Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Emerging HSCs require Jak2 and Pi3k signaling for proliferation and survival.
Embryonic HSCs are unaffected by the JAK2V617F mutation.
Abstract
The regulation of hematopoietic stem cell (HSC) emergence during development provides important information about the basic mechanisms of blood stem cell generation, expansion, and migration. We set out to investigate the role that cytokine signaling pathways play in these early processes and show here that the 2 cytokines interleukin 3 and thrombopoietin have the ability to expand hematopoietic stem and progenitor numbers by regulating their survival and proliferation. For this, they differentially use the Janus kinase (Jak2) and phosphatidylinositol 3-kinase (Pi3k) signaling pathways, with Jak2 mainly relaying the proproliferation signaling, whereas Pi3k mediates the survival signal. Furthermore, using Jak2-deficient embryos, we demonstrate that Jak2 is crucially required for the function of the first HSCs, whereas progenitors are less dependent on Jak2. The JAK2V617F mutation, which renders JAK2 constitutively active and has been linked to myeloproliferative neoplasms, was recently shown to compromise adult HSC function, negatively affecting their repopulation and self-renewal ability, partly through the accumulation of JAK2V617F-induced DNA damage. We report here that nascent HSCs are resistant to the JAK2V617F mutation and show no decrease in repopulation or self-renewal and no increase in DNA damage, even in the presence of 2 mutant copies. More importantly, this unique property of embryonic HSCs is stably maintained through ≥1 round of successive transplantations. In summary, our dissection of cytokine signaling in embryonic HSCs has uncovered unique properties of these cells that are of clinical importance.
Introduction
Adult-repopulating hematopoietic stem cells (HSCs) are first detected at embryonic day (E)10.5 in the mouse aorta-gonads-mesonephros (AGM) region, where they are thought to emerge from the ventral endothelium of the dorsal aorta.1-3 Relatively little is known about how this is mediated by the microenvironment and, more specifically, which soluble factors act on nascent HSCs to regulate their emergence, survival, expansion, and migration.2 Understanding these complex processes and applying this knowledge to the recreation of the right conditions in vitro to facilitate the de novo generation and expansion of HSCs would be of immense clinical value.
For this reason, our group previously carried out gene expression screens that resulted in the identification of novel positive and negative regulators of emerging HSCs.4 These included Igf2,4 Dlk1,5 and catecholamines that are secreted from the codeveloping sympathetic nervous system.6 Additional soluble factors that have been shown by other groups to be important for HSC production in the AGM include Bmp4,7 interleukin 1 (Il1),8 Il3,9 Hedgehog,10 retinoic acid,11 and nitric oxide.12,13
Discovering the sources of these factors also allows for identification of the cells that contribute to the developing HSC niche.2 These supportive cells are polarized to the ventral side of the AGM, encompassing the area of the developing gut,10,14 and include mesenchymal cells underneath the aorta.7,15 Cells of the sympathetic nervous system were shown to be part of the niche,6 and there may also be important signals derived from endothelial cells and hematopoietic cells that are in close contact with emerging HSCs. In fact, inflammatory response signals released from primitive innate immune cells were recently demonstrated to play an important role in regulating HSC production.16-19
A functional annotation enrichment analysis of the differentially expressed genes identified in our previous expression screen4 has shown that components of cytokine signaling pathways are enriched among the genes upregulated in the AGM at the peak of HSC production, thus prompting us to investigate whether cytokine signaling plays a role in AGM hematopoiesis. Among the cytokines tested, we found that Il3 and thrombopoietin (Thpo) enhanced hematopoietic progenitor (HP) and HSC production in the AGM and that this was mediated via the Janus kinase (Jak)-signal transducer and activator of transcription (Stat) and phosphatidylinositol 3-kinase (Pi3k) signaling pathways. Furthermore, although these cytokine pathways are known to also regulate adult hematopoiesis, we demonstrate here that there are differences in the response of nascent HSCs compared with adult HSCs to aberrant, disease-associated cytokine signaling. These findings are relevant to the understanding and treatment of myeloproliferative disorders.
Methods
Mice
Wild-type C57BL/6J, Jak2 heterozygous knockout (Jak2+/fl),20 and heterozygous JAK2V617F-expressing (Jak2+/VF)21 mice were crossed for embryo production. The morning of plug detection was considered day 0. All animals were housed according to institutional guidelines, and procedures were performed following UK Home Office regulations.
AGM explants
AGMs were cultured on Durapore filters (Millipore, Watford, UK) with M5300 culture medium (Stem Cell Technologies, Cambridge, UK) containing 10−6 M hydrocortisone (Sigma-Aldrich, Gillingham, UK). The following reagents or their respective diluents (control) were added to the medium at the start of the culture: Il2, Il3, Il6, Thpo (all from PeproTech, London, UK), erythropoietin (Epo; R&D Systems, Abingdon, UK), Jak2 inhibitor (TG101348; Charnwood Molecular, Loughborough, UK), Pi3k inhibitor (LY294002; Promega, Southampton, UK), and mitogen-activated protein kinase (Mapk) pathway (Mek) inhibitor (U0126; Promega). After 3 days, AGMs were dissociated by collagenase treatment (Sigma-Aldrich; 0.125% in phosphate-buffered saline). At least 3 AGMs were used per data point per individual experiment.
Colony-forming assays
Cells were plated in M3434 Methocult (Stem Cell Technologies) in triplicate at 20 000, 50 000, and 100 000 cells per plate. Colonies were scored and counted after 7 days.
Proliferation and apoptosis assays
To assess proliferation, 10 μM 5-bromo-2′-deoxyuridine (BrdU; BD, Oxford, UK) was added during the last 16 hours of culture. Dissociated cells were stained with CD45-phycoerythrin (PE) and ckit-allophycocyanin (APC) (eBioscience, Hatfield, UK), permeabilized, and stained with a BrdU-fluorescein isothiocyanate (FITC) antibody (BD). For the apoptosis analysis, ckit+CD45+ cells were stained with Annexin V-PacificBlue (BioLegend, London, UK) and 7-aminoactinomycin D (7AAD) (Invitrogen, Paisley, UK). Cells were analyzed on a CyAn ADP analyzer (Beckman Coulter, High Wycombe, UK) with FlowJo software (Tree Star, Ashland, OR).
Repopulation assays
Cell preparations were injected intravenously into irradiated (9.5 Gy) recipients that differed in CD45 isoforms. After 4 months, donor contribution to the peripheral blood was analyzed by flow cytometry using CD45.1-PE and CD45.2-FITC (eBioscience). Mice with a donor cell contribution of ≥5% were considered to be positive for repopulation. Multilineage analysis was carried out with B220-APC, CD3-PacificBlue (both BD), Mac1-APC, and Gr1-PacificBlue (both from eBioscience). Peripheral blood counts were performed on an ABC blood counter (Woodley, Bolton, UK).
Immunohistochemistry
Cryosections from paraformaldehyde-fixed embryos were stained with anti–CD34-biotin (BD), Stat5 (Santa Cruz, Heidelberg, Germany), Thpo-biotin (Peprotech), and Ki67 (Novacastra, Milton Keynes, UK), followed by anti–rabbit-Alexa555, anti–rabbit-Alexa488 (all from Life Technologies), and streptavidin-Cy5 (Jackson Immunoresearch, West Grove, PA). Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assays were performed using an ApopTag Red In Situ Apoptosis Detection Kit (Millipore). Sections were mounted with Vectashield (Vector Laboratories, Peterborough, UK), and images were obtained on a Zeiss Axio Imager fitted with a Hammatsu Flash 4 V2 sCMOS camera and analyzed with ZEN software.
DNA damage analysis
Donor Lin−Sca1+ckit+ (LSK) cells were sorted from bone marrow (BM) of recipients using the following antibodies: CD45.2-BDhorizonV500 (donor); CD3-APC, Ter119-APC, F4/80-APC, Nk1.1-APC, Gr1-APC, B220-APC, and CD19-APC (lineage depletion); and Sca1-PB and ckit-APCeF780 (BD, eBioscience). Sorted cells were spotted onto microscope slides, fixed with 4% paraformaldehyde, permeabilized with 0.15% Triton X-100, and blocked overnight with 1% bovine serum albumin. They were stained with an antibody to γ-H2A.X (Millipore), followed by anti-mouse Alexa488 (Life Technologies), and mounted with ProLong Gold antifade with 4,6 diamidino-2-phenylindole (Molecular Probes, Loughborough, UK). Foci were counted using an Axio Imager (Zeiss) attached to a Hammamatsu Flash 4 V2 sCMOS camera with ZEN software.
Gene expression analysis
Samples for semiquantitative reverse transcriptase-polymerase chain reaction (RT-PCR) were placed in Trizol (Invitrogen), RNA was extracted, DNase treated and reverse transcribed with Superscript II (Invitrogen). For quantitative RT-PCR, a miRNeasy micro kit (Qiagen, Manchester, UK) was used, and cDNA was generated with an iScript Advanced cDNA Synthesis kit (BioRad, Hemel Hempstead, UK). RNA from sorted cell populations was extracted with a PicoPure RNA Isolation Kit (Thermo Scientific, Lutterworth, UK). Semiquantitative PCR reactions were run on a Peltier Thermal Cycler, and quantitative real-time PCR (qPCR) reactions as triplicates on a LightCycler 480 (Roche, Burgess Hill, UK). qRT-PCR data are expressed as difference of expression (2ΔCT) relative to β-actin. Primer sequences are provided in supplemental Table 1, available on the Blood Web site.
RT Profiler PCR arrays
Wild-type AGM and BM HSCs were sorted using anti–CD34-FITC (BD), CD45-FITC (eBioscience), CD45-PE (eBioscience), ckit-APC (BioLegend), CD48-APC (Cambridge Bioscience, Cambridge, UK), CD150-PacificBlue (Cambridge Bioscience), and EPCR-PE (eBioscience). Mature blood cells were excluded from the BM sample using the Mouse Hematopoietic Progenitor Cell Enrichment Cocktail (Stem Cell Technologies). Cells were sorted straight into TriReagent (Sigma-Aldrich), and RNA was extracted using the miRNeasy Micro Kit (Qiagen). cDNA was synthesized using the RT2 PreAMP cDNA Synthesis Kit (Qiagen) and amplified with the Jak/Stat Signaling Pathway Pre-Amp PCR Master Mix (Qiagen). The cDNA was quantified using the RT2 SYBR Green Mastermix on Jak/Stat Signaling Pathway RT2 Profiler 96-Well PCR Arrays (Qiagen; PAMM-039ZC-2) and analyzed using the 7900 HT Fast Real-Time PCR System (Applied Biosystems, Paisley, UK). Results were standardized through thresholding of the data using the internal controls, and the data were globally normalized. Using the online RT data analysis tool (SABiosciences, Manchester, UK), genes with P < .05 and fold change >2 were identified.
Data analysis
Data were analyzed with GraphPad Prism, and statistical significance was calculated using the Student t test, except for repopulation data, which were assessed through the Mann-Whitney test.
Results
Il3 and Thpo promote hematopoietic progenitor cell production
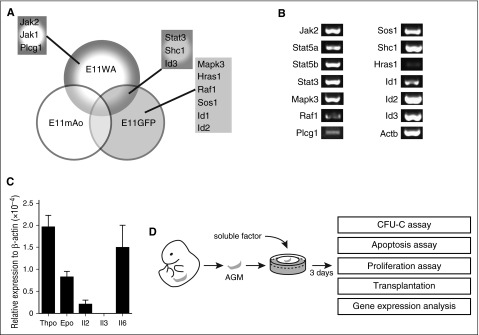
In our previous expression screen, we identified genes that are upregulated in the E11 dorsal aorta (E11WA), the middle region of the E11 dorsal aorta where HSCs are located (E11mAo), and in the HSC-enriched E11 Ly6A-GFP+ fraction (E11GFP).4 A functional enrichment analysis using the DAVID software indicated that components of cytokine signaling pathways were enriched in our data sets. These included the Il2, Il3, Il6, Epo, Thpo, Stat3, and extracellular signal-regulated kinase 1/extracellular signal-regulated kinase 2 Mapk signaling pathways, and the components were Hras1, Id1, Id2, Id3, Jak1, Jak2, Mapk3, Plcg1, Raf1, Shc1, Sos1, and Stat3 (Figure 1A; supplemental Table 2).4 We confirmed their expression by RT-PCR analysis, although Hras1 expression was low (Figure 1B).
Figure 1.
Cytokine signaling pathways are active in the AGM. (A) Components of cytokine signaling pathways found to be upregulated in the E11 dorsal aorta (E11WA), in E11 Ly6A-GFP+ cells (E11GFP), and in the middle part of the dorsal aorta (E11mAo) by expression profiling. (B) Confirmation of signaling component expression by semiquantitative RT-PCR analysis in the AGM region. (C) Analysis of cytokine expression in the E11 AGM by qPCR; n = 3. (D) Schematic diagram of experimental setup.
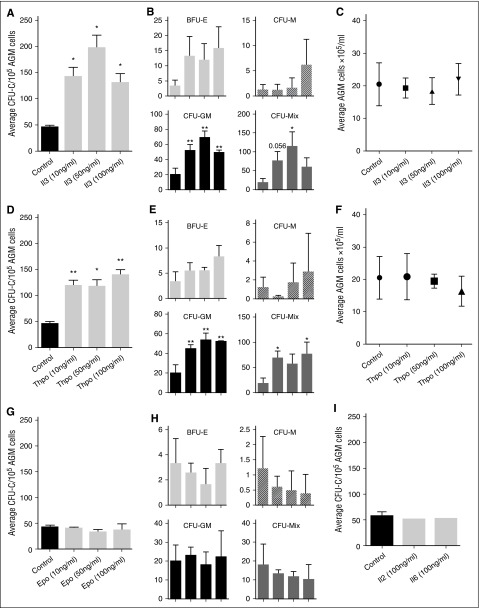
The upregulation of these genes in the E11 AGM region suggested that cytokine signaling may play a role in AGM hematopoiesis. Indeed, we found that Thpo, Epo, Il2, and Il6 were expressed in the AGM, with Thpo and Il6 showing the highest level of expression (Figure 1C). Il3 was undetectable by RT-PCR, as reported previously.9 To test whether they could influence AGM hematopoiesis, we added these cytokines to AGM explant cultures (Figure 1D). Addition of as little as 10 ng/mL Il3 resulted in a 3.1-fold enhanced colony formation (Figure 2A). Addition of 50 ng/mL resulted in a fourfold increase, whereas 100 ng/mL did not give a further increase. Analysis of the specific colony types indicated that Il3 had the strongest effect on the most immature colony types: colony-forming unit (CFU)-Mix and CFU-GM (granulocyte, monocyte) (Figure 2B). The overall number of AGM cells remained unaltered, suggesting that the effect of Il3 was hematopoiesis specific (Figure 2C).
Figure 2.
Il3 and Thpo expand hematopoietic progenitor cells. (A) Recombinant mouse Il3 was added to AGM explant cultures at the indicated concentrations. Three days later, cells were plated in methylcellulose, and total colonies were counted after 7 days. n = 3. (B) Total colonies separated into the different colony types. (C) Average number of total AGM cells obtained after 3 days of explant culture in the presence or absence of Il3. (D) Recombinant mouse Thpo was added to AGM explant cultures at the indicated concentrations. Three days later, cells were plated in methylcellulose, and total colonies were counted after 7 days. n = 3. (E) Total colonies separated into the different colony types. (F) Average number of total AGM cells obtained after 3 days of explant culture in the presence or absence of Thpo. (G) Recombinant mouse Epo was added to AGM explant cultures at the indicated concentrations. Three days later, cells were plated in methylcellulose, and total colonies were counted after 7 days. n = 3. (H) Total colonies separated into the different colony types. (I) Recombinant mouse Il2 or Il6 was added to AGM explant cultures at 100 ng/mL. Three days later, cells were plated in methylcellulose, and total colonies were counted after 7 days. n = 3.
Thpo increased the total number of hematopoietic colonies by 2.6-fold at 10 ng/mL (Figure 2D), only slightly less than Il3. Adding 50 ng/mL did not result in a further increase, and 100 ng/mL only gave an increase of threefold compared with the control. As was observed for Il3, Thpo mainly expanded CFU-GM and CFU-Mix colonies (Figure 2E), and it did not affect the total number of AGM cells (Figure 2F).
We also added Epo, Il2, and Il6 to AGM explant cultures, but saw no effect (Figure 2G-I).
Il3 and Thpo elicit prosurvival and proproliferation responses
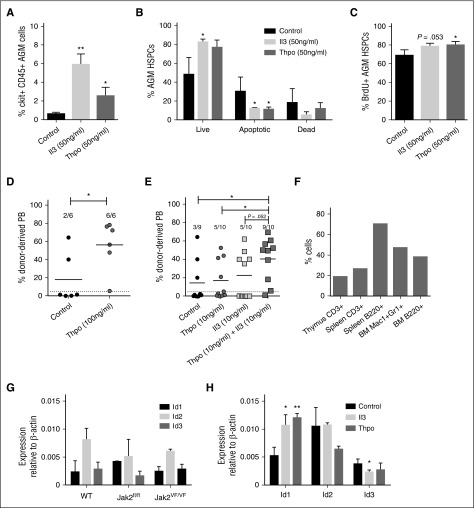
To shed light on the mechanism, we determined the number of apoptotic and proliferating cells within AGM hematopoietic stem and progenitor cells (HSPCs; ckit+CD45+). Il3 at 50 ng/mL produced an almost 10-fold increase in ckit+CD45+ cells, whereas the increase with the same concentration of Thpo amounted to 4.2-fold (Figure 3A). This is likely due to a significant decrease in early apoptotic cells (Figure 3B). In addition, we also detected an increase in BrdU+ proliferating HSPCs, which was significant for Thpo-treated cells, but just failed to reach significance (P = .053) for Il3 addition (Figure 3C).
Figure 3.
Il3 and Thpo have prosurvival and proproliferation effects and can expand HSCs. (A) The number of ckit+CD45+ AGM cells recovered after 3 days of explant culture in the presence or absence of Il3 or Thpo. (B) The percentage of live (7AAD−Annexin V−), early apoptotic (7AAD−Annexin V+), and dead (7AAD+Annexin V+) cells within the ckit+CD45+ population was determined. n = 3. (C) The percentage of proliferating cells that had incorporated BrdU during the last night of explant culture was determined within the ckit+CD45+ population. n = 3. (D) Repopulation levels of individual mice injected with AGM cells (0.1-0.3 embryo equivalents) explant-cultured in the presence or absence of Thpo. Dotted line represents 5% threshold. The number of positive mice out of total injected mice is indicated at the top. (E) Repopulation levels of individual mice injected with AGM cells explant-cultured in the presence or absence of Il3 and/or Thpo. Dotted line represents 5% threshold. The number of positive mice out of total injected mice is indicated at the top. (F) Multilineage analysis of donor cell contribution in one mouse highly repopulated with AGM cells cultured in the presence of Il3 and Thpo. (G) HSPCs (ckit+CD45+CD41intermediate) were sorted from uncultured AGMs of the indicated genotypes and analyzed by qPCR for Id gene expression. n = 3. (H) AGMs were cultured in the presence or absence of 100 ng/mL Il3 or Thpo, and ckit+CD45+CD41intermediate cells were sorted and then analyzed for the expression of Id1, Id2, or Id3 by quantitative real-time PCR analysis. n = 3. **P < .01, *P < .05.
Il3 and Thpo promote HSC production in an additive manner
To establish whether these cytokines were also able to expand HSCs, long-term repopulation assays were performed. Il3 was previously reported to expand AGM HSCs,9 whereas the effect of Thpo was unknown. Addition of 100 ng/mL Thpo to AGM explants resulted in 100% of the injected mice being repopulated, in contrast to one-third of recipients of control AGMs, with a significantly higher donor contribution in the peripheral blood (Figure 3D). To determine whether Il3 and Thpo can work additively, both cytokines were added at 10 ng/mL alone or in combination. Either cytokine on its own produced a slight increase from one-third to one-half of recipients being repopulated, whereas adding both cytokines resulted in 90% of repopulation (Figure 3E). Donor contribution was also higher in recipients of AGM cells pretreated with both cytokines compared with recipients of control or Thpo-only cells. The enhanced repopulation achieved with both cytokines was multilineage (Figure 3F). Although we cannot rule out that at least some of the effects of the cytokines may be mediated via responsive cells in the microenvironment, it is likely that Il3 and Thpo act directly on HSPCs because Il3 receptor expression was detected on phenotypic AGM HSPCs (CD34+ckit+),9 and transcripts for the Thpo receptor Mpl were found in intra-aortic clusters.22
Within our data set were 3 downstream target genes: Id1, Id2, and Id3 (Figure 1A). These can be activated through a number of stimuli including bone morphogenic proteins via Smads, C/EBPβ, T-cell receptor signaling via the Ras/MAPK pathway, and cytokines.23 Id1 and Id3 are subject to transient expression during development following stimulation, whereas Id2 mRNA is found more ubiquitously. Id1 is essential for adult HSC maintenance24,25 and is a downstream target of Il3 and Jak2-Stat5 signaling.23,26,27 Id2 and Id3 perform functions at later stages in hematopoiesis.28,29 All 3 are expressed in AGM HSPCs (Figure 3G). To determine whether AGM HSC expansion could be linked to one of these target genes, AGMs were cultured with no cytokine, Il3, or Thpo, and HSC-enriched populations (ckit+CD45+CD41intermediate)30,31 were analyzed for Id gene expression. Figure 3H shows that only Id1 is significantly upregulated by Il3 and Thpo, which suggests that Id1 may also be crucial for AGM HSC expansion and survival.
Jak2 and Pi3k signaling is required for hematopoietic progenitor production
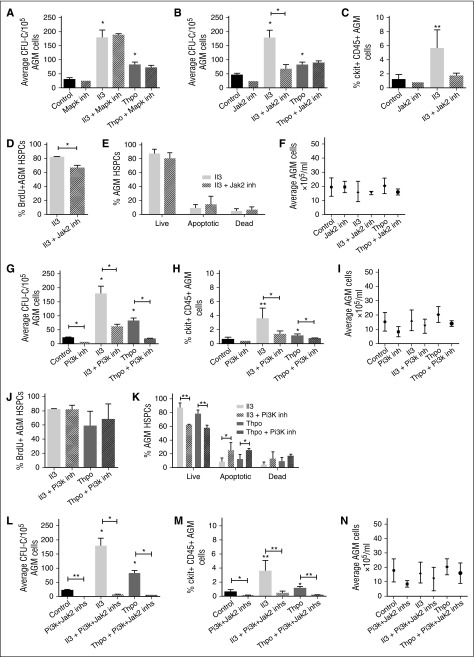
We next investigated the downstream signaling pathways of Il3 and Thpo. Components of the Jak-Stat and Mapk pathways were enriched in our data set (Figure 1A). In addition, Il3 and Thpo are known to signal through the Pi3k pathway. We tested whether inhibitors specific to these pathways can block the effects of Il3 and Thpo. Inhibitor concentrations were chosen based on published IC50 values and test experiments. Addition of a Mapk pathway inhibitor (U0126) had no effect on CFU-C numbers (Figure 4A), suggesting that the Mapk pathway is not essential for HSPC maintenance or expansion.
Figure 4.
Il3 and Thpo signal through the Jak2 and/or Pi3k pathways. AGMs were cultured as explants in the absence or presence of Il3 or Thpo (50 ng/mL) and/or (A) Mapk inhibitor (U0126; 5 μM), (B) Jak2 inhibitor (TG101348; 3 μM), (G) Pi3k inhibitor (LY294002; 14 μM), or (L) Jak2 inhibitor + Pi3k inhibitor for 3 days and then plated in methylcellulose. Colonies were scored 7 days later. n = 3. Alternatively, (C,H,M) the percentage of ckit+CD45+ at the end of the culture was determined, and the percentage of (D,J) BrdU+ cells or (E,K) live, apoptotic, and dead cells within this population. (F,I,N) The total number of cells recovered at the end of the each explant culture was also counted. n = 3; **P < .01, *P < .05.
We next added a Jak2 inhibitor (TG101348) and noted that this decreased CFU-C output in the absence of cytokines, albeit without quite reaching significance (Figure 4B). Interestingly, it blocked Il3-mediated CFU-C expansion, but did not interfere with Thpo signaling. Furthermore, it only reduced the number of BrdU+ HSPCs in the presence of Il3 (Figure 4C-D), whereas the number of live cells was unaffected (Figure 4E), suggesting that Jak2 mediates the proproliferation effect of Il3, but not the prosurvival signal. The overall number of AGM cells did not change, ruling out general toxicity (Figure 4F).
Finally, we tested a Pi3k inhibitor (LY294002) and observed that it reduced HP production even in the absence of cytokines, pointing to an essential role in AGM hematopoiesis (Figure 4G). Furthermore, the inhibitor also abrogated HP expansion induced by Il3 and Thpo (Figure 4G-H). Total AGM cell numbers were not significantly altered (Figure 4I). The Pi3k inhibitor did not interfere with the proproliferation effect of Il3 and Thpo (Figure 4J); however, it significantly decreased the number of live cells and increased the number of preapoptotic cells, thus blocking their prosurvival signal (Figure 4K). Taken together, these observations suggest that Jak2 and Pi3k perform independent functions downstream of Il3 and Thpo. We therefore added both inhibitors together to the explant cultures. This dramatically reduced hematopoietic colony output in the presence and absence of cytokines (Figure 4L) and also decreased the number of HSPCs (Figure 4M) to an extent that it precluded further analysis. The strong effect of the 2 inhibitors together was still hematopoiesis specific, as total AGM cell numbers were not significantly altered (Figure 4N). The fact that inhibition of both pathways had such a profound effect even in the absence of exogenous cytokines suggests that there may be an endogenous source of Il3 and Thpo. Indeed, expression of Il3 in the AGM in the proximity of HSPCs has previously been reported,9 and we detected patches of Thpo protein in the ventral subaortic mesenchyme (supplemental Figure 1A).
Jak2 is essential for emerging HSCs
Because we saw a slight decrease in colony formation with the Jak2 inhibitor in the absence of cytokines (Figure 4B) and detected Stat5, the main target of Jak2 in hematopoietic cells, in endothelial cells of the aorta and in intra-aortic hematopoietic clusters (Figure 5A), we decided to further investigate whether Jak2 is required for AGM hematopoiesis.
Figure 5.
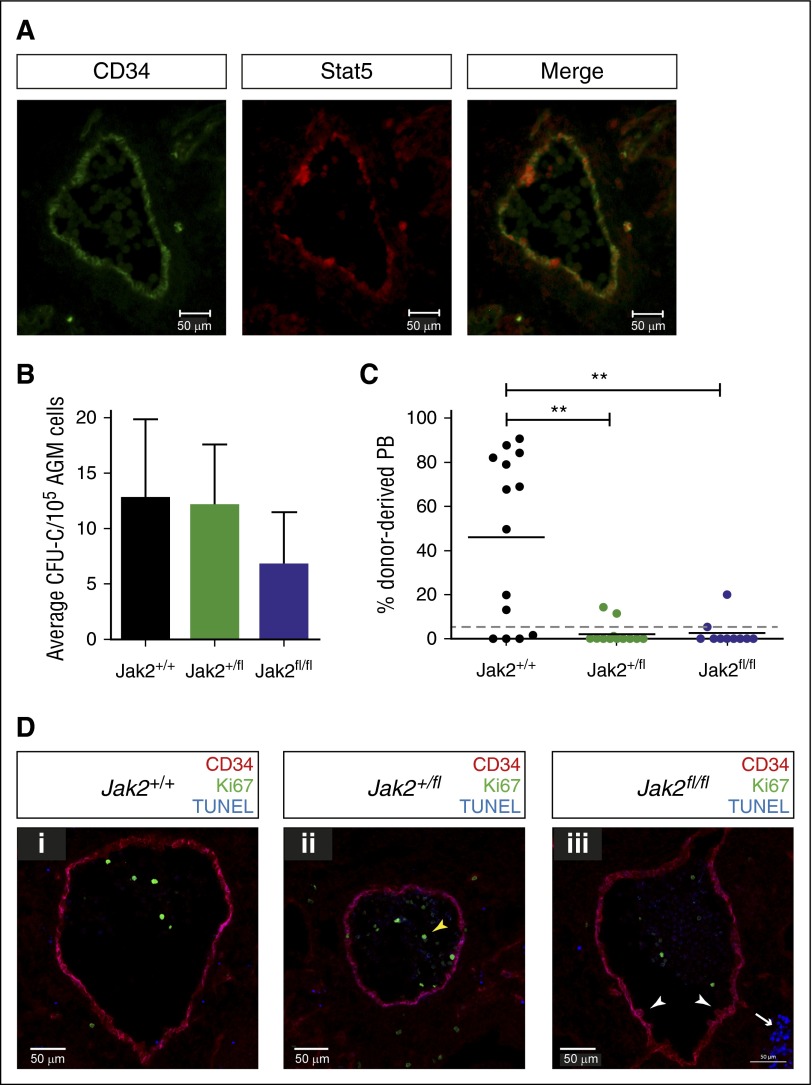
Jak2 signaling is required for HSC production in the AGM. (A) Cryosections (10 μM) were prepared from E11.5 embryos and stained with antibodies against CD34 and total Stat5 as indicated (ventral down). Pictures were taken with a Zeiss AxioSkop2 wide-field microscope (objective 20×/045 NA) fitted with a Zeiss AxioCam MRc5, and images were analyzed with the Zeiss AxioVision software. (B) E11.5 AGM cells from embryos with the indicated genotypes were directly plated in methylcellulose, and colonies were counted 7 days later. n = 3 for Jak2+/+; n = 4 for Jak2+/fl; n = 3 for Jak2fl/fl. (C) E11.5 AGM cells from embryos with the indicated genotypes were directly transplanted as 1 embryo equivalent, and donor cell contribution to the peripheral blood of the recipients was determined at 4 months. Dotted line represents 5% threshold. Fourteen recipients for Jak2+/+; 13 recipients for Jak2+/fl; 10 recipients for Jak2fl/fl. **P < .01. (D) Cryosections (10 μM) were prepared from (i) E11.5 Jak2+/+, (ii) Jak2+/fl, and (iii) Jak2fl/fl embryos and stained with antibodies against CD34 and Ki67, together with TUNEL staining as indicated (ventral down). n = 2 for each genotype; 16 to 21 sections were analyzed per genotype. Pictures were taken with a Zeiss Axio Imager Microscope (objective 40×) fitted with a Hammatsu Flash 4 V2 sCMOS camera, and images were analyzed with the Zen software.
We used Jak2-deficient mice, in which a mutant version of human JAK2, JAK2V617F, is knocked into the mouse Jak2 locus, creating a null allele.20 Human JAK2V617F is not expressed as this requires the presence of Cre recombinase. Thus, the genotypes Jak2+/fl and Jak2fl/fl represent heterozygous and homozygous knockout mice, respectively. We first tested the ability of uncultured Jak2-deficient AGMs to form hematopoietic colonies and observed that colony formation was only slightly reduced (Figure 5B). We next transplanted uncultured Jak2-deficient AGMs. Strikingly, removal of one or both copies of Jak2 dramatically decreased the number of repopulated mice and overall repopulation levels, demonstrating that Jak2 is essential for HSC production and/or maintenance in the AGM (Figure 5C). To investigate this further, we stained sections from E11.5 embryos with antibodies against CD34 and Ki67 to reveal hematopoietic clusters and proliferating cells, respectively. We also performed TUNEL assays to detect apoptotic cells (Figure 5D). The results indicate that HSPC formation proceeds normally in the absence of Jak2, as hematopoietic clusters were still detected in Jak2fl/fl embryos (white arrowheads in Figure 5Diii) and did not differ in numbers between genotypes. There was also no change in Id gene expression in enriched HSCs isolated from Jak2fl/fl AGMs compared with wild-type cells (Figure 3G). However, because there are very few HSCs present in an AGM at any one time and many of the clusters contain progenitor cells, changes in HSC numbers may be too subtle to detect. Very few proliferating cells were found in and around the aorta, as reported previously.5 These were mostly located within the circulation (yellow arrowhead in Figure 5Dii), with numbers and localization being comparable between the 3 genotypes. We found virtually no apoptotic cells in and around the aorta apart from clusters of cells in the subaortic mesenchyme (white arrow in Figure 5Diii), which is in agreement with previous results5 and did not differ between genotypes. The combined in vitro and in vivo data, therefore, demonstrate a role for Jak2 in HSC function following their emergence in the AGM.
AGM HSCs are resistant to the effects of the JAK2V617F mutation
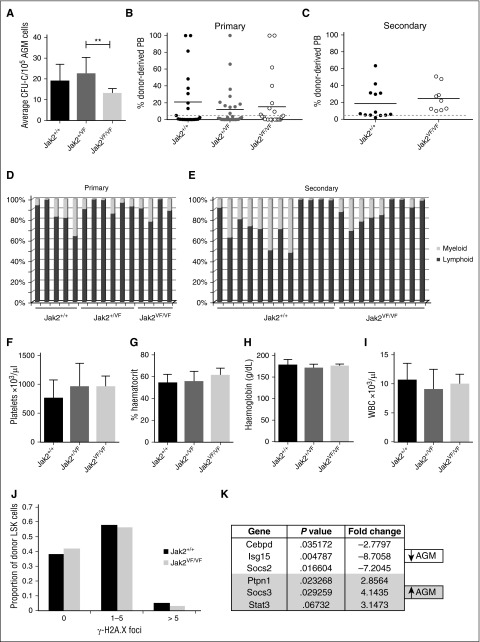
The JAK2V617F mutation is detected in the majority of patients with myeloproliferative neoplasms32 and creates a constitutively active JAK2 kinase. The effect of the JAK2V617F mutation on HSCs has been studied20,21,33-37 and was shown to compromise HSC function in one model,20,34 a defect that is even more severe when homozygous.21 Using the model, in which JAK2V617F is expressed from the mouse endogenous Jak2 locus following recombination with Stella-Cre,21 we set out to determine how constitutively active JAK2 would affect embryonic HSPCs. Expression of 1 copy of JAK2V617F did not affect AGM HP numbers, whereas the homozygous JAK2V617F AGMs showed a slight reduction in colony output (Figure 6A). Contrary to the results obtained with adult HSCs,20,21,34 similar percentages of mice were repopulated with wild-type (Jak2+/+; 43%), heterozygous (Jak2+/VF; 36%), and homozygous (Jak2VF/VF; 38%) AGMs, and the overall repopulation levels in the peripheral blood were not significantly different (Figure 6B). Similarly, expression of Id gene transcript was comparable between wild-type and Jak2VF/VF AGM HSPCs (Figure 3G). Due to an impairment of self-renewal in adult JAK2V617F-expressing HSCs, secondary transplantations gave rise to markedly reduced repopulation.20,34 It was thus striking to see that BM cells from primary recipients of homozygous AGM cells performed just as well in secondary repopulation assays as BM cells from primary recipients of wild-type AGM cells (Figure 6C). These results not only suggest that AGM HSCs are completely resistant to the JAK2V617F mutation but that they also retain this resistance after 4 months in an adult microenvironment.
Figure 6.
AGM HSCs are unaffected by the JAK2V617F mutation. (A) E11.5 AGM cells from embryos with the indicated genotypes were directly plated in methylcellulose, and colonies were counted 7 days later. n = 5 for Jak2+/+; n = 9 for Jak2+/VF; n = 4 for Jak2VF/VF. **P < .01. (B) E11.5 AGM cells from embryos with the indicated genotypes were directly transplanted as 1 embryo equivalent, and donor cell contribution to the peripheral blood of the recipients was determined at 4 months. Dotted line represents 5% threshold. Twenty-one recipients for Jak2+/+; 28 recipients for Jak2+/VF; 24 recipients for Jak2VF/VF. (C) Secondary transplants were performed with total BM cells from 3 primary recipients of Jak2+/+ AGM cells and 2 primary recipients of Jak2VF/VF AGM cells. Two to 3 million total BM cells were injected per secondary recipient with the amount adjusted to the repopulation levels in the primary recipient. Donor cell contribution to the peripheral blood was determined at 4 months after transplantation. Dotted line represents 5% threshold. Thirteen recipients for Jak2+/+; 9 recipients for Jak2VF/VF. Donor contribution within individual (D) primary and (E) secondary recipients was analyzed with respect to myeloid and lymphoid proportion. (F-I) Blood counts were performed on primary recipients at 4 months after transplantation. Error bars indicate standard deviation. (J) Donor LSK cells were sorted from secondary recipients of Jak2+/+ and Jak2VF/VF AGMs and stained for γ-H2A.X foci. The number of foci were counted in 200 to 300 cells per genotype. n = 2. (K) cDNA was prepared from sorted wild-type AGM HSCs (ckit+CD34+CD45+) and wild-type BM HSCs (CD45+CD48−CD150+EPCR+) and analyzed for the expression of Jak-Stat pathway components using the Qiagen Jak/Stat Signaling Pathway RT2 Profiler PCR Array. Differentially expressed genes with P < .05 and a fold change >2 are shown. n = 3.
It has been reported that JAK2V617F-expressing adult HSCs are prone to enhanced lineage bias34 and may in some cases show poor myeloid engraftment.21 Multilineage analysis of primary or secondary recipients of wild-type or JAK2V617F-expressing AGM HSCs revealed no obvious differences (Figure 6D-E).
Transplantability of JAK2V617F-associated disease has been demonstrated20,21,35; however, no changes in platelets, hematocrit, hemoglobin, or white blood cell counts were observed in recipients of heterozygous or homozygous AGMs, even when repopulated at high levels, demonstrating that embryonic HSCs do not initiate a myeloproliferative disease (Figure 6F-I).
The presence of the JAK2V617F mutation leads to increased DNA damage in HSCs, as measured by an approximately fourfold increase in γ-H2A.X foci,34,35 which may partly explain their compromised function. We investigated whether AGM HSCs are more resistant to JAK2V617F-induced DNA damage to explain their resilience to the presence of this mutation. Indeed, even after having undergone serial transplantation, AGM-derived HSCs homozygous for JAK2V617F did not accumulate any more γ-H2A.X–positive foci than their wild-type counterparts (Figure 6J).
We reasoned that the difference in sensitivity to the JAK2V617F mutation may be due to AGM HSCs expressing a different set of Jak-Stat signaling components. We therefore analyzed Jak-Stat pathway-related gene expression in sorted wild-type AGM and BM HSCs using the Qiagen Jak-Stat signaling pathway array. Figure 6K summarizes the genes that were found to be differentially expressed. Socs3 stands out as an obvious candidate as it was shown to inhibit JAK2V617F,38 and its promoter displays hypermethylation in some patients with myeloproliferative neoplasms.39,40 It will be our main target for future mechanistic studies.
Discussion
Cytokine signaling regulates cell proliferation, survival, and differentiation at all stages of the adult hematopoietic tree. Our results extend this to the early stages of definitive hematopoiesis, where we found Il3 and Thpo to be able to expand AGM HSCs and HPs by increasing HSPC proliferation and survival. A proproliferation and prosurvival effect of Il3 on ckit+ AGM cells was demonstrated previously,9 thus supporting our findings. Limiting dilution transplantations also showed that Il3 can expand HSC numbers.9 Considering that Thpo is required for adult HSC maintenance and expansion after transplantation41 and that we found it to increase HSPC proliferation and survival without bias toward a particular lineage, it is likely that Thpo can also increase AGM HSCs, although limiting dilution transplantation assays should be performed. The effect of Thpo on its own had not been investigated in AGM cells before even though it is routinely included in reaggregation studies that serve to reveal pre-HSC potential in the AGM31; however, Mpl−/− AGMs, lacking the Thpo receptor, had shown a slightly reduced HSPC number, delayed production of HSCs,22 and contained a higher percentage of apoptotic cells.42 This mild effect of Mpl deletion on AGM hematopoiesis is somewhat surprising in light of our results following Thpo addition, but may be explained by a partial compensation by intact Il3 signaling in the Mpl−/− mice.
Our inhibitor studies demonstrate that the Pi3k pathway is crucially important for AGM hematopoiesis and for relaying the Il3 and Thpo signal, whereas Jak2 only appears to be required for Il3 signaling. AGM hematopoiesis is almost completely abrogated when Jak2 and Pi3k are blocked simultaneously.
We also analyzed AGM hematopoiesis in Jak2 knockout mice. As had been observed in the inhibitor studies, HPs were only mildly affected by the absence of Jak2. At this stage of development, most of the progenitors found in uncultured AGMs are still yolk sac-derived and will not have been the product of HSC differentiation. This suggests that Jak2 is largely dispensable for yolk sac hematopoiesis. Recently, an essential role for Jak2 in adult HSC maintenance and self-renewal has been demonstrated.43,44 We now show that Jak2 is also crucially required for the function of the first HSCs that emerge in the AGM. Their generation and survival appear normal; however, they are compromised in their repopulation and self-renewal ability.
We also investigated what impact an overactive Jak2 kinase would have on AGM HSPCs. The V617F mutation in the human JAK2 protein results in a constitutively active kinase that has been associated with the development of myeloproliferative neoplasm. Several genetic mouse models have been generated,20,37,45-49 some of which have addressed the impact of the mutation on adult HSCs. The Li 2010 model presented with fewer phenotypic HSCs that were compromised in their reconstitution ability and self-renewal and had accumulated DNA damage,20,34 whereas the Mullaly 2010 model observed no impact on HSC competitiveness despite an altered cycling status.36,37 In the Marty 2010 model, an increase in HSC competitiveness was reported,33 whereas the Tiedt 2008 model showed a complex HSC phenotype with an increase of phenotypic long-term HSCs that were less competitive, expressed a short-term HSC gene signature, and presented with an increase in DNA damage.35 Using the Li et al and Kent et al models,20,21,34 in which a HSC defect had been observed, it was striking to see that nascent HSCs are completely unaffected by the JAK2V617F mutation, even when present in 2 copies. We previously reported other examples of how embryonic HSCs are differentially affected by certain mutations.2
Furthermore, the resilience to the effects of JAK2V617F is maintained in secondary transplantations, at which point adult JAK2V617F-expressing HSCs have almost become exhausted.20,34 This suggests that embryonic HSCs maintain at least part of their unique properties during the 4 months they reside in the adult BM niche prior to the secondary transplantation. AGM HSCs were also resistant to JAK2V617F-induced DNA damage. It will be interesting to see if this is a general property of embryonic HSCs or specific to JAK2V617F-induced DNA damage.
In an attempt to decipher how AGM HSCs escape the damaging effects of constitutively active Jak2, we determined whether any Jak-Stat signaling components are differentially expressed in AGM HSCs vs adult BM HSCs. Of the 6 differentially expressed genes, Socs3 stands out as a potential candidate because, together with Socs1, it is considered to be the most potent inhibitor of cytokine signaling. Unlike other Socs family members, Socs1 and Socs3 can directly inhibit the catalytic activity of Jak via a KIR motif that is unique to these 2 proteins.50 Importantly, it was demonstrated by in vitro assays that wild-type and mutant JAK2 are equally inhibited by SOCS338; however, additional processes such as promoter methylation, hyperphosphorylation, and the presence of certain cytokine receptors may interfere with SOCS3-mediated inhibition of JAK2V617F in vivo.39,40,51
We also found Stat3 to be expressed at higher levels in AGM HSCs. Interestingly, a Stat3-deficient background was reported to enhance Jak2V617F-associated disease in a mouse model.52 An upregulation of Stat3 may therefore make AGM HSCs more resilient to the effects of JAK2V617F. Furthermore, Stat3 was also shown to be required for the induction of Socs3,52 suggesting that higher levels of Stat3 result in upregulation of Socs3. It will be important to investigate these differences between embryonic and adult HSCs as these are not only relevant to human diseases, but may also allow for the exploitation of the unique properties of embryonic HSCs, such as proliferation without exhaustion and resistance to DNA damage, for adult HSC manipulation.
Acknowledgments
The authors thank the Cambridge Institute for Medical Research flow cytometry team, in particular Dr Reiner Schulte and Michal Maj, for sorting services, the microscopy team, Matthew Gratian and Mark Bowen, for technical support and advice, and the staff of the animal facility for assistance with animal maintenance and experimentation. The authors also thank Tina Hamilton and Dean Pask for help with animal experiments.
Core facilities are supported by strategic award WT100140 and equipment grant 093026. This work was funded by a Medical Research Council studentship (M.I.M.), a Kay Kendall Leukaemia Fund intermediate fellowship (KKL276) (K.O.), a Bloodwise Bennett senior fellowship (10015) (K.O.), and a British Society for Haematology Early Stage Investigator fellowship (K.O.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.I.M. carried out the majority of the experiments under the supervision of K.O.; W.A.B. conducted the RT Profiler PCR arrays and performed the immunohistochemistry on Jak2-mutant embryos with the assistance of S.W.P.C.; C.K. and G.K. provided assistance with the mouse experiments, genotyping, and analysis of transplantation results; C.K. performed immunohistochemistry, cell sorting, and qPCR; S.R.F. performed qPCR reactions; J.L. assisted with the experiments involving Jak2-deficient and JAK2V617F-expressing mouse lines and with the DNA damage analysis; A.R.G. provided crucial reagents and advice and assisted with the editing of the manuscript; and K.O. designed and supervised the study, assisted with the experiments, carried out the DNA damage analysis, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Katrin Ottersbach, MRC Centre for Regenerative Medicine, Scottish Centre for Regenerative Medicine, Edinburgh bioQuarter, 5 Little France Dr, University of Edinburgh, Edinburgh EH16 4UU, United Kingdom; e-mail: katrin.ottersbach@ed.ac.uk.
References
- 1.Ciau-Uitz A, Monteiro R, Kirmizitas A, Patient R. Developmental hematopoiesis: ontogeny, genetic programming and conservation. Exp Hematol. 2014;42(8):669–683. doi: 10.1016/j.exphem.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Mirshekar-Syahkal B, Fitch SR, Ottersbach K. Concise review: From greenhouse to garden: the changing soil of the hematopoietic stem cell microenvironment during development. Stem Cells. 2014;32(7):1691–1700. doi: 10.1002/stem.1680. [DOI] [PubMed] [Google Scholar]
- 3.Swiers G, Rode C, Azzoni E, de Bruijn MF. A short history of hemogenic endothelium. Blood Cells Mol Dis. 2013;51(4):206–212. doi: 10.1016/j.bcmd.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mascarenhas MI, Parker A, Dzierzak E, Ottersbach K. Identification of novel regulators of hematopoietic stem cell development through refinement of stem cell localization and expression profiling. Blood. 2009;114(21):4645–4653. doi: 10.1182/blood-2009-06-230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirshekar-Syahkal B, Haak E, Kimber GM, et al. Dlk1 is a negative regulator of emerging hematopoietic stem and progenitor cells. Haematologica. 2013;98(2):163–171. doi: 10.3324/haematol.2012.070789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitch SR, Kimber GM, Wilson NK, et al. Signaling from the sympathetic nervous system regulates hematopoietic stem cell emergence during embryogenesis. Cell Stem Cell. 2012;11(4):554–566. doi: 10.1016/j.stem.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durand C, Robin C, Bollerot K, Baron MH, Ottersbach K, Dzierzak E. Embryonic stromal clones reveal developmental regulators of definitive hematopoietic stem cells. Proc Natl Acad Sci USA. 2007;104(52):20838–20843. doi: 10.1073/pnas.0706923105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orelio C, Haak E, Peeters M, Dzierzak E. Interleukin-1-mediated hematopoietic cell regulation in the aorta-gonad-mesonephros region of the mouse embryo. Blood. 2008;112(13):4895–4904. doi: 10.1182/blood-2007-12-123836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robin C, Ottersbach K, Durand C, et al. An unexpected role for IL-3 in the embryonic development of hematopoietic stem cells. Dev Cell. 2006;11(2):171–180. doi: 10.1016/j.devcel.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Peeters M, Ottersbach K, Bollerot K, et al. Ventral embryonic tissues and Hedgehog proteins induce early AGM hematopoietic stem cell development. Development. 2009;136(15):2613–2621. doi: 10.1242/dev.034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanda B, Ditadi A, Iscove NN, Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 2013;155(1):215–227. doi: 10.1016/j.cell.2013.08.055. [DOI] [PubMed] [Google Scholar]
- 12.Adamo L, Naveiras O, Wenzel PL, et al. Biomechanical forces promote embryonic haematopoiesis. Nature. 2009;459(7250):1131–1135. doi: 10.1038/nature08073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.North TE, Goessling W, Peeters M, et al. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137(4):736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taoudi S, Medvinsky A. Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc Natl Acad Sci USA. 2007;104(22):9399–9403. doi: 10.1073/pnas.0700984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richard C, Drevon C, Canto PY, et al. Endothelio-mesenchymal interaction controls runx1 expression and modulates the notch pathway to initiate aortic hematopoiesis. Dev Cell. 2013;24(6):600–611. doi: 10.1016/j.devcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espín-Palazón R, Stachura DL, Campbell CA, et al. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell. 2014;159(5):1070–1085. doi: 10.1016/j.cell.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Q, Zhang C, Wang L, et al. Inflammatory signaling regulates hematopoietic stem and progenitor cell emergence in vertebrates. Blood. 2015;125(7):1098–1106. doi: 10.1182/blood-2014-09-601542. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Esain V, Teng L, et al. Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 2014;28(23):2597–2612. doi: 10.1101/gad.253302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawamiphak S, Kontarakis Z, Stainier DY. Interferon gamma signaling positively regulates hematopoietic stem cell emergence. Dev Cell. 2014;31(5):640–653. doi: 10.1016/j.devcel.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Spensberger D, Ahn JS, et al. JAK2 V617F impairs hematopoietic stem cell function in a conditional knock-in mouse model of JAK2 V617F-positive essential thrombocythemia. Blood. 2010;116(9):1528–1538. doi: 10.1182/blood-2009-12-259747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Kent DG, Godfrey AL, et al. JAK2V617F homozygosity drives a phenotypic switch in myeloproliferative neoplasms, but is insufficient to sustain disease. Blood. 2014;123(20):3139–3151. doi: 10.1182/blood-2013-06-510222. [DOI] [PubMed] [Google Scholar]
- 22.Petit-Cocault L, Volle-Challier C, Fleury M, Péault B, Souyri M. Dual role of Mpl receptor during the establishment of definitive hematopoiesis. Development. 2007;134(16):3031–3040. doi: 10.1242/dev.001818. [DOI] [PubMed] [Google Scholar]
- 23.Ling F, Kang B, Sun XH. Id proteins: small molecules, mighty regulators. Curr Top Dev Biol. 2014;110:189–216. doi: 10.1016/B978-0-12-405943-6.00005-1. [DOI] [PubMed] [Google Scholar]
- 24.Jankovic V, Ciarrocchi A, Boccuni P, DeBlasio T, Benezra R, Nimer SD. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc Natl Acad Sci USA. 2007;104(4):1260–1265. doi: 10.1073/pnas.0607894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry SS, Zhao Y, Nie L, Cochrane SW, Huang Z, Sun XH. Id1, but not Id3, directs long-term repopulating hematopoietic stem-cell maintenance. Blood. 2007;110(7):2351–2360. doi: 10.1182/blood-2007-01-069914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leeanansaksiri W, Wang H, Gooya JM, et al. IL-3 induces inhibitor of DNA-binding protein-1 in hemopoietic progenitor cells and promotes myeloid cell development. J Immunol. 2005;174(11):7014–7021. doi: 10.4049/jimmunol.174.11.7014. [DOI] [PubMed] [Google Scholar]
- 27.Wood AD, Chen E, Donaldson IJ, et al. ID1 promotes expansion and survival of primary erythroid cells and is a target of JAK2V617F-STAT5 signaling. Blood. 2009;114(9):1820–1830. doi: 10.1182/blood-2009-02-206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji M, Li H, Suh HC, Klarmann KD, Yokota Y, Keller JR. Id2 intrinsically regulates lymphoid and erythroid development via interaction with different target proteins. Blood. 2008;112(4):1068–1077. doi: 10.1182/blood-2008-01-133504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kee BL, Rivera RR, Murre C. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-beta. Nat Immunol. 2001;2(3):242–247. doi: 10.1038/85303. [DOI] [PubMed] [Google Scholar]
- 30.Robin C, Ottersbach K, Boisset JC, Oziemlak A, Dzierzak E. CD41 is developmentally regulated and differentially expressed on mouse hematopoietic stem cells. Blood. 2011;117(19):5088–5091. doi: 10.1182/blood-2011-01-329516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taoudi S, Gonneau C, Moore K, et al. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell. 2008;3(1):99–108. doi: 10.1016/j.stem.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Levine RL, Gilliland DG. Myeloproliferative disorders. Blood. 2008;112(6):2190–2198. doi: 10.1182/blood-2008-03-077966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasan S, Lacout C, Marty C, et al. JAK2V617F expression in mice amplifies early hematopoietic cells and gives them a competitive advantage that is hampered by IFNα. Blood. 2013;122(8):1464–1477. doi: 10.1182/blood-2013-04-498956. [DOI] [PubMed] [Google Scholar]
- 34.Kent DG, Li J, Tanna H, et al. Self-renewal of single mouse hematopoietic stem cells is reduced by JAK2V617F without compromising progenitor cell expansion. PLoS Biol. 2013;11(6):e1001576. doi: 10.1371/journal.pbio.1001576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundberg P, Takizawa H, Kubovcakova L, et al. Myeloproliferative neoplasms can be initiated from a single hematopoietic stem cell expressing JAK2-V617F. J Exp Med. 2014;211(11):2213–2230. doi: 10.1084/jem.20131371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullally A, Bruedigam C, Poveromo L, et al. Depletion of Jak2V617F myeloproliferative neoplasm-propagating stem cells by interferon-α in a murine model of polycythemia vera. Blood. 2013;121(18):3692–3702. doi: 10.1182/blood-2012-05-432989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mullally A, Lane SW, Ball B, et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell. 2010;17(6):584–596. doi: 10.1016/j.ccr.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varghese LN, Ungureanu D, Liau NP, et al. Mechanistic insights into activation and SOCS3-mediated inhibition of myeloproliferative neoplasm-associated JAK2 mutants from biochemical and structural analyses. Biochem J. 2014;458(2):395–405. doi: 10.1042/BJ20131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fourouclas N, Li J, Gilby DC, et al. Methylation of the suppressor of cytokine signaling 3 gene (SOCS3) in myeloproliferative disorders. Haematologica. 2008;93(11):1635–1644. doi: 10.3324/haematol.13043. [DOI] [PubMed] [Google Scholar]
- 40.Torun D, Nevruz O, Akyol M, et al. Methylation of SOCS3 in Myeloproliferative Neoplasms and Secondary Erythrocytosis/Thrombocythemia. Turk J Haematol. 2013;30(1):13–18. doi: 10.4274/tjh.98474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian H, Buza-Vidas N, Hyland CD, et al. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1(6):671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 42.Fleury M, Petit-Cocault L, Clay D, Souyri M. Mpl receptor defect leads to earlier appearance of hematopoietic cells/hematopoietic stem cells in the Aorta-Gonad-Mesonephros region, with increased apoptosis. Int J Dev Biol. 2010;54(6-7):1067–1074. doi: 10.1387/ijdb.103104mf. [DOI] [PubMed] [Google Scholar]
- 43.Akada H, Akada S, Hutchison RE, Sakamoto K, Wagner KU, Mohi G. Critical role of Jak2 in the maintenance and function of adult hematopoietic stem cells. Stem Cells. 2014;32(7):1878–1889. doi: 10.1002/stem.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grisouard J, Hao-Shen H, Dirnhofer S, Wagner KU, Skoda RC. Selective deletion of Jak2 in adult mouse hematopoietic cells leads to lethal anemia and thrombocytopenia. Haematologica. 2014;99(4):e52–e54. doi: 10.3324/haematol.2013.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akada H, Yan D, Zou H, Fiering S, Hutchison RE, Mohi MG. Conditional expression of heterozygous or homozygous Jak2V617F from its endogenous promoter induces a polycythemia vera-like disease. Blood. 2010;115(17):3589–3597. doi: 10.1182/blood-2009-04-215848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marty C, Lacout C, Martin A, et al. Myeloproliferative neoplasm induced by constitutive expression of JAK2V617F in knock-in mice. Blood. 2010;116(5):783–787. doi: 10.1182/blood-2009-12-257063. [DOI] [PubMed] [Google Scholar]
- 47.Shide K, Shimoda HK, Kumano T, et al. Development of ET, primary myelofibrosis and PV in mice expressing JAK2 V617F. Leukemia. 2008;22(1):87–95. doi: 10.1038/sj.leu.2405043. [DOI] [PubMed] [Google Scholar]
- 48.Tiedt R, Hao-Shen H, Sobas MA, et al. Ratio of mutant JAK2-V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood. 2008;111(8):3931–3940. doi: 10.1182/blood-2007-08-107748. [DOI] [PubMed] [Google Scholar]
- 49.Xing S, Wanting TH, Zhao W, et al. Transgenic expression of JAK2V617F causes myeloproliferative disorders in mice. Blood. 2008;111(10):5109–5117. doi: 10.1182/blood-2007-05-091579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babon JJ, Lucet IS, Murphy JM, Nicola NA, Varghese LN. The molecular regulation of Janus kinase (JAK) activation. Biochem J. 2014;462(1):1–13. doi: 10.1042/BJ20140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hookham MB, Elliott J, Suessmuth Y, et al. The myeloproliferative disorder-associated JAK2 V617F mutant escapes negative regulation by suppressor of cytokine signaling 3. Blood. 2007;109(11):4924–4929. doi: 10.1182/blood-2006-08-039735. [DOI] [PubMed] [Google Scholar]
- 52.Yan D, Jobe F, Hutchison RE, Mohi G. Deletion of Stat3 enhances myeloid cell expansion and increases the severity of myeloproliferative neoplasms in Jak2V617F knock-in mice. Leukemia. 2015;29(10):2050–2061. doi: 10.1038/leu.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]