Abstract
While WHO grade I meningiomas are considered benign, patients with WHO grade III meningiomas have very high mortality. The principles underlying tumor progression in meningioma are largely unknown yet a detailed understanding of these mechanisms will be required for effective management of patients with these high-grade, lethal tumors. We present a case of an intraventricular meningioma that at first presentation displayed remarkable morphologic heterogeneity – comprised of distinct regions independently fulfilling histopathologic criteria for WHO grade I, II and III designations. The lowest-grade regions had classic meningothelial features while the highest grade regions were markedly dedifferentiated. While progression in meningiomas is generally observed during recurrence following radiation and systemic medical therapies the current case offers us a snapshot into histologic progression and intratumor heterogeneity in a native, pre-treatment context. Using whole exome sequencing (WES) and high resolution array comparative genomic hybridization (aCGH) we observe marked genetic heterogeneity between the various areas. Notably, in the higher grade regions we find increased aneuploidy with progressive loss of heterozygosity, the emergence of mutations in the TERT promoter and compromise of ARID1A. These findings provide new insights into intratumoral heterogeneity in the evolution of malignant phenotypes in anaplastic meningiomas and potential pathways of malignant progression.
Keywords: meningioma, anaplastic, progression, intratumoral heterogeneity, swi/snf complex, chromatin remodeling
Introduction
Meningiomas are the most common primary tumors of the central nervous system, with 10–15% following an aggressive course with rapid growth, invasion into adjacent structures, and rapid recurrence even after radical resection and adjuvant treatment. These meningiomas are classified as World Health Organization (WHO) grade II or III and are associated with premature mortality [1]. Despite emerging understanding of the genetic changes underlying meningioma formation [2–10], the factors that drive malignant progression in meningioma remain poorly understood [11–13]. We encountered a case of a unique untreated intraventricular meningioma, which allowed us to investigate molecular factors associated with tumor progression and dedifferentiation.
Materials and Methods
Pathologic examination
Histopathologic diagnosis and tumor purity >80% was confirmed by review of the H&E stained sections by two neuropathologists (S.S., M.A.). This study was approved by the human subject institutional review board of the Dana-Farber/Brigham and Women’s Cancer Center.
Immunohistochemistry and RNAscope
Immunohistochemistry was performed using commercially available antibodies following standard automated and manual protocols on paraffin-embedded formalin-fixed tissue. The slides were incubated with antibodies for MIB-1 (Ki-67, Dako, Cat# M7240), epithelial membrane antigen (EMA, Dako, Cat# M0613 clone: E29) and ARID1A (monoclonal ARID1A/BAF250a, Cat# sc-32761X, Santa Cruz) and developed using 3,3′-diaminobenzidine (DAB) as the chromagen (Sigma Chemical Co., St. Louis, MO), before counterstaining with hematoxylin. Appropriate positive and negative controls were used throughout. In situ hybridization of TERT mRNA transcripts was performed using the RNAscope® assay with Probe-Hs-TERT (Cat# 605511, Advanced Cell Diagnostics, USA), following manufacturer’s protocols [10,14].
Genomic analysis
Array-based comparative genomic hybridization (aCGH) was performed using a stock 1×1M Agilent SurePrint G3 Human CGH Microarray chip in a CLIA-certified laboratory. A minimum of 1.3μg DNA, corresponding to approximately 6×1mm FFPE punches, was obtained for each specimen. Genomic DNA isolated from FFPE blocks was hybridized with a commercial reference DNA sample representing a pool of individuals with normal karyotypes (Promega, Madison, WI). The array platform contains 963,029 probes spaced with a 2.1kb overall median probe spacing and a 1.8kb probe spacing in RefSeq genes across the human genome. A genomic imbalance is reported when a minimum of eight consecutive probes, corresponding to approximately 14–16kb, show an average log2 ratio above +0.25 or below −0.5.
Whole-exome sequencing was performed on DNA isolated from each histopathologically distinct area, and matched germline DNA, to a mean depth of 80X, and analysis proceeded as previously described [8,15]. Raw sequencing data was processed using the Picard tools pipeline and the Genome Analysis Toolkit (GATK) [16,17]. Mutation analysis for single nucleotide variants (SNV) was performed using MuTect v1.1.4 [18], indel calling was performed using the GATK SomaticIndelDetector tool; SNVs and indels were annotated using Oncotator. To analyze somatic copy number alterations from whole exome data, we used the ReCapSeg algorithm, which assesses homologue-specific copy ratios (HSCRs) from segmental estimates of multipoint allelic copy-ratios at heterozygous loci incorporating the statistical phasing software (BEAGLE) and population haplotype panels (HAPMAP3) [19–21].
Results
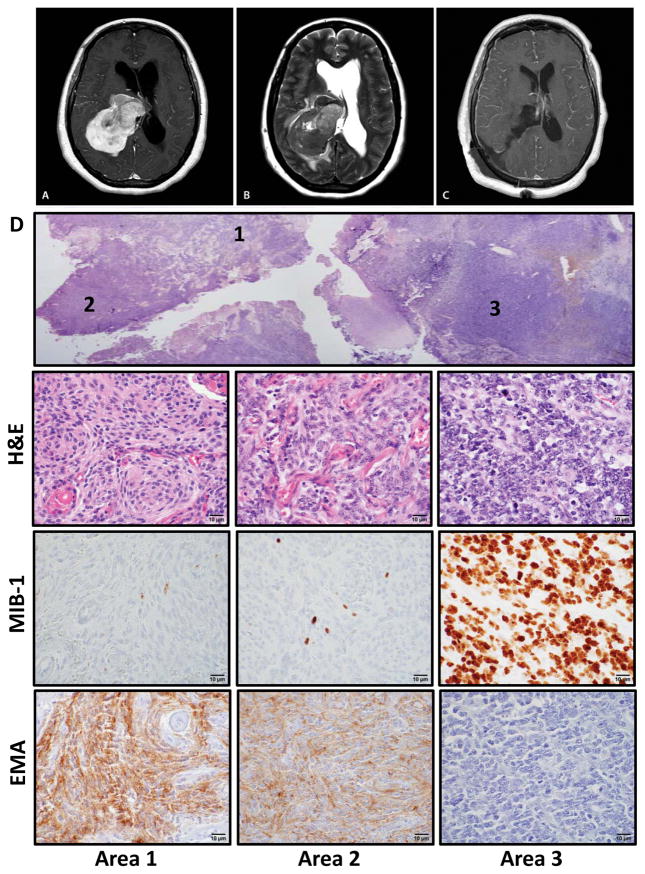
A 48-year-old woman presented with five days of headache and gait instability, prompting imaging that revealed a 6.9×4.7×5cm heterogeneously contrast-enhancing mass, centered within the atrium of the right lateral ventricle, causing obstructive hydrocephalus (Fig. 1A–B). A right parietal craniotomy was performed with gross total resection of the tumor (Fig. 1C). The patient’s symptoms improved post-operatively and she remains alive two years following the resection.
Figure 1.
Intratumoral heterogeneity within a high-grade meningioma. (A) T1-weighted gadolinium-enhanced axial MRI demonstrating a heterogeneously appearing contrast-avid tumor within the atrium of the right lateral ventricle, with (B) significant peritumoral edema and associated hydrocephalus on the T2-weighted axial MRI. (C) Post-operative T1-weighted axial MRI demonstrating complete resection. (D) Section of tumor, with 3 distinct appearing histologic regions (designated areas 1, 2, 3). Immunohistochemistry for epithelial membrane antigen (EMA) and MIB-1 (Ki-67). Scale bar, 10 μm.
The histopathology was notable for the concurrent presence of regions displaying classic features of low-grade meningothelial meningioma (WHO grade I), others with features of atypical WHO grade II meningioma, and areas harboring cells with a pronounced increase in nucleocytoplasmic ratio, frequent mitosis and loss of discernable features of meningothelial differentiation – hallmarks of dedifferentiation in a WHO grade III anaplastic meningioma (Fig. 1D). Consistent with this histologic progression, the MIB-1 proliferation index ranged from <1% to >90% in corresponding grade I versus grade III regions (Fig. 1D). Immunohistochemistry (IHC) for epithelial membrane antigen (EMA), a marker of meningothelial differentiation, also showed EMA expression in the low-grade regions that was entirely lost in the highest-grade regions, consistent with the morphologic dedifferentiation (Fig. 1D). To better understand the molecular underpinnings of the histologic heterogeneity within this tumor, we analyzed DNA extracted from core punches from each of these three regions using high-resolution array-based comparative genomic hybridization (aCGH), Sanger sequencing of the TERT promoter, and whole-exome sequencing (Fig. 2).
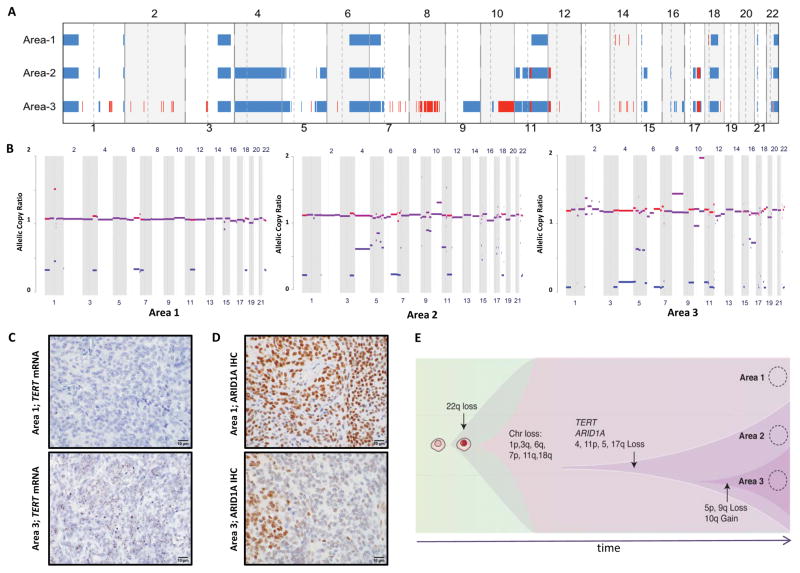
Figure 2.
Genetic heterogeneity within a high-grade meningioma. (A) High-resolution aCGH was performed on DNA extracted from core punches taken from area 1 (WHO grade I), area 2 (WHO grade II), and area 3 (WHO grade III), using 1×1M Agilent SurePrint G3 Human CGH Microarray chip [3,30]. Red indicates gain and blue indicates loss. (B) Copy number analysis from exome-sequencing data of the three areas shown as allelic copy ratios. Red indicates gain and blue indicates loss. (C) In situ hybridization of TERT mRNA transcripts using a TERT-specific RNAscope probe in FFPE sections of the meningioma samples [10,14]. (D) Immunohistochemistry for ARID1A. (E) Schematic representation of clonal evolution leading to dedifferentiation and intratumoral heterogeneity. Scale bar, 10 μm.
The aCGH data showed partial single-copy loss of chromosomes 1p, 3q, 6q, 7p, 11q, 18q (monosomy 18), and 22q (monosomy 22) in all three areas (Fig. 2A). The atypical and anaplastic regions both harbored an expanded loss of chromosome 11q, additional unique losses of 4, 11p, and regions of 5, 15q, and 17q that were not detected in the grade I areas. Expansion of 5p and 9q losses was distinct to the grade III area, as were gains on chromosome 8 and 10q. Copy-number analysis of the whole-exome sequencing data showed a similar pattern of losses and further revealed progressive loss of heterozygosity across the three regions and evidence for subclonal chromosomal losses in Area 2 that become clonal in Area 3 (Fig. 2B).
Because whole-exome sequencing does not cover the promoter region of the TERT gene, which has been reported to be mutated in meningiomas with histologic progression [2], we performed Sanger sequencing of this region. We found wild-type sequences in the grade I area but the C228T mutation in both the grade II and III areas. Using RNAscope in situ hybridization [14], we found that TERT mRNA is low in grade I areas and sharply increased in higher-grade areas (Fig. 2C). The whole-exome sequencing data revealed a low frequency of other mutations in the three samples with the notable presence of an ARID1A frameshift deletion (p.LHH2017fs) in the grade II and III areas (Supplementary Table 1). Coupled with single-copy loss of chromosome 1p, the frameshift would lead to a sharp decrease in ARID1A protein levels. Indeed, ARID1A levels are markedly reduced in the high-grade areas on IHC (Fig. 2D).
Discussion
The mutation and copy-number data reveal a concentric, rather than partially overlapping, pattern of alterations; nearly all mutations and copy number alterations in the areas with atypical histopathology were also observed in the sample with higher-grade anaplastic features. This suggests a common clonal origin to the distinct areas. Notably, this tumor was newly diagnosed, without prior surgical or chemo-radiation treatment. It is interesting to speculate that the de novo molecular heterogeneity observed in our case reflects a set of clonal sweeps where each ensuing clone consists of a subclone of the previous dominant clone (Fig. 2E). One might imagine that therapeutic interventions such as chemotherapy and radiation may precipitate a different pattern in which multiple surviving subclones compete for dominance.
The shared pattern of single-copy loss, involving chromosomes 1p, 3q, 6q, 18q, and 22q, is consistent with previously reported cytogenetic changes in high-grade meningiomas [22]. We found these alterations even in the areas with grade I histologic features, which had a low proliferative index and intact EMA expression, suggesting that additional alterations can be needed for histologic progression. Non-contiguous losses on 15q and 17q and the unique whole-arm loss of 9q observed only in the anaplastic region contrasts with previously observed gains of 9q, 15q, and 17q in some high-grade meningiomas and reflects the current incomplete understanding of the specific oncogenic drivers within these broad regions [13,22]. No CDKN2A (9p21.3) deletions, a frequent finding amongst aggressive meningiomas [13,23], were observed.
The differences in mutational profiles between the low (grade I) and high-grade (grades II and III) areas are intriguing. Mutations in the TERT promoter and in ARID1A were most notable. Telomerase activation is well-recognized as a feature of malignant meningiomas [24], with a recent report that TERT promoter mutations are specific to meningiomas which recur displaying histologic progression [2]. The observation of differential TERT expression within regions of the same tumor that had not been treated further supports the role for increased TERT in the malignant progression of meningioma. Moreover, sequencing studies have revealed that the genes encoding subunits of mammalian SWI/SNF complexes are mutated in over 20% of all human cancers [25–27]. ARID1A is important in ensuring correct lineage progression and in limiting self-renewal [28]. It is interesting to propose that concomitant activation of TERT and compromise of the SWI/SNF complex or other chromatin remodeling complexes through a range of genetic or epigenetic events [29] may synergize in allowing meningioma cells to develop high-grade malignant phenotypes [12] and dedifferentiate.
Supplementary Material
Genes identified to have mutations or indels across areas 1, 2, and 3, from whole-exome sequencing, as analyzed by MuTect. Allelic fractions are indicated when a mutation or indel was present; or blank if no somatic alteration was determined. Copy number gain (red) or loss (blue) are also highlighted as corresponding to the region of the chromosome where the gene is located.
Acknowledgments
We are grateful to Marian Slaney and Sebastian Valentino for assistance with histology, Terri Woo for assistance with immunohistochemistry and Marc L. Listewnik for cytogenetic data support for technical assistance. The work is supported by 35-1041 grant from King Abdulaziz City for Science and Technology (KACST), Saudi Arabia (M.A.A.), the Brain Science Foundation (S.S.) and the Jared Branfman Sunflowers for Life Fund for Pediatric Brain and Spinal Cancer Research (S.S.).
Footnotes
Conflict of interest: The authors declare no conflicts of interest with respect to this study.
References
- 1.Wiemels J, Wrensch M, Claus EB. Epidemiology and etiology of meningioma. J Neurooncol. 2010;99:307–314. doi: 10.1007/s11060-010-0386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goutagny S, Nault JC, Mallet M, Henin D, Rossi JZ, Kalamarides M. High Incidence of Activating TERT Promoter Mutations in Meningiomas Undergoing Malignant Progression. Brain Pathol. 2014;24:184–189. doi: 10.1111/bpa.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abedalthagafi MS, Merrill PH, Bi WL, et al. Angiomatous meningiomas have a distinct genetic profile with multiple chromosomal polysomies including polysomy of chromosome 5. Oncotarget. 2014;5:10596–10606. doi: 10.18632/oncotarget.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith MJ, O’Sullivan J, Bhaskar SS, et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nature genetics. 2013;45:295–298. doi: 10.1038/ng.2552. [DOI] [PubMed] [Google Scholar]
- 5.Sahm F, Bissel J, Koelsche C, et al. AKT1E17K mutations cluster with meningothelial and transitional meningiomas and can be detected by SFRP1 immunohistochemistry. Acta neuropathologica. 2013;126:757–762. doi: 10.1007/s00401-013-1187-5. [DOI] [PubMed] [Google Scholar]
- 6.Reuss DE, Piro RM, Jones DT, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta neuropathologica. 2013;125:351–358. doi: 10.1007/s00401-013-1093-x. [DOI] [PubMed] [Google Scholar]
- 7.Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339:1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nature genetics. 2013;45:285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aavikko M, Li SP, Saarinen S, et al. Loss of SUFU function in familial multiple meningioma. American journal of human genetics. 2012;91:520–526. doi: 10.1016/j.ajhg.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Z, Abedalthagafi M, Aizer AA, et al. Increased expression of the immune modulatory molecule PD-L1 (CD274) in anaplastic meningioma. Oncotarget. 2014 doi: 10.18632/oncotarget.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang HW, Kim TM, Song SS, et al. Alternative splicing of CHEK2 and codeletion with NF2 promote chromosomal instability in meningioma. Neoplasia. 2012;14:20–28. doi: 10.1593/neo.111574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kliese N, Gobrecht P, Pachow D, et al. miRNA-145 is downregulated in atypical and anaplastic meningiomas and negatively regulates motility and proliferation of meningioma cells. Oncogene. 2013;32:4712–4720. doi: 10.1038/onc.2012.468. [DOI] [PubMed] [Google Scholar]
- 13.Choy W, Kim W, Nagasawa D, et al. The molecular genetics and tumor pathogenesis of meningiomas and the future directions of meningioma treatments. Neurosurgical focus. 2011;30:E6. doi: 10.3171/2011.2.FOCUS1116. [DOI] [PubMed] [Google Scholar]
- 14.Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brastianos PK, Taylor-Weiner A, Manley PE, et al. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nature genetics. 2014;46:161–165. doi: 10.1038/ng.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International HapMap C. Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. American journal of human genetics. 2009;85:847–861. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riemenschneider MJ, Perry A, Reifenberger G. Histological classification and molecular genetics of meningiomas. Lancet neurology. 2006;5:1045–1054. doi: 10.1016/S1474-4422(06)70625-1. [DOI] [PubMed] [Google Scholar]
- 23.Bostrom J, Meyer-Puttlitz B, Wolter M, et al. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. The American journal of pathology. 2001;159:661–669. doi: 10.1016/S0002-9440(10)61737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon M, Park TW, Leuenroth S, Hans VH, Loning T, Schramm J. Telomerase activity and expression of the telomerase catalytic subunit, hTERT, in meningioma progression. Journal of neurosurgery. 2000;92:832–840. doi: 10.3171/jns.2000.92.5.0832. [DOI] [PubMed] [Google Scholar]
- 25.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer discovery. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan B, Wang TL, Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer research. 2011;71:6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dykhuizen EC, Hargreaves DC, Miller EL, et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature. 2013;497:624–627. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eroglu E, Burkard TR, Jiang Y, et al. SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell. 2014;156:1259–1273. doi: 10.1016/j.cell.2014.01.053. [DOI] [PubMed] [Google Scholar]
- 29.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Craig JM, Vena N, Ramkissoon S, et al. DNA fragmentation simulation method (FSM) and fragment size matching improve aCGH performance of FFPE tissues. PLoS One. 2012;7:e38881. doi: 10.1371/journal.pone.0038881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes identified to have mutations or indels across areas 1, 2, and 3, from whole-exome sequencing, as analyzed by MuTect. Allelic fractions are indicated when a mutation or indel was present; or blank if no somatic alteration was determined. Copy number gain (red) or loss (blue) are also highlighted as corresponding to the region of the chromosome where the gene is located.