell. 2012;23(18):3663–72. 10.1091/mbc.E12-06-0438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shimi T, Kittisopikul M, Tran J, et al. : Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell. 2015;26(22):4075–86. 10.1091/mbc.E15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stahley SN, Warren MF, Feldman RJ, et al. : Super-Resolution Microscopy Reveals Altered Desmosomal Protein Organization in Tissue from Patients with Pemphigus Vulgaris. J Invest Dermatol. 2016;136(1):59–66. 10.1038/JID.2015.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hu S, Tee YH, Kabla A, et al. : Structured illumination microscopy reveals focal adhesions are composed of linear subunits. Cytoskeleton (Hoboken). 2015;72(5):235–45. 10.1002/cm.21223 [DOI] [PubMed] [Google Scholar]
- 74. Case LB, Baird MA, Shtengel G, et al. : Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat Cell Biol. 2015;17(7):880–92. 10.1038/ncb3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kanchanawong P, Shtengel G, Pasapera AM, et al. : Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468(7323):580–4. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu J, Wang Y, Goh WI, et al. : Talin determines the nanoscale architecture of focal adhesions. Proc Natl Acad Sci U S A. 2015;112(35):E4864–73. 10.1073/pnas.1512025112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guillaume E, Comunale F, Do Khoa N, et al. : Flotillin microdomains stabilize cadherins at cell-cell junctions. J Cell Sci. 2013;126(Pt 22):5293–304. 10.1242/jcs.133975 [DOI] [PubMed] [Google Scholar]
- 78. Oldenburg J, van der Krogt G, Twiss F, et al. : VASP, zyxin and TES are tension-dependent members of Focal Adherens Junctions independent of the α-catenin-vinculin module. Sci Rep. 2015;5: 17225. 10.1038/srep17225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Westin L, Reuss M, Lindskog M, et al. : Nanoscopic spine localization of Norbin, an mGluR5 accessory protein. BMC Neurosci. 2014;15:45. 10.1186/1471-2202-15-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sonnen KF, Schermelleh L, Leonhardt H, et al. : 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1(10):965–76. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wynne DJ, Funabiki H: Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J Cell Biol. 2015;210(6):899–916. 10.1083/jcb.201506020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Duleh SN, Welch MD: WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken). 2010;67(3):193–206. 10.1002/cm.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hell SW: Far-field optical nanoscopy. Science. 2007;316(5828):1153–8. 10.1126/science.1137395 [DOI] [PubMed] [Google Scholar]
- 84. Klar TA, Hell SW: Subdiffraction resolution in far-field fluorescence microscopy. Opt Lett. 1999;24(14):954–6. 10.1364/OL.24.000954 [DOI] [PubMed] [Google Scholar]
- 85. Hofmann M, Eggeling C, Jakobs S, et al. : Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci U S A. 2005;102(49):17565–9. 10.1073/pnas.0506010102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hein B, Willig KI, Hell SW: Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proc Natl Acad Sci U S A. 2008;105(38):14271–6. 10.1073/pnas.0807705105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Westphal V, Lauterbach MA, Di Nicola A, et al. : Dynamic far-field fluorescence nanoscopy. New J Phys. 2007;9:435 10.1088/1367-2630/9/12/435 [DOI] [Google Scholar]
- 88. Westphal V, Rizzoli SO, Lauterbach MA, et al. : Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320(5873):246–9. 10.1126/science.1154228 [DOI] [PubMed] [Google Scholar]
- 89. D'Este E, Kamin D, Göttfert F, et al. : STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep. 2015;10(8):1246–51. 10.1016/j.celrep.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 90. Urban NT, Willig KI, Hell SW, et al. : STED nanoscopy of actin dynamics in synapses deep inside living brain slices. Biophys J. 2011;101(5):1277–84. 10.1016/j.bpj.2011.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Reimold C, Defeu Soufo HJ, Dempwolff F, et al. : Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology. Mol Biol Cell. 2013;24(15):2340–9. 10.1091/mbc.E12-10-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rak GD, Mace EM, Banerjee PP, et al. : Natural killer cell lytic granule secretion occurs through a pervasive actin network at the immune synapse. PLoS Biol. 2011;9(9):e1001151. 10.1371/journal.pbio.1001151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tamarit B, Bugault F, Pillet AH, et al. : Membrane microdomains and cytoskeleton organization shape and regulate the IL-7 receptor signalosome in human CD4 T-cells. J Biol Chem. 2013;288(12):8691–701. 10.1074/jbc.M113.449918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Shutova MS, Spessott WA, Giraudo CG, et al. : Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Curr Biol. 2014;24(17):1958–68. 10.1016/j.cub.2014.07.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ashour DJ, Pelka B, Jaaks P, et al. : The catalytic domain of inositol-1,4,5-trisphosphate 3-kinase-a contributes to ITPKA-induced modulation of F-actin. Cytoskeleton (Hoboken). 2015;72(2):93–100. 10.1002/cm.21208 [DOI] [PubMed] [Google Scholar]
- 96. Gad AK, Rönnlund D, Spaar A, et al. : Rho GTPases link cellular contractile force to the density and distribution of nanoscale adhesions. FASEB J. 2012;26(6):2374–82. 10.1096/fj.11-195800 [DOI] [PubMed] [Google Scholar]
- 97. Yang TT, Hampilos PJ, Nathwani B, et al. : Superresolution STED microscopy reveals differential localization in primary cilia. Cytoskeleton (Hoboken). 2013;70(1):54–65. 10.1002/cm.21090 [DOI] [PubMed] [Google Scholar]
- 98. Oddoux S, Zaal KJ, Tate V, et al. : Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements. J Cell Biol. 2013;203(2):205–13. 10.1083/jcb.201304063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rothmeier E, Pfaffinger G, Hoffmann C, et al. : Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog. 2013;9(9):e1003598. 10.1371/journal.ppat.1003598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rathje LS, Nordgren N, Pettersson T, et al. : Oncogenes induce a vimentin filament collapse mediated by HDAC6 that is linked to cell stiffness. Proc Natl Acad Sci U S A. 2014;111(4):1515–20. 10.1073/pnas.1300238111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rönnlund D, Gad AK, Blom H, et al. : Spatial organization of proteins in metastasizing cells. Cytometry A. 2013;83(9):855–65. 10.1002/cyto.a.22304 [DOI] [PubMed] [Google Scholar]
- 102. Kayser J, Grabmayr H, Harasim M, et al. : Assembly kinetics determine the structure of keratin networks. Soft Matter. 2012;8:8873–8879. 10.1039/c2sm26032h [DOI] [Google Scholar]
- 103. Depreux FF, Puckelwartz MJ, Augustynowicz A, et al. : Disruption of the lamin A and matrin-3 interaction by myopathic LMNA mutations. Hum Mol Genet. 2015;24(15):4284–95. 10.1093/hmg/ddv160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bhattacharya R, Gonzalez AM, Debiase PJ, et al. : Recruitment of vimentin to the cell surface by beta3 integrin and plectin mediates adhesion strength. J Cell Sci. 2009;122(Pt 9):1390–400. 10.1242/jcs.043042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gonzales M, Weksler B, Tsuruta D, et al. : Structure and function of a vimentin-associated matrix adhesion in endothelial cells. Mol Biol Cell. 2001;12(1):85–100. 10.1091/mbc.12.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Frigault MM, Lacoste J, Swift JL, et al. : Live-cell microscopy - tips and tools. J Cell Sci. 2009;122( Pt 6):753–67. 10.1242/jcs.033837 [DOI] [PubMed] [Google Scholar]
- 107. Schneckenburger H, Weber P, Wagner M, et al. : Light exposure and cell viability in fluorescence microscopy. J Microsc. 2012;245(3):311–8. 10.1111/j.1365-2818.2011.03576.x [DOI] [PubMed] [Google Scholar]
- 108. Wäldchen S, Lehmann J, Klein T, et al. : Light-induced cell damage in live-cell super-resolution microscopy. Sci Rep. 2015;5: 15348. 10.1038/srep15348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Vigers GP, Coue M, McIntosh JR: Fluorescent microtubules break up under illumination. J Cell Biol. 1988;107(3):1011–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Fernández-Martín L, Marcos-Ramiro B, Bigarella CL, et al. : Crosstalk between reticular adherens junctions and platelet endothelial cell adhesion molecule-1 regulates endothelial barrier function. Arterioscler Thromb Vasc Biol. 2012;32(8):e90–102. 10.1161/ATVBAHA.112.252080 [DOI] [PubMed] [Google Scholar]
- 111. Guizetti J, Schermelleh L, Mäntler J, et al. : Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331(6024):1616–20. 10.1126/science.1201847 [DOI] [PubMed] [Google Scholar]
- 112. Luo W, Yu CH, Lieu ZZ, et al. : Analysis of the local organization and dynamics of cellular actin networks. J Cell Biol. 2013;202(7):1057–73. 10.1083/jcb.201210123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Versaevel M, Braquenier JB, Riaz M, et al. : Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Sci Rep. 2014;4: 7362. 10.1038/srep07362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Komis G, Mistrik M, Samajová O, et al. : Dynamics and organization of cortical microtubules as revealed by superresolution structured illumination microscopy. Plant Physiol. 2014;165(1):129–48. 10.1104/pp.114.238477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zobel T, Bogdan S: A high resolution view of the fly actin cytoskeleton lacking a functional WAVE complex. J Microsc. 2013;251(3):224–31. 10.1111/jmi.12020 [DOI] [PubMed] [Google Scholar]
- 116. Burnette DT, Shao L, Ott C, et al. : A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J Cell Biol. 2014;205(1):83–96. 10.1083/jcb.201311104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Indra I, Hong S, Troyanovsky R, et al. : The adherens junction: a mosaic of cadherin and nectin clusters bundled by actin filaments. J Invest Dermatol. 2013;133(11):2546–54. 10.1038/jid.2013.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hookway C, Ding L, Davidson MW, et al. : Microtubule-dependent transport and dynamics of vimentin intermediate filaments. Mol Biol Cell. 2015;26(9):1675–86. 10.1091/mbc.E14-09-1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Shroff H, Galbraith CG, Galbraith JA, et al. : Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5(5):417–23. 10.1038/nmeth.1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wilmes S, Staufenbiel M, Lisse D, et al. : Triple-color super-resolution imaging of live cells: resolving submicroscopic receptor organization in the plasma membrane. Angew Chem Int Ed Engl. 2012;51(20):4868–71. 10.1002/anie.201200853 [DOI] [PubMed] [Google Scholar]
- 121. Mudrakola HV, Zhang K, Cui B: Optically resolving individual microtubules in live axons. Structure. 2009;17(11):1433–41. 10.1016/j.str.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chazeau A, Mehidi A, Nair D, et al. : Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J. 2014;33(23):2745–64. 10.15252/embj.201488837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rossier O, Octeau V, Sibarita JB, et al. : Integrins β 1 and β 3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat Cell Biol. 2012;14(10):1057–67. 10.1038/ncb2588 [DOI] [PubMed] [Google Scholar]
- 124. Tatavarty V, Das S, Yu J: Polarization of actin cytoskeleton is reduced in dendritic protrusions during early spine development in hippocampal neuron. Mol Biol Cell. 2012;23(16):3167–77. 10.1091/mbc.E12-02-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tatavarty V, Kim EJ, Rodionov V, et al. : Investigating sub-spine actin dynamics in rat hippocampal neurons with super-resolution optical imaging. PLoS One. 2009;4(11):e7724. 10.1371/journal.pone.0007724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ding JB, Takasaki KT, Sabatini BL: Supraresolution imaging in brain slices using stimulated-emission depletion two-photon laser scanning microscopy. Neuron. 2009;63(4):429–37. 10.1016/j.neuron.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Li Q, Wu SS, Chou KC: Subdiffraction-limit two-photon fluorescence microscopy for GFP-tagged cell imaging. Biophys J. 2009;97(12):3224–8. 10.1016/j.bpj.2009.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Moneron G, Hell SW: Two-photon excitation STED microscopy. Opt Express. 2009;17(17):14567–73. 10.1364/OE.17.014567 [DOI] [PubMed] [Google Scholar]
- 129. Gould TJ, Myers JR, Bewersdorf J: Total internal reflection STED microscopy. Opt Express. 2011;19(14):13351–7. 10.1364/OE.19.013351 [DOI] [PubMed] [Google Scholar]
- 130. Gustafsson MG, Shao L, Carlton PM, et al. : Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94(12):4957–70. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Shao L, Kner P, Rego EH, et al. : Super-resolution 3D microscopy of live whole cells using structured illumination. Nat Methods. 2011;8(12):1044–6. 10.1038/nmeth.1734 [DOI] [PubMed] [Google Scholar]
- 132. Strauss MP, Liew AT, Turnbull L, et al. : 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 2012;10(9):e1001389. 10.1371/journal.pbio.1001389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Huang B, Jones SA, Brandenburg B, et al. : Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008;5(12):1047–52. 10.1038/nmeth.1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Xu K, Babcock HP, Zhuang X: Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods. 2012;9(2):185–8. 10.1038/nmeth.1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Harke B, Ullal CK, Keller J, et al. : Three-dimensional nanoscopy of colloidal crystals. Nano Lett. 2008;8(5):1309–13. 10.1021/nl073164n [DOI] [PubMed] [Google Scholar]
- 136. Böhm U, Hell SW, Schmidt R: 4Pi-RESOLFT nanoscopy. Nat Commun. 2016;7: 10504. 10.1038/ncomms10504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Shelden E, Knecht DA: Mutants lacking myosin II cannot resist forces generated during multicellular morphogenesis. J Cell Sci. 1995;108(Pt 3):1105–15. [DOI] [PubMed] [Google Scholar]
- 138. Wu PH, Giri A, Sun SX, et al. : Three-dimensional cell migration does not follow a random walk. Proc Natl Acad Sci U S A. 2014;111(11):3949–54. 10.1073/pnas.1318967111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Cella Zanacchi F, Lavagnino Z, Perrone Donnorso M, et al. : Live-cell 3D super-resolution imaging in thick biological samples. Nat Methods. 2011;8(12):1047–9. 10.1038/nmeth.1744 [DOI] [PubMed] [Google Scholar]
- 140. Gao L, Shao L, Higgins CD, et al. : Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell. 2012;151(6):1370–85. 10.1016/j.cell.2012.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Winter PW, York AG, Nogare DD, et al. : Two-photon instant structured illumination microscopy improves the depth penetration of super-resolution imaging in thick scattering samples. Optica. 2014;1(3):181–91. 10.1364/OPTICA.1.000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Bartesaghi A, Merk A, Banerjee S, et al. : 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348(6239):1147–51. 10.1126/science.aab1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Phillipp F: Advances in High-Resolution Transmission Electron Microscopy. Materials Transactions JIM. 1998;39(9):888–902. 10.2320/matertrans1989.39.888 [DOI] [Google Scholar]