Abstract
A phylogeny of anthropophilic and zoophilic anopheline mosquito species was constructed, using the nuclear internal transcribed spacer 2 (ITS2) and mitochondrial cytochrome oxidase subunit I (COI) genes. The ITS2 alignment, typically difficult due to its noncoding nature and large size variations, was aided by using predicted secondary structure, making this phylogenetically useful gene more amenable to investigation. This phylogeny is unique in explicitly including zoophilic, non-vector anopheline species in order to illustrate their relationships to malaria vectors. Two new, cryptic species, Anopheles funestus-like and Anopheles rivulorum-like, were found to be present in Zambia for the first time. Sequences from the D3 region of the 28S rDNA suggest that the Zambian An. funestus-like may be a hybrid or geographical variant of An. funestus-like, previously reported in Malawi. This is the first report of An. rivulorum-like sympatric with An. rivulorum (Leeson), suggesting that these are separate species rather than geographic variants.
Keyword Index: Anopheles, phylogeny, ITS2, COI, Zambia
INTRODUCTION
The genus Anopheles contains over 400 species (Harbach 2004), of which 30–40 are vectors for human malaria. The Anopheles genus is subdivided into six subgenera, Anopheles, Cellia, Kerteszia, Lophopodomyia, Nyssorhynchus, and Stethomyia. All Anopheles in sub-Saharan Africa are grouped in Anopheles and Cellia (Gillies and DeMeillon 1968). The Anopheles subgenus is further divided into the Angusticorn and Laticorn Series. The Cellia subgenus is divided into the Cellia, Myzomyia, Neocellia, Pyretophorus, Neomyzomyia, and Paramyzomyia Series.
Most phylogenetic work on the Anopheles genus has either focused on higher-level taxonomy (Krzywinski et al. 2001, Sallum et al. 2002, Sallum et al. 2000) or on elucidating relationships between major malaria vectors and their closely related taxa (Anthony et al. 1999, Garros et al. 2004, Marshall et al. 2005), see (Harbach 2004) for review. In contrast, this study encompasses primary, secondary, and nonvector anophelines present in Zambia and southern Africa.
For the purposes of this study, we included sympatric anopheline species present in southern Zambia (Table 1) (Anopheles gambiae Giles, An. arabiensis Patton, An. quadriannulatus Theobald species A, An. pharoensis Theobald, An. squamosus Theobald, An. rufipes Gough, An. maculipalpis Giles, An. pretoriensis Theobald, An. coustani sensu lato Laveran, An. theileri Edwards, Anopheles rivulorum Leeson, An. leesoni Evans, An. funestus sensu strictu Giles, An. rivulorum-like, An. funestus-like), as well as species from South Africa (Anopheles vaneedeni Gillies & Coetzee, Anopheles parensis Gillies) that are closely related but with limited range. An. minimus Theobald species A from Thailand was included due to its close relatedness to An. leesoni. ITS2 sequences were downloaded from GenBank for An. minimus Theobald species C, An. rivuloum-like from Burkina Faso, and An. funestus-like from Malawi.
Table 1.
Behavioral and vectorial characteristics of species in this study. Major vectors are marked in bold and “potential secondary” indicates that the species has been shown to be capable of transmitting Plasmodium but is unlikely to be a vector due to mainly zoophilic feeding behavior.
| Species | anthropophilic vs zoophilic | endophilic vs. exophilic | vector status |
|---|---|---|---|
| An. arabiensis | variable | variable | major vector |
| An. coustani | variable, mainly zoophilic | mainly exophilic | potential secondary |
| An. funestus | highly anthropophilic | endophilic | major vector |
| An. funestus-like | potentially zoophilic | unknown | unknown |
| An. gambiae s.s. | highly anthropophilic | endophilic | major vector |
| An. leesoni | zoophilic | exophilic | nonvector |
| An. longipalpis | zoophilic | frequently endophilic | nonvector |
| An. maculipalpis | zoophilic | exophilic | nonvector |
| An. minimus | highly anthropophilic | endophilic | major vector |
| An. parensis | zoophilic | exophilic | nonvector |
| An. pharoensis | mainly zoophilic | mainly exophilic | potential secondary |
| An. pretoriensis | zoophilic | exophilic | nonvector |
| An. quadriannulatus | zoophilic | exophilic | nonvector |
| An. rivulorum | zoophilic | exophilic | potential secondary |
| An. rivulorum-like | potentially variable | unknown | unknown |
| An. rufipes | zoophilic | exo- | potential secondary |
| An. squamosus | mainly zoophilic | mainly exophilic | potential secondary |
| An. theileri | zoophilic | exophilic | nonvector |
| An. vaneedeni | zoophilic | exophilic | potential secondary |
Anopheles gambiae, An. arabiensis, and An. quadriannulatus belong to the An. gambiae s.l. complex (Pyretophorus Series) and all have a wide distribution throughout Africa, with An. gambiae restricted to wetter areas, while An. arabiensis is more tolerant to drought (Gillies and DeMeillon 1968). An. gambiae is highly anthropophilic and one of the most important malaria vectors in sub-Saharan Africa (Gillies and DeMeillon 1968, White et al. 1972). An. arabiensis can vary in anthropophilicity, depending on locale, and has been shown to be primarily anthropophilic in southern Zambia (Fornadel et al. 2010a, Kent et al. 2007) and is the primary malaria vector in that region. An. quadriannulatus, although closely related to these two highly efficient vectors, is almost entirely zoophilic and therefore does not vector malaria. An. quadriannulatus is now known to be composed of two species, A and B. In Macha, only An. quadriannulatus A has been collected, therefore An. quadriannulatus B was not included.
Anopheles pharoensis and An. squamosus (Cellia Series) have mainly been ignored as vectors due to the fact that they are considered zoophilic. However, dissections of An. squamosus have yielded Plasmodium sporozoites in Tanzania (Gillies 1964) and Zimbabwe (Gillies and DeMeillon 1968), and they have been shown to bite humans in Zambia (Fornadel et al. 2010b). An. pharoensis has been shown to bite humans in high numbers and have been implicated as a major vector in Egypt (Barber and Rice 1937) and some areas of Cameroon (Antonio-Nkondjio et al. 2006), and as a secondary vector in Senegal (Dia et al. 2008). Its variation in vector status and behavior, as well as chromosomal inversion studies, suggests that An. pharoensis may be a species complex (Miles et al. 1983).
Anopheles rufipes, An. maculipalpis, and An. Pretoriensis (Neocellia Series), are almost entirely zoophilic Because An. rufipes occasionally enters households and bites people, and due to a small number of positive sporozoite dissections, it has been considered a secondary vector (Gillies and DeMeillon 1968). Anopheles coustani s.l. (Anopheles subgenus, Laticorn series) is outside the Cellia subgenus and is therefore used here as an outgroup. It has highly variable behavior, with few collected in human landing catches in Cameroon (Antonio-Nkondjio et al. 2006), Kenya (Mbogo et al. 1995), and Senegal (Dia et al. 2008), but increased anthropophily reported in Ethiopia (Taye et al. 2006), South Africa (Coetzee 1983), Mozambique (Mendis et al. 2000), and Zambia (Fornadel et al. 2010b). Additionally, specimens infected with Plasmodium malariae have been found in Cameroon (Antonio-Nkondjio et al. 2006). Like An. pharoensis, its variability in behavior prompted studies which indicated that An. coustani s.l. includes at least two species, An. coustani sensu strictu and An. crypticus (Coetzee 1994).
The Funestus Group, in the Myzomyia series, includes the Rivuloum Subgroup (An. rivulorum, An. rivulorum-like, An. brucei, An. fuscivenosus, all Afrotropical), the Minimus Subgroup (An. minimus A, C, and E, An. leesoni, An. fluviatilis, An. flavirostris, all Asian except An. leesoni), and the Funestus Subgroup (An. funestus s.s., Anopheles vaneedeni, Anopheles parensis, and An. aruni, all Afrotropical) (Garros et al. 2005, Harbach 2004). These species are morphologically similar as adults, and adults in the Funestus Subgroup are indistinguishable (Gillies and DeMeillon 1968), but the most common species are easily distinguished by PCR diagnostic (Koekemoer et al. 2002).
Despite their morphological similarity, mosquitoes in the Funestus Group have variable host preferences and vector competence. The anthropophilic An. funestus s.s. is a highly competent malaria vector, rivaling An. gambiae ss. as a major vector in Africa. An. rivulorum (Wilkes et al. 1996) and An. vaneedeni (De Meillon et al. 1977) are capable of Plasmodium transmission but are primarily zoophilic and therefore considered secondary vectors, and all other members of the complex are zoophilic (Cohuet et al. 2003). An. minimus s.l. is anthropophilic and one of the primary malaria vectors in Southeast Asia (Harrison 1980), but the related An. leesoni is primarily zoophilic and a non-vector (Gillies and DeMeillon 1968).
Anopheles theileri is in the Wellcomei Group, Myzomyia Series, and is almost entirely zoophilic (Gillies and DeMeillon 1968). An. longipalpis, also in the Myzomyia Series, includes two cryptic species, Type A and Type C (Koekemoer et al. 2009), and Type C was used in this study. Anopheles longipalpis is almost entirely zoophilic (Gillies and DeMeillon 1968) but can be highly endophilic, making it easily mistaken for the vector An. funestus s.s. (Kent et al. 2007). Additionally, two cryptic species, Anopheles rivulorum-like and Anopheles funestus-like, have been discovered based upon divergent ITS2 sequences (Cohuet et al. 2003, Spillings et al. 2009).
Specimens of Anopheles rivulorum-like have only been found in West Africa, particularly Cameroon (Cohuet et al. 2003) and Burkina Faso (Hackett et al. 2000), while An. rivulorum is confined to southern Africa. The vector status of An. rivulorum-like is unknown. Prior to this, there have been no reports of these two species in the same location, however, here we document that they are sympatric in southern Zambia. Anopheles funestus-like was recently identified in Malawi, where it is sympatric with An. funestus s.s., but experiments have definitively shown the two species to be separate (Spillings et al. 2009). In Macha, An. funestus s.s. was collected prior to a drought in 2004–2005 (Kent et al. 2007), after which no specimens were collected for several years and it was presumed that the species was locally extinct. In 2008–2009, several specimens of both An. funestus s.s. and An. funestus-like were collected.
As part of ongoing entomological research in southern Zambia, our aim was to construct a phylogeny of anopheline species in southern Africa, including previously overlooked zoophilic and non-vector species. By including all species present in this geographic area, we hope to shed light on the evolution of characteristics such as anthropophilicity, endophily, adaption to human-altered environments, and vector competence for malaria.
MATERIALS AND METHODS
Study site and sample collection
The Johns Hopkins Malaria Research Institute field station in Macha, Zambia is located in the southern province at 16.39292° S, 26.79061° E at an elevation of approximately 1,100 m above sea level. The habitat around the field station, the Malaria Institute at Macha (MIAM), is Miombo woodland. Mosquitoes were collected by CDC light trap (Beier 2002, Fornadel et al. 2010a, Sudia and Chamberlain 1988), human landing collection (Service 1976), and by cattle-baited trap (Fornadel et al. 2010b) in four village areas within 10 km of the field station. All samples were identified morphologically (Gillies and Coetzee 1987) and packaged on silica gel for transport to Johns Hopkins. Samples from the An. gambiae complex were further molecularly identified by the diagnostic developed by Scott and others (Scott et al. 1993), and samples from the An. funestus group were identified by the diagnostic developed by Koekomer and others (Koekemoer et al. 2002). Blood meal host species were identified for samples of An. funestus s.s. and An. rivulorum-like by PCR (Fornadel and Norris 2008, Kent and Norris 2005). Samples of mosquito species not native to southern Zambia were obtained as follows: An. gambiae from the Keele strain maintained at the Johns Hopkins School of Public Health insectary, An. minimus A collected in Thailand, An. funestus, An. leesoni, An. rivulorum, and An. vaneedeni from South Africa were provided by Maureen Coetzee and Lizette Koekemoer. The ITS2 sequences of An. minimus C (accession no. AF 230462.1), An. funestus-like from Malawi (GenBank accession no. FJ438963), and An. rivulorum-like from Cameroon and Burkina Faso (GenBank accession no. AF210725.1) were downloaded from GenBank.
Gene targets used in phylogenetic analyses
The cytochrome c oxidase I subunit (COI) is a mitochondrial protein-coding gene that has often been used for anopheline phylogeny construction (Marshall et al. 2005, Mohanty et al. 2009, Sallum et al. 2002) due to its phylogenetic information content, reliable PCR amplification due to high copy number, and ease of alignment due to evolutionary constraints imposed by the triplet code and necessity of coding for a functional enzyme. The internal transcribed spacer 2 is a nuclear, non-coding gene that separates the 5.8S and 28S large subunit RNA genes on the ribosomal RNA precursor transcript. The rRNA genes are arrayed in tandem repeats of several thousand, and the flanking 5.8S and 28S genes provide highly conserved primer binding sites, facilitating PCR amplification. Despite the fact that there are thousands of copies of the ribosomal RNA cistron, the sequences are homogenized by concerted evolution, such that high quality gene sequences can be obtained (Coleman 2003, Hershkovitz 1999). Unlike protein-coding genes, the ITS2 is not constrained by amino acid coding and is therefore more variable than most coding genes used for phylogeny reconstruction, giving greater resolution (Coleman 2003, Schultz et al. 2005). A major drawback is the high sequence variation and differences in length that can make alignment of sequences from taxonomic levels above species difficult or impossible. However, during rRNA maturation, the ITS2 folds into a secondary structure necessary for ribosomal cistron processing (Venema and Tollervey 1999). This secondary structure is greatly conserved among eukaryotes (Coleman and Vacquier 2002, Mai and Coleman 1997, Michot et al. 1999, Schultz et al. 2005) and can be used to aid primary sequence alignment. This conserved secondary structure, which can be used to identify the proper secondary structure from predicted models, consists of four helix loop regions, of which helix II is highly conserved and helix III is the longest, a pyrimidine mismatch in the basal 7 nucleotide pairs of helix II, and a conserved UGGU sequence at the 5’ end of helix III (Schultz et al. 2005). The ITS2 has been widely used for phylogeny studies (Coleman 2003), especially in plants (Hershkovitz 1999), and has more recently been extended to insects (Wiemers et al. 2009, Young and Coleman 2004), but despite investigations into mosquito ITS2 sequence and structure (Paskewitz et al. 1993, Wesson et al. 1992) and use of ITS2 sequences for mosquito species identification (Cohuet et al. 2003, Garros et al. 2004, Koekemoer et al. 2002, Spillings et al. 2009), its phylogenetic use for Anopheles has been limited to the subspecies level (Dassanayake et al. 2008).
DNA extraction and sequencing
DNA was extracted from dried mosquito samples using a DNeasy® Blood and Tissue kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The variable internal transcribed spacer 2 region (ITS2) was amplified with primers from the flanking 5.8S and 28S genes, ITS2A (5’-TGT GAA CTG CAG GAC ACA T-3’) and ITS2B (5’-ACC CCC TGA ATT TAA GCA TA-3’) (Koekemoer et al. 2002). Because sequencing reactions with ITS2A frequently failed, some samples were amplified and sequenced using the novel primer ITS2B1 (5’-GTC CCT ACG TGC TGA GCT TC-3’). This primer binds further downstream in the 28S gene, such that the 3’ portions of products immediately upstream of the ITS2B binding site could be sequenced. Each 25 µl reaction contained 1X PCR buffer, 200µM each dNTP, 30 pmol each primer, two units Taq polymerase, and 1.0 µl DNA template. Products were amplified in a thermocycler (MJ Research, Watertown, MA, USA) using the following conditions: 2 min initial denaturation at 94° C, 40 cycles of 30 s at 94° C, 30 s at 50° C, and 40 at 72° C, and a 10 min final extension at 72° C. PCR product size ranged from 480 bp to 847 bp. An. squamosus and An. pharoensis (both Cellia Series) repeatedly failed to produce an ITS2 PCR product despite varied PCR conditions and the creation of new primers, and were therefore not included in the ITS2 tree.
An 877 bp fragment of the cytochrome oxidase subunit I (COI) gene was amplified with the primers COI (5’-TTG ATT TTT TGG TCA TCC AGA AGT-3’) and Cul-Rev (5’-TAG AGC TTA AAT TCA TTG CAC TAA TC-3’) (Oshaghi et al. 2006). PCR reaction mixtures were identical to the ITS2 reaction, with the following thermocycler conditions: 1 min initial denaturation at 94° C, 32 cycles of 1 min at 94° C, 1 min at 55° C, 2 min at 72° C, and a 7 min final extension at 72° C.
Domain 3 of the 28S rDNA (D3) was sequenced from samples of An. funestus-like from Zambia for comparison to published sequences of An. funestus s.s. and An. funestus-like from Malawi (GenBank accession no. DQ407757.1 and FJ843022, respectively) (Spillings et al. 2009). The product was amplified with the primers D3A (5’-GAC CCG TCT TGA AAC ACG GA-3’) and D3B (5’-TCG GAA GGA ACC AGC TAC TA-3’). Each 25 µl reaction contained 1X PCR buffer, 200 µM of each dNTP, 25 pmol of each primer, 2 U Taq polymerase, and 1 µl DNA template and was amplified using the following conditions: 3 min initial denaturation at 94° C, followed by 30 cycles of 94° C denaturation for 30 s, 63° C annealing for 40 s, 72° C extension for 40 s, and a 72° C final extension for 10 min.
Five µl of each PCR product was subjected to electrophoresis on a 2% agarose gel, stained with ethidium bromide, and visualized under UV illumination. The remainder of each successful PCR reaction was purified using a Qiaquick PCR prep kit (Qiagen, Valenca, CA). Products were sequenced in both directions using dye terminator chemistry on a 3730×1 DNA Analyzer (Applied Biosystems, Foster City, CA) at the Johns Hopkins University School of Medicine Sequencing and Synthesis Facility. Accession numbers for each sequence are given in Table 2.
Table 2.
Gene name, species, location collected, and GenBank accession numbers of samples used in phylogeny construction.
| Gene | Species | Location | Accession number |
|---|---|---|---|
| ITS2 | An. arabiensis | Zambia | JN994133 |
| ITS2 | An. coustani | Zambia | JN994134 |
| ITS2 | An. funestus | Zambia | JN994135 |
| ITS2 | An. funestus | South Africa | JN994136 |
| ITS2 | An. funestus-like | Zambia | JN994137 |
| ITS2 | An. gambiae | Keele strain colony | JN994138 |
| ITS2 | An. leesoni | Zambia | JN994139 |
| ITS2 | An. longipalpis upper band | Zambia | JN994140 |
| ITS2 | An. longipalpis lower band | Zambia | JN994141 |
| ITS2 | An. maculipalpis | Zambia | JN994142 |
| ITS2 | An. minimus C | Thailand | JN994143 |
| ITS2 | An. parensis | Zambia | JN994144 |
| ITS2 | An. pretoriensis | Zambia | JN994145 |
| ITS2 | An. quadriannulatus | Zambia | JN994146 |
| ITS2 | An. rivulorum-like | Zambia | JN994147 |
| ITS2 | An. rivulorum | South Africa | JN994148 |
| ITS2 | An. rivulorum | Zambia | JN994149 |
| ITS2 | An. rufipes | Zambia | JN994150 |
| ITS2 | An. theileri | Zambia | JN994151 |
| ITS2 | An. vaneedeni | South Africa | JN994152 |
| COI | An. arabiensis | Zambia | JN994153 |
| COI | An. coustani | Zambia | JN994154 |
| COI | An. funestus | Zambia | JN994155 |
| COI | An. funestus-like | Zambia | JN994156 |
| COI | An. gambiae s.s. | Keele strain colony | JN994157 |
| COI | An. leesoni | Zambia | JN994158 |
| COI | An. longipalpis | Zambia | JN994159 |
| COI | An. maculipalpis | Zambia | JN994160 |
| COI | An. minimus C | Thailand | JN994161 |
| COI | An. parensis | Zambia | JN994162 |
| COI | An. pharoensis | Zambia | JN994163 |
| COI | An. pretoriensis | Zambia | JN994164 |
| COI | An. quadriannulatus | Zambia | JN994165 |
| COI | An. rivulorum-like | Zambia | JN994166 |
| COI | An. rivulorum | South Africa | JN994167 |
| COI | An. rivulorum | Zambia | JN994168 |
| COI | An. rufipes | Zambia | JN994169 |
| COI | An. squamosus | Zambia | JN994170 |
| COI | An. theileri | Zambia | JN994171 |
| COI | An. vaneedeni | South Africa | JN994172 |
ITS2 PCR products from South African samples of An. leesoni, An. funestus, and An. rivulorum and COI PCR products from An. leesoni and An. funestus were 100% identical to their Zambian counterparts. They were therefore dropped from the alignment.
Alignment and phylogeny construction
The Clustal function in MEGA 4.0 (Tamura et al. 2007) was used to align the COI sequences and to trim the ends of sequences, and MEGA 4.0 was used to build phylogenies. For the Neighbor-Joining (NJ) tree, evolutionary distances were computed using the Maximum Composite Likelihood evolution method. The 1st+2nd+3rd+non-coding codon positions were used. Gaps and missing data were completely deleted from the dataset, leaving a total of 831 positions. The Maximum Parsimony (MP) tree was obtained using the Close-Neighbor-Interchange algorithm with a search level of 3, in which the initial trees were obtained with the random addition of sequences (ten replicates). The 1st+2nd+3rd+non-coding codon positions were used. Trees were rooted using An. coustani s.l. as an outgroup. Bootstrap tests with 1,000 replicates were performed for both trees. NJ and MP trees using translated COI protein sequences were constructed similarly but were poorly supported at most nodes (bootstrap value <50) and therefore not shown.
The 5.8S and 28S ends of the ITS2 were determined using the HMM Annotation tool on the ITS2 database website (Keller et al. 2009) at http://its2.bioapps.biozentrum.uni-wuerzburg.de, and sequences were trimmed to this length to remove the highly conserved fragments of the 5.8S and 28S rRNA genes that border the ITS2. Secondary structures were determined by custom homology modeling using the ITS2 database (Wolf et al. 2005) (http://its2.bioapps.biozentrum.uni-wuerzburg.de/cgi-bin/indexpl?custom.). Anopheles superpictus (GenBank accession number EU482198) was used as the template. For sequences that did not have significant similarity to An. superpictus, the An. pretoriensis secondary structure result was used as a template. ITS2 sequences were aligned using 4SALE (Seibel et al. 2006), which takes secondary structure into account during alignment. Phylogenetic trees were then constructed using the same NJ and MP methods described for COI, with the difference that gaps and missing data were deleted in only pairwise comparisons.
Because the ITS2 sequences of An. funestus-like from Malawi (GenBank accession no. FJ438963) and An. rivulorum-like from Cameroon and Burkina Faso (GenBank accession no. AF210725.1) are shorter than the sequences used in our phylogeny, separate alignments of these sequences with their Zambian counterparts were constructed in MEGA 4.0. Pairwise genetic distances were calculated as number of base substitutions per site, using the Maximum Composite Likelihood method and with gaps and missing data deleted only for pairwise comparisons. Additionally, the An. funestus-like Zambia D3 partial sequence was aligned with published An. funestus-like and An. funestus s.s. sequences using MEGA 4.0.
RESULTS
COI phylogenies
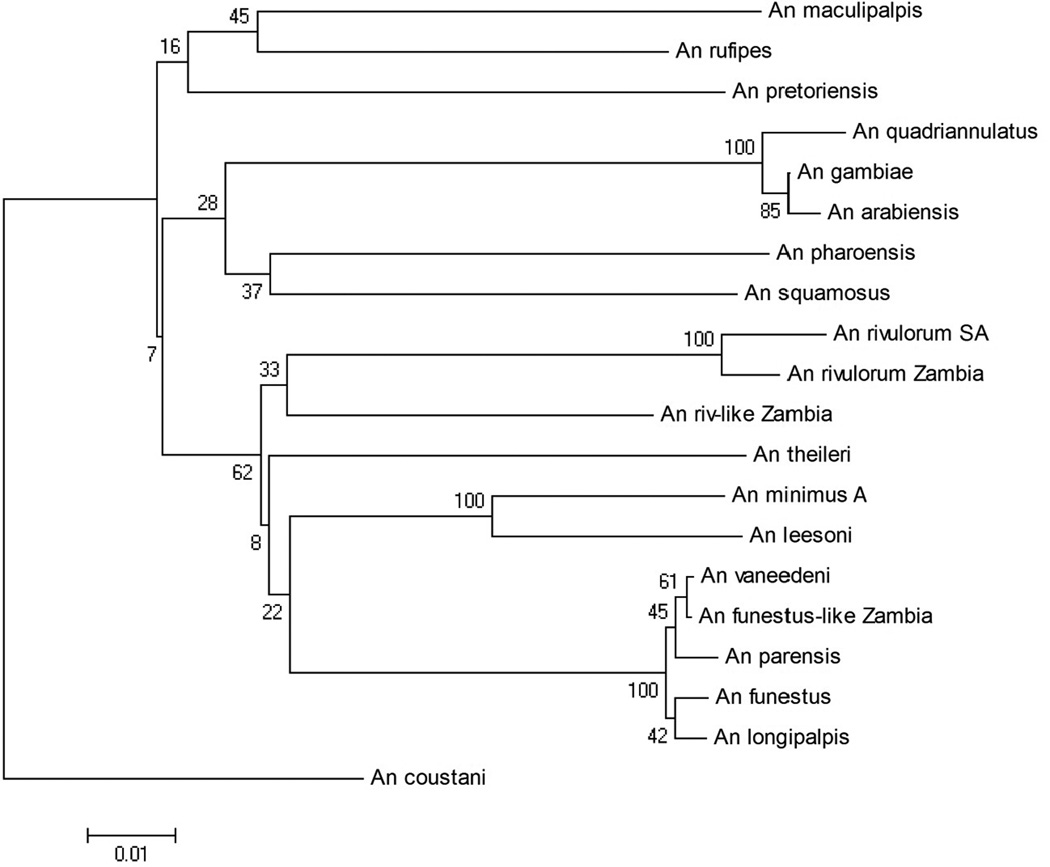
In the COI alignment, there were 831 positions in the dataset, 188 of which were parsimony informative. For the NJ tree, the optimum tree (with a branch length of 0.75231908) is shown (Figure 1) with bootstrap values at each node. This tree maintained all series as monophyletic groups, placing Cellia closest to Pyretophorus, and Neocellia as the outermost group. The Rivulorum and Minimus Subgroups within Myzomyia were monophyletic, An. theileri was placed within the Funestus Group, and An. longipalpis was placed within the Funestus Subgroup.
Figure 1.
Cytochrome oxidase sub unit I (COI) Neighbor-Joining tree, branch length = 0.75231908.
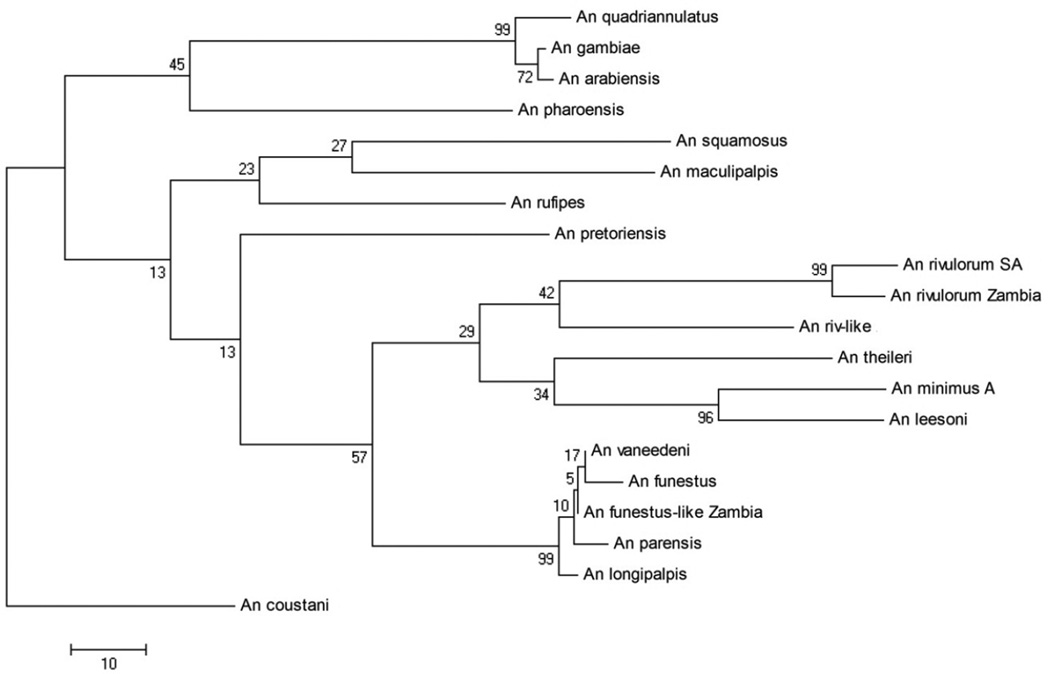
The Maximum Parsimony method found 12 most parsimonious trees, with a length of 639, one of which is shown (Figure 2). These 12 trees differed only in the internal arrangement of the clade containing An. vaneedeni, An. funestus s.s., An. funestus-like, An. parensis, and An. longipalpis. Six of the 12 trees included An. longipalpis within the Funestus Subgroup, while six put it outside the clade. This tree did not preserve the Cellia and Neocellia series but still put An. pharoensis closest to the An. gambiae complex. However, most higher-level nodes were poorly supported in both COI trees.
Figure 2.
Cytochrome oxidase subunit I (COI) Maximum Parsimony tree, one of 12 most parsimonious trees that differed in the arrangement of the Funestus Group clade.
ITS2 phylogenies
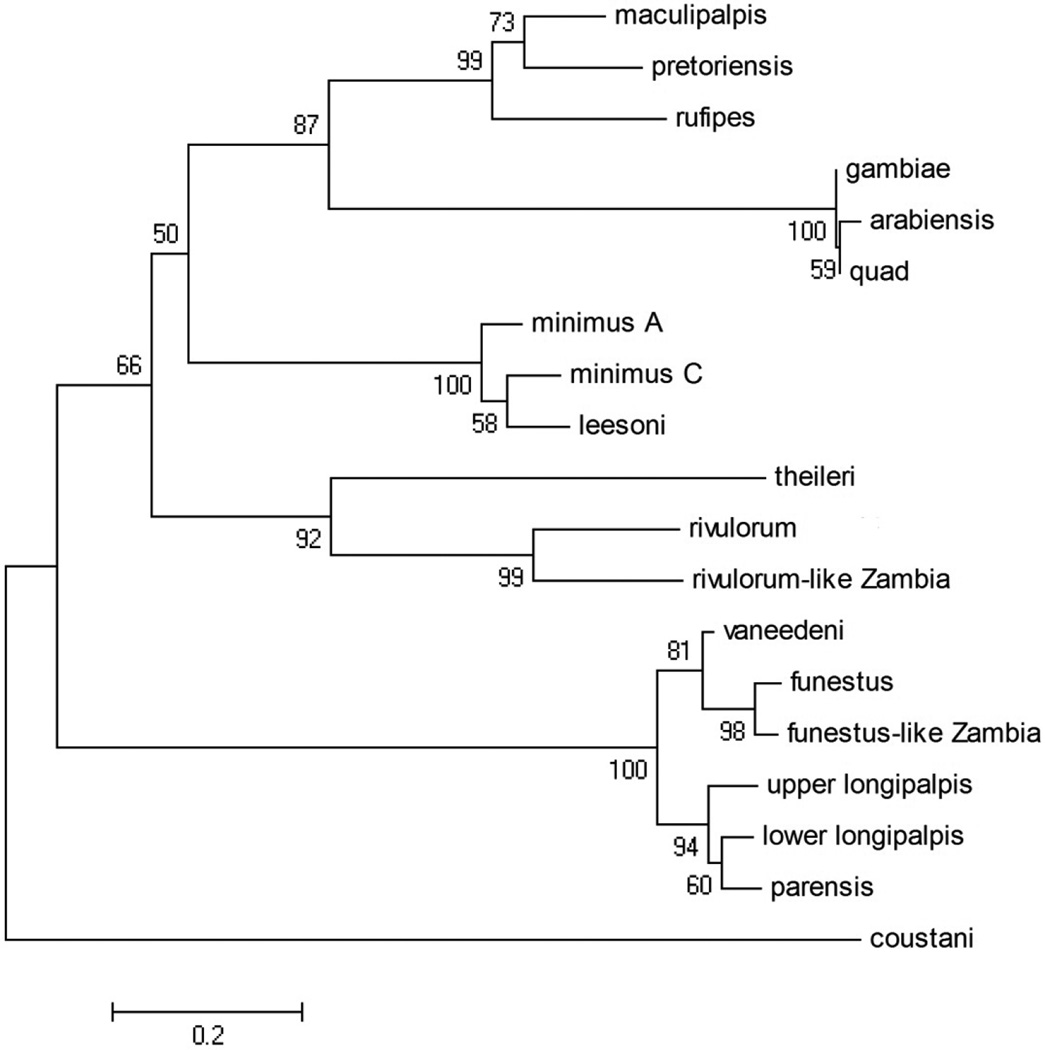
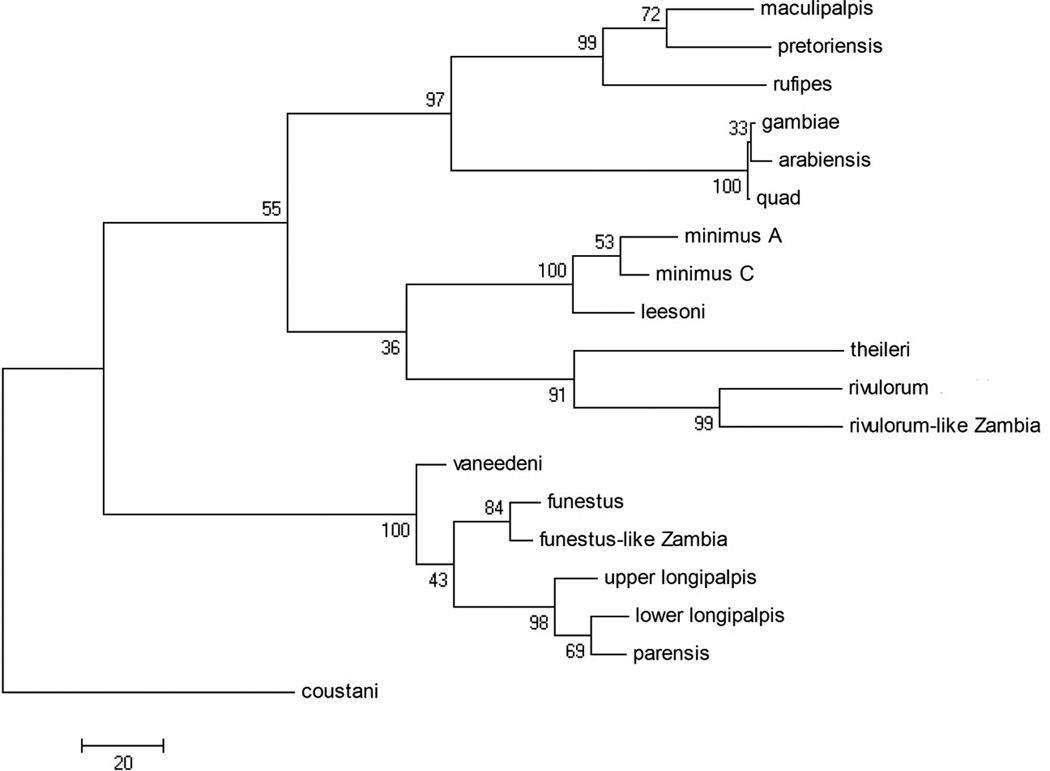
In the ITS2 alignment, there were 658 positions in the final dataset, of which 277 of which were parsimony informative. For the Neighbor-Joining method, the optimum tree (with a branch length of 5.06003986) is shown (Figure 3) with bootstrap values at each node. The Maximum Parsimony method found three most parsimonious trees, with a length of 902. These three trees differed only in the rearrangement of the An. gambiae complex clade. One of these trees is shown (Figure 4). The MP tree was in best agreement with the current classification of the Anopheles genus (Harbach 2004): the Neocellia Series (An. maculipalpis, An. pretoriensis, and An. rufipes) and Pyretophorus Series (An. gambiae complex) are monophyletic. However, the resulting phylogeny has the Myzomyia Series as paraphyletic, with An. minimus, An. leesoni, An. theileri, An. rivulorum, and An. rivulorum-like making up one clade, and An. vaneedeni, An. parensis, An. longipalpis, An. funestus and An. funestus-like making up the other. The Minimus and Rivulorum Subgroups are each monophyletic, and the Funestus Subgroup groups together but includes An. longipalpis. The ITS2 NJ tree is arranged nearly identically to the MP tree, with the exception that the Minimus and Rivulorum Subgroups are separate clades.
Figure 3.
Internal transcribed spacer 2 (ITS2) Neighbor-Joining tree, branch length = 5.06003986.
Figure 4.
Internal transcribed spacer 2 (ITS2) Maximum Parsimony tree, one of three most parsimonious trees that difered in the arrangement of the An. gambiae clade.
During the 2008–2009 rainy season, two specimens of An. funestus s.s., four specimens of An. funestus-like, one specimen of An. rivulorum, and 16 specimens of An. rivulorum-like were collected from nearby localities in southern Zambia. All were collected in CDC light traps inside houses, except for two An. rivulorum-like from cattle-baited traps and one from an indoor human landing catch. One An. funestus s.s. was blooded (human), as well as two An. funestus-like (one human, one goat), and five An. rivulorum-like (one human, one human+cow, one human+goat, two cows) (Table 3). Additionally, one archived An. funestus-like sample collected by pyrethroid spray catch in 2006 was identified by ITS2 sequencing and contained cow blood. All other specimens were unfed. ITS2 sequences were identical for all samples within each species.
Table 3.
An. funestus s.s., An. funestus-like, An. rivulorum, and An. rivulorum-like samples collected in Macha during the 2008–2009 rainy season. Locations in Universal Transverse Mercator grid 35L: Namwalinda (X 487921, Y8193087), Chidakwa (X 477263, Y8184855), Lupata (X475497, Y8188941), and regional collections (X495271, Y8192700).
| # collected | blood meals | location | |
|---|---|---|---|
| An. funestus s.s. | 2 | 1 human | Namwalinda |
| An. funestus-like | 5 | 1 cow | Namwalinda and Chidakwa |
| An. rivulorum | 3 | -- | Namwalinda |
| An. rivulorum-like | 20 | 1 human, 1 human+cow, 1 human+goat, 3 cows |
Namwalinda, Lupata, and Chidakwa |
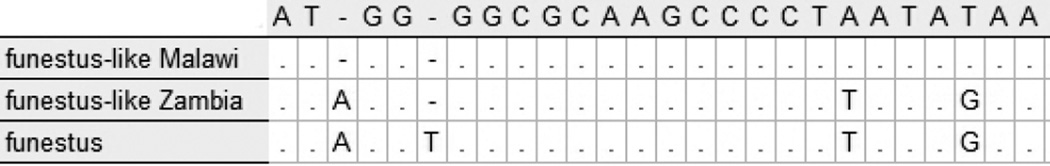
Although there was no ITS2 divergence within An. rivulorum and An. funestus s.s. species from Zambia and South Africa, the sequences of An. rivulorum-like and An. funestus-like from Zambia were divergent from their counterparts in Burkina Faso and Malawi, respectively. Out of 449 total basepairs, An. rivulorum-like Zambia had 15 basepair substitutions and a one bp insertion when compared to An. rivulorum-like Burkina Faso (Table 4). Of 740 total basepairs, An. funestus-like Zambia had eight substitutions and a four bp insertion when compared to An. funestus-like Malawi (Table 5). Interestingly, the D3 sequence of An. funestus-like Zambia was closest to that of An. funestus s.s., with only a single one bp deletion, vs a one bp insertion and two substitutions when compared to An. funestus-like Malawi (Figure 5).
Table 4.
Table of pairwise diferences for An. rivulorum and An. rivulorum-like ITS2 sequences. Number of base substitutions per site; analysis conducted using Maximum Composite Likelihood method. Gaps and missing data were deleted only in pairwise comparisons.
|
rivulorum-like Burkina Faso |
rivulorum-like Zambia |
rivulorum | |
|---|---|---|---|
| rivulorum-like Burkina Faso | --- | ||
| rivulorum-like Zambia | 0.035 | --- | |
| rivulorum | 0.165 | 0.170 | --- |
Table 5.
Table of pairwise diferences for An. funestus s.s. and An. funestus-like ITS2 sequences. Number of base substitutions per site; analysis conducted using Maximum Composite Likelihood method. Gaps and missing data were deleted only in pairwise comparisons.
|
funestus-like Malawi |
funestus-like Zambia |
funestus s.s. | |
|---|---|---|---|
| funestus-like Malawi | --- | ||
| funestus-like Zambia | 0.010 | --- | |
| funestus s.s. | 0.037 | 0.036 | --- |
Figure 5.
Variable section of Domain 3 of 28S for Malawi An. funestus-like, Zambian An. funestus-like, and An. funestus s.s.
DISCUSSION
A current theory suggests that the Anopheles ancestral phenotype is resistance to malaria (Riehle et al. 2006, Rios-Vasquez et al. 2013). Using this assumption, previous work has shown independent evolution of anthropophily and Plasmodium vectorial capacity has occurred multiple times within major African vector species (Marshall et al. 2005, Kamali et al. 2012). By including zoophilic species, our phylogeny suggests that anthropophilicity and capacity to vector Plasmodium evolved independently at least three times within the subgenus Cellia, with An. gambiae s.s. and An. arabiensis; An. minimus A and C; and An. funestus s.s. clustering more closely with zoophilic species than with each other. Intermediate levels of anthropophilicity, and minor vector status, have evolved at least twice, in the clade containing An. pharoensis and An. squamosus, and in An. rivulorum. Alternatively, if the ability to transmit Plasmodium is the ancestral phenotype, this trait was lost at least three times.
Historically, the status of An. longipalpis has been disputed (Koekemoer et al. 2009). Despite variation in the arrangement of the Funestus Subgroup, all the trees placed An. longipalpis C within this clade, with the exception of half the COI MP trees that placed An. longipalpis C immediately outside the clade. This agrees with previous work (Koekemoer et al. 2009) showing that An. longipalpis C is more closely related to the Funestus Subgroup than to the Minimus Subgroup. Additionally, all trees placed An. theileri, a member of the Wellcomei Group, within the Funestus Group, indicating that either the Funestus Group is not monophyletic, or that the Wellcomei Group should be included within it.
For distal nodes, the ITS2 trees appeared to be more robust than the COI trees. The use of secondary structure greatly assisted in properly aligning sequences, making it possible to use this gene. The ITS2 contained more parsimony informative sites by length (277 of 658 bp vs 188 of 831 bp for COI) and provided trees with higher bootstrap values at most nodes. Unlike the COI NJ and MP trees, which were not highly similar, the ITS2 trees had nearly identical topology, indicating that the ITS2 was less sensitive to which method was used. However, as shown by bootstrap values at higher-level nodes, the more variable ITS2 was less accurate in determining basal tree topology. Both trees inaccurately placed An. gambiae and An. minimus groups together, despite morphological evidence that An. minimus and An. funestus are sister taxa (Marshall et al. 2005, Garros et al. 2005). Another limitation was the inability to PCR amplify the ITS2 from species in the Cellia Series.
An additional consideration is whether these trees may be affected by introgression. In the An. gambiae group, autosomal genes and inversions have introgressed between sibling species, complicating the construction of phylogenetic trees (Besansky et al. 2003, Besansky et al. 1994). However, the X chromosome, particularly the pericentromeric region which contains the rDNA and ITS2 locus, is more divergent and less susceptible to introgression (Besansky et al. 2003).
This is the first report in southern Africa of An. rivulorum-like, previously limited to West Africa (Cohuet et al. 2003). An. rivulorum and An. rivulorum-like are sympatric in Zambia and there is no evidence of hybridization, which lends more weight to the theory that An. rivulorum-like is a separate species. Specimens of An. rivulorum-like from Zambia all had identical ITS2 sequences but were divergent from ITS2 sequences from West Africa. However, they were clearly more closely related to An. rivulorum-like than to An. rivulorum and may simply be geographic variants. In Zambia, An. rivulorum-like appears to be at least somewhat anthropophilic, as they were collected in indoor CDC traps and human landing catches, and over half the blooded samples contained either human or mixed human/animal blood meals. However, it is unknown how many were collected in cattle-baited traps as our lab does not routinely conduct molecular species diagnostic assays to distinguish An. rivulorum-like from the very common An. longipalpis collected in cattle-baited traps. Because the closely related An. rivulorum is known to be a potential secondary vector, it remains possible that An. rivulorum-like could vector malaria.
Additionally, this is the first report of An. funestus-like outside of Malawi. Like An. rivulorum-like, all Zambian ITS2 and D3 sequences were identical. However, it is unclear if this is an An. funestus s.s. x An. funestus-like hybrid, or an An. funestus-like geographic variant. The ITS2 sequence is more closely related to that of An. funestus-like, but the D3 sequence is clearly closer to An. funestus s.s. Differences as small as 2–3 bp in the D3 region have been shown to differentiate between species (Singh et al. 2004, Spillings et al. 2009), indicating that Zambian An. funestus-like may be a separate species. In Zambia, An. funestus-like enters houses and at least occasionally feeds on humans. Because An. funestus s.s. is such an efficient malaria vector, it will be important to monitor An. funestus-like to determine if it is also a malaria vector.
This phylogeny, which includes all Anopheles species present in southern Zambia, as well as many from southern Africa, clarifies the relationships between the anthropophilic malaria vectors An. gambiae s.s., An. arabiensis, An. funestus s.s., and more zoophilic, secondary vector or non-vector species in the subgenus Cellia. This information is important to understanding of the evolution of mosquito behavioral characteristics such as zoophily, anthropophily, endo- and exophagy, and endo- and exophily. Many species, such as An. arabiensis, are highly plastic in their behavior, and exhibit variation across the spectrum of these characteristics. As intensification of vector control measures such as insecticide-treated bed nets and indoor residual spraying with insecticides targets anthropophilic, endophagic, and endophilic species, there is the potential for shifts in the behavior of these species. Understanding the phylogenetic basis for these behaviors will be vital to support evidence-based vector control.
Acknowledgments
The authors thank Rebekah Kading, Christen Fornadel, Maureen Coetzee, and Lizette Koekemoer for providing mosquito samples, as well as the MIAM entomology team, especially Shadreck Habbanti, Limonty Simubali, Harry Hamapumbu, and Petros Moono for invaluable assistance with collections. We would also like to thank the two anonymous reviewers for their insightful comments. This work was funded by an NIH T32 AI 007417 Training Grant to LCN, research support to DEN from NIH (ICEMR) U19 AI089680, and the Johns Hopkins Malaria Research Institute.
REFERENCES CITED
- Anthony TG, Harbach RE, Kitching IJ. Phylogeny of the Pyretophorus Series of Anopheles subgenus Cellia (Diptera: Culicidae) Syst. Entomol. 1999;24:193–205. [Google Scholar]
- Antonio-Nkondjio C, Kerah CH, Simard F, Awono-Ambene P, Chouaibou M, Tchuinkam T, Fontenille D. Complexity of the malaria vectorial system in Cameroon: contribution of secondary vectors to malaria transmission. J. Med. Entomol. 2006;43:1215–1221. doi: 10.1603/0022-2585(2006)43[1215:cotmvs]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Barber MA, Rice JB. A survey of malaria in Egypt. Am. J. Trop. Med. Hyg. 1937;17:413–436. [Google Scholar]
- Beier JC. Vector incrimination and entomological inoculation rates. Meth. Mol. Med. 2002;72:3–11. doi: 10.1385/1-59259-271-6:01. [DOI] [PubMed] [Google Scholar]
- Besansky NJ, Powell JR, Caccone A, Hamm DM, Scott JA, Collins EH. Molecular phylogeny of the Anopheles gambiae complex suggests genetic introgression between principal malaria vectors. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6885–6888. doi: 10.1073/pnas.91.15.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Kryzwinski J, Lehmann T, Simard F, Kern M, Mukabayire O, Fontenille D, Touré Y, Sagnon N. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M. Chromosomal and cross-mating evidence for two species within Anopheles (A.) coustani (Diptera: Culicidae) Syst. Entomol. 1983;8:137–141. [Google Scholar]
- Coetzee M. Anopheles crypticus, new species from South Africa is distinguished from Anopheles coustani (Diptera: Culicidae) Mosq. Syst. 1994;26:125–131. [Google Scholar]
- Cohuet A, Simard F, Toto JC, Kengne P, Coetzee M, Fontenille D. Species identification within the Anopheles funestus group of malaria vectors in Cameroon and evidence for a new species. Am. J. Trop. Med. Hyg. 2003;69:200–205. [PubMed] [Google Scholar]
- Coleman AW. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 2003;19:370–375. doi: 10.1016/S0168-9525(03)00118-5. [DOI] [PubMed] [Google Scholar]
- Coleman AW, Vacquier VD. Exploring the phylogenetic utility of ITS sequences for animals: a test case for abalone (Haliotis) J. Mol. Evol. 2002;54:246–257. doi: 10.1007/s00239-001-0006-0. [DOI] [PubMed] [Google Scholar]
- Dassanayake RS, Gunawardene YINS, Nissanka BDD, De Silva K. ITS-2 secondary structures and phylogeny of Anopheles culicifacies species. Bioinformation. 2008;2:456–460. doi: 10.6026/97320630002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meillon B, Van Eeden GJ, Coetzee L, Coetzee M, Meiswinkel R, Du Toit CLN, Hansford CF. Observations on a species of the Anopheles funestus subgroup, a suspected exophilic vector of malaria parasites in northeastern Transvaal, South Africa. Mosq. News. 1977;37:657–661. [Google Scholar]
- Dia I, Konate L, Samb B, Sarr JB, Diop A, Rogerie F, Faye M, Riveau G, Remoue F, Diallo M, Fontenille D. Bionomics of malaria vectors and relationship with malaria transmission and epidemiology in three physiographic zones in the Senegal River Basin. Acta Trop. 2008;105:145–153. doi: 10.1016/j.actatropica.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Fornadel CM, Norris DE. Increased endophily by the malaria vector Anopheles arabiensis in southern Zambia and identification of digested blood meals. Am. J. Trop. Med. Hyg. 2008;79:876–880. [PMC free article] [PubMed] [Google Scholar]
- Fornadel CM, Norris LC, Norris DE. Centers for Disease Control light traps for monitoring Anopheles arabiensis human biting rates in an area with low vector density and high insecticide-treated bed net use. Am. J. Trop. Med. Hyg. 2010a;83:838–842. doi: 10.4269/ajtmh.2010.10-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornadel CM, Norris LC, Glass GE, Norris DE. Analysis of Anopheles arabiensis blood feeding behavior in southern Zambia during the two years after introduction of insecticide-treated bed nets. Am. J. Trop. Med. Hyg. 2010b;83:848–853. doi: 10.4269/ajtmh.2010.10-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garros C, Harbach RE, Manguin S. Morphological assessment and molecular phylogenetics of the Funestus and Minimus groups of Anopheles (Cellia) J. Med. Entomol. 2005;42:522–536. doi: 10.1093/jmedent/42.4.522. [DOI] [PubMed] [Google Scholar]
- Garros C, Koekemoer LL, Coetzee M, Coosemans M, Manguin S. A single multiplex assay to identify major malaria vectors within the African Anopheles funestus and the Oriental An. minimus groups. Am. J. Trop. Med. Hyg. 2004;70:583–590. [PubMed] [Google Scholar]
- Gillies M, Coetzee M. A Supplement to the Anophelinae of Africa South of the Sahara. Johannesburg: South African Institute for Medical Research; 1987. [Google Scholar]
- Gillies MT. The role of secondary vectors of malaria in north-east Tanganyika. Trans. R. Soc. Trop. Med. Hyg. 1964;58:154–158. doi: 10.1016/0035-9203(64)90004-5. [DOI] [PubMed] [Google Scholar]
- Gillies MT, DeMeillon B. The Anophelinae South of the Sahara (Ethiopian Zoological Region) Johannesburg: South African Institute for Medical Research; 1968. [Google Scholar]
- Hackett BJ, Gimnig J, Guelbeogo W, Costantini C, Koekemoer LL, Coetzee M, Collins FH, Besansky NJ. Ribosomal DNA internal transcribed spacer (ITS2) sequences differentiate Anopheles funestus and An. rivulorum, and uncover a cryptic taxon. Insect. Mol. Biol. 2000;9:369–374. doi: 10.1046/j.1365-2583.2000.00198.x. [DOI] [PubMed] [Google Scholar]
- Harbach RE. The classification of genus Anopheles (Diptera: Culicidae): a working hypothesis of phylogenetic relationships. Bull. Entomol. Res. 2004;94:537–553. doi: 10.1079/ber2004321. [DOI] [PubMed] [Google Scholar]
- Harrison BA. Medical entomology studies-XIII. The Myzomyia Series of Anopheles (Cellia) in Thailand, with emphasis on intra-interspecific variations (Diptera: Culicidae) Contrib Am. Entomol. Inst. 1980;17:195. [Google Scholar]
- Hershkovitz MA. Ribosomal DNA sequences and angiosperm systematics. In: Hollingsworth PM, editor. Molecular Systematics and Plant Evolution. Taylor and Francis; 1999. pp. 268–326. [Google Scholar]
- Kamali M, Xia A, Zhijian T, Sharakov I. A new chromosomal phylogeny supports the repeated origin of vectorial capacity in malaria mosquitoes of the Anopheles gambiae Complex. PLoS Path. 2012;8(10):el002960. doi: 10.1371/journal.ppat.1002960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Schleicher T, Schultz J, Muller T, Dandekar T, Wolf M. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430:50–7. doi: 10.1016/j.gene.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Kent RJ, Norris DE. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am. J. Trop. Med. Hyg. 2005;73:336–342. [PMC free article] [PubMed] [Google Scholar]
- Kent RJ, Thuma PE, Mharakurwa S, Norris DE. Seasonality, blood feeding behavior, and transmission of Plasmodium falciparum by Anopheles arabiensis after an extended drought in southern Zambia. Am. J. Trop. Med. Hyg. 2007;76:267–274. [PMC free article] [PubMed] [Google Scholar]
- Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am. J. Trop. Med. Hyg. 2002;66:804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- Koekemoer LL, Misiani EA, Hunt RH, Kent RJ, Norris DE, Coetzee M. Cryptic species within Anopheles longipalpis from southern Africa and phylogenetic comparison with members of the An. funestus group . Bull. Entomol. Res. 2009;99:41–49. doi: 10.1017/S0007485308006123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski J, Wilkerson RC, Besansky NJ. Toward understanding Anophelinae (Diptera, Culicidae) phylogeny: insights from nuclear single-copy genes and the weight of evidence. Syst. Biol. 2001;50:540–556. [PubMed] [Google Scholar]
- Mai JC, Coleman AW. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J. Mol. Evol. 1997;44:258–271. doi: 10.1007/pl00006143. [DOI] [PubMed] [Google Scholar]
- Marshall JC, Powell JR, Caccone A. Short report: Phylogenetic relationships of the anthropophilic Plasmodium falciparum malaria vectors in Africa. Am. J. Trop. Med. Hyg. 2005;73:749–752. [PubMed] [Google Scholar]
- Mbogo CN, Snow RW, Khamala CP, Kabiru EW, Ouma JH, Githure JI, Marsh K, Beier JC. Relationships between Plasmodium falciparum transmission by vector populations and the incidence of severe disease at nine sites on the Kenyan coast. Am. J. Trop. Med. Hyg. 1995;52:201–206. doi: 10.4269/ajtmh.1995.52.201. [DOI] [PubMed] [Google Scholar]
- Mendis C, Jacobsen JL, Gamage-Mendis A, Bule E, Dgedge M, Thompson R, Cuamba N, Barreto J, Begtrup K, Sinden RE, Hogh B. Anopheles arabiensis and An. funestus are equally important vectors of malaria in Matola coastal suburb of Maputo, southern Mozambique. Med. Vet. Entomol. 2000;14:171–180. doi: 10.1046/j.1365-2915.2000.00228.x. [DOI] [PubMed] [Google Scholar]
- Michot B, Joseph N, Mazan S, Bachellerie JP. Evolutionarily conserved structural features in the ITS2 of mammalian pre-rRNAs and potential interactions with the snoRNA U8 detected by comparative analysis of new mouse sequences. Nucleic Acids Res. 1999;27:2271–82. doi: 10.1093/nar/27.11.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles SJ, Green CA, Hunt RH. Genetic observations on the taxon Anopheles (Cellia) pharoensis Theobald (Diptera: Culicidae) J. Trop. Med. Hyg. 1983;86:153–157. [PubMed] [Google Scholar]
- Mohanty A, Swain S, Kar SK, Hazra RK. Analysis of the phylogenetic relationship of Anopheles species, subgenus Cellia (Diptera: Culicidae) and using it to define the relationship of morphologically similar species. Infect Genet Evol. 2009;9:1204–1224. doi: 10.1016/j.meegid.2009.06.021. [DOI] [PubMed] [Google Scholar]
- Oshaghi MA, Yaaghoobi F, Abaie MR. Pattern of mitochondrial DNA variation between and within Anopheles stephensi (Diptera: Culicidae) biological forms suggests extensive gene flow. Acta Trop. 2006;99:226–233. doi: 10.1016/j.actatropica.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Paskewitz SM, Wesson DM, Collins FH. The internal transcribed spacers of ribosomal DNA in five members of the Anopheles gambiae species complex. Insect Mol. Biol. 1993;2:247–257. doi: 10.1111/j.1365-2583.1994.tb00144.x. [DOI] [PubMed] [Google Scholar]
- Riehle MM, Markianos K, Niare O, Xu J, Li J, Touré AM, Podiougou B, Odoul F, Diawara S, Diallo M, Coulibaly B, Ouatara A, Kruglyak L, Traoure SF, Vernick KD. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312:577–579. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]
- Rios-Velasquez CM, Martins-Campos KM, Simoes RC, Izzo T, dos Santos EV, Pessoa FAC, Lima JBP, Monteiro WM, Secundino NEC, Lacerda MVG, Tadei WP, Pimenta PEP. Experimental infection of key Anopheles species from the Brazilian Amazon. Malar. J. 2013;12:460. doi: 10.1186/1475-2875-12-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallum MAM, Schultz TR, Foster PG, Aronstein K, Wirtz RA, Wilkerson RC. Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Syst. Entomol. 2002;27:361–382. [Google Scholar]
- Sallum MAM, Schultz TR, Wilkerson RC. Phylogeny of Anophelinae (Diptera Culicidae) based on morphological characters. Ann. Entomol. Soc. Am. 2000;93:745–775. [Google Scholar]
- Schultz J, Maisel S, Gerlach D, Muller T, Wolf M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA. 2005;11:361–364. doi: 10.1261/rna.7204505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Brogdon WG, Collins EH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- Seibel PN, Muller T, Dandekar T, Schultz J, Wolf M. 4SALE--a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformat. 2006;7:498. doi: 10.1186/1471-2105-7-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service MW. Mosquito Ecology: Field Sampling Methods. Elsevier Applied Science; 1976. [Google Scholar]
- Singh OP, Chandra D, Nanda N, Raghavendra K, Sunil S, Sharma SK, Dua VK, Subbarao SK. Differentiation of members of the Anopheles fluviatilis species complex by an allele-specific polymerase chain reaction based on 28S ribosomal DNA sequences. Am. J. Trop. Med. Hyg. 2004;70:27–32. [PubMed] [Google Scholar]
- Spillings BL, Brooke BD, Koekemoer LL, Chiphwanya J, Coetzee M, Hunt RH. A new species concealed by Anopheles funestus Giles, a major malaria vector in Africa. Am. J. Trop. Med. Hyg. 2009;81:510–515. [PubMed] [Google Scholar]
- Sudia WD, Chamberlain RW. Battery-operated light trap, an improved model. In: Sudia WD, Chamberlain RW, editors. J. Am. Mosq. Contr. Assoc. Vol. 4. 1988. pp. 536–538. [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Taye A, Hadis M, Adugna N, Tilahun D, Wirtz RA. Biting behavior and Plasmodium infection rates of Anopheles arabiensis from Sille, Ethiopia. Acta Trop. 2006;97:50–54. doi: 10.1016/j.actatropica.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae . Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Wesson DM, Porter CH, Collins FH. Sequence and secondary structure comparisons of ITS rDNA in mosquitoes (Diptera: Culicidae) Mol. Phylogenet. Evol. 1992;1:253–269. doi: 10.1016/1055-7903(92)90001-w. [DOI] [PubMed] [Google Scholar]
- White GB, Magayuka SA, Boreham PEL. Comparative studies on sibling spcies of the Anopheles gambiae Giles complex (Dipt. Culicidae): binomics and vectorial activity of species A and species B at Segera, Tanzania. Bull. Entomol. Res. 1972;62:295–317. [Google Scholar]
- Wiemers M, Keller A, Wolf M. ITS2 secondary structure improves phylogeny estimation in a radiation of blue butterflies of the subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus) BMC Evol. Biol. 2009;9:300. doi: 10.1186/1471-2148-9-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes TJ, Matola YG, Charlwood JD. Anopheles rivulorum, a vector of human malaria in Africa. Med. Vet. Entomol. 1996;10:108–110. doi: 10.1111/j.1365-2915.1996.tb00092.x. [DOI] [PubMed] [Google Scholar]
- Wolf M, Achtziger M, Schultz J, Dandekar T, Muller T. Homology modeling revealed more than 20,000 rRNA internal transcribed spacer 2 (ITS2) secondary structures. RNA. 2005;11:1616–1623. doi: 10.1261/rna.2144205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young I, Coleman AW. The advantages of the ITS2 region of the nuclear rDNA cistron for analysis of phylogenetic relationships of insects: a Drosophila example. Mol. Phylogenet. Evol. 2004;30:236–242. doi: 10.1016/s1055-7903(03)00178-7. [DOI] [PubMed] [Google Scholar]