Abstract
Gene editing enables the site-specific modification of the genome. These technologies have rapidly advanced such that they have entered common use in experimental hematology to investigate genetic function. In addition, genome editing is becoming increasingly plausible as a treatment modality to rectify genetic blood disorders and improve cellular therapies. Genome modification typically ensues from site-specific double-strand breaks and may result in a myriad of outcomes. Even single-strand nicks and targeted biochemical modifications that do not permanently alter the DNA sequence (epigenome editing) may be powerful instruments. In this review, we examine the various technologies, describe their advantages and shortcomings for engendering useful genetic alterations, and consider future prospects for genome editing to impact hematology.
Introduction
The immense potential of site-specific genomic modification has been appreciated since the advent of molecular genetics. Prior to the availability of engineerable targeted nucleases, homologous recombination could be instigated in mammalian cells by the presence of extrachromosomal donor templates, including small oligonucleotides.1-4 The uses of triple helix forming oligonucleotides as triggers and viral genomes such as adeno-associated virus (AAV) as donors for homologous recombination in the absence of additional nucleases continue to demonstrate promise as effective means of targeted genome modification.5,6 However, the relatively low rates of gene correction that can typically be realized by these approaches has both limited research and deterred clinical applications.7 Following the introduction of a double-strand break (DSB) at a locus, rates of homologous recombination increase by roughly 3 orders of magnitude.8-10 Thus, the discovery and subsequent development of targeted nucleases has provided important novel mechanisms for site-specific homologous recombination. In addition, targeted nucleases may be used to produce additional sequence-specific genetic and epigenetic outcomes that may be exploited to gain knowledge of genome function and produce desirable alterations.11
Each of the targeted nucleases described below contains 2 major functional moieties, the first of which is specific DNA recognition. The various platforms differ based on the biochemical nature of the recognition (by protein or RNA), by the modularity of recognition (whether 1, 2, or more components are involved), the size of the recognition domain (and its attendant challenges to cellular delivery), the ease with which the interaction may be engineered to recognize a variety of target sequences, and the specificity of such recognition. The second function is endonucleic DNA cleavage. The nature of both the cleaving enzyme (monomeric vs multimeric organization, size) and the resultant cleavage (blunt-ended, staggered-ended, single-stranded nick) are distinguishing characteristics.
A number of recent reviews have covered the fundamental discoveries and engineering breakthroughs that have yielded the current toolkit of genome editing reagents.12-15 Here we provide a brief overview of the major classes of targeted nucleases and then focus on considerations and opportunities for the widespread application of these tools for laboratory research, as well as clinical application to ameliorate hematologic disorders.
Genome editing tools
Meganucleases
Meganucleases (also called homing endonucleases) were the first targeted nuclease described.16 These 20- to 37-kDa (∼0.6-1 kb) sequence-specific nucleases are active as monomers and named based on their ability to recognize relatively long (14-40 bp) target sequences.17 Meganucleases are distinguished from restriction endonucleases by their much larger target site, whose occurrence may be as rare as a single instance per genome.17
Applications of meganucleases are limited by the low frequency of target site presence at most genes, thus essentially preventing the use of naturally occurring forms for functional genomics or therapeutic development.18,19 Meganucleases may be engineered by both rational structure-guided design and high-throughput screening to recognize diverse targets, including those within human genes, although this remains a nontrivial task.18,20 In addition, the tendency of meganucleases to tolerate some target sequence degeneracy may limit their specificity.21
Zinc-finger nucleases
Zinc-finger nucleases (ZFNs) are chimeric nucleases that consist of individual zinc-finger protein (ZFP) motifs, each of which is capable of specifically recognizing 3 to 4 bases of DNA.22 Linking multiple ZFPs together allows for the targeting of extended stretches of DNA (typically 9-18 bp). The coupling of a FokI nuclease (which cleaves without sequence specificity but requires dimerization) to each of a pair of monomers allows the composite dimeric enzyme to introduce a site-specific DSB.23 Each ZFN target site is composed of 2 ZFP binding sites on either side of a spacer region of 5 to 7 bp within which the dimerized FokI cleaves.24 Although the need for dimerization allows for increased target site specificity (due to the increased binding site size), wild-type FokI can still homodimerize and lead to undesired breaks. FokI domains containing mutations (ELD/KKR) in the dimerization interface result in increased specificity via obligate heterodimerization.25-27 Each ZFN monomer is encoded by a roughly 1.2 kb (∼45 kDa) gene, a pair of which need to be delivered to target cells to execute genome editing. Like meganucleases, the requirement for de novo design and testing of ZFN target sequence recognition is a barrier to their widespread use.
Transcription activator–like effector nucleases
Similar to ZFNs, transcription activator–like effector nucleases (TALENs) are chimeric nucleases composed of an engineerable DNA-binding domain and a FokI nuclease. The DNA binding domain consists of an array of individual TALE repeats composed of 34 amino acid modules that determine DNA specificity based merely on the sequence of the 12th and 13th residues (so-called repeat variable diresidues [RVDs]).23 Like ZFNs, TALENs are typically designed to act as obligate heterodimers to increase specificity. Thus, active TALENs consist of a pair of monomers (2.5-3.0 kb or 90-110 kDa) separated by a 12- to 20-bp spacer region that introduce a DSB on FokI dimerization.24
TALEN design and assembly is more straightforward than that of ZFNs as each RVD targets a single DNA base and multiple RVDs can be stitched together in essentially any order. Online resources are available to assist with design and cloning from preformed modules.15 Hybrid nucleases consisting of the easily engineerable TALE binding domain and the site-specific meganuclease head (so-called megaTALs) may offer additional specificity, affinity, and potentially simplified cellular delivery because they are active as ∼75-kDa (∼2-kb) monomers.28
Clustered regularly interspaced short palindromic repeat/Cas
Clustered regularly interspaced short palindromic repeat (CRISPR) sequences are a defining component of a prokaryotic adaptive immune system. From these loci, bacteria express genomically encoded RNAs to guide nuclease cleavage to matching sequences of invading phage and plasmid DNA.29,30 The widely studied type II system from Streptococcus pyogenes consists of 3 components: (1) the nuclease Cas9, containing both RuvC and HNH nuclease domains, which produces blunt DSBs; (2) a DNA-binding CRISPR RNA (crRNA), which includes a 20-nt guide RNA (gRNA) sequence with precise complementarity to its DNA target; and (3) an auxiliary trans-activating crRNA (tracrRNA) bridging the crRNA to Cas9. Recognition of a target site by SpCas9 depends on the presence of a protospacer adjacent motif (PAM) sequence NGG immediately downstream of the gRNA target sequence. Cleavage occurs 3 bp upstream of the PAM.31
The most common implementation of CRISPR genome editing in eukaryotic cells relies on a 2-component system consisting of SpCas9 (∼160 kDa, ∼4.2 kb) and a single chimeric guide RNA (sgRNA) of ∼110 nt that subserves the recognition and structural functions of the crRNA and tracrRNA.32
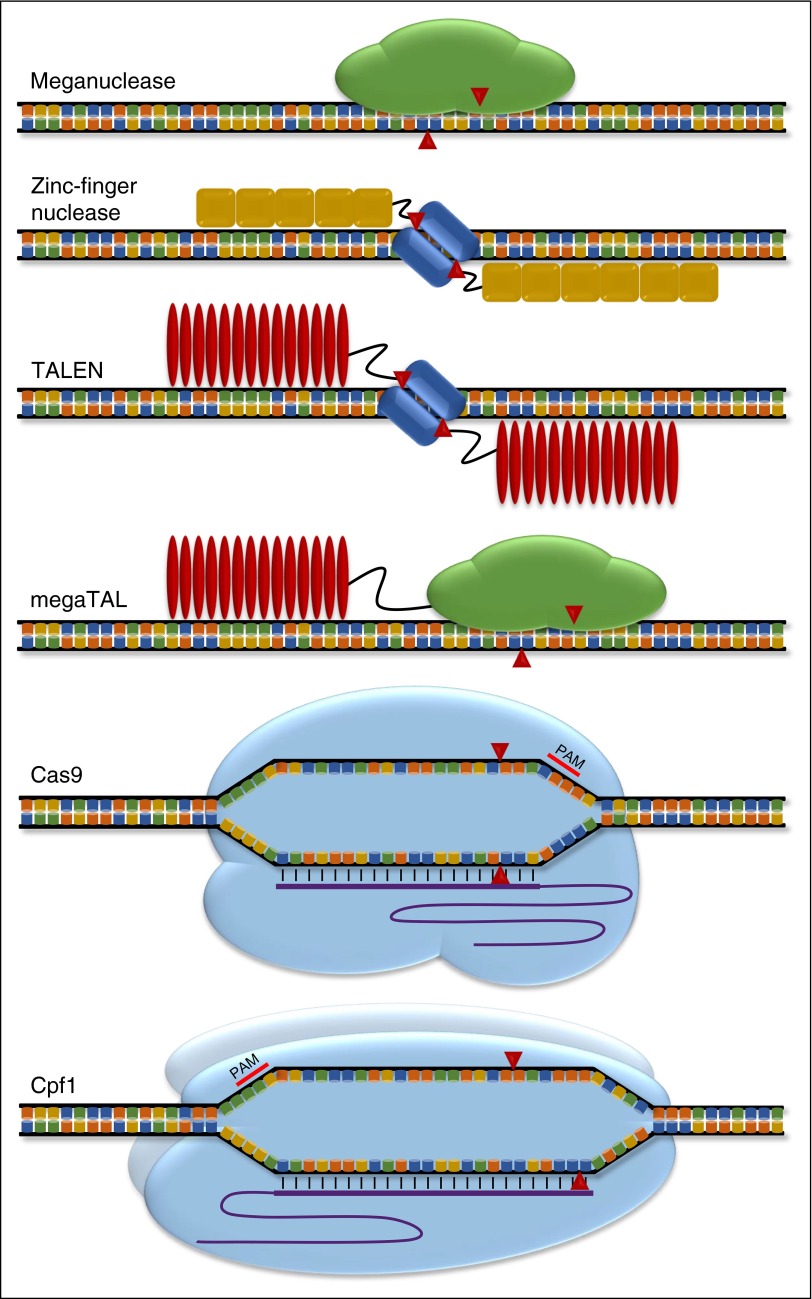
The discrete rules underlying SpCas9/sgRNA genome editing belie the extensive diversity of CRISPR/Cas systems. Several naturally occurring Cas9 orthologs have been adapted for genome editing of human cells, and many more are likely to be discovered.33-35 For example, the smaller size of the Staphylococcus aureus Cas9 (∼120 kDa, ∼3.3 kb) compared with SpCas9 may provide advantages in terms of cellular delivery, but its restriction by a different PAM sequence (NNGRRT) may limit SaCas9 to a somewhat narrower set of genomic targets.33 Engineered variants of SaCas9 and SpCas9 with novel PAM recognition sequences broaden the repertoire of possible genomic targets.36,37 Cpf1, a member of a distantly related CRISPR nuclease family, differs from SpCas9 in that it uses a single gRNA without tracrRNA, has a different PAM sequence (TTN), appears to act as a dimer rather than monomer, and introduces a staggered rather than blunt DSB.35 This multitude of CRISPR/Cas variants provides scientists with a rapidly expanding toolkit with extensive versatility (Figure 1).
Figure 1.
Targeted nucleases. Schematic of genome editing systems. DNA recognition and cleavage components for each nuclease shown. ZFP motifs shown as yellow boxes. TALE RVDs shown as red ovals. FokI shown as blue rectangles. Single guide RNA shown as purple lines. Red arrowheads indicate cleavage points for each enzyme. Cpf1 illustrated as dimer.
Molecular outcomes of genome editing
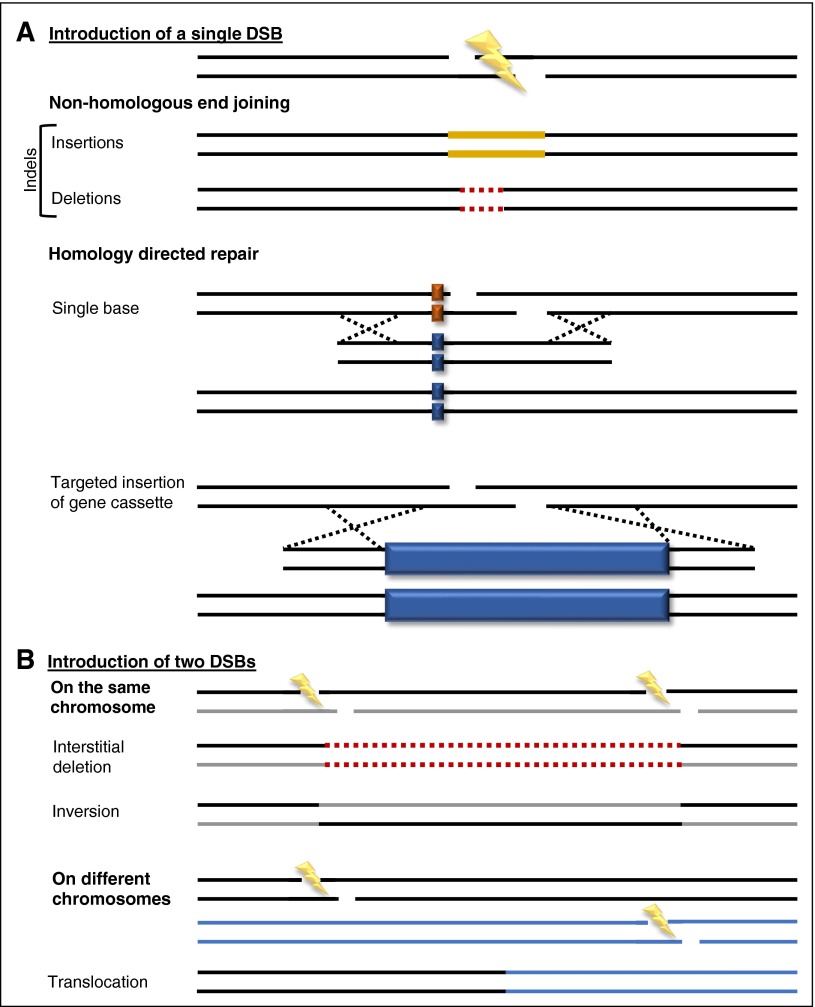
Following introduction of a DSB in the genome, cellular repair prevents disastrous chromosomal catastrophes. The varied repair responses may be simplified into 2 major pathways, each of which relies on a large number of host factors: nonhomologous end joining (NHEJ) and homology-directed repair (HDR). NHEJ involves reuniting the 2 broken ends of the chromosome in a process that often leads to small insertions or deletions (so-called indels) at the cleavage site. These small “scars” left behind by indel mutations may result in functional disruption of essential target sequences, such as producing missense or frameshift mutations or interruption of splice sites or transcription factor binding sites24 (Figure 2A). The simultaneous introduction of 2 DSBs some distance away from each other on the same chromosome may result in large interstitial deletions and inversions or in translocations if on different chromosomes38-42 (Figure 2B).
Figure 2.
Molecular outcomes of genome editing. Multiple possible repair outcomes of (A) 1 or (B) 2 DSBs in the genome. Schematics of indels, gene insertion, and gene correction are shown. Solid gold lines, inserted bases; dashed red lines, deleted bases; solid dark blue boxes, gene cassettes. Outcomes depicted are not exhaustive, and other outcomes, such as insertion of a gene cassette via NHEJ, are possible.78,79
In contrast, HDR depends on a donor template for repair. In “natural” HDR, following genotoxic injury, homologous sister chromatids serve as template for precise repair in replicating cells. In the case of genome editing, an extrachromosomal donor sequence may be used to integrate sequences of choice adjacent to induced DSBs (or even to single-strand breaks, as may be produced by nickase mutants of Cas9). Because expression of HDR pathway components is limited to the S/G2 phase of the cell cycle, only dividing cells are competent for this type of repair.43-45
Genome editing as a research tool
A fundamental goal of human genetics is to relate genotype to phenotype. Genome editing provides a powerful means to rapidly generate novel alleles to determine the function of coding and noncoding sequences. Given its robustness and ease of use, CRISPR/Cas9 has quickly become an indispensable tool for many experimental hematologists to evaluate gene function. The ability to create null alleles on demand appears at least as powerful a paradigm shift as preceding RNA interference technology for functional genomics. Various manifestations of genome editing as a research methodology include the production of knockout and knock-in alleles in hematopoietic cell lines and in hematopoietic stem cells transplanted to mice, to model leukemia-associated translocations, and to produce various animal models of genetic modification in both the zygote and restricted to somatic lineages.40,46-51
The control of Cas9 specificity merely by the nature of the 20-nt gRNA sequence makes the generation and interrogation of large-scale sgRNA libraries straightforward by massively parallel oligonucleotide synthesis and sequencing, respectively. With increased specificity and efficiency over RNAi screens, CRISPR-based forward genetic screens may be performed by introduction to cells of genome-wide libraries of sgRNAs as mutagens, cells with a given phenotype retrieved, and then the mutagens of interest easily identified.52,53 These approaches have been successfully applied to identify unexpected dependencies for various infectious diseases and cancers.54-56
Systematic CRISPR approaches not only provide an opportunity to identify genes of interest, but also delineate key coding and non-coding sequences. Dense sgRNA libraries targeted throughout coding sequences can identify functional domains within a gene, such as druggable dependencies in hematologic malignancy.57 Methodical perturbation of regulatory sequences, such as the fetal hemoglobin-associated erythroid-specific enhancer of BCL11A58 by CRISPR/Cas9 and ZFN/TALEN mutagenesis, have identified critical regulatory sequences that themselves may serve as therapeutic targets.59,60
In contrast to conventional genome editing, which implies permanent modification of genomic target sequences, genome editing tools may be repurposed to engender potent biological outcomes without mutagenesis. For example, catalytically inactive mutants of Cas9 (dCas9) may be guided to proximal promoters or coding sequences to block transcription.61 Linking the site-specific binding domains of targeted nucleases to gene regulatory domains (along with disabled nuclease function) may allow for potent modulation of gene expression to characterize mechanisms of transcriptional regulation as well as redirect cellular phenotype.62 For example, fusion of dCas9 to a repressor such as the Kruppel-associated box promotes repressive histone modifications such as histone H3K9 trimethylation and targeted gene silencing.62 Pairing TALEs or dCas9 with chromatin regulators such as LSD1 histone demethylase can result in targeted epigenetic regulation of noncoding elements and help define relationships between genes and distal regulatory elements.63-65
In addition to gene repression, gene activation can also be achieved using modified targeted nuclease platforms. Coupling dCas9 to individual positive transcriptional regulators such as the VP64 transactivation domain or the core region of the histone acetyltransferase p300 results in robust transcription of target genes, and higher-order clusters of activation domains may serve as even more potent activators.66-68 Targeting of such dCas9-based transcriptional activators or repressors to a large set of gene promoters with sgRNA libraries permits genome-wide screens.69,70
Therapeutic considerations
Repair mode
The ultimate cure for a genetic disorder would be to revert the disease mutation back to normal sequence. Conditions for which a single or predominant mutation underlies the disease would seem to be most amenable to this approach (because a single targeting strategy could be broadly applicable). In addition, a point mutation might be simpler to correct than a larger scale rearrangement. For example, sickle cell disease, with its high incidence and characteristic causative β-globin adenine-to-thymine transversion, would seem an auspicious target for gene correction. In fact, HDR-based genome editing correction of the sickle cell disease point mutation has been achieved in multiple cell types, including induced pluripotent stem cells,71-73 although the rates of editing that could be achieved in long-term hematopoietic stem cells (HSCs) have thus far remained below the level of therapeutic relevance.74
For other conditions with numerous mutations of an affected gene, like the varied mutations of IL2RG underlying X-linked severe combined immunodeficiency (X-SCID), a more universal strategy might be attractive, such as the targeted integration of an entire gene cassette.75,76 One strategy would be to target a gene cassette including autonomous regulatory elements to a universal “safe harbor” locus. Another option would be to target a minimal gene cassette to an endogenous locus. Targeting to intronic sequences allows splicing of a promoterless transgene downstream of the upstream exons and retains the endogenous gene promoter and distal regulatory elements for gene control.75 This strategy could be particularly advantageous for conditions in which endogenous gene regulation is imperative. For example, CD40 ligand (CD40L), whose deficiency results in combined immunodeficiency, is typically only transiently expressed on activated T cells. Constitutive expression of CD40L in T cells of CD40L-deficient mice results in lymphoproliferative disorder, suggesting the potential risks of nonendogenous gene regulation.77 Although targeted integration often insinuates HDR repair, NHEJ pathways may also be harnessed to achieve targeted integration, which may be particularly important in nondividing cells.78-80 Even if a mutant gene were effectively repaired or replaced, an additional challenge may be to avoid immune responses against the novel gene product.81
Although gene correction might seem the most intuitive approach to therapeutic genome editing, in fact the first clinical trial using targeted nucleases in human patients has relied on NHEJ-based genetic disruption rather than gene repair. One advantage of this strategy is that NHEJ tends to be a more active repair pathway compared with HDR, particularly in quiescent cells.82 Another benefit of gene disruption may be that modulation of a disease-modifying pathway could produce a more universal remedy for the disease compared with individual mutation-specific corrections. The first-in-human genome editing trial used ZFNs in autologous T cells to target the HIV5 coreceptor CCR5,83 inspired by individuals naturally resistant to HIV infection due to CCR5 deficiency, as well as the remarkable case of an HIV-positive recipient of an allogeneic CCR5-deficient hematopoietic stem cell transplant (HSCT) whose HIV became undetectable without antiretroviral therapy.66,84
The indels left behind following NHEJ repair may be useful not only for disrupting coding sequences but also noncoding regulatory sequences such as the erythroid-specific enhancer of BCL11A for the β-hemoglobinopathies59,60 or DMD splice sites to promote exon skipping in muscular dystrophy.85 Paired DSBs may also result in desirable outcomes, such as interstitial deletions of mutant exons in DMD85-87 or reversion of large chromosomal inversions underlying hemophilia A.44
Even epigenetic editing approaches could be considered for therapeutic utility, such as forced chromatin looping for the β-hemoglobinopathies to redirect interactions with the powerful globin enhancer locus control region away from mutant β-globin toward compensatory γ-globin.88,89 To the degree that persistent expression of the artificial DNA binding factor would be required to maintain therapeutic gene expression, the kiss-and-run promise of therapeutic genome editing would be lost, and the challenges of long-term stable and safe expression would be similar to those facing conventional gene therapy.90
Target cell
The ideal somatic target cell for therapeutic gene editing depends on the specific clinical situation. In general, the goal is for a durable, potentially even lifelong (ie, curative), benefit from gene editing. The ability to access and manipulate the appropriate target cell may determine the feasibility of the endeavor. Limiting edits to disease-associated cells may minimize risk of adverse clinical effects of genetic perturbation of cell types unconnected to the disease, although the nature of this risk depends on the particular perturbation.
For many hematologic conditions, the most relevant cell type to edit would be the HSC, the rare self-renewing cells atop the hematopoietic hierarchy. Certainly for genetic disorders of the HSC itself (such as inherited bone marrow failure disorders like Fanconi anemia or dyskeratosis congenita), modification of this cell would be required for a salutary effect. For monogenic disorders of downstream lineages, such as of erythrocytes (eg, hemoglobinopathies91) or granulocytes (eg, chronic granulomatous disease92,93), modification of the HSC appears to be required to achieve a renewing source of corrected cells. Engineering HSCs may even have the advantage to confer exceptional properties (ie, more ameliorating than mere healthy cells) to downstream progeny. For example, supraphysiologic expression of arylsulfatase A in corrected microglia derived from autologous HSCs in metachromatic leukodystrophy appears to confer benefits beyond that of allogenic HSCT due to cell nonautonomous cross-correction.94 For all HSC-based therapies, in addition to any risks intrinsic to the gene editing itself, risks of isolating the cells from the appropriate source and preparative conditioning therapy must be considered.
One caveat to engineering hematopoiesis is that most knowledge of HSC function is based on cellular capacity to reconstitute ablated animals and patients. Emerging studies of steady-state hematopoiesis suggest that a large fraction of hematopoietic output derives from long-lived lineage-restricted progenitor cells,95-97 raising the question of whether modifying appropriate lineage-restricted progenitors could have durable therapeutic utility under specific conditions.
For inherited disorders of lymphocytes (eg, immunodeficiencies), modification of long-lived T cells might sometimes be adequate, although in some conditions, T cells may be absent. The results of gene therapy for X-SCID suggest that modification of a barely measurable fraction of HSCs can result in robust (although sometimes oligoclonal) T-cell reconstitution, reflecting the intense selective advantage for rescued T cells in certain immunodeficiencies.98 Another consideration is the number of rescued cells required to provide a therapeutic advantage. For example, in CD40L deficiency, allogeneic HSCT with mixed chimerism may be curative.99 Therefore, in this type of condition, correction of merely a subset of HSCs or even T cells might be expected to be of therapeutic value.
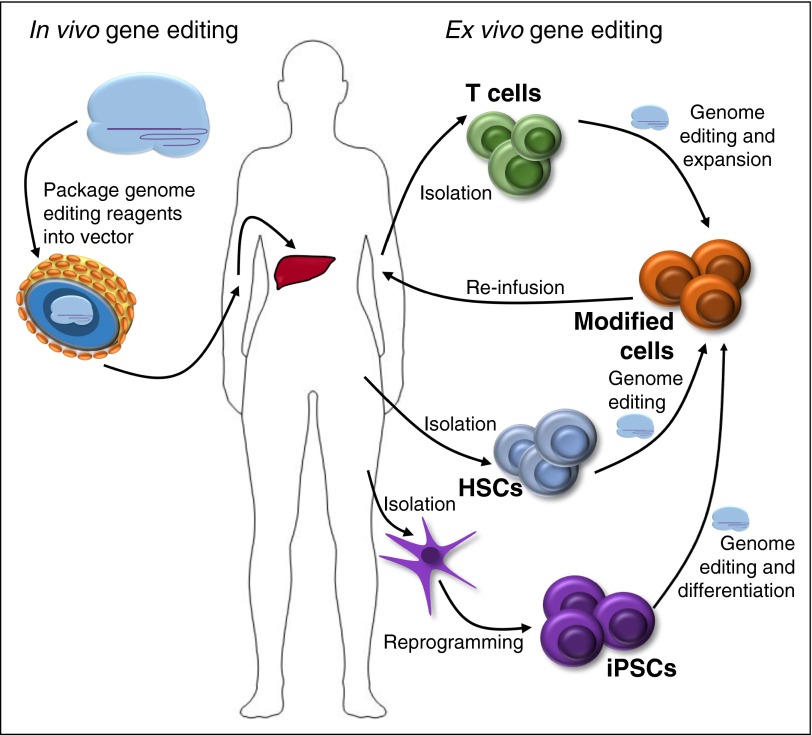
Isolated T-cell modification may be desirable in various adoptive immunotherapy applications. A technical advantage may be that T cells can be easier to expand and manipulate ex vivo compared with HSCs100 (Figure 3). Also T-cell editing could have a theoretical safety advantage with respect to myeloproliferative risk. Allogeneic T-cell therapies could be useful in a wide variety of immune, infectious, and malignant conditions. Autologous T cells with enforced expression of chimeric antigen receptors (CARs)101-103 recognizing tumor-associated antigens have shown remarkable clinical responses; however, the current approaches require expensive, labor-intensive, time-sensitive autologous cell processing. Gene disruption of the endogenous T-cell receptor (TCR) might allow for production of allogeneic CAR T cells while avoiding risk of suboptimal or graft-versus-host responses.104-107 Recently, a report of a single remarkable case with molecular remission following treatment with TALEN TCR-edited allogeneic CAR T cells (with subsequent allogeneic HSCT) for multiply relapsed B-cell acute lymphoblastic leukemia has generated great excitement.108
Figure 3.
Therapeutic genome editing. Schematic of delivery strategies and target cells for in vivo and ex vivo genome editing. iPSCs, induced pluripotent stem cells.
Other long-lived hematopoietic cells in addition to HSCs might be considered as targets for therapeutic gene editing. For example, congenital pulmonary alveolar proteinosis results from absence of pulmonary alveolar macrophages due to granulocyte-macrophage colony-stimulating factor deficiency. In mouse models, transplantation of gene corrected or healthy pulmonary alveolar macrophages can result in disease reversal for ≥1 year,109 suggesting that tissue macrophages can be genetically manipulated, long-lived, and biologically potent under particular clinical circumstances.
A variety of hematologic conditions might benefit from gene editing of nonhematopoietic lineages, chiefly the inherited bleeding disorders. Hepatocytes are both the frequent target for various genetic therapies110 and the natural source of synthesis of various coagulation factors that may be congenitally deficient such as factor IX in hemophilia B. However, expression of factor IX from cells besides hepatocytes (eg, myocytes, megakaryocytes111,112) and expression of factor VIII from hepatocytes113 (rather than endogenous endothelial cells) may be restorative in the hemophilias. These results suggest that recapitulating the endogenous cell source for secreted factors may be less important than other considerations such as ease and safety of delivery and achievable level of transgene expression.
Finally, genome editing of pluripotent stem cells has a number of advantages, including the ability to select rare desired mutations and to extensively characterize clonal on-target and off-target effects, including by whole genome sequencing.114,115 The major challenge is to develop efficient protocols to produce hematopoietic stem cells or other clinically relevant cellular outputs.116 Irrespective of the target cell, a successful clinical gene editing approach must consider the fraction of the tissue that must be corrected to achieve therapeutic benefit. This may depend on the degree of selective advantage of effector cell relative to target cell (eg, survival advantage to be expected for erythrocytes and erythroid precursors deriving from corrected HSCs in hemoglobinopathy or for B-lymphocytes derived from corrected HSCs in X-linked agammaglobulinemia117).
Delivery
Robust cellular delivery may be the limiting factor for therapeutic genome editing. Most strategies involve just a transient burst of nuclease expression with the rationale that after desired edits have been produced, nucleases serve no productive role. Meganucleases and megaTALs need only the expression of 1 protein, ZFNs and TALENs require the expression of 2 monomers, and the CRISPR/Cas9 system depends on simultaneous expression of both a protein (Cas9) and RNA (sgRNA) to yield an active ribonucleoprotein complex in target cells.31
For genome editing of hematopoietic cells, most attention has focused on ex vivo delivery. TALENs have proven difficult to deliver via AAV or lentiviral vectors due to their large size and highly repetitive sequences.118 Delivery of SpCas9 via AAV is particularly challenging given that the gene itself nearly outstrips the viral genome cargo limit, leaving very little room for regulatory elements, sgRNA cassettes, or homology donor sequences. Smaller orthologs such as SaCas9 appear better suited for AAV delivery.33 Concerns of low-level AAV integration could become particularly relevant in the setting of clinical delivery of active nucleases.119 Although widely tropic integrase-defective lentiviral vectors might appear a logical approach, these are hindered by relatively low gene expression levels compared with integrating counterparts.120-122
Transient delivery of targeted nucleases to primary hematopoietic cells has been most successfully achieved by electroporation.75,123-125 Interestingly, electroporation of mRNA has reduced risk of integration and appears to result in reduced cellular toxicity compared with DNA. Delivery of in vitro transcribed ZFN mRNA to HSCs and T cells via electroporation may be compatible with clinical-scale genome editing.60,83,126,127 CRISPR/Cas9 may be delivered to both HSCs and T cells via electroporation of ribonucleoprotein complexes of Cas9 protein and sgRNA.123
Alternative delivery platforms such as osmocytosis, cell-penetrating peptides, cationic lipids, and microfluidic devices have been proposed, but their efficacy and lack of toxicity for primary hematopoietic cells remain to be determined.128-131
In contrast to the focus on ex vivo delivery for hematopoietic cells, in vivo delivery has been investigated particularly for therapeutic genome editing of nonhematopoietic tissues. For example, intravenous delivery by AAV of ZFNs and a donor cassette targeting the factor IX locus allows for the targeted integration and therapeutic production of factor IX in hemophilia B mice.80,132 An alternate strategy is to use AAV delivery of ZFNs and a promoterless donor cassette to target expression to the first intron of the albumin locus. Recent proof-of-principle for this approach has been demonstrated to ameliorate both factor VIII and factor IX deficiencies in relevant mouse models.133
In addition, 3 groups recently showed that AAV delivery of Cas9 and a pair of sgRNAs could delete a mutant exon, resulting in therapeutic exon skipping in mouse models of Duchenne muscular dystrophy.85-87 Collectively, these studies demonstrate proof-of-principle of systematic delivery of nucleases to target genome editing in liver and muscle, each of which might serve as appropriate sites of synthesis in coagulation factor deficiency.
In addition to viral vectors, nonviral delivery, particularly to the liver, may be considered for in vivo genome editing. Already hydrodynamic injection of CRISPR/Cas9 plasmids has been shown to correct the metabolic liver disease hereditary tyrosinemia in mice.134 Other forms of nonviral in vivo delivery, such as ultrasound with microbubbles, cationic lipid complexes, and DNA nanoparticles, have been investigated.135-137 Given the pace of advances, even in vivo delivery of genome editing reagents targeted directly to hematopoietic cells including HSCs seems conceivable. In contrast to their typical use via ex vivo transduction, lentiviral and nonlentiviral vectors may directly transduce HSCs following in vivo intraosseous or intravenous delivery.138,139 One issue that in vivo delivery might face is development of immune responses against the targeted nucleases themselves (ie, bacterially derived nucleases in the case of CRISPR), which could restrict the potential for successive therapies.
Editing efficiency and specificity
The ideal genome edit would occur with high efficiency at the desired (“on-target”) site and with few if any consequences at other (“off-target”) genetic loci.140 Both efficiency and specificity of editing may depend on the nature of the nuclease platform itself, features of the genomic target site, and the mechanisms by which the nucleases are delivered to the target cells.
Various claims have been made about the relative superiority of numerous platforms.141 Overall, it is unclear whether differences observed in individual studies are true class effects or related to the individual loci, target sequences, cell types, and delivery modalities. Particularly given the numerous permutations of CRISPR/Cas systems, it is difficult to generalize. For an individual nuclease, its level of expression appears to be a critical determinant of efficiency.142 Cell type–specific chromatin context may play an important role in determining accessibility and subsequent modification of a given nuclease at a target site.143,144
Typically alleles repaired by NHEJ exceed those repaired by HDR following introduction of DSBs, particularly in nondividing cells. A variety of strategies have been used to promote HDR compared with NHEJ outcomes, including pharmacologic or genetic inhibition of NHEJ, augmentation of HDR, and synchronization of cell cycle.45,145,146 Some have suggested that the unique 3′ overhang generated by a meganuclease head (including by megaTALs) may enhance the rate of HDR relative to FokI or Cas9 cleavage.100
Gene therapy trials with integrating retroviral vectors targeting HSCs have demonstrated both clinical efficacy and severe adverse events due to vector integration-mediated oncogenesis, although newer-generation lentiviral vectors appear to substantially reduce risk of insertional mutagenesis.147-149 In contrast to conventional gene therapy, which counts as success thousands of unique semirandom vector integration events with many distributed near active genes, genome editing raises the theoretical potential for seamless repair, with only the desired genetic modification. Numerous strategies have been described to minimize risks of off-target mutagenesis in response to targeted nuclease exposure.
The first step is to design reagents with the lowest possible risk for off-target cleavages. Target sequences with exact or close matches in the genome in addition to the on-target site carry the greatest risk for off-target cleavage because targeted nuclease DNA recognition may be somewhat promiscuous. For CRISPR gRNAs, the so-called seed sequences proximal to the PAM are particularly intolerant of mismatches.
Second, once any close matching sequences (possible off-target sites) are identified, their cleavage may be closely monitored experimentally.150-152 However, the ability to predict off-target sites may be imperfect, indicating the importance of systematic approaches for unbiased assessment of off-target cleavage.153,154 Whole-genome sequencing may be considered the ultimate unbiased technique, but the sensitivity is relatively low and thus unable to exclude rare off-target effects in populations of cells. A number of methods for systematic off-target identification have been developed, each with its own caveats in terms of false positives and false negatives and ability to detect events in therapeutically relevant cellular contexts.33,143,155-157 Also the basal rate of DSBs and mutagenesis that exists in normal somatic tissues158 including HSCs,159 must be taken into account.
In addition to design of a highly unique recognition sequence, several other technical approaches may promote specificity of genome editing, including simply minimizing the duration and degree of expression of the targeted nuclease. ZFNs may be engineered at both their DNA recognition domains and linker and dimerization domains to maximize specficity.160 For megaTALs, additional TALE RVD recognition modules may improve effective specificity.100 For CRISPR/Cas systems, a number of technical strategies have been explored, including short gRNAs (of 17-18 nt), nickase mutants of Cas9 coupled to paired gRNAs targeting opposing strands, dCas9-FokI dimers for enforced heterodimerization, mutant Cas9 coupled to a programmable DNA binding domain such as ZFP or TALE,161 and engineered variants of Cas9 that reduce its interaction strength with the gRNA:target DNA heteroduplex,162,163 each of which appear to improve specificity.
For therapeutic translation, these approaches need to balance a favorable portfolio of cellular delivery, on-target efficiency, and off-target specificity, as measured under clinically relevant conditions.
Safety
In contrast to the above molecular considerations, the ultimate utility of therapeutic genome editing approaches will be judged based on clinical safety and efficacy. It is important to note that unlike insertional mutagenesis, which is widely described as a mechanism of tumorigenesis, examples of biologically relevant nuclease off-target genotoxicity under conditions that would approximate therapeutic genome editing are vanishingly scarce. The vast majority of off-target cleavages would be expected to result in neutral indels at noncoding sequences. Therefore, rational nuclease design ought to prioritize avoiding cleavage within critical sequences, such as within recurrently mutated tumor suppressor genes. Extrapolating from regulatory requirements for conventional gene therapy, evaluation for any biological aberrations of target cells, such as propensity for clonal outgrowth or tumorigenesis in vitro or in animals, will justifiably receive emphasis rather than reliance on merely molecular outcomes.
Conclusions
Genome (and epigenome) editing offers the opportunity to prospectively alter DNA (and associated chromatin) to investigate fundamental genome function and to ameliorate disease. The fast pace of technological advancements in these areas appears poised to accelerate knowledge of gene regulation and genome biology. Multiple preclinical and early stage clinical programs are underway, initially using ZFNs, TALENs, and megaTALs, with CRISPR/Cas-based therapies swiftly following. The continued improvement of the delivery and specificity of these reagents promises to hasten clinical implementation of genuine precision medicine.
Acknowledgments
M.D.H. is supported by the National Institutes of Health (NIH), National Heart, Lung, and Blood Institute grant T32HL007574 and D.E.B. by the NIH, National Institute of Diabetes and Digestive and Kidney Diseases grant K08DK093705 (Career Development Award), the Doris Duke Charitable Foundation, the Charles H. Hood Foundation, the American Society of Hematology, the Borough’s Wellcome Fund, and Cooley’s Anemia Foundation.
Authorship
Contribution: M.D.H. and D.E.B. wrote and edited the manuscript.
Conflict-of-interest disclosure: D.E.B. is an inventor on a patent application related to therapeutic genome editing of BCL11A, served as a consultant to Editas Medicine and CRISPR Therapeutics, and received research support from Biogen. M.D.H. declares no competing financial interests.
Correspondence: Daniel E. Bauer, Boston Children’s Hospital, 1 Blackfan Circle, Karp RB 07007A, Boston, MA 02115; e-mail: bauer@bloodgroup.tch.harvard.edu.
References
- 1.Cole-Strauss A, Yoon K, Xiang Y, et al. Correction of the mutation responsible for sickle cell anemia by an RNA-DNA oligonucleotide. Science. 1996;273(5280):1386-1389. [DOI] [PubMed]
- 2.Igoucheva O, Alexeev V, Yoon K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 2001;8(5):391–399. doi: 10.1038/sj.gt.3301414. [DOI] [PubMed] [Google Scholar]
- 3.Olsen PA, Randøl M, Luna L, Brown T, Krauss S. Genomic sequence correction by single-stranded DNA oligonucleotides: role of DNA synthesis and chemical modifications of the oligonucleotide ends. J Gene Med. 2005;7(12):1534–1544. doi: 10.1002/jgm.804. [DOI] [PubMed] [Google Scholar]
- 4.Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244(4910):1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 5.Reza F, Glazer PM. Therapeutic genome mutagenesis using synthetic donor DNA and triplex-forming molecules. Methods Mol Biol. 2015;1239:39–73. doi: 10.1007/978-1-4939-1862-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barzel A, Paulk NK, Shi Y, et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517(7534):360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goncz KK, Prokopishyn NL, Abdolmohammadi A, et al. Small fragment homologous replacement-mediated modification of genomic beta-globin sequences in human hematopoietic stem/progenitor cells. Oligonucleotides. 2006;16(3):213–224. doi: 10.1089/oli.2006.16.213. [DOI] [PubMed] [Google Scholar]
- 8.Osman F, Subramani S. Double-strand break-induced recombination in eukaryotes. Prog Nucleic Acid Res Mol Biol. 1998;58:263–299. doi: 10.1016/s0079-6603(08)60039-2. [DOI] [PubMed] [Google Scholar]
- 9.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91(13):6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23(24):5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilton IB, Gersbach CA. Enabling functional genomics with genome engineering. Genome Res. 2015;25(10):1442–1455. doi: 10.1101/gr.190124.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096-1258096. [DOI] [PubMed]
- 13.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 15.Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15(5):321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 16.Hafez M, Hausner G. Homing endonucleases: DNA scissors on a mission. Genome. 2012;55(8):553–569. doi: 10.1139/g2012-049. [DOI] [PubMed] [Google Scholar]
- 17.Pâques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. 2007;7(1):49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 18.Smith J, Grizot S, Arnould S, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34(22):e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva G, Poirot L, Galetto R, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11(1):11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grizot S, Smith J, Daboussi F, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37(16):5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Ulge UY, Hovde BT, Doyle LA, Monnat RJ., Jr Comprehensive homing endonuclease target site specificity profiling reveals evolutionary constraints and enables genome engineering applications. Nucleic Acids Res. 2012;40(6):2587–2598. doi: 10.1093/nar/gkr1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott CT. The zinc finger nuclease monopoly. Nat Biotechnol. 2005;23(8):915–918. doi: 10.1038/nbt0805-915. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Pinera P, Ousterout DG, Gersbach CA. Advances in targeted genome editing. Curr Opin Chem Biol. 2012;16(3-4):268–277. doi: 10.1016/j.cbpa.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25(7):786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 26.Doyon Y, Vo TD, Mendel MC, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8(1):74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 27.Miller JC, Holmes MC, Wang J, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 28.Boissel S, Jarjour J, Astrakhan A, et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42(4):2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1(1):7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 31.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816-821. [DOI] [PMC free article] [PubMed]
- 32.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou Z, Zhang Y, Propson NE, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110(39):15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleinstiver BP, Prew MS, Tsai SQ, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33(12):1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Canver MC, Bauer DE, Dass A, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem. 2014;289(31):21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunet E, Simsek D, Tomishima M, et al. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci USA. 2009;106(26):10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piganeau M, Ghezraoui H, De Cian A, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23(7):1182–1193. doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee HJ, Kweon J, Kim E, Kim S, Kim J-S. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22(3):539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park C-Y, Kim DH, Son JS, et al. Functional correction of large factor VIII gene chromosomal inversions in hemophilia A patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17(2):213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7(18):2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32(12):3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orthwein A, Noordermeer SM, Wilson MD, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528(7582):422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Heckl D, Kowalczyk MS, Yudovich D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32(9):941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ablain J, Durand EM, Yang S, Zhou Y, Zon LIA. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell. 2015;32(6):756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun. 2014;5:3964. doi: 10.1038/ncomms4964. [DOI] [PubMed] [Google Scholar]
- 51.Buechele C, Breese EH, Schneidawind D, et al. MLL leukemia induction by genome editing of human CD34+ hematopoietic cells. Blood. 2015;126(14):1683–1694. doi: 10.1182/blood-2015-05-646398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue H-Y, Ji L-J, Gao A-M, et al. CRISPR-Cas9 for medical genetic screens: applications and future perspectives. J Med Genet. 2015;53(2):91-97. [DOI] [PubMed]
- 53.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16(5):299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 56.Shen B, Zhang W, Zhang J, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11(4):399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 57.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33(6):661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bauer DE, Kamran SC, Lessard S, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Canver MC, Smith EC, Sher F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vierstra J, Reik A, Chang K-H, et al. Functional footprinting of regulatory DNA. Nat Methods. 2015;12(10):927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thakore PI, D’Ippolito AM, Song L, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12(12):1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mendenhall EM, Williamson KE, Reyon D, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013;31(12):1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kearns NA, Pham H, Tabak B, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12(5):401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hilton IB, D’Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zalatan JG, Lee ME, Almeida R, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160(1-2):339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chavez A, Scheiman J, Vora S, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12(4):326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159(3):647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sebastiano V, Maeder ML, Angstman JF, et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29(11):1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun N, Zhao H. Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng. 2014;111(5):1048–1053. doi: 10.1002/bit.25018. [DOI] [PubMed] [Google Scholar]
- 73.Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118(17):4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoban MD, Cost GJ, Mendel MC, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125(17):2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Genovese P, Schiroli G, Escobar G, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510(7504):235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lombardo A, Genovese P, Beausejour CM, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25(11):1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 77.Brown MP, Topham DJ, Sangster MY, et al. Thymic lymphoproliferative disease after successful correction of CD40 ligand deficiency by gene transfer in mice. Nat Med. 1998;4(11):1253–1260. doi: 10.1038/3233. [DOI] [PubMed] [Google Scholar]
- 78.Cristea S, Freyvert Y, Santiago Y, et al. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng. 2013;110(3):871–880. doi: 10.1002/bit.24733. [DOI] [PubMed] [Google Scholar]
- 79.Maresca M, Lin VG, Guo N, Yang Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013;23(3):539–546. doi: 10.1101/gr.145441.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anguela XM, Sharma R, Doyon Y, et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood. 2013;122(19):3283–3287. doi: 10.1182/blood-2013-04-497354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sack BK, Herzog RW, Terhorst C, Markusic DM. Development of Gene Transfer for Induction of Antigen-specific Tolerance. Mol Ther Methods Clin Dev. 2014;1:14013. doi: 10.1038/mtm.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mohrin M, Bourke E, Alexander D, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7(2):174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hütter G, Nowak D, Mossner M, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360(7):692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 85.Long C, Amoasii L, Mireault AA, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2015 doi: 10.1126/science.aad5725. 351(6271):400-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tabebordbar M, Zhu K, Cheng JKW, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2015;351(6271):407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2015;351(6271):403-407. [DOI] [PMC free article] [PubMed]
- 88.Deng W, Lee J, Wang H, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149(6):1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deng W, Rupon JW, Krivega I, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158(4):849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526(7573):351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 91.Hoban MD, Orkin SH, Bauer DE. Genetic treatment of a molecular disorder: gene therapy approaches to sickle cell disease. Blood. 2016;127(7):839-848. [DOI] [PMC free article] [PubMed]
- 92.Zou J, Sweeney CL, Chou BK, et al. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011;117(21):5561–5572. doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kang EM, Malech HL. Gene therapy for chronic granulomatous disease. Methods Enzymol. 2012;507:125–154. doi: 10.1016/B978-0-12-386509-0.00007-7. [DOI] [PubMed] [Google Scholar]
- 94.Biffi A, Montini E, Lorioli L, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341(6148):1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 95.Sun J, Ramos A, Chapman B, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514(7522):322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Busch K, Klapproth K, Barile M, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518(7540):542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 97.Paul F, Arkin Y, Giladi A, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163(7):1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 98.Mukherjee S, Thrasher AJ. Gene therapy for PIDs: progress, pitfalls and prospects. Gene. 2013;525(2):174–181. doi: 10.1016/j.gene.2013.03.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sacco MG, Ungari M, Catò EM, et al. Lymphoid abnormalities in CD40 ligand transgenic mice suggest the need for tight regulation in gene therapy approaches to hyper immunoglobulin M (IgM) syndrome. Cancer Gene Ther. 2000;7(10):1299–1306. doi: 10.1038/sj.cgt.7700232. [DOI] [PubMed] [Google Scholar]
- 100.Sather BD, Romano Ibarra GS, Sommer K, et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci Transl Med. 2015;7(307):307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grupp SA, Kalos M, Barrett D, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maude SL, Frey N, Shaw PA, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Torikai H, Reik A, Liu PQ, et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119(24):5697–5705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Torikai H, Reik A, Soldner F, et al. Toward eliminating HLA class I expression to generate universal cells from allogeneic donors. Blood. 2013;122(8):1341–1349. doi: 10.1182/blood-2013-03-478255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Provasi E, Genovese P, Lombardo A, et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat Med. 2012;18(5):807–815. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Poirot L, Philip B, Schiffer-Mannioui C, et al. Multiplex genome edited T-cell manufacturing platform for “off-the-shelf” adoptive T-cell immunotherapies. Cancer Res. 2015;75(18):3853–3864. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
- 108.Qasim W, Jal Amrolia P, Samarashinghe S, et al. First clinical application of Talen engineered universal CAR19 T cells in B-ALL [abstract]. Blood. 2015;126(23). Abstract 2046. [Google Scholar]
- 109.Suzuki T, Arumugam P, Sakagami T, et al. Pulmonary macrophage transplantation therapy. Nature. 2014;514(7523):450–454. doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.High KA, Anguela XM. Adeno-associated viral vectors for the treatment of hemophilia. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv475. 35(R1):R36-R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arruda VR, Stedman HH, Haurigot V, et al. Peripheral transvenular delivery of adeno-associated viral vectors to skeletal muscle as a novel therapy for hemophilia B. Blood. 2010;115(23):4678–4688. doi: 10.1182/blood-2009-12-261156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang G, Shi Q, Fahs SA, Kuether EL, Walsh CE, Montgomery RR. Factor IX ectopically expressed in platelets can be stored in alpha-granules and corrects the phenotype of hemophilia B mice. Blood. 2010;116(8):1235–1243. doi: 10.1182/blood-2009-11-255612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McIntosh J, Lenting PJ, Rosales C, et al. Therapeutic levels of FVIII following a single peripheral vein administration of rAAV vector encoding a novel human factor VIII variant. Blood. 2013;121(17):3335–3344. doi: 10.1182/blood-2012-10-462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miyaoka Y, Chan AH, Judge LM, et al. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat Methods. 2014;11(3):291–293. doi: 10.1038/nmeth.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tenzen T, Zembowicz F, Cowan CA. Genome modification in human embryonic stem cells. J Cell Physiol. 2010;222(2):278–281. doi: 10.1002/jcp.21948. [DOI] [PubMed] [Google Scholar]
- 116.Vo LT, Daley GQ. De novo generation of HSCs from somatic and pluripotent stem cell sources. Blood. 2015;125(17):2641–2648. doi: 10.1182/blood-2014-10-570234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kerns HM, Ryu BY, Stirling BV, et al. B cell-specific lentiviral gene therapy leads to sustained B-cell functional recovery in a murine model of X-linked agammaglobulinemia. Blood. 2010;115(11):2146–2155. doi: 10.1182/blood-2009-09-241869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Holkers M, Maggio I, Liu J, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41(5):e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nault J-C, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47(10):1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- 120.Nightingale SJ, Hollis RP, Pepper KA, et al. Transient gene expression by nonintegrating lentiviral vectors. Mol Ther. 2006;13(6):1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 121.Pelascini LPL, Janssen JM, Gonçalves MAFV. Histone deacetylase inhibition activates transgene expression from integration-defective lentiviral vectors in dividing and non-dividing cells. Hum Gene Ther. 2013;24(1):78–96. doi: 10.1089/hum.2012.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Joglekar AV, Stein L, Ho M, Hoban MD, Hollis RP, Kohn DB. Dissecting the mechanism of histone deacetylase inhibitors to enhance the activity of zinc finger nucleases delivered by integrase-defective lentiviral vectors. Hum Gene Ther. 2014;25(7):599–608. doi: 10.1089/hum.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hendel A, Bak RO, Clark JT, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33(9):985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mandal PK, Ferreira LMR, Collins R, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15(5):643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Papapetrou EP, Zoumbos NC, Athanassiadou A. Genetic modification of hematopoietic stem cells with nonviral systems: past progress and future prospects. Gene Ther. 2005;12(Suppl 1):S118–S130. doi: 10.1038/sj.gt.3302626. [DOI] [PubMed] [Google Scholar]
- 126.Wang J, Exline CM, DeClercq JJ, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol. 2015;33(12):1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beane JD, Lee G, Zheng Z, et al. Clinical scale zinc finger nuclease-mediated gene editing of PD-1 in tumor infiltrating lymphocytes for the treatment of metastatic melanoma. Mol Ther. 2015;23(8):1380–1390. doi: 10.1038/mt.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.D’Astolfo DS, Pagliero RJ, Pras A, et al. Efficient intracellular delivery of native proteins. Cell. 2015;161(3):674–690. doi: 10.1016/j.cell.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 129.Han X, Liu Z, Jo MC, et al. CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci Adv. 2015;1(7):e1500454. doi: 10.1126/sciadv.1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu J, Gaj T, Yang Y, et al. Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells. Nat Protoc. 2015;10(11):1842–1859. doi: 10.1038/nprot.2015.117. [DOI] [PubMed] [Google Scholar]
- 131.Ramakrishna S, Kwaku Dad A-B, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24(6):1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li H, Haurigot V, Doyon Y, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475(7355):217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sharma R, Anguela XM, Doyon Y, et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126(15):1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Matsui H, Fujimoto N, Sasakawa N, et al. Delivery of full-length factor VIII using a piggyBac transposon vector to correct a mouse model of hemophilia A. PLoS One. 2014;9(8):e104957. doi: 10.1371/journal.pone.0104957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Omata D, Negishi Y, Suzuki R, Oda Y, Endo-Takahashi Y, Maruyama K. Nonviral gene delivery systems by the combination of bubble liposomes and ultrasound. Adv Genet. 2015;89:25–48. doi: 10.1016/bs.adgen.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 136.Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sun W, Ji W, Hall JM, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54(41):12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang X, Shin SC, Chiang AFJ, et al. Intraosseous delivery of lentiviral vectors targeting factor VIII expression in platelets corrects murine hemophilia A. Mol Ther. 2015;23(4):617–626. doi: 10.1038/mt.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Burtner CR, Beard BC, Kennedy DR, et al. Intravenous injection of a foamy virus vector to correct canine SCID-X1. Blood. 2014;123(23):3578–3584. doi: 10.1182/blood-2013-11-538926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Corrigan-Curay J, O’Reilly M, Kohn DB, et al. Genome editing technologies: defining a path to clinic. Mol Ther. 2015;23(5):796–806. doi: 10.1038/mt.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39(21):9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Joglekar AV, Hollis RP, Kuftinec G, Senadheera S, Chan R, Kohn DB. Integrase-defective lentiviral vectors as a delivery platform for targeted modification of adenosine deaminase locus. Mol Ther. 2013;21(9):1705–1717. doi: 10.1038/mt.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu X, Scott DA, Kriz AJ, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32(7):670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Knight SC, Xie L, Deng W, et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350(6262):823-826. [DOI] [PubMed]
- 145.Yu C, Liu Y, Ma T, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16(2):142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 148.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kohn DB. Eliminating SCID row: new approaches to SCID. Hematology Am Soc Hematol Educ Program. 2014;2014(1):475–480. doi: 10.1182/asheducation-2014.1.475. [DOI] [PubMed] [Google Scholar]
- 150.Perez EE, Wang J, Miller JC, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26(7):808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pattanayak V, Guilinger JP, Liu DR. Determining the specificities of TALENs, Cas9, and other genome-editing enzymes. Methods Enzymol. 2014;546:47–78. doi: 10.1016/B978-0-12-801185-0.00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gabriel R, Lombardo A, Arens A, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29(9):816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 155.Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33(2):179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim D, Bae S, Park J, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237–243, 1, 243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 158.Lodato MA, Woodworth MB, Lee S, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350(6256):94-98. [DOI] [PMC free article] [PubMed]
- 159.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39(1):381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bolukbasi MF, Gupta A, Wolfe SA. Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat Methods. 2015;13(1):41–50. doi: 10.1038/nmeth.3684. [DOI] [PubMed] [Google Scholar]
- 162.Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84-88. [DOI] [PMC free article] [PubMed]
- 163.Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]