Abstract
The sense of smell collects vital information about the environment by detecting a multitude of chemical odorants. Breadth and sensitivity are provided by a huge number of chemosensory receptor proteins, including more than 1,400 olfactory receptors (ORs). Organizing the sensory information generated by these receptors so that it can be processed and evaluated by the central nervous system is a major challenge. This challenge is overcome by monogenic and monoallelic expression of OR genes. The single OR expressed by each olfactory sensory neuron determines the neuron’s odor sensitivity and the axonal connections it will make to downstream neurons in the olfactory bulb. The expression of a single OR per neuron is accomplished by coupling a slow chromatin-mediated activation process to a fast negative-feedback signal that prevents activation of additional ORs. Singular OR activation is likely orchestrated by a network of interchromosomal enhancer interactions and large-scale changes in nuclear architecture.
Keywords: stochastic expression, interchromosomal interactions, chromatin, enhancer, allelic exclusion, nuclear architecture
INTRODUCTION: THE LOGIC OF THE OLFACTORY SYSTEM
The sense of smell provides vital information about the surrounding environment. An odor may indicate that food or a predator is nearby, or it may carry social information about conspecifics. This information is conveyed by a vast diversity of chemical compounds, referred to as odorants, which are detected by the olfactory system. How an odorant is perceived depends on its physical and chemical features, such as its molecular size and shape, and the presence of chemical functional groups (Haddad et al. 2008, Saito et al. 2009). Real-world odors, such as the smell of wine or a rose, may be a complex mixture of tens or hundreds of different odorants (Aycı et al. 2005, Aznar et al. 2001), so the olfactory system must be able to discern differences among complex mixtures of odorants. This discriminatory power can be vitally important, for example, in determining whether food is fresh or spoiled. Thus, the task of the olfactory system is to take in a complex mixture of odorants, generate a response that reflects the composition of this mixture, and relay this information to the brain in an organized manner. The olfactory system is remarkably good at this task. Even the human olfactory system, which is relatively reduced in size and receptor diversity compared with those of most mammals, is nonetheless able to discriminate an extraordinary number of different smells (Bushdid et al. 2014).
Odorant Detection
The vast diversity of odorants detected by the olfactory system is mirrored by a large repertoire of chemosensory proteins. The olfactory system detects odorants with G protein–coupled receptors (GPCRs) expressed on the surface of sensory neurons. The olfactory GPCRs belong to several different families: the olfactory receptors (ORs), trace amine–associated receptors (TAARs), type-1 and type-2 vomeronasal receptors (V1Rs and V2Rs), and, in rodents, the formyl peptide receptors (FPRs). In mice, these four families include more than 2,000 receptor genes. Although several hundred of these genes are nonfunctional pseudogenes, at least 1,300 code for full-length receptors (Clowney et al. 2011, Ishii & Mombaerts 2011, Liberles & Buck 2006, Rodriguez et al. 2002). Together, these chemosensory proteins account for the ability of the olfactory system to detect odorants from the environment.
The ORs compose the largest family of olfactory chemosensory receptors in mammals. Mice have 1,431 OR genes, of which 1,075 are intact (Buck & Axel 1991, Clowney et al. 2011). There are two subfamilies of ORs: 160 type I ORs that are related to ORs found in fish and other vertebrates, and 1,271 mammal-specific type II ORs. Type I ORs are all encoded by a large gene cluster located on mouse chromosome 7, whereas the type II OR genes are arranged in clusters that are scattered across most chromosomes. Both types of ORs are expressed by the olfactory sensory neurons (OSNs) of the main olfactory epithelium (MOE), which is the largest structure of the olfactory system. Although the odorant ligands recognized by most ORs remain unknown, in vitro and in vivo studies have recently been making rapid progress toward identification of pairings of odorants and ORs (McClintock et al. 2014, Nara et al. 2011). Odorant–OR pairing is combinatorial: each odorant activates multiple ORs, and higher concentrations of an odorant activate additional ORs (Malnic et al. 1999, McClintock et al. 2014, Nara et al. 2011). Most ORs appear to be tuned toward specific odorants, or odorants with a specific physicochemical feature, but some ORs are broadly tuned and recognize many, structurally diverse odorants (Grosmaitre et al. 2009, Nara et al. 2011).
Other families of olfactory GPCRs detect specialized types of odorants. The TAARs are a family of GPCRs that have high sensitivity to amine odorants (Dewan et al. 2013, Liberles & Buck 2006, Pacifico et al. 2012, Zhang et al. 2013). Fourteen of the 15 TAAR genes are expressed by a small number of OSNs in the MOE that do not express ORs. Olfactory TAARs detect several behaviorally important odorants, including death-associated diamines such as cadaverine and β-phenylethylamine, an amine odorant that is enriched in the urine of carnivores (Dewan et al. 2013, Ferrero et al. 2011, Hussain et al. 2013).
V1Rs and V2Rs are expressed by vomeronasal sensory neurons (VSNs) in the vomeronasal organ (VNO), a structure located at the base of the nasal cavity (Dulac & Axel 1995, Herrada & Dulac 1997, Matsunami & Buck 1997, Ryba & Tirindelli 1997). V1Rs and V2Rs are expressed by distinct populations of VSNs within the VNO; V1Rs are expressed by apically located type I VSNs, and V2Rs are expressed by basally located type II VSNs. These receptors primarily detect pheromones, which are chemical signals sent between conspecifics (Halpern 1987). The deletion of a large V1R cluster results in mating deficits (Del Punta et al. 2002), whereas mutations that affect vomeronasal receptor signaling alter mating behaviors and aggression (Kimchi et al. 2007, Stowers et al. 2002), supporting the role of these genes in social interactions. In rodents, the VNO also includes neurons expressing GPCRs of the FPR family. Five of the seven FPR genes are expressed in the VNO; the remaining members are expressed in the immune system (Liberles et al. 2009, Rivière et al. 2009). It has been proposed that FPRs recognize pathogen-associated odorants, such as formylated signal peptides from bacteria (Bufe et al. 2012, 2015; Rivière et al. 2009).
The Sorting of Odorant Information
In aggregate, the sensory neurons of the olfactory system express thousands of receptors, each of which may recognize many odorants with varying affinity. How is this information gathered and organized? Great progress has been made in understanding how this is achieved for ORs. Central to this process is monogenic and monoallelic OR expression: Each OSN expresses only one allele of one OR gene. This review examines how the choice of a single OR for expression serves to organize olfactory information. We also examine how singular OR expression is achieved. We focus on OR choice in mice, but similar mechanisms appear to control OR choice across vertebrates (Ferreira et al. 2014, Mori et al. 2000, Ngai et al. 1993).
OLFACTORY RECEPTOR CHOICE DEFINES OLFACTORY SENSORY NEURON IDENTITY
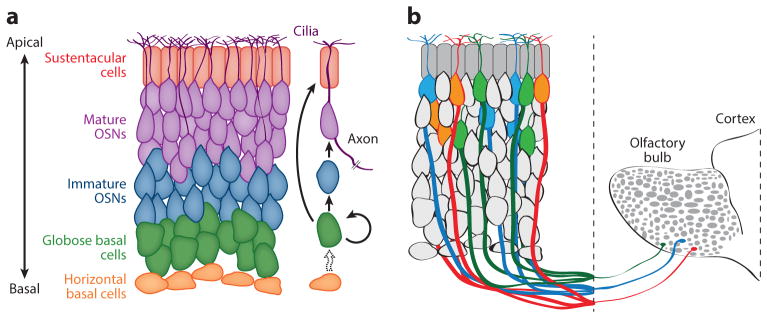
The MOE is lined with neurogenic, pseudostratified epithelium (Figure 1a), which is home to OSNs and various non-neuronal cell types, including supporting sustentacular cells. In addition, the MOE is home to two basally located stem cell populations: a cycling population of globose basal cells (GBCs), which give rise to OSNs and sustentacular cells, and a quiescent population of horizontal basal cells (HBCs), which can regenerate the GBCs. OSNs have variable but finite life spans, and are continuously regenerated from the cycling GBC population (Kondo et al. 2010). Mature OSNs reside toward the apical side of the tissue and extend dendritic cilia into the nasal lumen, providing a surface for OR proteins to bind odorants from the environment. The axon of each OSN projects from the basal surface of the neuron. Axons from many OSNs fasciculate into bundles, cross the cribriform plate separating the nasal cavity from the brain, and extend, without branching, into the olfactory bulb. Olfactory axons project to specialized structures in the olfactory bulb called glomeruli. In these glomeruli, olfactory axons form synapses with dendrites from mitral and tufted neurons of the olfactory bulb. After initial processing in the olfactory bulb, olfactory information is relayed to higher brain regions, such as the piriform cortex and the medial amygdala (Sosulski et al. 2011).
Figure 1.
Structure of the olfactory system. (a) Pseudostratified organization of the principal cell types of the main olfactory epithelium. Mitotically active globose basal cells (GBCs) differentiate into sustentacular cells and olfactory sensory neurons to replace cells lost to cell death. Quiescent horizontal basal cells differentiate into GBCs as needed (dotted arrow), particularly in the case of injury. For clarity, most olfactory sensory neuron (OSN) dendrites and axons are not shown. (b) OSNs expressing three different olfactory receptors (ORs) ( green, orange, and blue). Neurons expressing each OR are scattered throughout the tissue. Axons from neurons expressing the same OR converge onto the same target glomerulus (ovals) in the olfactory bulb.
The large repertoire of OR genes gives rise to diversity among the OSNs of the MOE. Each OSN expresses only one of the approximately 1,100 functional OR genes (Chess et al. 1994, Clowney et al. 2012). Moreover, only one allele of the chosen OR gene is expressed (Chess et al. 1994, Shykind et al. 2004). The identity of the OR expressed by an OSN defines its odorant sensitivity (Bozza et al. 2002), although some variability in odorant sensitivity and response kinetics has been reported for OSNs expressing the same OR (Grosmaitre et al. 2006). As a result, monogenic OR choice effectively defines the odorant sensitivity of approximately 1,100 distinct OSN cell types. Monoallelic expression increases this diversity further in cases in which there are functional differences between the two alleles of an OR.
The expression of an OR is a critical checkpoint in OSN maturation. OSNs that cannot activate an olfactory receptor due to a mutation affecting OR choice become arrested in an immature state (Lyons et al. 2013). These cells continue to express immature markers and fail to express markers of mature OSNs, such as Adcy3. This arrest can be bypassed by the ectopic expression of an OR from a transgene with an inducible promoter.
RNA in situ hybridization studies have shown that individual ORs are expressed within constrained zones of the MOE (Ressler et al. 1993, Strotmann et al. 1992, Vassar et al. 1993). Early analyses proposed that the MOE is divided into four zones and that each OR is expressed within one of these four zones, but a more comprehensive follow-up study suggested that zones of expression are specific to each OR (Miyamichi et al. 2005). Within its zone of expression, an OR is expressed in a punctate, seemingly stochastic pattern.
The axons of neurons expressing the same OR converge onto a specific pair of glomeruli in the olfactory bulb (Figure 1b) (Mombaerts et al. 1996, Ressler et al. 1994, Vassar et al. 1994). This targeting is partially determined by the graded expression of axon-guidance molecules along the dorsal–ventral axis of the MOE (Mori & Sakano 2011, Takeuchi et al. 2010). Because ORs are expressed in defined zones, OSNs expressing the same OR will have similar levels of these guidance molecules. However, axon targeting also depends on the identity of the OR expressed by each OSN. Even slight differences in the amino acid sequence of the OR protein may alter the pattern of axonal projection (Feinstein & Mombaerts 2004, Feinstein et al. 2004). This targeting has been shown to be influenced by the basal odorant-independent signaling activity of the OR protein (Nakashima et al. 2013). Basal activity varies among ORs and influences the expression of guidance molecules involved in anterior–posterior patterning, potentially providing a mechanism to couple OR identity to axon targeting. OR transgenes are sometimes expressed in a different zone from the endogenous OR, resulting in OR expression with a different set of guidance molecules and the projection of an axon to a different glomerulus (Vassalli et al. 2002). Remarkably, the expression of an OR transgene outside its normal zone can reroute axons expressing the matching endogenous OR (Ma et al. 2014, Tsai & Barnea 2014, Vassalli et al. 2002). These findings raise the possibility that the identity of the expressed OR directly contributes to targeting. OR protein is detectable on the OSN axons, which may allow the OR protein to function directly in axon guidance (Barnea et al. 2004, Feinstein et al. 2004, Strotmann et al. 2004).
The mechanisms governing OR choice are flexible and can accommodate a new OR. The β2-adrenergic receptor is the non-OR GPCR with the highest similarity to ORs, and it has several sequence features that are highly conserved among ORs (Feinstein et al. 2004, Nakashima et al. 2013). When the β2-adrenergic receptor is introduced into mice as a transgene with an OR promoter, it is expressed monogenically and monoallelically, like a normal OR (Feinstein et al. 2004). A similar outcome was obtained when targeted mutagenesis was used to replace the coding sequence of an endogenous OR with the coding sequence of the β2-adrenergic receptor (Nakashima et al. 2013, Omura et al. 2014). OSNs expressing the β2-adrenergic receptor target to discrete glomeruli, and these glomeruli show activity in response to a known β2-adrenergic receptor ligand, isoproterenol (Feinstein et al. 2004, Nakashima et al. 2013, Omura et al. 2014).
Each glomerulus pools information from a population of OSNs, all expressing the same OR. As a result of this organization, a given odorant at a given concentration will manifest as a particular pattern of glomerular activation, reflecting the degree to which it activates each OR (Mori & Sakano 2011). Odorant mixtures will activate glomeruli in a combinatorial fashion, and a given odorant mixture will generate a particular pattern of glomerular activation.
NEGATIVE FEEDBACK ENFORCES MONOGENIC AND MONOALLELIC EXPRESSION OF OLFACTORY RECEPTORS
The singular expression of ORs is enforced by negative feedback; the expression of one OR allele prevents the expression of all other ORs, including ORs that have identical regulatory sequences, such as the second allele of the chosen OR (Figure 2). This phenomenon is referred to as allelic exclusion.
Figure 2.
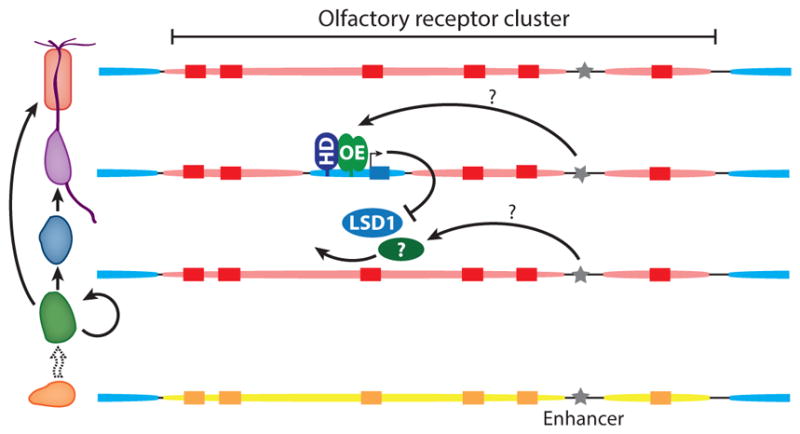
Epigenetic regulation of olfactory receptor (OR) genes. OR genes (rectangles) are embedded within cluster-wide chromatin domains. In horizontal basal cells, OR clusters are decorated with histone modifications ( yellow) characteristic of facultative heterochromatin (H3K9me2 and H4K20me2); flanking euchromatin is blue. In sustentacular cells and immature olfactory sensory neurons, OR clusters bear marks of constitutive heterochromatin (H3K9me3 and H4K20me3) (red ovals). OR activation involves the derepression of a single OR by the histone demethylase LSD1 and additional, unidentified chromatin-modifying enzymes. Following derepression, OR transcription is induced by the binding of O/E family (OE) and homeodomain (HD) transcription factors to OR promoters. OR transcription triggers a negative feedback mechanism that prevents derepression of additional ORs by downregulating LSD1. OR enhancer elements ( gray stars) are euchromatic in main olfactory epithelium tissue as a whole, but their chromatin state has not been studied in individual cell types. OR enhancers could contribute to OR activation by targeting OR derepression, by activating OR transcription following derepression, or both.
As described in the section Olfactory Receptor Choice Defines Olfactory Sensory Neuron Identity, each OSN expresses only one allele of one OR. Singular expression was originally proposed based on transcript cloning experiments performed on small pools of OSNs (Chess et al. 1994). After single-nucleotide polymorphisms that distinguish alleles of OR genes had been identified, it was shown that small pools of OSNs contained transcripts from one allele or the other, but not from both. Subsequently, monoallelic expression was confirmed by generating knock-in mice in which OSNs expressing the targeted OR also expressed a reporter gene from an internal ribosome entry site (IRES) element in the OR 3′ untranslated region (Feinstein & Mombaerts 2004, Serizawa et al. 2003, Shykind et al. 2004). By tagging each allele of an OR with a different reporter, individual OSNs have been shown to express only the maternal allele or the paternal allele (Shykind et al. 2004). Monogenic expression is more difficult to definitively establish because it is not feasible to test for coexpression of any given OR with every other OR. Single-cell reverse transcriptase polymerase chain reaction analyses (Clowney et al. 2012, Imai et al. 2006), numerous two-color in situ hybridization experiments, and lineage-tracing experiments (Shykind et al. 2004) all support the one-receptor-per-neuron rule. It remains possible that OSNs expressing a given OR may express low quantities of transcripts from other OR genes, or that a small fraction of OSNs may express more than one OR, at least before their terminal differentiation. A definitive test of these possibilities will require single-cell RNA-sequence profiling of a large number of OSNs expressing a variety of different ORs. However, the fact that OSNs expressing the same OR converge to the same glomerulus supports the idea that the vast majority of cells express a single receptor at functionally significant levels.
Singular OR expression has also been observed with OR transgenes. Transgenes encoding an OR and flanking sequences are frequently expressed in the MOE in a similar manner to endogenous ORs (Serizawa et al. 2000, 2003; Vassalli et al. 2002). OR transgenes are not coexpressed with the identical endogenous OR despite having the same regulatory sequences, and two differently tagged but otherwise identical transgenes are not coexpressed (Ebrahimi et al. 2000, Serizawa et al. 2000). Further, like endogenous ORs, OR transgenes are expressed monoallelically (Vassalli et al. 2002, Zhang et al. 2007).
Singular OR expression depends on a feedback-based mechanism that is triggered by translation of a functional OR. When a synthetic promoter is used to express an OR transgene prior to OR choice, the transgene is expressed broadly throughout the MOE, and OSNs that express the transgene do not express endogenous ORs (Fleischmann et al. 2008). In contrast, coexpression has been observed among OR genes that do not code for a functional OR protein (Feinstein et al. 2004, Lewcock & Reed 2004, Serizawa et al. 2003). OR feedback not only prevents OR coexpression but also stabilizes expression of the chosen OR allele. OSNs that express a given OR switch to a different OR at a low frequency (Shykind et al. 2004). However, switching occurs much more frequently if the OR gene is mutated to remove the coding sequence. Similarly, the expression of OR pseudogenes is unstable, presumably because they do not code for a functional receptor and, thus, cannot activate feedback. Finally, a proper feedback signal depends not only on an intact protein sequence but also on robust expression levels. In a knock-in mouse that expresses the rat I7 OR under the control of the OMP promoter, the majority of OSNs appear to coexpress an endogenous OR allele together with low, but functionally significant, levels of the transgenic I7 OR (Zhou & Belluscio 2012). The requirement for robust OR expression for the initiation of a feedback signal likely is a direct consequence of the fact that ORs elicit this feedback by inducing the unfolded protein response (UPR) (Dalton et al. 2013). The UPR senses the presence of unfolded proteins and coordinates a multipart response that includes expressing chaperones and slowing the rate of translation (Ron & Walter 2007). OR expression acts through this pathway by activating a kinase, Perk, that phosphorylates translation-initiation factor eIF2α (Dalton et al. 2013). eIF2α phosphorylation halts the translation of most transcripts but increases the translation of select transcripts that have short open reading frames in the 5′ untranslated region. One such transcript, activating transcription factor 5 (ATF5) (Ron & Walter 2007, Watatani et al. 2008), is transcribed very highly in the MOE, but is only translated in a small number of immature OSNs (Dalton et al. 2013). OR expression is required and sufficient to induce translation of ATF5, and ATF5 is required for OR feedback and OSN maturation (Dalton et al. 2013, Wang et al. 2012). Intriguingly, the loss of ATF5 does not lead to OR coexpression, but rather it leads to a phenotype similar to that seen with OR pseudogenes: unstable OR expression and frequent switching between ORs (Dalton et al. 2013).
Beyond the UPR, OR-activated G protein signaling has a role in maintaining feedback inhibition. The G protein alpha (Gα) subunit does not have a role in feedback inhibition because a constitutively active Gα subunit fails to repress ORs (Imai et al. 2006). However, recent evidence from zebrafish suggests that signaling through the beta–gamma subunit helps prevent the activation of additional ORs (Ferreira et al. 2014).
CHROMATIN-MEDIATED SILENCING OF OLFACTORY RECEPTOR GENES IS CENTRAL TO OLFACTORY RECEPTOR CHOICE
Allelic exclusion enforces singular, monoallelic OR expression by making OR choice a winner-takes-all process. The first OR to be transcribed and translated to a sufficient level induces the UPR and ATF5 translation. This stabilizes the expression of the chosen OR and prevents expression of additional ORs. But how does the initial OR choice occur?
Olfactory Receptor Promoters Are Simple Sequences Bound by Abundant Transcriptional Regulators
OR-like expression can be recapitulated with OR transgenes as small as 2.2 kb, and with as little as 161 base pairs (bp) of OR promoter sequence (Rothman et al. 2005, Vassalli et al. 2002, Zhang et al. 2007). OR promoter sequences are required for the expression of OR transgenes, and an OR promoter is sufficient to drive MOE-specific expression of a reporter transgene (Plessy et al. 2012; Rothman et al. 2005; Vassalli et al. 2002, 2011). These findings suggest that OR promoters contain sequences that are sufficient to recapitulate OR activation.
Analyses of OR promoters have identified shared regulatory sequences (Clowney et al. 2011; Michaloski et al. 2006; 2011; Plessy et al. 2012; Young et al. 2011). These studies have reliably identified O/E family motifs and homeodomain motifs in most OR promoter sequences (Figure 2). O/E and homeodomain motifs occur in characteristic locations within OR promoters; O/E motifs are usually located between 50 and 150 bp upstream of the transcriptional start site, whereas homeodomain sites tend to be found further upstream. Deletion and mutagenesis studies support a role for these elements in OR expression. The promoter of olfactory receptor 151 (Olfr151) contains both O/E and homeodomain sites, and deleting these sites reduces the expression of Olfr151 transgenes (Rothman et al. 2005). Similar mutations in the promoter of the endogenous Olfr151 gene reduce expression, although to a lesser extent than is observed for transgenes. This reduced effect may be due to the availability of other OR regulatory elements in the vicinity of the endogenous OR allele, or may reflect the failure of OR transgenes to recapitulate more nuanced aspects of endogenous OR loci, such as the chromatin state. Adding nine copies of a homeodomain sequence to the promoter of an OR transgene increases the frequency of transgene expression, further supporting a role for homeodomains in choice (Vassalli et al. 2011). An analysis of OR promoters has also determined that these sequences are much more A- and T-rich than most murine promoters and that they lack a peak of G- and C-rich sequences near the transcriptional start site (Clowney et al. 2011). These features, combined with O/E and homeodomain motifs, may help define a signature that distinguishes OR promoters from the promoters of other genes.
In contrast, sequence analysis has failed to shed light on the determinants of zonal OR expression. The propensity for OR transgenes to be expressed in a different zone from the endogenous alleles could be due to the absence of zone-specific cis-regulatory elements from the transgene. However, this could also be due to the influence of non-OR regulatory elements near the random integration site. Coding-sequence swap experiments have provided stronger evidence for the existence of zonal cis-regulatory elements: When the coding sequence of Olfr17 is replaced by the coding sequence of an OR expressed in different zones, the swapped OR is still expressed in the Olfr17 zone (Wang et al. 1998). However, no sequence has been identified that distinguishes the promoters of ORs expressed in different zones (Clowney et al. 2011). This search has been limited by the small number of ORs (approximately 100) for which the pattern of zonal expression has been characterized; more complete characterization of OR expression patterns may allow zonal regulatory sequences to be identified.
Transcription factors that bind O/E motifs and homeodomain sites are abundantly expressed in the olfactory epithelium and have been shown to regulate OR expression. O/E motifs are bound by the Olf/EBF family of transcription factors (Wang & Reed 1993, Wang et al. 1997, 2002). All four family members (Olf1–Olf4) can bind O/E motif sequences as homo- or heterodimers and can activate the expression of a reporter gene in a heterologous system, although Olf4 is a less potent activator than the other family members (Wang et al. 2002). All four Olfs are expressed in the MOE, and all appear to be at least partially redundant with one another. Olfactory epithelium development is normal in Olf1 knock-outs (Lin & Grosschedl 1995, Wang et al. 1997). In Olf2 and Olf3 knock-out animals, OSN axons fail to project to dorsal regions of the olfactory bulb, and compound mutants heterozygous for Olf2 and Olf3 exhibit a similar phenotype, suggesting that the overall dosage of Olf family members may be functionally important (Wang et al. 2004). The zinc-finger protein OAZ is a negative regulator of O/E proteins that is transiently expressed by cells in the process of differentiating into OSNs (Cheng & Reed 2007, Tsai & Reed 1997). Artificially sustaining OAZ expression has been shown to disrupt OSN maturation and result in the continued expression of immature markers (Cheng & Reed 2007). In such animals, OSN axon targeting was disrupted, and the two ORs that were examined showed altered patterns of expression.
Several homeodomain transcription factors are expressed in the MOE, but two, Lhx2 and Emx2, have been investigated in the context of OR expression on the basis of their ability to bind an OR-promoter homeodomain site in a yeast one-hybrid assay and their high level of expression (Hirota & Mombaerts 2004). In Lhx2 knock-out mice the differentiation of most OSNs arrests in an immature state, and the arrested cells fail to express ORs (Hirota & Mombaerts 2004, Kolterud et al. 2004). However, some OSNs in the dorsal MOE are able to differentiate to mature OSNs (Hirota et al. 2007, Kolterud et al. 2004), all of which express type I OR genes (Hirota et al. 2007). OSNs that express type I ORs constitute a distinct subtype of OSNs (Bozza et al. 2009), so developmental differences, rather than a specific role for Lhx2 in OR choice, may account for this difference. Further, Lhx2 knock-out mice have fewer OSNs expressing type I ORs than wild-type mice, suggesting that Lhx2 contributes to differentiation in these cells. In contrast, Emx2 knockout mice have mature OSNs present throughout the MOE (McIntyre et al. 2008). However, these mice have fewer mature OSNs than do wild-type mice. For the most part, a commensurate reduction has been observed in the number of OSNs expressing each OR. However, in Emx2 knock-out mice some ORs are unaffected and others are expressed in more cells. This effect is seen for both type-I and type-II ORs, and there is not a clear relationship between the genomic clustering of OR genes and the change in expression: ORs that increase in expression are present in the same OR clusters as ORs that decrease or are unchanged in expression.
Examination of OR promoters suggests that they are bound by a small number of highly expressed common transcription factors. This arrangement seems ill suited to achieving singular OR expression, particularly because the chosen OR allele is transcribed very highly and comprises one of the most abundant protein-coding RNAs in OSNs. Somehow, the nonchosen ORs, which vastly outnumber the chosen OR, must be kept transcriptionally silent. This silencing is likely to be critical to OSN function; OR basal activity specifies axon targeting to the correct glomerulus in the olfactory bulb, so coexpression of additional ORs could lead to mistargeting that would disrupt the glomerular map (Clowney et al. 2012). Given the huge number of nonchosen ORs, even low basal expression could easily mask the activity of the chosen OR. How is silencing of nonchosen ORs accomplished? Remarkably, feedback inhibition does not require an OR promoter sequence. OR transgenes with synthetic promoters are also resistant to activation in mature OSNs, indicating that the OR coding sequence is sufficient for feedback inhibition to occur (Fleischmann et al. 2013, Nguyen et al. 2007).
The Chromatin State Governs Olfactory Receptor Expression
In the MOE, heterochromatin compacts and silences OR genes. Heterochromatin is a compact chromatin structure that is inaccessible to many transcription factors and the transcriptional machinery. Heterochromatin formation accompanies gene silencing in many developmental systems. Compaction into heterochromatin renders silenced genes refractory to future transcriptional activation, thus stabilizing cell fate decisions. OR genes exhibit biochemical hallmarks of compaction and heterochromatin formation in the MOE (Magklara et al. 2011). In most cases, developmentally regulated heterochromatin formation is directed by the Polycomb complex. Nucleosomes that have been compacted by Polycomb are decorated with a characteristic modification: histone H3 lysine 27 trimethylation (H3K27me3). Polycomb-silenced regions of the genome can be identified by chromatin immunoprecipitation using antibodies specific for this histone modification. However, OR genes are devoid of H3K27me3 in the olfactory epithelium, and instead bear histone modifications characteristic of constitutive heterochromatin (H3K9me3 and H4K20me3). The functional significance of OR compaction into H3K9me3- or H4K20me3-marked constitutive heterochromatin rather than H3K27me3-marked facultative heterochromatin has not been examined. However, this may provide opportunities for specialized regulatory mechanisms to target OR chromatin separately and in parallel to normal, developmentally regulated gene silencing by facultative heterochromatin formation. Consistent with this hypothesis, constitutive heterochromatin marks are abundant on other gene families characterized by monoallelic or combinatorial patterns of expression, including vomeronasal receptor genes, formyl-peptide receptors, and clustered protocadherins (Magklara et al. 2011).
Heterochromatin is not limited to OR genes, but rather H3K9me3 and H4K20me3 marks decorate genomic regions between and surrounding OR genes. The heterochromatic region roughly coincides with full OR clusters, as defined by boundary elements, such as binding sites of the zinc-finger protein CTCF (Magklara et al. 2011). These broad regions likely correspond to topological domains (Dixon et al. 2012) that contain OR regulatory elements that drive OR-like expression of the genes within. Consistent with this, a β-galactosidase reporter transgene that normally exhibits broad expression in mature OSNs instead exhibits sparse OR-like expression when inserted within the heterochromatic region near an endogenous OR (Magklara et al. 2011, Pyrski et al. 2001). Further, this transgene acquires OR-like properties that are characteristic of the nearby ORs, including monoallelic expression and dependence on Emx2 (Magklara et al. 2011).
Surprisingly, the chromatin-mediated silencing of OR genes precedes OR expression (Magklara et al. 2011). In HBCs, the multipotent progenitor cells of the MOE, OR genes are free of H3K9me3 and H4K20me3 (Figure 2). In these cells, OR clusters instead bear H3K9 and H4K20 dimethylated nucleosomes that are also observed in other tissues. H3K9me3 and H4K20me3 are present on OR genes in cells that are in the process of differentiating into OSNs but that do not yet express an OR, a stage marked by expression of an Ngn1-green fluorescent protein (GFP) reporter gene. These heterochromatic marks remain in mature OR-expressing neurons. However, heterochromatin marks are absent from the chosen OR allele; in cells sorted based upon their expression of an OR-IRES-GFP reporter, the chosen, labeled allele is free of silencing marks but H3K9me3 is present on the inactive, unlabeled allele and other OR genes. Further, the chosen OR bears histone modifications characteristic of transcription initiation (H3K4me3), indicating that it is embedded within transcriptionally active euchromatin.
OR expression requires derepression of a previously silenced OR gene. LSD1 is a histone demethylase with dual activities: It can completely demethylate H3K9me2, a mark associated with gene silencing, and it can also demethylate H3K4me2, which is associated with gene expression. The deletion of an LSD1 conditional allele prior to OR choice results in widespread loss of OR expression (Lyons et al. 2013). However, LSD1 is dispensable for OR expression after choice; the deletion of LSD1 with either an OR-IRES-Cre or a mature OSN-specific Cre (OMP-Cre) has no effect on OR expression. OR genes show hallmarks of LSD1-catalyzed H3K9 demethylation, suggesting that LSD1 acts directly on OR chromatin. LSD1 activity generates hydrogen peroxide, which can oxidize nearby DNA, thereby generating oxidized DNA bases, such as 8-oxoguanosine (8-oxoG). DNA immunoprecipitation experiments have demonstrated that OR genes exhibit high levels of 8-oxoG. This 8-oxoG likely results, at least in part, from LSD1 activity because over-expression of LSD1 from a transgene increases the amount of 8-oxoG present on ORs. If LSD1 acts by derepressing the chosen OR allele, then the chosen allele should exhibit high levels of 8-oxoG. Indeed, OSNs sorted by their expression of an OR-IRES-GFP reporter exhibit high levels specifically on the chosen OR. Notably, LSD1 does not act on trimethylated H3K9me3, meaning that some other protein must carry out the initial steps of OR gene derepression before LSD1 can act.
LSD1 is the target of the OR-dependent negative feedback signal. OR feedback induces expression of Adcy3, which leads to downregulation of LSD1 (Dalton et al. 2013, Lyons et al. 2013). LSD1 downregulation can be prevented by deleting Adcy3, by deleting components of the UPR, or by the ectopic expression of LSD1 from a transgene (Dalton et al. 2013, Lyons et al. 2013). Continued LSD1 expression might be expected to result in OR coexpression, but a switching phenotype is observed instead. Functional ORs are chosen and expressed, but only transiently. This switching state is reversible: The silencing of ectopic LSD1 expression results in a return to normal, stable OR expression within 3 weeks. Switching may result from the alternative role of LSD1 as a demethylase that targets histone modifications associated with transcription (H3K4me2). Thus, LSD1 has a dual role in regulating OR choice: desilencing the chosen OR to allow expression and promoting inactivation of ORs that fail to activate the OR feedback pathway. This latter function likely accounts for the unstable expression of OR pseudogenes and for the low rate of switching reported for functional ORs.
Does OR silencing have a function beyond ensuring singular, monoallelic OR expression? Remarkably, blocking the formation of H3K9me3-marked heterochromatin has a dramatic effect on the pattern of OR expression (Lyons et al. 2014). Blocking H3K9me3 formation has been accomplished by conditionally deleting two histone methyltransferases, G9a and GLP, that generate H3K9me2, a necessary precursor of H3K9me3. The deletion of these enzymes in the developing olfactory epithelium radically, and reproducibly, skews OR expression toward a small subset of ORs. The magnitude of this skew depends on the dosage of G9a and GLP: A single knock-out has a less severe skew than a compound heterozygous or single knock-out, which, in turn, is less severe than a double knock-out. No such phenotype has been observed when H4K20me3 formation is blocked, suggesting that H3K9me3 may be the more functionally important modification. It remains unclear what defines the particular subset of ORs that are favored in the G9a–GLP knock-out. It is possible that these ORs have unusually strong promoters or have unusual access to OR enhancers. Nonetheless, it appears that OR silencing levels the playing field, giving all ORs roughly equal opportunity to be chosen for expression regardless of the relative strength of their promoter. Furthermore, in G9a–GLP double knock-out mice, OR coexpression occurs at low but detectable frequencies among the few ORs that are expressed in a dominant fashion, supporting a role for this epigenetic silencing in the singularity of OR expression.
Does chromatin-based regulation suffice to generate the singularity of OR choice or does choice involve additional layers of control? Recent theoretical studies have shown that monoallelic OR choice can be modeled with mechanisms similar to those described above, namely OR derepression and the subsequent inhibition of the derepressing mechanism (Alsing & Sneppen 2013, Tan et al. 2013). The success of this simple activation–feedback mechanism depends on the ratio between the average time it takes to activate an OR and the time it takes for feedback inhibition to occur (Tan et al. 2013). Coexpression occurs if the activation of a second OR occurs shortly after the activation of the first—that is, before feedback inhibition can shut off the derepression mechanism. As a result, singular OR choice is favored by a fast feedback mechanism and a slow derepression mechanism. By estimating two of these parameters, a 2% rate of OR coexpression (Tian & Ma 2008) and a 1–2 h delay required for feedback inhibition, the authors estimated the third parameter: an OR activation rate of approximately 1 every 5–10 days (Tan et al. 2013). This estimate roughly agrees with the timing of OSN differentiation (Kondo et al. 2010) and the regeneration rate of OSNs following removal of the olfactory bulb (Gokoffski et al. 2010). Notably, this model predicts that failed feedback or sustained LSD1 expression will result in persistent OR switching, rather than in OR coexpression, which is in agreement with the findings of experimental manipulations (Dalton et al. 2013, Lyons et al. 2013, Tan et al. 2013). An appealing aspect of this model is that it can easily accommodate the variable rates of OR switching and of OR coexpression that have been reported for individual ORs or OR subfamilies. For example, a reduced ability to engage feedback inhibition or zonal variation in the level of LSD1 could account for the high rate of switching reported for ORs of the OR37 family (Bader et al. 2010, Strotmann et al. 2009).
Our understanding of the chromatin events surrounding OR choice fits the broad-scale patterns of OR choice remarkably well. However, many uncertainties remain that hint at additional layers of control. Most simply, more events must surround OR activation than LSD1-catalyzed desilencing. LSD1 catalyzes only demethylation of dimethylated H3K9; a different enzyme must carry out the initial demethylation of H3K9me3 to H3K9me2. Full demethylation of H3K9me3 could be carried out by a multiprotein complex containing LSD1 and a histone tri-demethylase (Wissmann et al. 2007), or the initial demethylation could be an independent event from subsequent derepression by LSD1. Theoretical modeling predicts that the initial demethylation is likely the slow, rate-limiting step in OR desilencing, so this could easily be a key regulatory step gating OR choice (Tan et al. 2013). Further, once OR derepression is complete, the OR must still be transcribed and translated to initiate feedback inhibition. It is unclear whether derepression is sufficient for expression or whether expression requires some additional activating mechanism. These regulatory events could also be coupled by multiprotein complexes containing LSD1 and other regulatory proteins. Our understanding of the molecular events surrounding OR switching are similarly limited; the demethylase that removes the first methyl group from H3K4me3 from the nonchosen OR has not been identified and the events that follow subsequent demethylation by LSD1 are also a mystery. In particular, although continued LSD1 expression is sufficient to induce switching, it is unclear how the switched OR is silenced: Is it returned to heterochromatin, or is it repressed by some other mechanism?
DOES NUCLEAR ORGANIZATION COORDINATE OLFACTORY RECEPTOR CHOICE?
Recent findings have suggested that interchromosomal chromatin interactions and large-scale changes in nuclear organization are involved in coordinating OR choice. Experimentally disrupting these interactions disrupts singular OR expression. However, it remains unclear whether these processes are directly involved in OR choice and, if so, what part they play.
Cis-regulatory enhancer elements are critical for the expression of several ORs. The first OR enhancer was identified by studying transgenes bearing a large insertion spanning several clustered OR genes (Serizawa et al. 2003). Mapping experiments with a variety of transgene constructs have identified a conserved region, named H, which is required for high-level expression of the linked ORs. When H is added to an OR transgene it drives expression of the transgene OR across a large portion of the MOE. Subsequently, a second OR enhancer, named P, was identified by its sequence similarity with the promoter of Olfr713 (Bozza et al. 2009). P is also able to drive broad expression of OR transgenes in the MOE. Similar to OR promoters, the H and P enhancers contain O/E motifs and homeodomain motifs, and these motifs are required for the enhancer activity of H (Bozza et al. 2009, Nishizumi et al. 2007). The deletion of either H or P reduces the expression of several nearby OR genes, but does not affect every gene in the surrounding OR cluster, nor does the deletion affect the expression of ORs outside the cluster (Fuss et al. 2007, Khan et al. 2011, Nishizumi et al. 2007).
Recently, 35 more candidate OR enhancers, named after Greek islands, have been identified on the basis of characteristic patterns of histone modifications and chromatin accessibility (Markenscoff-Papadimitriou et al. 2014). Using a high-throughput zebrafish transgene assay, 12 of these candidate enhancers have been shown to drive reporter-gene expression specifically in olfactory neurons. In mouse reporter transgenes, several of these were subsequently shown to drive broad expression in the MOE, and deletion of one of these enhancers, Lipsi, was shown to reduce the expression of nearby OR genes. Regulatory sequences within these enhancers were identified, on the basis that they were protected from digestion by deoxyribonuclease I (DNaseI), using high-throughput DNaseI sequencing and sequence capture to enrich for enhancer sequences. This analysis identified protected O/E and homeodomain motifs in the enhancers, and binding of Lhx2 to these sites was demonstrated by chromatin immunoprecipitation. This approach also identified several additional protected motifs in OR promoters. Among these were the motif bound by the ATF family, suggesting a possible link between ATF5-mediated feedback inhibition and enhancer activity, and a motif recognized by the transcription factor Bptf. The deletion of a Bptf conditional allele in the developing MOE dramatically reduces OR expression and arrests differentiating OSNs in an immature state.
OR enhancers make chromatin contacts with many OR promoters. Chromosome-conformation capture experiments have shown that the H enhancer contacts the promoter of Olfr1507, which requires H for its expression (Lomvardas et al. 2006). However, H also interacts with the promoters of many ORs that are unaffected by deleting H, including ORs on different chromosomes. These interchromosomal interactions have also been detected by DNA fluorescent in situ hybridization (DNA-FISH), which showed frequent colocalization of H with ORs from other chromosomes. Remarkably, simultaneous RNA- and DNA-FISH experiments have demonstrated that H frequently colocalizes with the transcribed OR allele. A similar pattern has been observed with the newly identified Greek island enhancers (Markenscoff-Papadimitriou et al. 2014). Fifteen of these enhancers have been shown to interact with the Olfr1507 promoter in OSNs expressing Olfr1507, and these interactions are much weaker or absent in OSNs that express other ORs. Yet, when one of these enhancers, Lipsi, was knocked out, Olfr1507 expression was unchanged.
These findings suggest that the chosen OR may interact with several OR enhancers and that individual enhancers, while required in cis, are redundant when acting in trans. Consistent with this model, multicolor DNA-FISH experiments have shown that multiple OR enhancers colocalize with Olfr1507 DNA simultaneously in OSNs expressing Olfr1507, but not in other OSNs (Markenscoff-Papadimitriou et al. 2014). Indeed, when OSNs are examined as a whole, a complex network of interactions among OR enhancers is detected. These interactions are not found in the non-neuronal sustentacular cells of the MOE, even though these sustentacular cells are continually born from the same stem cell pool as OSNs and have the same constitutive heterochromatin marks on OR genes (Magklara et al. 2011, Markenscoff-Papadimitriou et al. 2014). Remarkably, the deletion of Bptf not only disrupts OR expression but also greatly reduces interactions among OR enhancers, raising the possibility that enhancer trans interactions, although individually redundant, may nonetheless be required for OR expression (Markenscoff-Papadimitriou et al. 2014).
The formation of OR enhancer interactions over OSN differentiation occurs in the context of larger-scale changes in the organization of OR genes within the nucleus (Figure 3). By using a heterogeneous DNA-FISH probe that detects the vast majority of OR sequences, it has been shown that OR gene sequences are dispersed throughout a broad region of the nucleus in basal stem cells, but are condensed into a small number of compact foci in mature OSNs (Clowney et al. 2012). These foci include OR genes from multiple chromosomes. The formation of OR foci accompanies a larger movement of heterochromatin from the nuclear periphery to the center of the nucleus (Figure 3a). The OR foci are located at the edge of the larger heterochromatin foci (Armelin-Correa et al. 2014, Clowney et al. 2012). However, constitutive heterochromatin marks are neither sufficient nor absolutely required for the formation of OR foci. OR genes bear histone marks characteristic of constitutive heterochromatin in sustentacular cells, yet OR genes remain dispersed (Clowney et al. 2012, Magklara et al. 2011). OR genes are not heterochromatinized in G9a–GLP knock-out mice, but OR foci still form, although they are not as well organized as in wild-type OSNs (Lyons et al. 2014).
Figure 3.
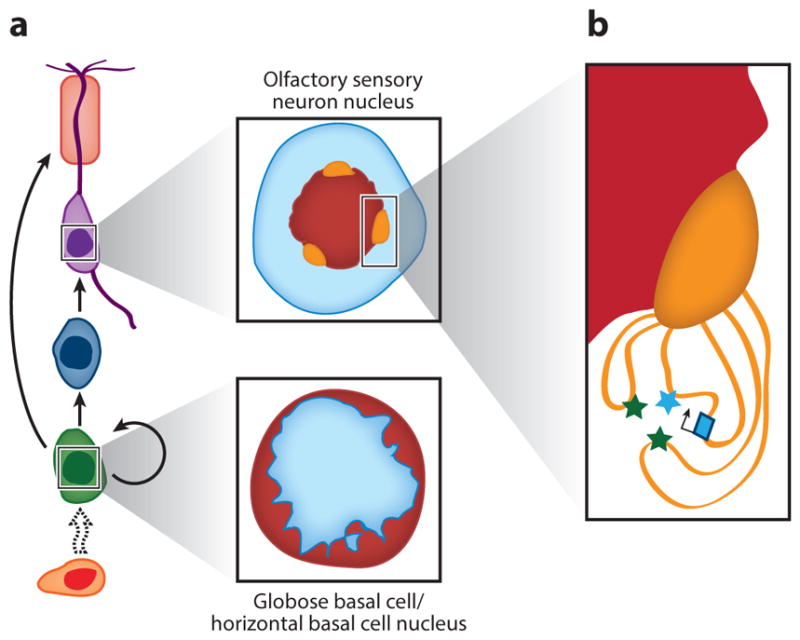
Nuclear organization in olfactory sensory neurons (OSNs). (a) OSN differentiation is accompanied by a reorganization of heterochromatin (red ) and euchromatin (blue) within the nucleus. Heterochromatin relocalizes from the nuclear periphery to the center of the nucleus. This reorganization is accompanied by a coalescence of (orange) OR genes into a few dense foci located at the edge of the larger heterochromatin focus. (b) The active OR allele (blue) is located outside these OR foci and colocalizes with OR enhancers from the same cluster (blue star) and from different clusters ( green stars).
The active OR allele, as determined by RNA-FISH, is located outside OR foci in euchromatic regions of the nucleus, which is consistent with the observation of euchromatin histone modifications on the active OR allele (Clowney et al. 2012, Magklara et al. 2011) (Figure 3b). Furthermore, although OR enhancer sequences are usually located within OR foci, colocalization of a pair of OR enhancers has been shown to preferentially occur outside OR foci (Markenscoff-Papadimitriou et al. 2014). These findings suggest that the active OR allele and multiple OR enhancers may occupy a separate, active compartment outside the repressive OR foci.
Evidence suggests that OR foci may contribute to OR silencing. The reorganization of OSN nuclei coincides with the downregulation of lamin b receptor (Lbr), which is an integral membrane protein that maintains heterochromatin at the nuclear periphery through interactions with the heterochromatin protein HP1β (Clowney et al. 2012, Ye & Worman 1996, Ye et al. 1997). The ectopic expression of Lbr in mature OSNs disrupts OR foci and results in heterochromatin remaining on the nuclear periphery throughout differentiation (Clowney et al. 2012). Ectopic Lbr expression does not affect the levels of constitutive heterochromatin marks on OR genes, but it does reduce the association of HP1β with ORs and increases the accessibility of ORs to DNaseI. Further, ectopic Lbr expression does not affect chromatin interactions within OR clusters in cis, for example, between H and Olfr1507, but strongly reduces the frequency of enhancer–enhancer and enhancer–promoter interactions in trans (Clowney et al. 2012, Markenscoff-Papadimitriou et al. 2014). This is accompanied by a strong reduction in OR expression and OR coexpression (Clowney et al. 2012). Thus, it appears that the complete repression of the nonchosen ORs and the singular, robust expression of a chosen OR allele are intimately linked processes that rely on epigenetic silencing and the nuclear aggregation of most OR alleles. These molecular events not only prevent transcription-factor binding to most OR promoters (K. Monahan, unpublished observations) and insulation of the nonchosen ORs from the transcriptional machinery, but also ensure that high levels of transcription factors will remain available to the chosen OR by preventing their sequestration to the thousands of shared binding sites found in the rest of the gene family. Furthermore, it appears that the aggregation of silent OR genes to distinct heterochromatic foci generates an architectural platform that promotes the frequent interchromosomal interaction of OR-linked distant enhancers that may be necessary for robust OR transcription.
SUMMARY AND FUTURE DIRECTIONS
Obtaining the widest receptive field possible while retaining ligand specificity and sensitivity is not trivial. The immune system, the only other mammalian system facing such an extreme challenge, evolved unique DNA-rearrangement-based mechanisms for the generation of a diverse repertoire of immunoglobulin and T cell receptor molecules. The olfactory system instead solved the problem by rapidly expanding the OR gene family to a remarkable number of genes, as has been recently highlighted by the identification of approximately 4,000 elephant ORs (Niimura et al. 2014). Because the sensitivity of this sensory system depends upon singular and robust OR expression, a multilayered regulatory mechanism evolved in parallel to coordinate the monogenic and monoallelic nature of OR transcription. An unusual nuclear organization likely coordinates the complete silencing of most ORs and the enhanced transcription of a single allele, and a feedback signal that silences the desilencer ensures that this singular selection will be maintained for the life of the neuron. Additional processes, such as the activity-dependent expression of a histone variant that affects the longevity of each neuron (Santoro & Dulac 2012), likely allow this seemingly stochastic process to adapt to environmental cues in a fashion reminiscent of the adaptive processes of the immune system.
Although progress has been made in our molecular understanding of this fascinating process, fundamental questions remain open. For example, it is not known if the widespread, interchromosomal interactions among distant enhancers and the transcriptionally active allele contribute to the transcription of this allele. And, furthermore, if the formation of a multienhancer complex is indeed necessary for OR gene activation, then why do multiple enhancers have to act in a coordinated fashion? Only when all of the protein components that govern the formation and function of this enhanceosome have been identified (Thanos & Maniatis 1995) will we be able to provide convincing answers to these questions. In any case, if cell type–specific nuclear architecture can influence the interchromosomal association of regulatory sequences with mechanistic consequences, then the principles uncovered in the case of OR choice could be applicable to every developmental system that seeks to generate cellular diversity.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Kevin Monahan, Email: kgm2108@columbia.edu.
Stavros Lomvardas, Email: sl682@columbia.edu.
LITERATURE CITED
- Alsing AK, Sneppen K. Differentiation of developing olfactory neurons analysed in terms of coupled epigenetic landscapes. Nucleic Acids Res. 2013;41(9):4755–64. doi: 10.1093/nar/gkt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armelin-Correa LM, Gutiyama LM, Brandt DYC, Malnic B. Nuclear compartmentalization of odorant receptor genes. PNAS. 2014;111(7):2782–87. doi: 10.1073/pnas.1317036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aycı F, Aydınlı M, Bozdemir ÖA, Tutaş M. Gas chromatographic investigation of rose concrete, absolute and solid residue. Flavour Fragr J. 2005;20(5):481–86. [Google Scholar]
- Aznar M, López R, Cacho JF, Ferreira V. Identification and quantification of impact odorants of aged red wines from Rioja. GC-olfactometry, quantitative GC-MS, and odor evaluation of HPLC fractions. J Agric Food Chem. 2001;49(6):2924–29. doi: 10.1021/jf001372u. [DOI] [PubMed] [Google Scholar]
- Bader A, Bautze V, Haid D, Breer H, Strotmann J. Gene switching and odor induced activity shape expression of the OR37 family of olfactory receptor genes. Eur J Neurosci. 2010;32(11):1813–24. doi: 10.1111/j.1460-9568.2010.07458.x. [DOI] [PubMed] [Google Scholar]
- Barnea G, O’Donnell S, Mancia F, Sun X, Nemes A, et al. Odorant receptors on axon termini in the brain. Science. 2004;304(5676):1468. doi: 10.1126/science.1096146. [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 2002;22(8):3033–43. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T, Vassalli A, Fuss S, Zhang J-J, Weiland B, et al. Mapping of class I and class II odorant receptors to glomerular domains by two distinct types of olfactory sensory neurons in the mouse. Neuron. 2009;61(2):220–33. doi: 10.1016/j.neuron.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65(1):175–87. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Bufe B, Schumann T, Kappl R, Bogeski I, Kummerow C, et al. Recognition of bacterial signal peptides by mammalian formyl peptide receptors: A new mechanism for sensing pathogens. J Biol Chem. 2015;290(12):7369–87. doi: 10.1074/jbc.M114.626747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, Schumann T, Zufall F. Formyl peptide receptors from immune and vomeronasal system exhibit distinct agonist properties. J Biol Chem. 2012;287(40):33644–55. doi: 10.1074/jbc.M112.375774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushdid C, Magnasco MO, Vosshall LB, Keller A. Humans can discriminate more than 1 trillion olfactory stimuli. Science. 2014;343(6177):1370–72. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LE, Reed RR. Zfp423/OAZ participates in a developmental switch during olfactory neurogenesis. Neuron. 2007;54(4):547–57. doi: 10.1016/j.neuron.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78(5):823–34. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151(4):724–37. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Magklara A, Colquitt BM, Pathak N, Lane RP, Lomvardas S. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Res. 2011;21(8):1249–59. doi: 10.1101/gr.120162.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155(2):321–32. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysocki CJ, et al. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature. 2002;419(6902):70–74. doi: 10.1038/nature00955. [DOI] [PubMed] [Google Scholar]
- Dewan A, Pacifico R, Zhan R, Rinberg D, Bozza T. Non-redundant coding of aversive odours in the main olfactory pathway. Nature. 2013;497(7450):486–89. doi: 10.1038/nature12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83(2):195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Ebrahimi FAW, Edmondson J, Rothstein R, Chess A. YAC transgene-mediated olfactory receptor gene choice. Dev Dyn. 2000;217:225–31. doi: 10.1002/(SICI)1097-0177(200002)217:2<225::AID-DVDY9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117(6):833–46. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117(6):817–31. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Ferreira T, Wilson SR, Choi YG, Risso D, Dudoit S, et al. Silencing of odorant receptor genes by G protein βγ signaling ensures the expression of one odorant receptor per olfactory sensory neuron. Neuron. 2014;81(4):847–59. doi: 10.1016/j.neuron.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DM, Lemon JK, Fluegge D, Pashkovski SL, Korzan WJ, et al. Detection and avoidance of a carnivore odor by prey. PNAS. 2011;108(27):11235–40. doi: 10.1073/pnas.1103317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Abdus-Saboor I, Sayed A, Shykind B. Functional interrogation of an odorant receptor locus reveals multiple axes of transcriptional regulation. PLOS Biol. 2013;11(5):e1001568. doi: 10.1371/journal.pbio.1001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Shykind BM, Sosulski DL, Franks KM, Glinka ME, et al. Mice with a “monoclonal nose”: perturbations in an olfactory map impair odor discrimination. Neuron. 2008;60(6):1068–81. doi: 10.1016/j.neuron.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130(2):373–84. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Gokoffski KK, Kawauchi S, Wu H-H, Santos R, Hollenbeck PLW, et al. Feedback regulation of neurogenesis in the mammalian olfactory epithelium: new insights from genetics and systems biology. In: Menini A, editor. Neurobiology of Olfaction. Boca Raton, FL: CRC; 2010. pp. 241–66. [PubMed] [Google Scholar]
- Grosmaitre X, Fuss SH, Lee AC, Adipietro KA, Matsunami H, et al. SR1, a mouse odorant receptor with an unusually broad response profile. J Neurosci. 2009;29(46):14545–52. doi: 10.1523/JNEUROSCI.2752-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmaitre X, Vassalli A, Mombaerts P, Shepherd GM, Ma M. Odorant responses of olfactory sensory neurons expressing the odorant receptor MOR23: a patch clamp analysis in gene-targeted mice. PNAS. 2006;103(6):1970–75. doi: 10.1073/pnas.0508491103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad R, Khan R, Takahashi YK, Mori K, Harel D, Sobel N. A metric for odorant comparison. Nat Methods. 2008;5(5):425–29. doi: 10.1038/nmeth.1197. [DOI] [PubMed] [Google Scholar]
- Halpern M. The organization and function of the vomeronasal system. Annu Rev Neurosci. 1987;10:325–62. doi: 10.1146/annurev.ne.10.030187.001545. [DOI] [PubMed] [Google Scholar]
- Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90(4):763–73. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. PNAS. 2004;101(23):8751–55. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota J, Omura M, Mombaerts P. Differential impact of Lhx2 deficiency on expression of class I and class II odorant receptor genes in mouse. Mol Cell Neurosci. 2007;34(4):679–88. doi: 10.1016/j.mcn.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Hussain A, Saraiva LR, Ferrero DM, Ahuja G, Krishna VS, et al. High-affinity olfactory receptor for the death-associated odor cadaverine. PNAS. 2013;110(48):19579–84. doi: 10.1073/pnas.1318596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314(5799):657–61. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Ishii T, Mombaerts P. Coordinated coexpression of two vomeronasal receptor V2R genes per neuron in the mouse. Mol Cell Neurosci. 2011;46(2):397–408. doi: 10.1016/j.mcn.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Khan M, Vaes E, Mombaerts P. Regulation of the probability of mouse odorant receptor gene choice. Cell. 2011;147(4):907–21. doi: 10.1016/j.cell.2011.09.049. [DOI] [PubMed] [Google Scholar]
- Kimchi T, Xu J, Dulac C. A functional circuit underlying male sexual behaviour in the female mouse brain. Nature. 2007;448:1009–14. doi: 10.1038/nature06089. [DOI] [PubMed] [Google Scholar]
- Kolterud A, Alenius M, Carlsson L, Bohm S. The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development. 2004;131(21):5319–26. doi: 10.1242/dev.01416. [DOI] [PubMed] [Google Scholar]
- Kondo K, Suzukawa K, Sakamoto T, Watanabe K, Kanaya K, et al. Age-related changes in cell dynamics of the postnatal mouse olfactory neuroepithelium: cell proliferation, neuronal differentiation, and cell death. J Comp Neurol. 2010;518(11):1962–75. doi: 10.1002/cne.22316. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. PNAS. 2004;101(4):1069–74. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442(7103):645–50. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Horowitz LF, Kuang D, Contos JJ, Wilson KL, et al. Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ. PNAS. 2009;106:9842–47. doi: 10.1073/pnas.0904464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376(6537):263–67. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126(2):403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154(2):325–36. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DB, Magklara A, Goh T, Sampath SC, Schaefer A, et al. Heterochromatin-mediated gene silencing facilitates the diversification of olfactory neurons. Cell Rep. 2014;9(3):884–92. doi: 10.1016/j.celrep.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Wu Y, Qiu Q, Scheerer H, Moran A, Yu CR. A developmental switch of axon targeting in the continuously regenerating mouse olfactory system. Science. 2014;344:194–97. doi: 10.1126/science.1248805. [DOI] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145(4):555–70. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96(5):713–23. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, Goh T, Murphy KK, et al. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 2014;159(3):543–57. doi: 10.1016/j.cell.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90(4):775–84. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Adipietro K, Titlow WB, Breheny P, Walz A, et al. In vivo identification of eugenol-responsive and muscone-responsive mouse odorant receptors. J Neurosci. 2014;34(47):15669–78. doi: 10.1523/JNEUROSCI.3625-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JC, Bose SC, Stromberg AJ, McClintock TS. Emx2 stimulates odorant receptor gene expression. Chem Senses. 2008;33(9):825–37. doi: 10.1093/chemse/bjn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloski JS, Galante PAF, Malnic B. Identification of potential regulatory motifs in odorant receptor genes by analysis of promoter sequences. Genome Res. 2006;16(9):1091–98. doi: 10.1101/gr.5185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloski JS, Galante PAF, Nagai MH, Armelin-Correa L, Chien M-S, et al. Common promoter elements in odorant and vomeronasal receptor genes. PLOS ONE. 2011;6(12):e29065. doi: 10.1371/journal.pone.0029065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25(14):3586–92. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, et al. Visualizing an olfactory sensory map. Cell. 1996;87(4):675–86. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Mori K, Sakano H. How is the olfactory map formed and interpreted in the mammalian brain? Annu Rev Neurosci. 2011;34(1):467–99. doi: 10.1146/annurev-neuro-112210-112917. [DOI] [PubMed] [Google Scholar]
- Mori T, Sakai M, Matsuoka I, Kurihara K. Analysis of promoter activity of 5′-upstream regions of zebrafish olfactory receptor genes. Biol Pharm Bull. 2000;23(2):165–73. doi: 10.1248/bpb.23.165. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Takeuchi H, Imai T, Saito H, Kiyonari H, et al. Agonist-independent GPCR activity regulates anterior-posterior targeting of olfactory sensory neurons. Cell. 2013;154(6):1314–25. doi: 10.1016/j.cell.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara K, Saraiva LR, Ye X, Buck LB. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci. 2011;31(25):9179–91. doi: 10.1523/JNEUROSCI.1282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngai J, Dowling MM, Buck L, Axel R, Chess A. The family of genes encoding odorant receptors in the channel catfish. Cell. 1993;72(5):657–66. doi: 10.1016/0092-8674(93)90395-7. [DOI] [PubMed] [Google Scholar]
- Nguyen MQ, Zhou Z, Marks CA, Ryba NJP, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131(5):1009–17. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 2014;24:1485–96. doi: 10.1101/gr.169532.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizumi H, Kumasaka K, Inoue N, Nakashima A, Sakano H. Deletion of the core-H region in mice abolishes the expression of three proximal odorant receptor genes in cis. PNAS. 2007;104(50):20067–72. doi: 10.1073/pnas.0706544105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura M, Grosmaitre X, Ma M, Mombaerts P. The β2-adrenergic receptor as a surrogate odorant receptor in mouse olfactory sensory neurons. Mol Cell Neurosci. 2014;58:1–10. doi: 10.1016/j.mcn.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifico R, Dewan A, Cawley D, Guo C, Bozza T. An olfactory subsystem that mediates high-sensitivity detection of volatile amines. Cell Rep. 2012;2(1):76–88. doi: 10.1016/j.celrep.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessy C, Pascarella G, Bertin N, Akalin A, Carrieri C, et al. Promoter architecture of mouse olfactory receptor genes. Genome Res. 2012;22(3):486–97. doi: 10.1101/gr.126201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrski M, Xu Z, Walters E, Gilbert DJ, Jenkins NA, et al. The OMP-lacZ transgene mimics the unusual expression pattern of OR-Z6, a new odorant receptor gene on mouse chromosome 6: Implication for locus-dependent gene expression. J Neurosci. 2001;21(13):4637–48. doi: 10.1523/JNEUROSCI.21-13-04637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73(3):597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79(7):1245–55. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Rivière S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459(7246):574–77. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Del Punta K, Rothman A, Ishii T, Mombaerts P. Multiple new and isolated families within the mouse superfamily of V1r vomeronasal receptors. Nat Neurosci. 2002;5(2):134–40. doi: 10.1038/nn795. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rothman A, Feinstein P, Hirota J, Mombaerts P. The promoter of the mouse odorant receptor gene M71. Mol Cell Neurosci. 2005;28(3):535–46. doi: 10.1016/j.mcn.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Ryba NJ, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19(2):371–79. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2(60):ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Dulac C. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. eLife. 2012;1:e00070. doi: 10.7554/eLife.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, et al. Mutually exclusive expression of odorant receptor transgenes. Nat Neurosci. 2000;3(7):687–93. doi: 10.1038/76641. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302(5653):2088–94. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, et al. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117(6):801–15. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Sosulski DL, Bloom ML, Cutforth T, Axel R, Datta SR. Distinct representations of olfactory information in different cortical centres. Nature. 2011;472(7342):213–16. doi: 10.1038/nature09868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L, Holy TE, Meister M, Dulac C, Koentges G. Loss of sex discrimination and male-male aggression in mice deficient for TRP2. Science. 2002;295(5559):1493–500. doi: 10.1126/science.1069259. [DOI] [PubMed] [Google Scholar]
- Strotmann J, Bader A, Luche H, Fehling HJ, Breer H. The patch-like pattern of OR37 receptors is formed by turning off gene expression in non-appropriate areas. Mol Cell Neurosci. 2009;41(4):474–85. doi: 10.1016/j.mcn.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Strotmann J, Levai O, Fleischer J, Schwarzenbacher K, Breer H. Olfactory receptor proteins in axonal processes of chemosensory neurons. J Neurosci. 2004;24(35):7754–61. doi: 10.1523/JNEUROSCI.2588-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann J, Wanner I, Krieger J, Raming K, Breer H. Expression of odorant receptors in spatially restricted subsets of chemosensory neurones. NeuroReport. 1992;3(12):1053–56. doi: 10.1097/00001756-199212000-00005. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Inokuchi K, Aoki M, Suto F, Tsuboi A, et al. Sequential arrival and graded secretion of Sema3F by olfactory neuron axons specify map topography at the bulb. Cell. 2010;141(6):1056–67. doi: 10.1016/j.cell.2010.04.041. [DOI] [PubMed] [Google Scholar]
- Tan L, Zong C, Xie XS. Rare event of histone demethylation can initiate singular gene expression of olfactory receptors. PNAS. 2013;110(52):21148–52. doi: 10.1073/pnas.1321511111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83(7):1091–100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Tian H, Ma M. Activity plays a role in eliminating olfactory sensory neurons expressing multiple odorant receptors in the mouse septal organ. Mol Cell Neurosci. 2008;38(4):484–88. doi: 10.1016/j.mcn.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai L, Barnea G. A critical period defined by axon-targeting mechanisms in the murine olfactory bulb. Science. 2014;344(6180):197–200. doi: 10.1126/science.1248806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, Reed RR. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J Neurosci. 1997;17(11):4159–69. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, Feinstein P, Mombaerts P. Homeodomain binding motifs modulate the probability of odorant receptor gene choice in transgenic mice. Mol Cell Neurosci. 2011;46(2):381–96. doi: 10.1016/j.mcn.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35(4):681–96. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nuñez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79(6):981–91. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74(2):309–18. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93(1):47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364(6433):121–26. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- Wang SS, Betz AG, Reed RR. Cloning of a novel Olf-1/EBF-like gene, O/E-4, by degenerate oligo-based direct selection. Mol Cell Neurosci. 2002;20(3):404–14. doi: 10.1006/mcne.2002.1138. [DOI] [PubMed] [Google Scholar]
- Wang SS, Lewcock JW, Feinstein P, Mombaerts P, Reed RR. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131(6):1377–88. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- Wang SS, Tsai RY, Reed RR. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci. 1997;17(11):4149–58. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-Z, Ou J, Zhu LJ, Green MR. Transcription factor ATF5 is required for terminal differentiation and survival of olfactory sensory neurons. PNAS. 2012;109(45):18589–94. doi: 10.1073/pnas.1210479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watatani Y, Ichikawa K, Nakanishi N, Fujimoto M, Takeda H, et al. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J Biol Chem. 2008;283(5):2543–53. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Müller JM, Greschik H, Fodor BD, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9(3):347–53. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272(23):14983–89. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–56. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- Young JM, Luche RM, Trask BJ. Rigorous and thorough bioinformatic analyses of olfactory receptor promoters confirm enrichment of O/E and homeodomain binding sites but reveal no new common motifs. BMC Genomics. 2011;12:561. doi: 10.1186/1471-2164-12-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Pacifico R, Cawley D, Feinstein P, Bozza T. Ultrasensitive detection of amines by a trace amine-associated receptor. J Neurosci. 2013;33(7):3228–39. doi: 10.1523/JNEUROSCI.4299-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-Q, Breer H, Strotmann J. Promotor elements governing the clustered expression pattern of odorant receptor genes. Mol Cell Neurosci. 2007;36(1):95–107. doi: 10.1016/j.mcn.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Belluscio L. Coding odorant concentration through activation timing between the medial and lateral olfactory bulb. Cell Rep. 2012;2(5):1143–50. doi: 10.1016/j.celrep.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]