Abstract
Despite nearly complete understanding of the genetics of the β-hemoglobinopathies for several decades, definitive treatment options have lagged behind. Recent developments in technologies for facile manipulation of the genome (zinc finger nucleases, transcription activator-like effector nucleases, or clustered regularly interspaced short palindromic repeats–based nucleases) raise prospects for their clinical application. The use of genome-editing technologies in autologous CD34+ hematopoietic stem and progenitor cells represents a promising therapeutic avenue for the β-globin disorders. Genetic correction strategies relying on the homology-directed repair pathway may repair genetic defects, whereas genetic disruption strategies relying on the nonhomologous end joining pathway may induce compensatory fetal hemoglobin expression. Harnessing the power of genome editing may usher in a second-generation form of gene therapy for the β-globin disorders.
Introduction
The β-hemoglobinopathies, namely sickle cell disease (SCD) and β-thalassemia, result from genetic mutations in the β-globin gene and are among the most common monogenic diseases in the world.1 SCD results from a nonsynonymous A to T mutation in codon 6 of the β-globin gene leading to a Glu-Val replacement,2,3 whereas β-thalassemias are caused by diverse point mutations or deletions.4-9 Treatment options are largely supportive. Transfusion and iron chelation are mainstays in the thalassemias whereas pain management, hydration, and hydroxyurea are used in SCD.10-16
The hemoglobin tetramer is composed of 2 α-like globin chains encoded by any of the 3 genes in the α-globin cluster on chromosome 16 and 2 β-like globin chains encoded from any of the 5 genes in the β-globin locus on chromosome 11. The expression of the 3 genes at the α-globin locus (ζ, α1, α2) and the 5 genes at the β-globin locus (ε, Gγ, Aγ, δ, β) are developmentally regulated. It has been appreciated for many years that levels of fetal hemoglobin (HbF; α2γ2), subject to developmental silencing in the months after birth, is a modifier of disease severity in patients with β-globin disorders.16-23 This protective effect of HbF has motivated the therapeutic strategy to reinduce its expression in adult life. Hydroxyurea, a cytotoxic agent that inhibits ribonucleotide reductase, induces HbF modestly through an unknown mechanism. However, it has dose-limiting myelosuppressive effects and some patients are nonresponders to therapy.10-13 Although bone marrow transplant (BMT) represents the sole established curative option for patients, its use is limited by donor availability and graft-versus-host disease (GVHD). A clinical trial has demonstrated successful gene addition of an antisickling form of β-globin to a transfusion-dependent βEβ0 thalassemia patient who gained transfusion independence as a result of gene transfer.24 Several additional somatic gene therapy trials for β-thalassemias and SCD are ongoing.25 Despite a deep understanding of molecular defects and gene control mechanisms, treatment options for the majority of patients remain limited.3
The emergence of designer nucleases for eukaryotic genome editing has ushered in an era of unprecedented control over the genome. The development of zinc finger nucleases (ZFNs),26-35 transcription activator-like (TAL) effector nucleases (TALENs),36-40 and meganucleases41-44 established genome editing as a valuable laboratory technique. The emergence of the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) nuclease system,45-49 which utilizes a single guide RNA (sgRNA) to direct the Cas9 nuclease for site-specific cleavage, has engendered tremendous excitement about potential clinical applications. The breakneck speed at which new variations on the general theme are developed is truly remarkable. Other Cas9-like systems include the CRISPR/Cpf1 nuclease platform,50 dimeric RNA-guided FokI nucleases,51,52 and use of Cas9s derived from a variety of prokaryotic species.53,54 It is unlikely that the discovery of novel CRISPR-based systems and Cas9-like nucleases capable of eukaryotic genome editing will end soon.55 The relative benefits of the newly developed CRISPR-based systems, ZFNs, and TALENs are still subject to debate. Although CRISPR-based systems are often cited as the most efficient,56 ZFNs are the only editing technology that has been brought thus far to a clinical trial. The CCR5 gene has been targeted by ZFNs in autologous CD4+ T cells from patients with HIV. The gene-modified cells were subsequently reinfused, which led to a decrease in the blood level of HIV in most patients.57 Notably, this study demonstrated that reinfusion of autologous genome-edited primary human cells could be achieved, well tolerated, and possibly lead to clinical benefit.
Genome-editing–based therapies rely on gene correction or disruption. Double-strand break (DSB) induction by an engineered nuclease is repaired by the endogenous repair pathways of homology-directed repair (HDR) or nonhomologous end joining (NHEJ).58 Genetic correction strategies exploit the HDR pathway to insert custom sequences into the genome through codelivery of an extrachromosomal repair template in conjunction with an engineered nuclease. The creation of a DSB improves HDR frequency.59 As such, wild-type (or customized) sequences can be provided as an extrachromosomal donor for repair following site-specific cleavage by the nuclease. In contrast, genetic disruption strategies rely on the NHEJ pathway following nuclease-induced DSB to produce local insertions/deletions (indels).47,48,58 Introduction of 2 engineered nucleases can result in targeted deletion or inversion, duplication, local indels at nuclease cleavage sites, or translocations/chromosomal rearrangements.60-73 Here, we review genome-editing approaches for genetic correction and disruption strategies for the β-hemoglobinopathies as summarized in Figure 1 and Table 1.
Figure 1.
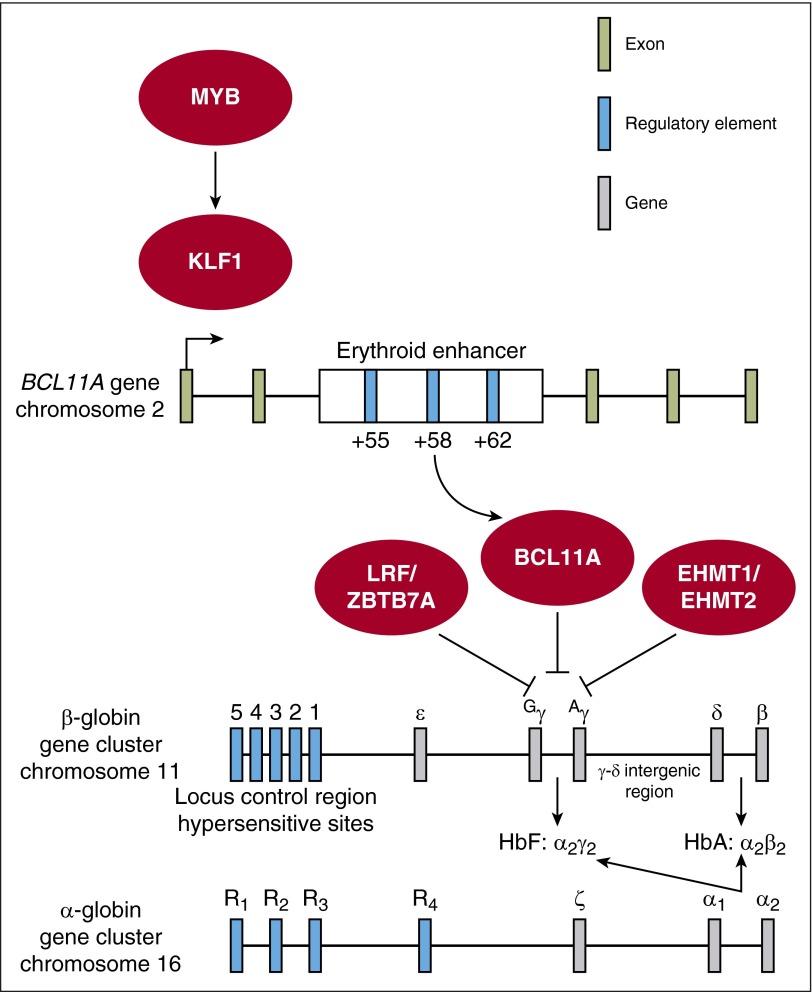
Network of potential targets for genome-editing–based therapy of the β-globin disorders. Therapeutic genome-editing strategies rely on genetic correction through the HDR pathway or genetic disruption through the NHEJ pathway. Genetic correction/repair strategies involve direct modification of the β-globin gene cluster through (1) correction of the sickle mutation in the β-globin gene or (2) insertion of the HPFH-associated single-nucleotide polymorphisms (SNPs) into the Gγ or Aγ promoters. Genetic disruption strategies involve targeted disruption of (1) BCL11A coding sequence, (2) the minimal critical sequences in the +58 DHS within the erythroid-specific BCL11A enhancer, (3) the HbF-associated sequences within the Aγ-δ intergenic region, or (4) other genes with a known role in γ-globin regulation such as MYB, KLF1, LRF/ZBTB7A, or EHMT1/EHMT2. HbA, adult hemoglobin.
Table 1.
Potential targets for therapeutic genome editing for the β-globin disorders
| Target | Repair strategy | Efficiency | Advantages/Disadvantages |
|---|---|---|---|
| Repair of SCD allele | HDR | Low-moderate | Single target allele; inadvertent generation of β-thalassemia alleles |
| Repair of β-thalassemia allele | HDR | Low-moderate | Heterogeneous target alleles |
| Recreation of nondeletional HPFH | HDR | Low-moderate | Inadvertent generation of γ-null alleles; identified HPFH patients support mutation tolerance/clinical benefit |
| Recreation of deletional HPFH | NHEJ | Low-moderate | Insufficient efficiency of targeted deletion; identified HPFH patients support mutation tolerance/clinical benefit |
| Other targets in β-globin cluster | NHEJ | — | Targets unknown |
| BCL11A | NHEJ | High | HSC/B-cell dysfunction due to BCL11A requirement; haploinsufficient patients have significant HbF induction |
| BCL11A enhancer | NHEJ | High | Erythroid-specific BCL11A loss; haploinsufficient patients have significant HbF induction |
| α-Globin | NHEJ | Moderate-high | Balance α-β chains; inadvertent generation of α-thalassemia cells |
| KLF1 | NHEJ | High | Broad role in cell proliferation and cellular development |
| MYB | NHEJ | High | Broad role in cell proliferation and cellular development |
| LRF/ZBTB7A | NHEJ | High | Broad role in cell proliferation and cellular development |
| EHMT1/EHMT2 | NHEJ | High | Role in hematopoiesis unknown |
| LIN28B pathway | NHEJ | High | Role in hematopoiesis unknown |
General strategies for SCD and β-thalassemia therapeutic genome editing
Correction of underlying genetic defects
The most appealing and theoretically straightforward application of genome editing for monogenic disorders is correction of a mutant DNA sequence and, in that manner, preservation of all intrinsic regulatory mechanisms acting on the gene of interest. Precise gene correction relies on HDR from an extrachromosomal template containing a wild-type gene sequence. Typically, the frequency of HDR is relatively low, and particularly low in CD34+ hematopoietic stem and progenitor cells (HSPCs) as discussed in the next section. However, gene correction strategies may benefit from mixed chimerism allogeneic transplant studies suggesting that low levels of chimerism can produce clinical benefit.74 Clinical development of such strategies requires optimizing efficiency and safety of correcting the sickle mutation, whereas the diverse spectrum of β-thalassemia point mutations and deletions necessitates optimization for each unique genetic target, a significant challenge for clinical translation.
Gene editing for reactivation of HbF in SCD and β-thalassemia
Elevated HbF is beneficial in SCD and β-thalassemia. Targets for manipulation include sequences lying within the β-globin cluster or within the genes encoding transcriptional regulators of globin gene expression (Figure 1; Table 1). Depending on the target, editing would rely on HDR or NHEJ. Suitability of each target relates to the ease with which the desired gene modifications can be generated and the extent to which the modifications reactivate HbF expression. In SCD, the goal is to induce sufficient HbF to prevent sickle hemoglobin (HbS) polymerization. In β-thalassemia, the aim is to replace deficient β-globin and thereby reduce globin chain imbalance.
Possible targets for editing
Genetic correction of the SCD and β-thalassemia mutations
Classical gene-targeting approaches have been used to repair the SCD mutation in embryonic stem cells,75 but this approach cannot be applied to CD34+ HSPCs due to low efficiency and the necessity to isolate and propagate faithful recombinants. Correction of genetic defects in cultured cells with an engineered nuclease and a donor repair template has been achieved for multiple disorders, including cystic fibrosis,76-79 Duchenne muscular dystrophy,80,81 ornithine transcarbamylase deficiency,82 hereditary tyrosinemia,83 and other diseases.71,84-87 Gene correction for SCD and β-thalassemia has also been accomplished in a laboratory setting.75,88-91 Of note, a recent study reported correction of an SCD allele at nearly 20% gene modification in CD34+ HSPCs upon delivery of a repair template via integration-deficient lentivirus or by DNA oligonucleotide electroporation in the presence of a β-globin–targeted ZFN. Similar levels of correction were observed in bone marrow cells isolated from SCD patients.88 Despite successful HDR in bulk cells in vitro, the levels of HDR were 10- to 20-fold reduced in the spleen and bone marrow of transplanted immunodeficient mice, suggesting that HDR within long-term engrafting hematopoietic stem cells (HSCs) was far less efficient than in downstream progenitors. Another study reported HDR rates of 17% to 43% at 2 genomic loci in fetal liver–derived or mobilized peripheral blood–derived cells via electroporation of ZFN messenger RNA (mRNA) in conjunction with an adeno-associated virus (AAV) donor repair template.92 These rates of HDR were maintained in vivo, suggesting the ability to perform HDR in primitive repopulating cells. Studies using AAV in conjunction with megaTALs,93 TAL effectors coupled to a sequence-specific homing endonuclease, demonstrated ∼14% rates of HDR in CD34+ HSPCs.94 Although megaTALs may enhance HDR through generation of 3′ DNA overhangs in HSPCs, the rate of HDR in repopulating HSCs has not been examined.
The relative efficiency of HDR vs NHEJ is critical to potential use of gene editing for gene correction. High rates of NHEJ-mediated indel formation are suboptimal for clinical translation of β-globin gene correction as the process creates the possibility of disruption of β-globin production and inadvertent generation of β-thalassemia alleles. Another consideration is that mutagenesis has also been observed in the highly homologous δ-globin gene in β-globin gene correction experiments, which may result in deletions and rearrangements affecting β-globin that may be difficult to detect by standard polymerase chain reaction (PCR)-based genotyping approaches.88
It is possible that small molecules that enhance HDR and/or inhibit NHEJ may improve the efficiency of gene correction within CD34+ HSPCs, so long as they do not impair cell engraftment capability.95-98 Another possibility is the use of asymmetric donor template DNA to enhance rates of HDR.99 NHEJ is the dominant pathway in G1, S, and G2 phases of the cell cycle, whereas HDR preferentially occurs during late S-phase and G2 phase when sister chromatid templates become available.100 Because HSCs, the rare long-term repopulating cells within CD34+ HSPC preparations, are largely quiescent, HDR is not favored. These observations are supported by the roles of BRCA1, PALB2, and BRCA2 in DSB repair. BRCA1 creates single-strand DNA through end resection and interacts with PALB2 to recruit BRCA2 and RAD51 to mediate HDR at sites of DSB. Identification of the cell cycle’s role in suppressing BRCA1 in the G1 phase supports the dominance of NHEJ repair in quiescent cells.101 However, restoration of the BRCA1-PALB2 interaction during the G1 phase can support HDR. Therefore, it may be possible to enhance HDR in quiescent HSCs through modulation of the BRCA1-PALB2-BRCA2 pathway.101 Moreover, 1 study demonstrated enhanced rates of HDR in HEK293T and nonhematopoietic primary cells through cell cycle synchronization to achieve nuclease-mediated cleavage during the optimal portions of the cell cycle for HDR.100 However, triggering proliferation in HSCs tends to impair their ultimate repopulating potential. Whether expansion of HSPC populations with small molecules such as SR1102,103 or UM171104 will allow for improved HDR efficiencies with concomitant retention of stem cell activity in vivo is as yet unknown.
Modification of the β-globin locus to recreate hereditary persistence of fetal hemoglobin
As would be anticipated from the existence of rare hereditary persistence of fetal hemoglobin (HPFH) alleles, genome-wide association studies (GWAS) have linked the β-globin cluster itself to HbF levels.105-110 This corroborated previous human genetic studies that identified HPFH patients with elevated HbF levels resulting from large deletions within the β-globin cluster.111-113 Re-creating the larger deletional HPFH alleles is impractical given their large size.60 However, opportunities may exist for targeting discrete regions of the β-globin gene cluster by NHEJ. Comparison of large deletions in the cluster that generate either HPFH or δβ-thalassemia phenotypes has implicated sequences in the Aγ-δ intergenic region as harboring silencers of γ-gene expression. Notably, study of 3 families with overlapping deletions in the β-globin cluster identified a 3.5-kb region between the Aγ and δ genes that may be essential for γ-globin repression. Additional indirect support was derived from chromatin immunoprecipitation–PCR experiments that suggest BCL11A binding within this region.111,114-117 At present, the optimal sequences in the cluster amenable for targeted deletion by editing and NHEJ have not been identified.
Several point mutations or small deletions in the Aγ or Gγ-globin gene promoters lead to persistence of HbF into adult life. HbF levels in heterozygotes with these nondeletional HPFH mutations may be as high as 30%.116,118-120 One of the strongest HPFH alleles (−175 T>C in the Aγ promoter) was recently created in cultured K562 cells with TALENs. Increased γ-globin production resulted, most likely through de novo generation of a TAL1-binding site that facilitated increased chromatin looping between the Aγ promoter and the locus control region.121 An HPFH allele with a small deletion in the Aγ promoter was re-created in CD34+ HSPCs with sgRNA and Cas9 expression, presumably due to microdeletion of a repeated sequence.122 Therapeutic genome editing to generate HPFH mutations is an attractive strategy as the effects of these mutations are known through study of families with these rare beneficial alleles. The approach, however, faces many of the same challenges as precise gene correction, given the apparent dominance of the NHEJ pathway at the expense of HDR efficiency in HSCs.
BCL11A targeting
BCL11A gene disruption
The GWAS-implicated transcription factor BCL11A is a validated repressor of HbF.105-110,114,123 Erythroid-lineage Bcl11a knockout in a mouse model of SCD led to pancellular HbF induction and phenotypic correction of a mouse model of SCD without perturbing other hematologic parameters.124 Haploinsufficient patients with microdeletions within the BCL11A locus have significant neurocognitive phenotypes as well as elevated HbF at levels near or above therapeutic thresholds.125,126 In principle, the genetic knockout of BCL11A by targeting BCL11A coding sequence in order to create frameshift null alleles represents a potential therapeutic strategy. Roles of BCL11A in nonhematopoietic lineages, including the neural lineage,127,128 pancreatic progenitors,129 and the breast epithelium,130 would not be problematic upon modification of BCL11A in autologous CD34+ HSPCs. However, this strategy is limited by extraerythroid roles of BCL11A in the hematopoietic system, including its requirement for B-cell development127,131-133 and HSC function.134,135 These roadblocks might be circumvented by use of erythroid-restricted expression of genome-editing components. A variation of this approach involves erythroid-specific, short hairpin RNA–mediated knockdown of BCL11A expression, which is under development as a gene-therapy strategy.136 Delivery of genome-editing tools stably to CD34+ HSPCs would be inadvisable due to potential insertional mutagenesis137 as well as elevated risk of off-target mutagenesis over time. Furthermore, the effects of long-term expression of ZFNs, TALENs, or CRISPR/Cas9 on CD34+ HSPCs are unknown.
BCL11A gene enhancer
Recent fine mapping of HbF-associated GWAS variants led to the identification of a developmental stage-specific, erythroid-restricted 12-kb region bearing a characteristic enhancer chromatin signature. This enhancer region is composed of 3 DNaseI hypersensitive sites (DHS), termed +55, +58, and +62 as their distance in kilobases from the BCL11A transcriptional start site. Deletion of the orthologous element in a murine erythroid cell line resulted in a complete loss of BCL11A at both the RNA and protein levels whereas expression was spared in a B-cell line with the same deletion.67 Subsequent deletion studies demonstrated a similar requirement for this element for BCL11A expression in human erythroid cells.138,139
BCL11A enhancer targeting has several distinct advantages over coding sequence disruption: (1) GWAS studies have demonstrated that variation in the BCL11A enhancer is associated with elevated HbF levels and is both common and well tolerated.67 (2) Targeted deletion of this element in a human erythroid cell line leads to loss of BCL11A expression and subsequent HbF induction nearly comparable to BCL11A null clones.138 (3) Targeted deletion of the murine +62 DHS within the Bcl11a erythroid enhancer results in delayed hemoglobin switching sparing expression in the brain and nonerythroid hematopoietic lineages.138 The +62 DHS knockout mice were viable and born in normal Mendelian ratios as compared with Bcl11a−/− knockout mice that are perinatal lethal likely due to neural defects.123,138 These results further highlight the erythroid specificity of this element in vivo.138 (4) Targeting the BCL11A enhancer has been shown to be better tolerated even within the erythroid lineage as compared with targeting the BCL11A coding sequence, suggesting a residual low level of BCL11A present after enhancer targeting is insufficient to repress γ-globin, but promotes cellular fitness.138,139
Therefore, an alternative approach to targeting BCL11A coding sequence might be targeted deletion of the 12-kb BCL11A erythroid enhancer.67 However, although targeted deletions from ∼1 kb to 1 Mb have been demonstrated to occur at an appreciable frequency, these are unlikely to occur at a sufficient frequency at clinical scale with current genome-editing technologies due to competing outcomes to deletion when using a dual nuclease strategy including scarring (multifocal indels), inversions, and duplications.47,48,60,68,140 Furthermore, the heterogeneous population of cells resulting from a dual nuclease strategy would be suboptimal for clinical translation.
Functional footprinting-informed targeting by ZFNs/TALENs within the BCL11A enhancer and comprehensive functional mapping of the BCL11A enhancer by CRISPR/Cas9-mediated saturating mutagenesis has revealed an “Achilles' heel” to the BCL11A enhancer within the +58 DHS.138,139 Disruption of this minimal functional sequence at the core of the DHS +58 by CRISPR/Cas9 or ZFNs/TALENs resulted in γ-globin induction comparable to targeting coding sequence in CD34+ HSPCs subject to erythroid differentiation conditions.138,139 The core region has been fine-mapped to an ∼20-bp region including a GATA1-binding motif which appears to be essential for BCL11A expression and subsequent HbF repression.138,139 As previously discussed, the erythroid specificity of the regulatory element would not require erythroid-specific expression of the genome-editing components, as would be necessary with a BCL11A coding sequence targeting approach. Taken together, targeting of the BCL11A enhancer at the functional core of +58 DHS in autologous CD34+ HSPCs followed by BMT represents a promising therapeutic strategy to induce HbF expression in patients with the β-globin disorders (Figure 2).
Figure 2.
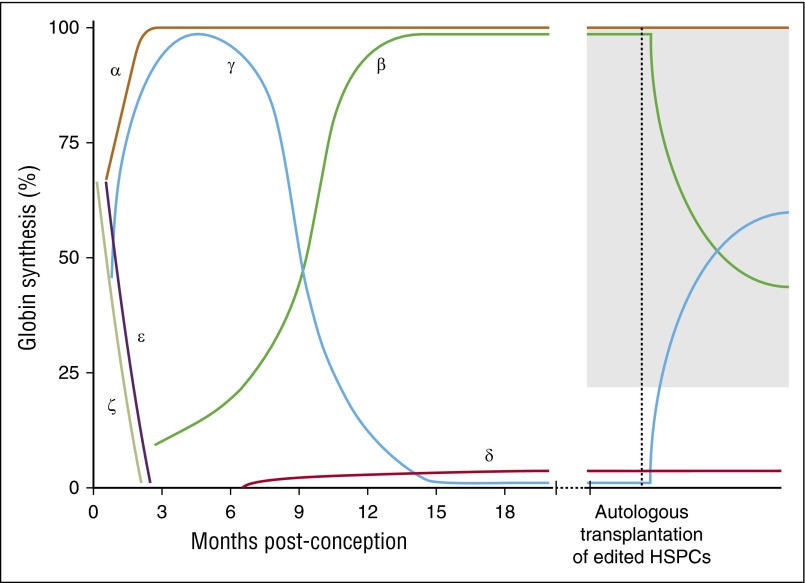
Reversal of hemoglobin switching to induce therapeutic levels of HbF. Reversal of hemoglobin switching can be accomplished through autologous bone marrow transplantation of genome-edited CD34+ HSPCs. The gray region indicates the hypothesized levels of HbF required for clinical benefit.
LRF/ZBTB7A gene disruption
Another transcription factor LRF/ZBTB7A (also referred to as Pokemon) has more recently been recognized as a major repressor of γ-globin.141 LRF-knockout mice exhibit elevated levels of the embryonic globin Hbb-βh1 with normal levels of Hbb-y. This contrasts from Bcl11a-null mice that exhibit elevation of both embryonic globins, Hbb-y > Hbb-βh1.123,124 Zbtb7a−/− mice are embryonic lethal due to anemia, whereas conditional knockout of Zbtb7a in adult mice leads to inefficient terminal erythropoiesis resulting in a mild macrocytic anemia.142 CRISPR/Cas9-mediated knockout of LRF in an erythroid cell line resulted in dramatic upregulation of γ-globin. LRF/BCL11A double knockout in this system was near additive and resulted in HbF of >90%, suggesting LRF’s role in γ-globin regulation is partially independent of BCL11A. Subsequent analysis demonstrated a mild delay in erythroid differentiation upon knockdown of LRF in primary human CD34+ HSPCs differentiated down the erythroid lineage with a corresponding induction of γ-globin.141 Although the effect of LRF loss on γ-globin is striking, the role of LRF in cell fate decisions in multiple hematopoietic lineages and its requirement for terminal erythropoiesis may limit its therapeutic potential.143
Reducing chain imbalance for β-thalassemias
The physiologic hallmark of β-thalassemia is globin chain imbalance, such that deficiency of β chains leads to precipitation of unstable, free α chains, membrane damage, hemolysis, and ineffective erythropoiesis.20,144-146 α-Thalassemia serves as a genetic modifier of β-thalassemia, as chain imbalance is reduced.146 This is supported by a milder disease course in patients with the co-inheritance of α-thalassemia and β-thalassemia.146-148 In principle, therefore, α-globin genes or their regulatory elements constitute potential targets for genome editing. Targeting an α-globin gene itself could result in a heterogeneous population of cells including those null for α-globin which might not support erythropoiesis. This approach could become a viable option if technological advancements allow for the precise control of the number of α-globin null alleles generated. However, with present editing tools, the inability to control the number of α-globin null alleles created through targeting α-globin coding sequences makes targeting known regulatory elements of α-globin or RNA interference–mediated knockdown of α-globin more attractive alternatives in this context.
Other potential therapeutic targets
Transcription factors KLF1 and MYB have previously been considered potential targets for HbF reactivation, but are not attractive due to their broad roles in cell proliferation and cellular development.112,113 Other genes such as EHMT1/EHMT2 and the LIN28B pathway have been implicated in the regulation of γ-globin; however, the selectivity of these targets and roles in hematopoiesis need further investigation.149-151
The platform: autologous bone marrow transplantation of genome-edited cells
Significant obstacles to wider use of BMT for cure of patients with β-globin disorders are the availability of compatible donors and risk of GVHD. Donor availability is particularly severe for SCD patients. The most persuasive rationale for therapeutic genome editing of β-globin disorders rests with the use of autologous CD34+ HSPCs as the cellular target. Through use of the patient’s own cells for therapy, donor availability and GVHD are avoided. As with more “conventional” somatic gene therapy with modified viruses, delivery of the requisite editing components to the target cells is the principal hurdle to be overcome in achieving clinical success.152 Delivery of therapeutic genes to CD34+ HSPCs has been accomplished with integrating and nonintegrating viral vectors (such as lentiviral, adenovirus, and AAV vectors), as well as physical methods (eg, electroporation).88,153-155 The optimal method for gene editing is currently unknown but is likely related to the specific technology used. High-efficiency delivery at clinical scale, roughly >108 CD34+ HSPCs cells, presents a practical challenge. However, recent studies have taken promising steps forward with electroporation of mRNA to CD34+ HSPCs at clinical scale (>1 × 108 cells).139,156 Robust cellular delivery is required for clinical translation of any envisioned therapeutic genome-editing approaches.
Genome editing is generally more difficult in primary cells as compared with immortal cell lines for reasons that are not entirely well understood, but may reflect inefficient delivery, diminished promoter activity of constructs, interferon responses, exonuclease activity, and host mechanisms of DNA repair.152 Electroporation of ZFNs, TALENs, and CRISPRs as DNA, RNA, and/or protein is an efficient delivery strategy to CD34+ HSPCs in a laboratory setting.88,92,140,152,157 For the CRISPR/Cas9 system, mRNA or ribonucleoprotein electroporation may obviate toxicity associated with DNA delivery, as well as yield higher rates of editing in cell lines and CD34+ HSPCs.100,152 The identification of novel Cas9 proteins isolated from diverse prokaryotes or other Cas9-like nucleases that are smaller than the widely used Streptococcus pyogenes–derived Cas9 may facilitate delivery efficiency, particularly for viral vectors.53 In addition, chemical modification of sgRNAs enhances editing efficiency in primary hematopoietic cells and CD34+ HSPCs.152
BMT poses risks to SCD and β-thalassemia patients beyond GVHD as reviewed in Lucarelli et al.158 Myeloablative or submyeloablative conditioning will be required to allow for engraftment of edited HSPCs. The inverse relationship between conditioning and engraftment rates suggests that myeloablative regimens maximize the likelihood of clinically beneficial engraftment rates. However, a myeloablative approach has elevated risk of BMT-associated morbidity and mortality.158 The optimal conditioning regimen for autologous BMT of genome-edited HSPCs requires investigation and may vary from patient to patient depending on the extent of end-organ damage resulting from vaso-occlusive events in SCD and/or iron overload in β-thalassemia patients. Given the risks associated with BMT, clinical guidelines for treatment may be stratified based on clinical disease severity for both SCD and β-thalassemia patients. BMT represents a viable therapeutic avenue in developed countries in spite of the associated risks, albeit the price of allogenic BMT exceeds $200 000 (in the United States).159 The extent of infrastructure and resources required for BMT restricts its wide use in developing countries,160,161 which have the highest prevalence of β-globin disorders.162
Steps to clinical translation
Although clinical translation of therapeutic genome editing for the β-hemoglobinopathies is appealing, several steps must be taken before the vision can become a reality. (1) Target selection, (2) delivery of editing reagents to HSCs, and (3) empirical testing of off-target potential must all be addressed and optimized. Targets that may be chosen for clinical development are summarized in Figure 1 and Table 1. (1) Strategies that rely on NHEJ are likely to be the first attempted using current technologies due to the dominance of NHEJ in quiescent HSCs and overall high efficiency of NHEJ as compared with HDR. At present, disruption of the core BCL11A enhancer sequences within the +58 DHS by NHEJ appears quite favorable in terms of potency of an effect on HbF expression and sparing of consequences for nonerythroid lineages. It may also be possible to combine an NHEJ-based approach with an HDR strategy or delivery of antisickling adult hemoglobin to enhance potential clinical benefit.163,164 (2) Transient delivery (electroporation or nonintegrating viral vectors) represents a safer alternative to stable integration of genome-editing components due to reduced risk of insertional mutagenesis and risk of off-target cleavage, as well as freedom from the uncertainty of long-term expression of genome-editing tools in CD34+ HSPCs. Transient delivery also necessitates high levels of on-target editing within a shorter window of time prior to loss of the genome-editing components through cell division. One possibility would be to enrich for edited cells prior to BMT of autologous cells,60 which could be further enhanced by strategies to expand HSCs ex vivo.102-104 (3) Off-target cleavages represent a legitimate concern for therapeutic genome editing. Newly developed techniques allow for unbiased genome-wide identification of off-target mutagenesis.165,166 Various methods have been reported that aim to enhance on-target vs off-target specificity. These include use of Cas9 nickase, truncated guides, dimeric RNA-guided FokI nucleases, and rationally engineered enhanced specificity Cas9.51,52,68,167-169 In addition, alternative RNA-directed nucleases (Cpf1) or modified Cas9 derivatives with reduced off-target cleavage potential appear to be steps toward “clean” editing reagents. It will be necessary to empirically test the optimized editing reagents for off-target cleavage potential and assess the associated risk of inappropriate DSBs within the genome. As methods to predict and detect off-target cleavages continue to improve, it may become possible to screen autologous genome-edited cells prior to BMT for possible pathogenic off-target mutations. “CD34+ humanized” mice, NOD-SCID-γ mice with bone marrow–engrafted human CD34+ HSPCs, can be used to evaluate the safety of genome-editing tools as these models can demonstrate multilineage reconstitution, self-renewal, and the ability to monitor leukemogenesis. However, due to the inability to model all human hematopoietic lineages, notably the erythroid lineage, and general limitations of chimera mouse models, humanized mice have limitations in assessing safety of genome-editing treatments in vivo. Furthermore, most off-target cleavages will exhibit a neutral effect on cellular fitness, whereas only rare off-target cleavages will be pathogenic, including induction of myelodysplastic syndrome or leukemic disorders. It will be important to enhance off-target cleavage prediction and detection methods to minimize risk of these rare pathogenic mutations.
One additional challenge for clinical development is harvesting CD34+ HSPCs for autologous stem cell transplantation from patients with β-globin disorders. Sufficient numbers of CD34+ HSPCs for BMT can be harvested from 2 sources, peripheral blood or bone marrow. Harvest of CD34+ HSPCs from peripheral blood is preferred due its minimal invasiveness and higher yield of CD34+ HSPCs following mobilization by granulocyte colony-stimulating factor (G-CSF).170 Use of G-CSF as a mobilizing agent is generally well tolerated for healthy adults and cancer patients. However, there are significant risks of G-CSF administration for patients with β-globin disorders. SCD patients have significant risk of vaso-occlusive events, acute chest syndrome, multiorgan system failure, and death,171 whereas β-thalassemia patients are susceptible to splenic rupture, hyperleukocytosis, and thrombosis.170 Plerixafor is an alternative mobilizing agent that may provide a safer option to G-CSF.170,172,173 Although the effect of plerixafor in SCD patients requires investigation, it has been shown to be safe and effective in both splenectomized and nonsplenectomized β-thalassemia patients. In contrast, although G-CSF was well tolerated in nonsplenectomized patients, it resulted in hyperleukocytosis and lower yield of CD34+ HSPCs as compared with plerixafor in splenectomized β-thalassemia patients.172 Combination of plerixafor with a reduced dose of G-CSF to avoid adverse effects has been shown to be superior to either agent alone.170,173 Therefore, plerixafor or combination G-CSF/plerixafor mobilization may provide a safe avenue for peripheral blood CD34+ HSPC harvests for β-thalassemia patients. Until acceptable protocols for mobilization of CD34+ HSPCs are established for SCD patients, traditional bone marrow harvesting may be required. It may be advisable as well to test ex vivo–editing efficiencies and maintenance of modified cells upon transfer into suitable immunodeficient mice for CD34+ HSPCs obtained by different methods to ensure optimization for clinical use.
Conclusions
The technological advances in genome manipulation are breathtaking in terms of the speed with which they have been reported in the past several years. The potential of genome-editing approaches for clinical benefit in the β-globin disorders is immense. Besides the choice of the editing platform and its delivery to repopulating cells within CD34+ HSPC harvests, a major factor in considering application to these conditions is the target sequences to be modified. If the goal is precise gene correction, the desired sequence alteration is clear. This strategy relies on HDR, and at the moment must await improved protocols for HDR in bona fide repopulating cells for clinical implementation.
Reactivation of HbF is an attractive approach, as it might be “one size fits all” in principle, suitable for both SCD and the β-thalassemias. The precise levels of pancellular HbF necessary for clinical benefit remain elusive, but is hypothesized to be ≥20% for SCD and likely somewhat higher in β-thalassemia (Figure 2). Due to the inability to precisely model HbF control experimentally, it may be difficult to assess the minimal threshold for clinical benefit in a laboratory setting. Furthermore, it is unlikely that HSCs undergoing therapeutic genome editing based on the strategies reviewed here will have a selective advantage in vivo. However, results from mixed chimerism allogeneic transplant demonstrate that low levels of chimerism can produce clinical benefit due to the survival advantage of normal red blood cells.74
Given the current state of genome-editing technologies, HbF induction mediated by NHEJ repair may provide a long-sought “silver bullet” for therapy. As such, harnessing the power of genome-editing tools may finally allow for therapeutic exploitation of the deep understanding of the genetics of hemoglobin and lead to a genome-editing–based therapeutic option for the β-hemoglobinopathies in the near future.
Acknowledgments
M.C.C. was supported by the National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases (F30DK 103359). S.H.O. was supported by the NIH National Heart, Lung, and Blood Institute (HL032259 and P01HL032262).
Authorship
Contribution: M.C.C. reviewed the literature; and M.C.C. and S.H.O. wrote the manuscript.
Conflict-of-interest disclosure: S.H.O. is an inventor on a patent related to this work. M.C.C. declares no competing financial interests.
Correspondence: Stuart H. Orkin, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: stuart_orkin@dfci.harvard.edu.
References
- 1.Bauer DE, Orkin SH. Hemoglobin switching’s surprise: the versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr Opin Genet Dev. 2015;33:62–70. doi: 10.1016/j.gde.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingram VM. A specific chemical difference between the globins of normal human and sickle-cell anaemia haemoglobin. Nature. 1956;178(4537):792–794. doi: 10.1038/178792a0. [DOI] [PubMed] [Google Scholar]
- 3.Orkin SH, Higgs DR. Medicine. Sickle cell disease at 100 years. Science. 2010;329(5989):291–292. doi: 10.1126/science.1194035. [DOI] [PubMed] [Google Scholar]
- 4.Orkin SH, Kazazian HH, Jr, Antonarakis SE, et al. Linkage of β-thalassaemia mutations and β-globin gene polymorphisms with DNA polymorphisms in human β-globin gene cluster. Nature. 1982;296(5858):627–631. doi: 10.1038/296627a0. [DOI] [PubMed] [Google Scholar]
- 5.Orkin SH, Kazazian HH., Jr The mutation and polymorphism of the human β-globin gene and its surrounding DNA. Annu Rev Genet. 1984;18:131–171. doi: 10.1146/annurev.ge.18.120184.001023. [DOI] [PubMed] [Google Scholar]
- 6.Wong C, Antonarakis SE, Goff SC, Orkin SH, Boehm CD, Kazazian HH., Jr On the origin and spread of β-thalassemia: recurrent observation of four mutations in different ethnic groups. Proc Natl Acad Sci USA. 1986;83(17):6529–6532. doi: 10.1073/pnas.83.17.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kazazian HH, Jr, Orkin SH, Antonarakis SE, et al. Molecular characterization of seven β-thalassemia mutations in Asian Indians. EMBO J. 1984;3(3):593–596. doi: 10.1002/j.1460-2075.1984.tb01853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonarakis SE, Kazazian HH, Jr, Orkin SH. DNA polymorphism and molecular pathology of the human globin gene clusters. Hum Genet. 1985;69(1):1–14. doi: 10.1007/BF00295521. [DOI] [PubMed] [Google Scholar]
- 9.Lie-Injo LE, Cai SP, Wahidijat I, et al. β-thalassemia mutations in Indonesia and their linkage to β haplotypes. Am J Hum Genet. 1989;45(6):971–975. [PMC free article] [PubMed] [Google Scholar]
- 10.Platt OS, Orkin SH, Dover G, Beardsley GP, Miller B, Nathan DG. Hydroxyurea enhances fetal hemoglobin production in sickle cell anemia. J Clin Invest. 1984;74(2):652–656. doi: 10.1172/JCI111464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letvin NL, Linch DC, Beardsley GP, McIntyre KW, Nathan DG. Augmentation of fetal-hemoglobin production in anemic monkeys by hydroxyurea. N Engl J Med. 1984;310(14):869–873. doi: 10.1056/NEJM198404053101401. [DOI] [PubMed] [Google Scholar]
- 12.Charache S, Terrin ML, Moore RD, et al. Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. N Engl J Med. 1995;332(20):1317–1322. doi: 10.1056/NEJM199505183322001. [DOI] [PubMed] [Google Scholar]
- 13.Platt OS. Hydroxyurea for the treatment of sickle cell anemia. N Engl J Med. 2008;358(13):1362–1369. doi: 10.1056/NEJMct0708272. [DOI] [PubMed] [Google Scholar]
- 14.Bunn HF. Pathogenesis and treatment of sickle cell disease. N Engl J Med. 1997;337(11):762–769. doi: 10.1056/NEJM199709113371107. [DOI] [PubMed] [Google Scholar]
- 15.Rees DC, Williams TN, Gladwin MT. Sickle-cell disease. Lancet. 2010;376(9757):2018–2031. doi: 10.1016/S0140-6736(10)61029-X. [DOI] [PubMed] [Google Scholar]
- 16.Rund D, Rachmilewitz E. β-thalassemia. N Engl J Med. 2005;353(11):1135–1146. doi: 10.1056/NEJMra050436. [DOI] [PubMed] [Google Scholar]
- 17.Platt OS, Brambilla DJ, Rosse WF, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330(23):1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 18.Platt OS, Thorington BD, Brambilla DJ, et al. Pain in sickle cell disease. Rates and risk factors. N Engl J Med. 1991;325(1):11–16. doi: 10.1056/NEJM199107043250103. [DOI] [PubMed] [Google Scholar]
- 19.Castro O, Brambilla DJ, Thorington B, et al. The Cooperative Study of Sickle Cell Disease. The acute chest syndrome in sickle cell disease: incidence and risk factors. Blood. 1994;84(2):643–649. [PubMed] [Google Scholar]
- 20.Musallam KM, Taher AT, Cappellini MD, Sankaran VG. Clinical experience with fetal hemoglobin induction therapy in patients with β-thalassemia. Blood. 2013;121(12):2199–2212. doi: 10.1182/blood-2012-10-408021. [DOI] [PubMed] [Google Scholar]
- 21.Galanello R, Sanna S, Perseu L, et al. Amelioration of Sardinian β0 thalassemia by genetic modifiers. Blood. 2009;114(18):3935–3937. doi: 10.1182/blood-2009-04-217901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson J. A study of sickling of young erythrocytes in sickle cell anemia. Blood. 1948;3(4):465–469. [PubMed] [Google Scholar]
- 23.Herman EC, Jr, Conley CL. Hereditary persistence of fetal hemoglobin. A family study. Am J Med. 1960;29:9–17. doi: 10.1016/0002-9343(60)90003-6. [DOI] [PubMed] [Google Scholar]
- 24.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467(7313):318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoban MD, Orkin SH, Bauer DE. Genetic treatment of a molecular disorder: gene therapy approaches to sickle cell disease. Blood. 2016;127(7):839–848. doi: 10.1182/blood-2015-09-618587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith J, Berg JM, Chandrasegaran S. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999;27(2):674–681. doi: 10.1093/nar/27.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161(3):1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300(5620):764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 30.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300(5620):763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 31.Urnov FD, Miller JC, Lee Y-L, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 32.Moehle EA, Rock JM, Lee Y-L, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases [published correction appears in Proc Natl Acad Sci USA. 2007;104(14):6090]. Proc Natl Acad Sci USA. 2007;104(9):3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 34.Miller JC, Holmes MC, Wang J, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Haurigot V, Doyon Y, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475(7355):217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 37.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 38.Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, Huang S, Jiang WZ, et al. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39(1):359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 41.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19(1):7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith J, Grizot S, Arnould S, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34(22):e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva G, Poirot L, Galetto R, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11(1):11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thierry A, Dujon B. Nested chromosomal fragmentation in yeast using the meganuclease I-Sce I: a new method for physical mapping of eukaryotic genomes. Nucleic Acids Res. 1992;20(21):5625–5631. doi: 10.1093/nar/20.21.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 46.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai SQ, Wyvekens N, Khayter C, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wyvekens N, Topkar VV, Khayter C, Joung JK, Tsai SQ. Dimeric CRISPR RNA-guided FokI-dCas9 nucleases directed by truncated gRNAs for highly specific genome editing. Hum Gene Ther. 2015;26(7):425–431. doi: 10.1089/hum.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10(11):1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33(6):661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12(4):393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bibikova M, Carroll D, Segal DJ, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21(1):289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Canver MC, Bauer DE, Dass A, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem. 2014;289(31):21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blasco RB, Karaca E, Ambrogio C, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Reports. 2014;9(4):1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 64.Xiao A, Wang Z, Hu Y, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41(14):e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta A, Hall VL, Kok FO, et al. Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 2013;23(6):1008–1017. doi: 10.1101/gr.154070.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20(1):81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bauer DE, Kamran SC, Lessard S, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity [published correction appears in Cell. 2013;155(2):479-480]. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andrey G, Kraft K, Geuer S, et al. Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in mice. Cell Reports. 2015;10:833–839. doi: 10.1016/j.celrep.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 71.Park C-Y, Kim DH, Son JS, et al. Functional correction of large factor VIII gene chromosomal inversions in hemophilia A patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17(2):213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 72.Li J, Shou J, Guo Y, et al. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol. 2015;7(4):284–298. doi: 10.1093/jmcb/mjv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L, Jia R, Palange NJ, et al. Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS One. 2015;10(3):e0120396. doi: 10.1371/journal.pone.0120396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andreani M, Testi M, Gaziev J, et al. Quantitatively different red cell/nucleated cell chimerism in patients with long-term, persistent hematopoietic mixed chimerism after bone marrow transplantation for thalassemia major or sickle cell disease. Haematologica. 2011;96(1):128–133. doi: 10.3324/haematol.2010.031013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang JC, Ye L, Kan YW. Correction of the sickle cell mutation in embryonic stem cells. Proc Natl Acad Sci USA. 2006;103(4):1036–1040. doi: 10.1073/pnas.0510177103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwank G, Koo B-K, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 77.Wu Y, Liang D, Wang Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13(6):659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 78.Firth AL, Menon T, Parker GS, et al. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Reports. 2015;12(9):1385–1390. doi: 10.1016/j.celrep.2015.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lokody I. Genetic therapies: correcting genetic defects with CRISPR-Cas9. Nat Rev Genet. 2014;15(2):63. doi: 10.1038/nrg3656. [DOI] [PubMed] [Google Scholar]
- 80.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345(6201):1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang Y, Wang L, Bell P, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34(3):334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin H, Song C-Q, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu Y, Zhou H, Fan X, et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25(1):67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Osborn MJ, Gabriel R, Webber BR, et al. Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum Gene Ther. 2015;26(2):114–126. doi: 10.1089/hum.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang C-W, Lai Y-S, Westin E, et al. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Reports. 2015;12(10):1668–1677. doi: 10.1016/j.celrep.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 87.Flynn R, Grundmann A, Renz P, et al. CRISPR-mediated genotypic and phenotypic correction of a chronic granulomatous disease mutation in human iPS cells. Exp Hematol. 2015;43(10):838–848. doi: 10.1016/j.exphem.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoban MD, Cost GJ, Mendel MC, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125(17):2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118(17):4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun N, Zhao H. Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng. 2014;111(5):1048–1053. doi: 10.1002/bit.25018. [DOI] [PubMed] [Google Scholar]
- 91.Xie F, Ye L, Chang JC, et al. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24(9):1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang J, Exline CM, DeClercq JJ, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol. 2015;33(12):1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boissel S, Jarjour J, Astrakhan A, et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42(4):2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sather BD, Romano Ibarra GS, Sommer K, et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci Transl Med. 2015;7(307):307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu C, Liu Y, Ma T, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16(2):142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pinder J, Salsman J, Dellaire G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43(19):9379–9392. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33(5):538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chu VT, Weber T, Wefers B, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33(5):543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 99.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34(3):339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 100.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Orthwein A, Noordermeer SM, Wilson MD, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528(7582):422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329(5997):1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wagner JE, Jr, Brunstein CG, Boitano AE, et al. Phase I/II trial of StemRegenin-1 expanded umbilical cord blood hematopoietic stem cells supports testing as a stand-alone graft. Cell Stem Cell. 2016;18(1):144–155. doi: 10.1016/j.stem.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fares I, Chagraoui J, Gareau Y, et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science. 2014;345(6203):1509–1512. doi: 10.1126/science.1256337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Menzel S, Garner C, Gut I, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39(10):1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 106.Uda M, Galanello R, Sanna S, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci USA. 2008;105(5):1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lettre G, Sankaran VG, Bezerra MA, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci USA. 2008;105(33):11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nuinoon M, Makarasara W, Mushiroda T, et al. A genome-wide association identified the common genetic variants influence disease severity in β0-thalassemia/hemoglobin E. Hum Genet. 2010;127(3):303–314. doi: 10.1007/s00439-009-0770-2. [DOI] [PubMed] [Google Scholar]
- 109.Solovieff N, Milton JN, Hartley SW, et al. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5′ olfactory receptor gene cluster. Blood. 2010;115(9):1815–1822. doi: 10.1182/blood-2009-08-239517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bhatnagar P, Purvis S, Barron-Casella E, et al. Genome-wide association study identifies genetic variants influencing F-cell levels in sickle-cell patients. J Hum Genet. 2011;56(4):316–323. doi: 10.1038/jhg.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sankaran VG, Xu J, Byron R, et al. A functional element necessary for fetal hemoglobin silencing. N Engl J Med. 2011;365(9):807–814. doi: 10.1056/NEJMoa1103070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sankaran VG, Orkin SH. The switch from fetal to adult hemoglobin. Cold Spring Harb Perspect Med. 2013;3(1):a011643. doi: 10.1101/cshperspect.a011643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bauer DE, Kamran SC, Orkin SH. Reawakening fetal hemoglobin: prospects for new therapies for the β-globin disorders. Blood. 2012;120(15):2945–2953. doi: 10.1182/blood-2012-06-292078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 115.Bank A. Regulation of human fetal hemoglobin: new players, new complexities. Blood. 2006;107(2):435–443. doi: 10.1182/blood-2005-05-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Forget BG. Molecular basis of hereditary persistence of fetal hemoglobin. Ann N Y Acad Sci. 1998;850:38–44. doi: 10.1111/j.1749-6632.1998.tb10460.x. [DOI] [PubMed] [Google Scholar]
- 117.Chakalova L, Osborne CS, Dai Y-F, et al. The Corfu deltabeta thalassemia deletion disrupts γ-globin gene silencing and reveals post-transcriptional regulation of HbF expression. Blood. 2005;105(5):2154–2160. doi: 10.1182/blood-2003-11-4069. [DOI] [PubMed] [Google Scholar]
- 118.Comi P, Giglioni B, Ottolenghi S, et al. Globin chain synthesis in single erythroid bursts from cord blood: studies on gamma leads to beta and G gamma leads to A gamma switches. Proc Natl Acad Sci USA. 1980;77(1):362–365. doi: 10.1073/pnas.77.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ottolenghi S, Nicolis S, Taramelli R, et al. Sardinian G gamma-HPFH: a T----C substitution in a conserved “octamer” sequence in the G gamma-globin promoter. Blood. 1988;71(3):815–817. [PubMed] [Google Scholar]
- 120.Martin DI, Tsai S-F, Orkin SH. Increased γ-globin expression in a nondeletion HPFH mediated by an erythroid-specific DNA-binding factor. Nature. 1989;338(6214):435–438. doi: 10.1038/338435a0. [DOI] [PubMed] [Google Scholar]
- 121.Wienert B, Funnell APW, Norton LJ, et al. Editing the genome to introduce a beneficial naturally occurring mutation associated with increased fetal globin. Nat Commun. 2015;6:7085. doi: 10.1038/ncomms8085. [DOI] [PubMed] [Google Scholar]
- 122.Traxler E, Yao Y, Li C, et al. Genome editing recreates hereditary persistence of fetal hemoglobin in primary human erythroblasts [abstract]. Blood. 2015;126(23). Abstract 640. [Google Scholar]
- 123.Sankaran VG, Xu J, Ragoczy T, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460(7259):1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu J, Peng C, Sankaran VG, et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334(6058):993–996. doi: 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Funnell APW, Prontera P, Ottaviani V, et al. 2p15-p16.1 microdeletions encompassing and proximal to BCL11A are associated with elevated HbF in addition to neurologic impairment. Blood. 2015;126(1):89–93. doi: 10.1182/blood-2015-04-638528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Basak A, Hancarova M, Ulirsch JC, et al. BCL11A deletions result in fetal hemoglobin persistence and neurodevelopmental alterations. J Clin Invest. 2015;125(6):2363–2368. doi: 10.1172/JCI81163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.John A, Brylka H, Wiegreffe C, et al. Bcl11a is required for neuronal morphogenesis and sensory circuit formation in dorsal spinal cord development. Development. 2012;139(10):1831–1841. doi: 10.1242/dev.072850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kuo TY, Chen CY, Hsueh YP. Bcl11A/CTIP1 mediates the effect of the glutamate receptor on axon branching and dendrite outgrowth. J Neurochem. 2010;114(5):1381–1392. doi: 10.1111/j.1471-4159.2010.06852.x. [DOI] [PubMed] [Google Scholar]
- 129.Benitez CM, Qu K, Sugiyama T, et al. An integrated cell purification and genomics strategy reveals multiple regulators of pancreas development. PLoS Genet. 2014;10(10):e1004645. doi: 10.1371/journal.pgen.1004645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khaled WT, Choon Lee S, Stingl J, et al. BCL11A is a triple-negative breast cancer gene with critical functions in stem and progenitor cells. Nat Commun. 2015;6:5987. doi: 10.1038/ncomms6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu P, Keller JR, Ortiz M, et al. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4(6):525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 132.Yu Y, Wang J, Khaled W, et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 2012;209(13):2467–2483. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin Y, Zhang Q, Zhang HM, et al. Transcription factor and miRNA co-regulatory network reveals shared and specific regulators in the development of B cell and T cell. Sci Rep. 2015;5:15215. doi: 10.1038/srep15215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Powers AN, Satija R. Single-cell analysis reveals key roles for Bcl11a in regulating stem cell fate decisions. Genome Biol. 2015;16:199. doi: 10.1186/s13059-015-0778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tsang JCH, Yu Y, Burke S, et al. Single-cell transcriptomic reconstruction reveals cell cycle and multi-lineage differentiation defects in Bcl11a-deficient hematopoietic stem cells. Genome Biol. 2015;16(1):178. doi: 10.1186/s13059-015-0739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guda S, Brendel C, Renella R, et al. miRNA-embedded shRNAs for lineage-specific BCL11A knockdown and hemoglobin F induction. Mol Ther. 2015;23(9):1465–1474. doi: 10.1038/mt.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118(9):3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Canver MC, Smith EC, Sher F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vierstra J, Reik A, Chang K-H, et al. Functional footprinting of regulatory DNA. Nat Methods. 2015;12(10):927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mandal PK, Ferreira LMR, Collins R, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15(5):643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Masuda T, Wang X, Maeda M, et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016;351(6270):285–289. doi: 10.1126/science.aad3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Maeda T, Ito K, Merghoub T, et al. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev Cell. 2009;17(4):527–540. doi: 10.1016/j.devcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lunardi A, Guarnerio J, Wang G, Maeda T, Pandolfi PP. Role of LRF/Pokemon in lineage fate decisions. Blood. 2013;121(15):2845–2853. doi: 10.1182/blood-2012-11-292037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Musallam KM, Sankaran VG, Cappellini MD, Duca L, Nathan DG, Taher AT. Fetal hemoglobin levels and morbidity in untransfused patients with β-thalassemia intermedia. Blood. 2012;119(2):364–367. doi: 10.1182/blood-2011-09-382408. [DOI] [PubMed] [Google Scholar]
- 145.Wilber A, Hargrove PW, Kim Y-S, et al. Therapeutic levels of fetal hemoglobin in erythroid progeny of β-thalassemic CD34+ cells after lentiviral vector-mediated gene transfer. Blood. 2011;117(10):2817–2826. doi: 10.1182/blood-2010-08-300723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mettananda S, Gibbons RJ, Higgs DR. α-Globin as a molecular target in the treatment of β-thalassemia. Blood. 2015;125(24):3694-3701. [DOI] [PMC free article] [PubMed]
- 147.Kan YW, Nathan DG. Mild thalassemia: the result of interactions of alpha and beta thalassemia genes. J Clin Invest. 1970;49(4):635–642. doi: 10.1172/JCI106274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thein SL. Genetic modifiers of the β-haemoglobinopathies. Br J Haematol. 2008;141(3):357–366. doi: 10.1111/j.1365-2141.2008.07084.x. [DOI] [PubMed] [Google Scholar]
- 149.Renneville A, Van Galen P, Canver MC, et al. EHMT1 and EHMT2 inhibition induces fetal hemoglobin expression. Blood. 2015;126(16):1930–1939. doi: 10.1182/blood-2015-06-649087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Krivega I, Byrnes C, de Vasconcellos JF, et al. Inhibition of G9a methyltransferase stimulates fetal hemoglobin production by facilitating LCR/γ-globin looping. Blood. 2015;126(5):665–672. doi: 10.1182/blood-2015-02-629972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee YT, de Vasconcellos JF, Yuan J, et al. LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood. 2013;122(6):1034–1041. doi: 10.1182/blood-2012-12-472308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hendel A, Bak RO, Clark JT, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33(9):985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nienhuis AW, Persons DA. Development of gene therapy for thalassemia. Cold Spring Harb Perspect Med. 2012;2(11):a011833. doi: 10.1101/cshperspect.a011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nienhuis AW. Development of gene therapy for blood disorders: an update. Blood. 2013;122(9):1556–1564. doi: 10.1182/blood-2013-04-453209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Papapetrou EP, Zoumbos NC, Athanassiadou A. Genetic modification of hematopoietic stem cells with nonviral systems: past progress and future prospects. Gene Ther. 2005;12(suppl 1):S118–S130. doi: 10.1038/sj.gt.3302626. [DOI] [PubMed] [Google Scholar]
- 156.Urnov FD, Reik A, Vierstra J, et al. Clinical-scale genome editing of the human BCL11A erythroid enhancer for treatment of the hemoglobinopathies [abstract].; Blood; 2015. Abstract 204. [Google Scholar]
- 157.Buechele C, Breese EH, Schneidawind D, et al. MLL leukemia induction by genome editing of human CD34+ hematopoietic cells. Blood. 2015;126(14):1683–1694. doi: 10.1182/blood-2015-05-646398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lucarelli G, Isgrò A, Sodani P, Gaziev J. Hematopoietic stem cell transplantation in thalassemia and sickle cell anemia. Cold Spring Harb Perspect Med. 2012;2(5):a011825. doi: 10.1101/cshperspect.a011825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cupit MC, Duncan C, Savani BN, Hashmi SK. Childhood to adult transition and long-term follow-up after blood and marrow transplantation. Bone Marrow Transplant. 2016;51(2):176–181. doi: 10.1038/bmt.2015.228. [DOI] [PubMed] [Google Scholar]
- 160.Faulkner LB. Setting up low-risk bone marrow transplantation for children with thalassemia may facilitate pediatric cancer care. South Asian J Cancer. 2013;2(3):109–112. doi: 10.4103/2278-330X.114098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mahmoud H, El-Haddad A, Fahmy O, et al. Hematopoietic stem cell transplantation in Egypt. Bone Marrow Transplant. 2008;42(suppl 1):S76–S80. doi: 10.1038/bmt.2008.136. [DOI] [PubMed] [Google Scholar]
- 162.Piel FB, Hay SI, Gupta S, Weatherall DJ, Williams TN. Global burden of sickle cell anaemia in children under five, 2010-2050: modelling based on demographics, excess mortality, and interventions. PLoS Med. 2013;10(7):e1001484. doi: 10.1371/journal.pmed.1001484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zuccato C, Breda L, Salvatori F, et al. A combined approach for β-thalassemia based on gene therapy-mediated adult hemoglobin (HbA) production and fetal hemoglobin (HbF) induction. Ann Hematol. 2012;91(8):1201–1213. doi: 10.1007/s00277-012-1430-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Breda L, Rivella S, Zuccato C, Gambari R. Combining gene therapy and fetal hemoglobin induction for treatment of β-thalassemia. Expert Rev Hematol. 2013;6(3):255–264. doi: 10.1586/ehm.13.24. [DOI] [PubMed] [Google Scholar]
- 165.Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33(2):179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yannaki E, Karponi G, Zervou F, et al. Hematopoietic stem cell mobilization for gene therapy: superior mobilization by the combination of granulocyte-colony stimulating factor plus plerixafor in patients with β-thalassemia major. Hum Gene Ther. 2013;24(10):852–860. doi: 10.1089/hum.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fitzhugh CD, Hsieh MM, Bolan CD, Saenz C, Tisdale JF. Granulocyte colony-stimulating factor (G-CSF) administration in individuals with sickle cell disease: time for a moratorium? Cytotherapy. 2009;11(4):464–471. doi: 10.1080/14653240902849788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yannaki E, Papayannopoulou T, Jonlin E, et al. Hematopoietic stem cell mobilization for gene therapy of adult patients with severe β-thalassemia: results of clinical trials using G-CSF or plerixafor in splenectomized and nonsplenectomized subjects. Mol Ther. 2012;20(1):230–238. doi: 10.1038/mt.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Karponi G, Psatha N, Lederer CW, et al. Plerixafor+G-CSF-mobilized CD34+ cells represent an optimal graft source for thalassemia gene therapy. Blood. 2015;126(5):616–619. doi: 10.1182/blood-2015-03-629618. [DOI] [PMC free article] [PubMed] [Google Scholar]