Abstract
Background
Anaplasma marginale is a well-known cattle pathogen of tropical and subtropical world regions. Even though, this obligate intracellular bacterium has been reported in other host species different than bovine, it has never been documented in Myrmecophaga tridactyla (giant anteater) or Hippocamelus antisense (taruca), which are two native endangered species.
Methods
Samples from two sick wild animals: a Myrmecophaga tridactyla (blood) and a Hippocamelus antisense (blood and serum) were studied for the presence of A. marginale DNA through msp5 gene fragment amplification. Further characterization was done through MSP1a tandem repeats analysis and MLST scheme and the genetic relationship among previously characterized A. marginale sequences were studied by applying, eBURST algorithm and AMOVA analysis.
Results
Anaplasma marginale DNA was identified in the Myrmecophaga tridactyla and Hippocamelus antisense samples. Through molecular markers, we identified an identical genotype in both animals that was not previously reported in bovine host. The analysis through eBURST and AMOVA revealed no differentiation between the taruca/anteater isolate and the bovine group.
Conclusions
In the present publication we report the identification of A. marginale DNA in a novel ruminant (Hippocamelus antisense) and non-ruminant (Myrmecophaga tridactyla) host species. Genotyping analysis of isolates demonstrated the close relatedness of the new isolate with the circulation population of A. marginale in livestock. Further analysis is needed to understand whether these two hosts contribute to the anaplasmosis epidemiology.
Keywords: Anaplasma marginale, Wild species, Taruca, Giant anteater
Background
Anaplasma marginale is an obligate intracellular pathogen from the phylum Proteobacteria, class alpha Proteobacteria, order Rickettsiales, and family Anaplasmataceae. Anaplasma marginale is known as an intraerythrocytic pathogen that causes moderate to severe hemolytic anemia, jaundice and hemoglobinuria without hemoglobinemia in cattle [1], although, other host species different than bovine were reported to be infected by this bacteria. In previous publications, Mazama gouazoubira, Blastocerus dichotomus, Syncerus caffer, Bubalus bubalis, Oryx gazella, Camelus dromedarius, Kobus vardonii and Aepyceros melampus have been described as A. marginale hosts [2–7]. The economic losses generated by anaplasmosis are not only associated with morbidity and mortality in cattle, but also with a lower weight gain rate, lower milk production, abortions and treatment costs. The identification of new hosts, the study of the genotypes associated to wild species and their interactions with genotypes frequently found in livestock could improve the complete understanding of the eco-epidemiology of anaplasmosis, and will be beneficial for surveillance and disease control. In the present study, we describe the identification of A. marginale DNA in two wild species (Myrmecophaga tridactyla and Hippocamelus antisensis) where this pathogen has not been previously reported. After molecular diagnosis of A. marginale by amplifying a fragment of the msp5 gene, both isolates were characterized applying two different molecular markers. Through MSP1a tandem repeats analysis and the A. marginale MLST scheme we were able to characterize the genotypes and study their geographical distribution. Results suggest that A. marginale could be circulating in Myrmecophaga tridactyla and Hippocamelus antisensis and that there seems to be directionality in the transmission of certain genotypes from cattle to these wild species.
Methods
Case reports
Case 1
In May 2013, as part of a reintroduction program carried out in Corrientes province, Argentina, a 20 days old female of giant anteater (Myrmecophaga tridactyla) (Xenarthra, Myrmecophagidae) had been released from illegal trafficking and transferred from Santiago del Estero province (Argentina) to the rescue center in Corrientes province (Argentina) (Fig. 1). One year later, a blood smear examination showed intraerythrocytic structures suggestive of A. marginale (Fig. 2). Finally, on 28 October 2014, the anteater died and venous blood samples were remitted to our laboratory for A. marginale molecular diagnosis.
Fig. 1.
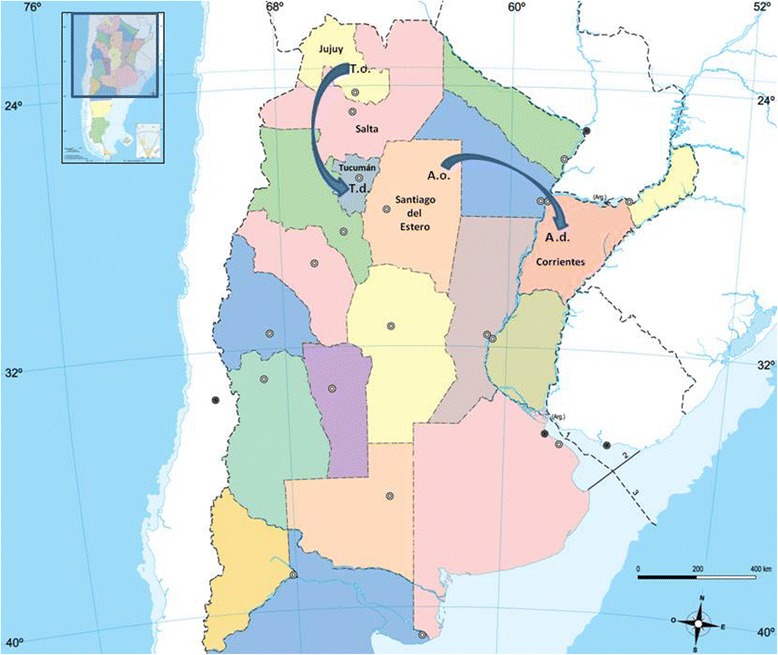
Map representing the north of Argentina and the taruca and the giant anteater movements. T.o: Taruca origin, T.d: Taruca destination, A.o: anteater origin and A.d: anteater destination
Fig. 2.
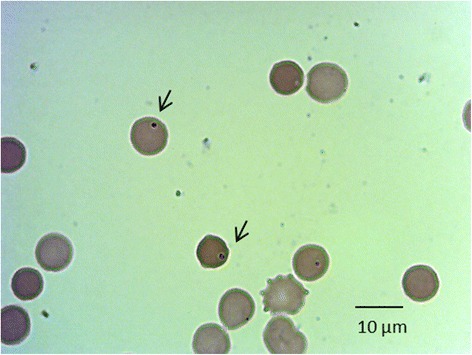
Blood smear from Myrmecophaga tridactyla. The arrows point out spherical inclusions suggestive of A. marginale. May Grunwald-Giemsa-Giemsa, 100× oil immersion
Case 2
In October 2012, a nine month old north Andean deer “taruca” (Hippocamelus antisensis) (Cetartiodactyla, Cervidae) arrived to the “Reserva Experimental de Flora y Fauna de Horco Molle” in Tucuman province (Argentina) as a result of a wildlife rescue procedure. The original location of the cervid was a forest area in Jujuy province (Argentina) (Fig. 1). The cervid underwent a pre-surgical analysis and a significant low PCV value was detected (17 %) in the absence of bleeding history or bloody feces. Intraerythrocytic structures suggestive of A. marginale were observed after blood smear examination (Fig. 3) and blood samples were remitted to our laboratory for A. marginale molecular diagnosis. Also, a frozen stored serum sample that has been taken previously (in May 2012) was sent as a background sample. In March 2014 the cervid died as a result of an infected myiasis in the head and subsequent sepsis.
Fig. 3.
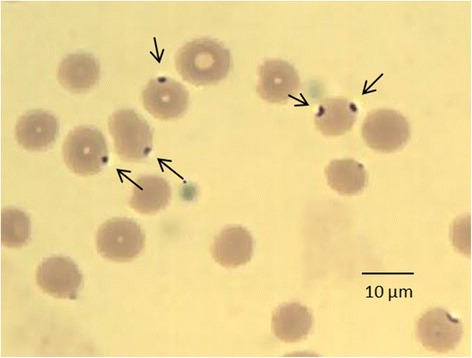
Blood smear from Hippocamelus antisensis. The arrows point out spherical inclusions suggestive of A. marginale. May Grunwald-Giemsa-Giemsa, 100× oil immersion
In both cases, samples were collected for routine diagnostic purposes following institutional guidelines. Capture and transit permits (Reference numbers: 1140 and 000336) were obtained from the provincial government through Natural Resources Agency of Corrientes and Tucuman, respectively.
Samples and genomic DNA isolation
Blood samples from the taruca and the giant anteater (one per animal) and a serum sample from the taruca were received and analyzed in our laboratory for A. marginale identification. The genomic DNA extraction from blood and serum samples was performed by phenol/chloroform method and a standard ethanol precipitation [8].
PCR assays for A. marginale
The first assay was based on the amplification of a fragment from a single copy gene that encodes the outer major surface protein MSP5 from A. marginale. Reaction and amplification conditions were carried out as described by Torioni de Echaide et al. [9]. For further isolate characterization two molecular marker schemes were employed. First, msp1α gene was amplified and sequenced for identifying the number and type of tandem repeats in the 5’region of the gene, according to the protocol described by [10]. Secondly, a multilocus sequence type (MLST) scheme based on the allelic polymorphism of seven A. marginale housekeeping genes (dnaA, ftsZ, lipA, groEl, recA, secY and sucB) was applied [11]. The nucleotide sequence of the primers employed and the resulting amplicon size are shown in Table 1.
Table 1.
Primers employed for A. marginale identification and characterization
| Gene | Product | Primer sequence | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| msp5 | major surface protein MSP5 | F: 5' GCATAGCCTCCCCCTCTTTC 3' | 548 | [9] |
| R: 5' TCCTCGCCTTGCCCCTCAGA 3' | ||||
| F: 5' TACACGTGCCCTACCGACTTA 3' | 345 | |||
| R: 5' TCCTCGCCTTGCCCCTCAGA 3' | ||||
| msp1α | major surface protein MSP1a | F: 5'GCATTACAACGCAACGCTTGAG 3' | 84–87 bp tandem repeats | [10] |
| R: 5'GCTTTACGCCGCCGCCTGCGCC 3' | ||||
| dnaA | Chromosomal replication initiation protein | F: 5' GTTCATAAGCGGGAAGGACA 3' | 512 | [11] |
| R: 5' CTTGTCTCGGTCTGGCTAGG 3' | ||||
| ftsZ | Cell division protein FtsZ | F: 5' CCTGACCACCAATCCGTATC 3' | 575 | |
| R: 5' CCCGTATGAAGCACCGTATC 3' | ||||
| groEl | Chaperonin GroEL | F: 5' AGCATAAAGCCCGAGGAACCTT 3' | 699 | |
| R: 5' GCCGAGCATGTCCTTCCTTCTG 3' | ||||
| lipA | Lipoyl synthase | F: 5' TGTGGATAGGGACGACCTTC 3' | 538 | |
| R: 5' AAAGTCATCCTCAGCGTGGT 3' | ||||
| recA | Recombinase A | F: 5' GGGCGGTAACTGTGCTTTTA 3' | 579 | |
| R: 5' ACGCCCATGTCGACTATCTC 3' | ||||
| secY | Preprotein translocase subunit SecY | F: 5' TTCACGCTGCTAGCCCTAAT 3' | 501 | |
| R: 5' TACGAGGGAAATGCCGTTAC 3' | ||||
| sucB | Dihydrolipoamide acetyltransferase component | F: 5' GAGATAGCATCTCCGGTTGC 3' | 808 | |
| R: 5' CTCCCCTGGCCTTTTTACTC 3' |
Molecular markers analysis
Multiple alignments of the msp1α sequences obtained for the taruca and the anteater isolates were performed using the Clustal W2 software (EMBL-EBI, Wellcome Trust Genome Campus, Hinxton, Cambridgeshire, UK). After the alignment, nucleotide sequences were translated to amino acid sequences in order to identify the tandem repeats. The repeat patterns were compared to those previously published [12, 13].
For automate MLST-DNA sequence editing and analysis, a custom-designed bioinformatic “Galaxy MLST-Pipeline” has been employed (http://bioinformatica.inta.gov.ar/galaxy/) [11]. The information obtained from this pipeline was then used for inferring relatedness between allelic profiles through the eBURST algorithm [14]. The sequence type (ST) from 74 A. marginale strains was retrieved from the MLST-Pipeline database (MLST-DB) (http://bioinformatica.inta.gov.ar/mlst/) for the study of the genotype diversity and distribution of the new isolates. This database includes information relating to strains from cattle of diverse geographic origins (USA, Puerto Rico, Italy, South Africa, Mexico, Colombia, Cuba, Brazil, Argentina and Uruguay). Applying the PHYLOViZ program [15], a triple locus variant (TLV) criteria was employed for the clonal complexes (CC) construction, meaning that those related genotypes that differ in up to three genes from the founder genotype will be arranged in the same CC. Also the full MLST was run; this option links all STs through the construction of a Minimum Spanning Tree and specifying the number of alleles that are different between each pair of STs. Also, the Arlequin 3.1 program [16] was employed to assess how much of the total genetic variation (measured by ST) was partitioned between the isolates through AMOVA [17]).
Results
Diagnosis and genotyping of A. marginale
Samples from the taruca (blood and serum) and the giant anteater (blood), tested for A. marginale with the msp5 specific assay, were PCR-positive (Fig. 4). After confirming the presence of A. marginale DNA, both isolates were characterized from blood samples by the msp1α marker and the MLST scheme. The MSP1a tandem repeat profile of the taruca and the anteater displayed the same genotype. The MSP1a profile consisted on two tandem repeats (repeat 13 and repeat 27) that were previously found in other genotypes, but were not previously reported in this combination, thus resulting in a new MSP1a genotype (Table 2).
Fig. 4.
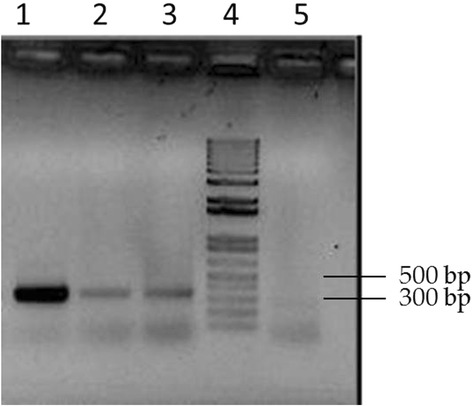
Agarose gel electrophoresis of msp5 gene amplification. 1 % agarose. Lane 1: positive control; Lane 2: anteater sample; Lane 3: taruca sample; Lane 4: Molecular marker (1 Kb plus); Lane 5: negative control
Table 2.
Amino acid sequence for MSP1a tandem repeats found for the taruca and the giant anteater A. marginale isolates
| Repeat | Encoded sequence | Number of repeats found |
|---|---|---|
| 13 | TDSSSASGQQQESSVLSQSDQASTSSQLG | 1 |
| 27 | ADSSSASGQQQESSVLSQSDQASTSSQLG | 1 |
For the MLST scheme, both isolates were assigned the same ST. Each one of the seven alleles was already registered in the MLST-DB, though the combination of alleles retrieved a new ST that resulted unique for the taruca and the anteater A. marginale isolate (Table 3). Sequences from msp1α and from the seven MLST genes were deposited in GenBank (submission ID: 1892187).
Table 3.
Alleles found for each gene and the resulting MLST scheme ST
| ST | dnaA | ftsZ | groEl | lipA | recA | secY | sucB |
|---|---|---|---|---|---|---|---|
| 69 | 3 | 3 | 2 | 2 | 13 | 4 | 4 |
Genetic relationship among isolates
Applying eBURST algorithm with a TLV criteria, a unique CC was found. USA isolates were arranged together around ST 4 (Puerto Rico). The rest of the CC was comprised of STs from diverse Latin-American countries (Argentina, Brazil, Uruguay, Mexico, Cuba and Colombia) and two STs from Italia (STs 39 and 40), all of them arranged around the ST 11 (Argentina). In particular, ST 69, which corresponds to the taruca and the giant anteater isolates, was established as a link between one ST from Brazil (ST 34) and two STs from northwest Argentina (STs 20 and 44), specifically from Salta, a neighboring province of Jujuy (Figs. 1 and 2). The South African ST (ST 33), two STs from Italy (STs 38 and 41) and one from Argentina (ST 27) remained as singletons (not integrated to the CC). The full MLST run showed the two main parts of the CC linked through ST 54 from Cuba (Fig. 5). Thus, we considered the genotypes that differ in up to two genes from the founder genotype (double locus variant criteria) to come up with a CC comprised of STs from Latin-American countries and the taruca/anteater isolates (STs linked by 1 and 2, Fig. 5). We compute the pairwise population comparisons of genetic differentiation between populations (FST), and no differentiation was shown between the taruca/anteater isolate and the bovine group (FST = -0.04699), revealing that most of the genetic diversity was observed among individuals within populations.
Fig. 5.
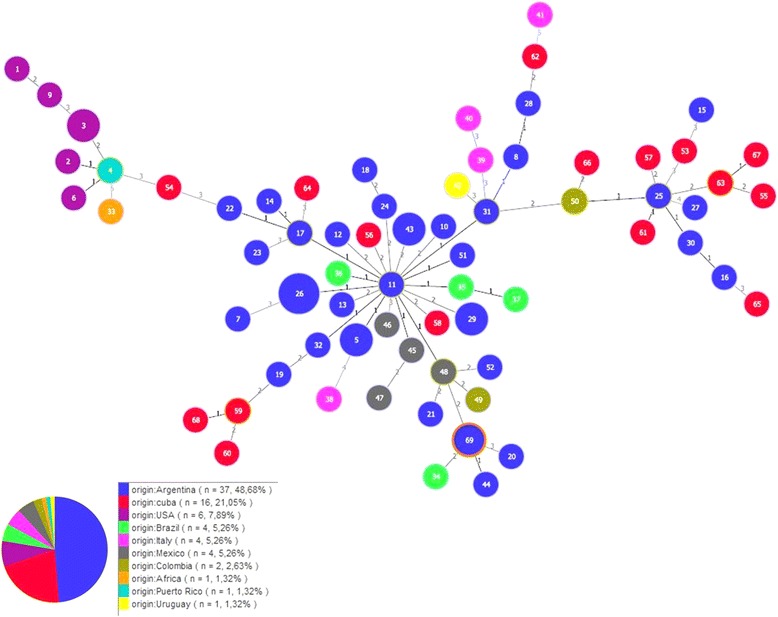
eBURST representation of genetic relationship among isolates. ST 69 is highlighted with an orange circle
Discussion
The comprehensive study of the anaplasmosis epidemiology needs the identification of all the involved factors, including susceptible, carrier and potential reservoir hosts. Herein, the evidence of A. marginale presence in the taruca and the giant anteater samples was supported not only by smear examination but remarkably by the PCR amplification of nine different gene sequences: one gene fragment currently used for diagnosis (msp5) and eight for molecular typing (msp1a and the seven neutral fragments from MLST scheme). Notably, the genotyping data revealed the presence of the same A. marginale isolate in both wild species.
Relevant information that arises from the results obtained here is the presence of A. marginale DNA in the taruca serum sample. Although serum samples are not ideally suited for genomic DNA extraction, especially from intraerythrocytic agents, we could manage to obtain a sufficient amount of A. marginale DNA for PCR detection. This fact probably has to do with the presence of the dense (infective) forms of A. marginale, which are able to survive outside the erythrocyte before parasitizing another cell [1]. The presence of A. marginale DNA in the taruca serum is an important fact since this sample was obtained two years before the blood sample, when the cervid was in its original location in Jujuy province, indicating that the taruca was already infected when moved to Horco Molle in Tucuman province.
Even though A. marginale is the most prevalent tick-borne disease in livestock worldwide and studies of its impact are mainly focused in domestic cattle. [1], this bacteria has also been detected in blood from other wild mammalian ruminant species such as Mazama gouazoubira, Blastocerus dichotomus, Syncerus caffer, Bubalus bubalis, Oryx gazella, Camelus dromedarius, Kobus vardonii and Aepyceros melampus [2–7, 18]. To date, there are no reports describing the presence of A. marginale in a taruca (Hippocamelus antisensis) and/or giant anteater (Myrmecophaga tridactyla), two native endangered species. Moreover, the finding in the giant anteater is the first demonstration of A. marginale in a non-ruminant species. Other Anaplasma species, A. phagocytophilum, has been found in a great diversity of hosts including human. This species has been considered a generalist pathogen with highly adaptive strategies that enables this bacterium to infect a wide range of hosts [18]. Recently, employing the groEL gene as a molecular marker Jahfari et al. [19] could discriminate four different A. phagocytophilum ecotypes with significantly different host ranges and zoonotic potential; these findings suggest that certain genotypes may be adapted to specific host species. It is likely that, this ‘wide host adaptation’ ability may be a common feature to other Anaplasma species, and this could explain why A. marginale is found in other host species different than cattle.
In Argentina the enzootic area for A. marginale, extends from the northern boundary of the country to the parallel 33 ° S. The main vector implicated in anaplasmosis transmission is Rhipicephalus microplus, an Ixodidae tick distributed through the north of the country. While not in anteater, there are reports of R. microplus parasitizing various cervid species [20, 21]. Also, other tick genus has been implicated as A. marginale vectors [22]. In this sense, Amblyomma cajennense, could be a potential A. marginale vector [23] and was previously reported parasitizing cattle [21, 24], giant anteater [24–26] and cervids [20, 21], highlighting the importance of following up its role as a source of infection of A. marginale for domestic and wild species. Moreover, A. cajennense is well distributed through Jujuy, Tucuman and Santiago del Estero province in Argentina [27]. In addition to ticks, other hematophagous insects could cause mechanical transmission of A. marginale [1]. In fact, Tabanus spp. is well distributed over South America and may act as a mechanical vector between cattle and new hosts species. In the two cases reported in this study, no ticks or other hematophagous arthropod were found parasitizing the taruca nor the giant anteater neither arriving at rescue centers or during their stay there.
Additionally we provide information regarding epidemiological analysis through genotyping of both isolates and comparing them with cattle genotypes reported to date. In this sense, the ST 69 found in both wild species resulted in a new variant consisting of seven alleles previously described in cattle from Argentina and other world regions [11]. Similarly, the two tandem repeats found through the MSP1a marker (repeat 13 and 27) were previously published for A. marginale isolates in cattle from Argentina, Mexico, Brazil and South Africa [12], but, as they had never been found together, their combination resulted in a novel genotype.
As we have previously reported [11], there is a broad genetic diversity of A. marginale isolates in bovine host. Moreover, the chance of finding the same genotype in cattle samples is scarce (Fig. 5 and [12]). Identical isolates have only been detected in samples collected in the same herd at the same time point (ST 26, 29 and 43). However, taken together, the information obtained from the msp1α genotype, the eBURST and AMOVA analysis highlights that, although different, the newly emerged genotype found in wild species is not separated from the rest of genotypes found in bovines. This finding might suggest that transmission of the new A. marginale isolate from cattle to accidental hosts could have arisen by means of a better transmission vector-competent isolate or by the emergence of a favored variant with an improved fitness for wider hosts. Further evidence is needed to support this hypothesis, mainly through the characterization of other A. marginale isolates from a greater number of wild host species.
To our knowledge, this is the first demonstration of A. marginale DNA in a taruca and a giant anteater. Moreover, for the taruca, A. marginale was detected over time, since the bacterium DNA was present in a serum sample and in a blood sample collected two years after.
Since the anteater and the taruca sustained other pathogens simultaneously, we are unable to affirm that A. marginale has infected or was the cause of death of the animals. Even though DNA from A. marginale has been detected from both taruca and anteater blood samples, whether these two hosts species contribute to the anaplasmosis epidemiology needs further investigation.
Conclusions
Our study demonstrated the presence of A. marginale DNA in two up to now non-acknowledged host species: Hippocamelus antisense and Myrmecophaga tridactyla. Moreover, this is the first report of A. marginale in a non-ruminant host. The sequence analysis of the molecular markers revealed that the same genotype was present in both the cervid and the anteater, and further analysis demonstrated the close relatedness of the new isolate with the circulation population of A. marginale in livestock. On one hand, these results could be an evidence of anaplasmosis as a threat for wild life and additionally could represent a sign of alternative mammalian reservoir for A. marginale. The acceptance of this hypothesis is worth an epidemiological base search and analysis.
Acknowledgements
We thank Dr. Verónica Lia for valuable suggestions.
Funding
This work was financed by grants from the Argentine Ministry of Science and Technology (PICT 0932) and INTA PNBIO (1131043 and 1131032). Salaries were provided by CONICET (ECG, LLA., SW and MF).
Authors’ contributions
ECG carried out part of the molecular genetic studies, ran the eBURST analysis and drafted the manuscript. SF carried out part of the molecular genetic studies and helped to draft the manuscript. MO participated in the design of the study and revised the manuscript critically. JPM carried out the giant anteater sampling and drafted the part of the manuscript referred to it clinical case. EC carried out the taruca sampling and drafted the part of the manuscript referred to it clinical case. JF participated in the sequence alignments. LLA carried out the protein sequence alignment and its analysis. MP carried out the nucleotide sequence alignment and its analysis. CB participated in the eBURST run and in the analysis of it results. VP carried out part of the PCR amplifications. SEW participated in the design of the study and revised the manuscript critically. MDF conceived of the study and gave the final approval of the version to be published. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
In both cases, samples were sent to the laboratory for routine diagnostic purposes following institutional guidelines. Capture and transit permits (Reference numbers: 1140 and 000336) were obtained from the provincial government through Natural Resources Agency of Corrientes and Tucuman, respectively.
References
- 1.Kocan KM, de la Fuente J, Guglielmone AA, Meléndez RD. Antigens and alternatives for control of Anaplasma marginale infection in cattle antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin Microbiol Rev. 2003;16:698–712. doi: 10.1128/CMR.16.4.698-712.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eygelaar D, Jori F, Mokopasetso M, Sibeko KP, Collins NE, Vorster I, Troskie M, Oosthuizen MC. Tick-borne haemoparasites in African buffalo (Syncerus caffer) from two wildlife areas in Northern Botswana. Parasit Vectors. 2015;8:1–11. doi: 10.1186/s13071-014-0627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudan V, Sharma RL, Borah MK. Subclinical anaplasmosis in camel (Camelus dromedarius) and its successful therapeutic management. J Parasit Dis. 2014;38:163–5. doi: 10.1007/s12639-012-0206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashraf QU a, Khan AU, Khattak RM, Ali M, Shaikh RS, Ali M, Iqbal F. A report on the high prevalence of Anaplasma sp. in buffaloes from two provinces in Pakistan. Ticks Tick Borne Dis. 2013;4:395–8. [DOI] [PubMed]
- 5.Silveira JAG, Rabelo EML, Ribeiro MFB. Molecular detection of tick-borne pathogens of the family Anaplasmataceae in Brazilian brown brocket deer (Mazama gouazoubira, Fischer,1814) and marsh deer (Blastocerus dichotomus, Illiger, 1815) Transbound Emerg Dis. 2012;59:353–60. doi: 10.1111/j.1865-1682.2011.01278.x. [DOI] [PubMed] [Google Scholar]
- 6.Tonetti N, Berggoetz M, Rühle C, Pretorius AM, Gern L. Ticks and tick-borne pathogens from wildlife in the Free State Province, South Africa. J Wildl Dis. 2009;45:437–46. doi: 10.7589/0090-3558-45.2.437. [DOI] [PubMed] [Google Scholar]
- 7.Munang’andu HM, Siamudaala VM, Munyeme M, Nalubamba KS. Detection of parasites and parasitic infections of free-ranging wildlife on a game ranch in Zambia: A challenge for disease control. J Parasitol Res. 2012;35(11-12):1–8. doi: 10.1155/2012/296475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor: N. Y. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 9.Torioni de Echaide S, Knowles DP, McGuire, Travis C, Palmer GH, Suarez CE, Mcelwain TF. Detection of cattle naturally infected with Anaplasma marginale in a region of endemicity by nested PCR and a competitive enzyme-linked immunosorbent assay using detection of cattle naturally infected with Anaplasma marginale in a region of endemicity. J Clin Microbiol. 1998;36:777–82. [DOI] [PMC free article] [PubMed]
- 10.De la Fuente J, Van Den Bussche RA, Kocan KM. Molecular phylogeny and biogeography of North American isolates of Anaplasma marginale (Rickettsiaceae: Ehrlichieae) Vet Parasitol. 2001;97:65–76. doi: 10.1016/S0304-4017(01)00378-8. [DOI] [PubMed] [Google Scholar]
- 11.Guillemi EC, Ruybal P, Lia V, Gonzalez S, Lew S, Zimmer P, Lopez Arias L, Rodriguez JL, Rodriguez SY, Frutos R, Wilkowsky SE, Farber MD. Development of a multilocus sequence typing scheme for the study of Anaplasma marginale population structure over space and time. Infect Genet Evol. 2015;30:186–94. doi: 10.1016/j.meegid.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 12.De la Fuente J, Ruybal P, Mtshali MS, Naranjo V, Shuqing L, Mangold AJ, Rodríguez SD, Jiménez R, Vicente J, Moretta R, Torina A, Almazán C, Mbati PM, de Echaide ST, Farber M, Rosario-Cruz R, Gortazar C, Kocan KM. Analysis of world strains of Anaplasma marginale using major surface protein 1a repeat sequences. Vet Microbiol. 2007;119:382–90. doi: 10.1016/j.vetmic.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Ruybal P, Moretta R, Perez A, Petrigh R, Zimmer P, Alcaraz E, Echaide I, Torioni de Echaide S, Kocan KM, de la Fuente J, Farber M. Genetic diversity of Anaplasma marginale in Argentina. Vet Parasitol. 2009;162:176–80. [DOI] [PubMed]
- 14.Feil EJ, Enright MC. Analyses of clonality and the evolution of bacterial pathogens. Curr Opin Microbiol. 2004;7:308–13. doi: 10.1016/j.mib.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Francisco AP, Bugalho M, Ramirez M, Carriço JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics. 2009;10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Biol Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 17.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–91. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum - a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol. 2013;3:1–33. doi: 10.3389/fcimb.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahfari S, Coipan EC, Fonville M, van Leeuwen AD, Hengeveld P, Heylen D, Heyman P, van Maanen C, Butler CM, Földvári G, Szekeres S, van Duijvendijk G, Tack W, Rijks JM, van der Giessen J, Takken W, van Wieren SE, Takumi K, Sprong H. Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasit Vectors. 2014;7:365. doi: 10.1186/1756-3305-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Da Silveira J a G, Rabelo ÉML, Ribeiro MFB. Detection of Theileria and Babesia in brown brocket deer (Mazama gouazoubira) and marsh deer (Blastocerus dichotomus) in the State of Minas Gerais, Brazil. Vet Parasitol. 2011;177:61–6. doi: 10.1016/j.vetpar.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-Vivas RI, Miller RJ, Ojeda-Chi MM, Rosado-Aguilar J a, Trinidad-Martínez IC, de Pérez León AA. Acaricide and ivermectin resistance in a field population of Rhipicephalus microplus (Acari: Ixodidae) collected from red deer (Cervus elaphus) in the Mexican tropics. Vet Parasitol. 2014;200:179–88. doi: 10.1016/j.vetpar.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 22.De la Fuente J, Kocan KM, Blouin EF, Zivkovic Z, Naranjo V, Almazán C, Esteves E, Jongejan F, Daffre S, Mangold AJ. Functional genomics and evolution of tick-Anaplasma interactions and vaccine development. Vet Parasitol. 2010;167:175–86. doi: 10.1016/j.vetpar.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Da Silva JB, da Fonseca AH, Barbosa JD. Molecular characterization of Anaplasma marginale in ticks naturally feeding on buffaloes. Infect Genet Evol. 2015;35:38–41. doi: 10.1016/j.meegid.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Garcia MV, Silva DC Da, Almeida RFC De, Cunha RC, Matias J, Barros JC, Andreotti R, Szabó MPJ. Environmentally associated ticks (Acari: Ixodidae) in Campo Grande, Mato Grosso do Sul, Brazil. Rev Bras Parasitol veterinária. 2013;22:124–8. [DOI] [PubMed]
- 25.Bechara GH, Szabó MP., Almeida Filho WV, Bechara JN, Pereira RJG, Garcia JE, MCP. Ticks associated with Armadillo (Euphractus sexcinctus) and Anteater (Myrmecophaga tridactyla) of Emas National Park, State of Goias, Brazil. Ann NY Acad Sci. 2002;969:290–3. [DOI] [PubMed]
- 26.Labruna MB, de Paula CD, Lima TF, Sana DA. Ticks (Acari: Ixodidae) on wild animals from the Porto-Primavera Hydroelectric power station area, Brazil. Mem Inst Oswaldo Cruz. 2002;97:1133–6. doi: 10.1590/S0074-02762002000800012. [DOI] [PubMed] [Google Scholar]
- 27.Guglielmone AA, Nava S. (Acari: Ixodidae): Distribución Y Hospedadores. RIA. 2006;35:133–53. [Google Scholar]


