Abstract
Background
Leucine-rich repeat receptor-like kinases (LRR-RLKs) represent a large class of proteins in regulating plant development and immunity. The LRR-RLK XA21 confers resistance to the bacterial disease caused by the pathogen of Xanthomonas oryzae pv. oryzae (Xoo). Several XA21 binding proteins have been characterized, however the early events governing XA21 signaling have not been fully elucidated.
Results
Here we report the identification of one LRR-RLK gene (XIK1) whose expression is induced rapidly upon the infection with the pathogen of Xoo. Expression pattern analysis reveals that XIK1 is preferentially expressed in reproductive leaves and panicles, and that expression is associated with plant development. By using RNA interference (RNAi), we silenced the expression of XIK1 in rice with Xa21 and found that reduced expression of XIK1 compromised disease resistance mediated by XA21. In addition, we found that the expression of the downstream marker genes of pathogen associated molecular pattern (PAMP) triggered immunity (PTI) in rice was compromised in Xa21 plants silenced for XIK1.
Conclusion
Our study reveals that the LRR-RLK gene XIK1 is Xoo-responsive and positively regulates Xa21-mediated disease resistance.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-014-0034-1) contains supplementary material, which is available to authorized users.
Keywords: LRR-RLK, Xa21, Disease resistance, Xanthomonas oryzae pv.oryzae
Background
Receptor-Like Kinases (RLKs) represent one of the largest protein families in plants, with more than 600 members in Arabidopsis (Shiu and Bleecker 2001) and 1000 members in rice (Shiu et al. 2004). A typical RLK protein contains extracellular structure, and transmembrane and kinase domains (Greeff et al. 2012). Based on the variation of the N-terminal domains, RLKs are divided into more than 44 subclasses (Gish and Clark 2011). The leucine-rich repeat (LRR)-RLK subclass is the largest, with 165 members in Arabidopsis (Shiu and Bleecker 2001) and 292 members in rice (Shiu et al. 2004). The LRR domain is a 24 residues-containing motif rich for leucine or other hydrophobic amino acids. Each LRR-RLK contains one or more LRRs to form a pocket used for binding to various ligands (Kolade et al. 2006; Gish and Clark 2011).
Previous studies have revealed that several LRR-RLKs are involved in plant development and immunity. For examples, the Arabidopsis BRI1, CLAVATA1, ERECTA1 (Clark et al. 1993; Torii et al. 1996; Clark et al. 1997; Li and Chory 1997; Jinn et al. 2000), rice BRI1 (OsBRI1) (Li and Chory 1997), and Petunia inflate PRK1 (Mu et al. 1994), regulate plant growth and development. The Arabidopsis flagellin receptor FLS2, the EF-Tu receptor EFR (Gomez-Gomez and Boller 2000; Zipfel et al. 2006), and the rice protein XA21 (Song et al. 1995) mediate immunity during plant-microbe interaction.
Xa21 encodes a protein of rice LRR-RLK XII subclass and confers resistance against diverse strains of Xoo (Song et al. 1995). Although the expression of Xa21 is quite stable during rice development, XA21-mediated disease resistance is development-dependent (Mazzola et al. 1994). Previous studies have shown that several XA21 binding proteins (XBs) regulate XA21-mediated immunity. These XBs belong to different protein families: such as RING finger ubiquitin ligase (XB3), transcriptional factor OsWRKY62 (XB10), protein phosphatase 2C (XB15), ATPase (XB24), endoplasmic reticulum chaperone (BiP3) and PANK protein (XB25) (Chen et al. 2010; Peng et al. 2008; Park et al. 2010; Wang et al. 2006; Park et al. 2008; Jiang et al. 2013). Recently, we also reported that OsSERK2, the rice ortholog of BAK1, regulates XA21-mediated immunity in a mechanism distinct from that of BAK1 in regulation of FLS2- and EFR-mediated immunity in Arabidposis (Chen et al. 2014b). Even with these advances, the mechanism of XA21-mediated immunity is still largely unknown.
Here, we characterized XIK1 (LOC_Os02g34790), which encodes a LRR-RLK protein, through analysis of the Rice Oligonucleotide array database (ROAD: http://www.ricearray.org/). We found that the expression of XIK1 was slightly induced upon the inoculation with Xoo in Kitaake but more strongly in Xa21 plants. We found that XIK1 is ubiquitously expressed in different rice tissues and preferentially in leaves and panicles. Transgenic Xa21 plants silenced for XIK1 exhibit compromised disease resistance to Xoo. We also found that the expression of XIK1 is increasing during rice development, which might be the reason why XA21-mediated disease resistance is developmentally dependent. In addition, we found that the expression of marker genes for pathogen associated molecular pattern (PAMP) triggered immunity (PTI) is reduced in Xa21 plants silenced for XIK1. Taken together, our study reveals that the expression of XIK1 is Xoo-responsive and that XIK1 positively regulates XA21-mediated immunity.
Results
Identification of XIK1
LRR-RLK is a large subgroup of the rice RLK family, of which, XA21, XA3/XA26 and OsSERK2 have been reported to regulate plant immunity against Xoo, the causal agent of bacterial leaf blight. To determine if any other LRR-RLK is involved in the immune response against Xoo, we analyzed global transcriptional analysis on the expression pattern of rice LRR-RLK genes to search those with Xoo-responsiveness using the data available from the Rice Oligonucleotide array database (http://www.ricearray.org/). We identified eight LRR-RLK genes whose transcriptional expressions were rapidly (at 2 h post inoculation) up-regulated upon Xoo inoculation (Additional file 1: Figure S1). We then selected one gene, LOC_Os02g34790, for further study. The protein encoded by LOC_Os02g34790 contains a typical signal peptide at the N-terminal, 18 repeated LRR motif and a kinase domain at the C-terminal (Figure 1). The phylogenetic tree analysis reveals that this protein belongs to LRR II subfamily (Hwang et al. 2011). We then named this gene as Xoo-induced kinase 1 (XIK1).
Figure 1.

Prediction of the amino acid sequence and protein structure of the LRR-RLK XIK1. The sequence from amino acid residue 1 to 26 is predicted as a signal peptide. The extra-cellular region with 17 LRRs consists of the amino acid residues from 28 to 422; the amino acids characterizing the LRR motifs are marked in dark. The transmembrane region consisting of amino acid residues from 486 to 509 are underlined with single solid line. The intracellular kinase domain consisting of the amino acid residues from 550 to 802, with the conserved amino acid residues is indicated by double lines. The structure of XIK1 was predicted according to the structure of XA21 reported previously (G. L. Wang et al. 1996; Xiang et al. 2006).
The expression pattern of XIK1
We performed quantitative RT-PCR to measure the transcription levels of XIK1 in the rice tissues, including root, stem, leaf and panicle at the productive stage. We found that XIK1 was ubiquitously expressed in all of the tissues with a preference in leaves and panicles (Figure 2A). We then measured the expression of XIK1 in leaves collected from different developmental stages of rice and found that the expression of XIK1 increases during the development of rice (Figure 2B). These results suggest that XIK1 may function in both tissue- and development-dependent manners in rice.
Figure 2.
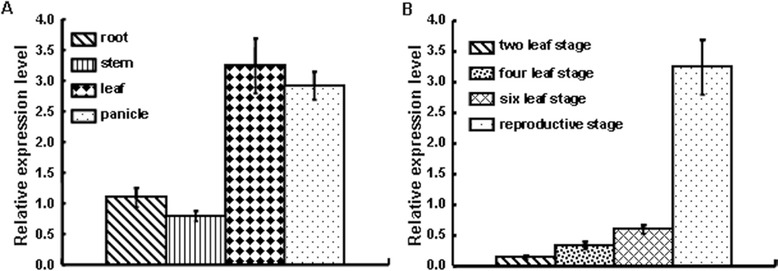
The expressions pattern of XIK1 . (A). qRT-PCR was performed on cDNA synthesized from RNA samples extracted from rice cultivar Kitaake reproductive stage tissues. Expression of XIK1 was normalized to the expression of the ubiquitin5 reference gene. Error bars indicating standard deviation (SD) obtained from three technical replicates. Three independent biological experiments were repeated with the similar results. (B). Determination of the transcriptional expression of XIK1 in leaves at different developmental stages as indicated. Gene expression of XIK1 was normalized to the expression of the ubiquitin5 reference gene. Error bars indicating SD obtained from three technical replicates. Three independent biological experiments were repeated with the similar results.
The expression of XIK1 in Xa21 plants is induced by Xoo
As the microarray data of XIK1 indicates that the expression of XIK1 in Nipponbare is responsive to Xoo, we tested whether it could also be induced in Xa21 plants (Peng et al. 2008). For this purpose, we collected the Xa21 rice leaves post the inoculation with Xoo or H2O (mock-treatment) and measured the expression of XIK1 using qRT-PCR. We found that the expression of XIK1 was almost 1.5 folds of that obtained from mock treatment in Xa21 resistant lines (Figure 3). This result suggests that the expression of XIK1 in Xa21 plants is induced upon the inoculation with Xoo.
Figure 3.
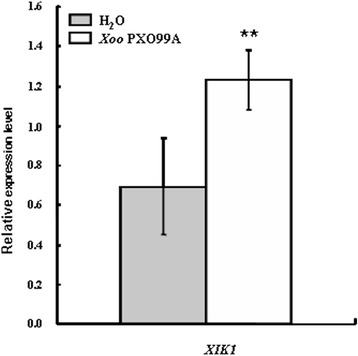
XIK1 is rapidly induced by Xoo inoculation. The expression of XIK1 is induced within 2 h after Xoo inoculation. The ** indicate significant differences (P<0.05) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
Silencing of XIK1 compromises the XA21-mediated resistance to Xoo
Because the LRR-RLK XA21 functions as a pattern recognition receptor (PRR) to confer resistance to Xoo and XIK1 is rapidly induced by Xoo inoculation in Xa21 plants, we presumed that XIK1 regulates XA21-mediated immunity. To test this hypothesis, we introduced the XIK1Ri construct into ProA-Xa21 homozygous rice lines to silence the expression of XIK1 in Xa21 stable transgenic plants. ProA-Xa21 homozygous rice lines confer full resistance to the Xoo strain PXO99A (Chen et al. 2010). We obtained 12 independent double transgenic RNAi lines carrying both Xa21 and XIK1Ri and named them Xa21XIK1Ri (XXIK1Ri) lines. Three representative lines with reduced expression levels of XIK1 and stable expression level of the XIK1 homolog LOC_Os02g34750 were found (Additional file 2: Figure S2). Compared with that in the wild type ProA-Xa21 plants, the expression of XIK1 is reduced and is about 40%, 45% and 80% in the three XXIK1RNAi lines, XXIK1Ri-1, XXIK1Ri-3 and XXIK1Ri-8, respectively. Expression of LOC_Os02g34750, with the highest identity (80.1%) of cDNA sequence to LOC_Os02g34790, was not altered (Additional file 2: Figure S2). We inoculated the T0 generations of these transgenic lines, and observed that the Xa21 plants silenced for XIK1 exhibited compromised disease resistance to the Xoo strain PXO99A compared with the wild type ProA-Xa21 plants (Additional file 3: Figure S3A-C). We then analyzed the disease resistance of plants from T1 generations after inoculation with PXO99A. We found that the T1 plants segregated with partial resistance in some segregants and typical full resistance in others. The double transgenic plants carrying Xa21 and silenced for XIK1 are partially resistant, showing typical long water-soaked lesions. The segregants carrying Xa21 but lacking of XIK1Ri confer full resistance to Xoo as do the wild type Xa21 plants (Additional file 3: Figure S3D). The Xa21 plants are highly resistant and the Kitaake plants are highly susceptible to the Xoo strain PXO99A as expected.
To characterize the effect of XIK1 on XA21-mediated disease resistance, we performed more detailed investigations using the plants (XXIK1Ri) homozygous for both Xa21 and XXIK1Ri derived respectively from three of the double transgenic lines. After we verified that the expression of XIK1 was reduced whereas the expression of Xa21 was not obviously changed in these RNAi lines (Additional file 4: Figure S4), we inoculated the plants with the Xoo strain PXO99AZ. At 14 days post inoculation (DPI), all of these three XXIK1Ri lines displayed longer lesions than the Xa21 control (Figure 4A-E). The average lesion length of the line XXIK1Ri-1 with the lowest expression level of XIK1 is about 4.29 ± 0.69 cm, with the longest lesion length among these three represent XIK1 silence lines. Meanwhile, XXIK1Ri-8 with the highest expression of XIK1 showed the shortest lesion length (3.29 ± 0.72) cm among these three XIK1Ri lines. The positive control Xa21 plants and the negative control Kitaake plants respectively show the average lesion length of 0.26 ± 0.10 cm for the full resistance and 7.33 ± 0.10 cm for the full susceptibility as expected (Figure 4F). These results suggest that these RNAi transgenic plants exhibit partial resistance, showing typical longer water-soaked lesions than ProA-Xa21 plants but shorter than Kitaake plants.
Figure 4.
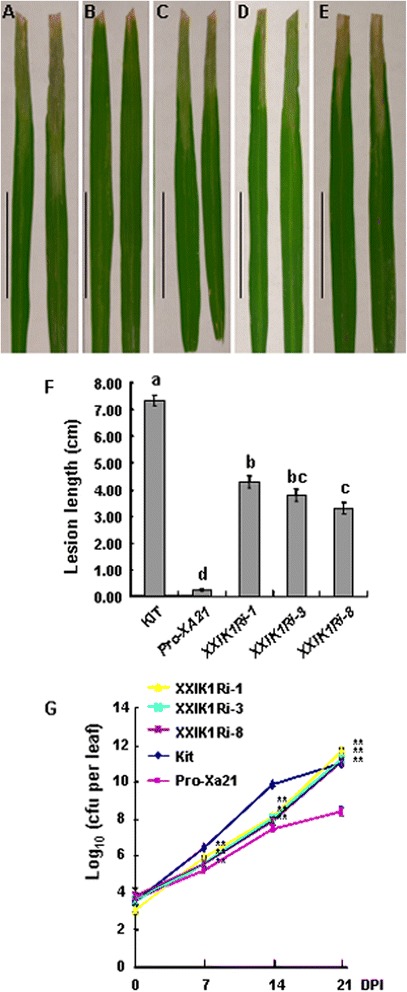
Determination of disease resistance of ProA-Xa21 plants silenced for XIK1. Six week-old plants were inoculated with the Xoo strain PXO99A. The ProA-Xa21 and Kitaake (Kit) were used as resistant and susceptible controls, respectively. Photograph depicts the representative leaves from plants at 14 DPI. The leaves from Kitaake displayed the full susceptibility to Xoo (A) whereas the leaves from ProA-Xa21 displayed full resistance to Xoo (B). The panels C, D, and E show the representative leaves from the three transgenic lines, XXIK1Ri-1, XXIK1Ri-3 and XXIK1Ri-8, respectively. All photographs were taken at 14 DPI. Bars= 5cm. (F), Lesion lengths of XXIK1Ri after inoculation with Xoo. Lesion length was measured at 14 DPI. Graph shows average lesion length ± SD of at least 20 leaves from 10 independent plants homozygous for both Xa21 and XIK1Ri. The letters indicate significant differences (P<0.05) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. (G), Bacterial cell numbers accumulated in ProA-Xa21 plants silenced for XIK1 after inoculation with Xoo. Bacterial cell numbers were counted at day 0, 7, 14, 21 post inoculations. Each data point represents the average ± SD of six leaves from two independent plants. The ** indicate significant differences (P<0.05) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. These experiments were repeated at least three times with similar results.
We then quantified bacteria accumulation in plant leaves after inoculation with Xoo at 0, 7, 14 and 21 DPI. We found that, at 21 DPI, the bacterial numbers in XXIK1Ri lines were almost 100 fold (4.8×1011 for XXIK1Ri-1, 2.0×1011 for XXIK1Ri-3 and 1.4×1011 for XXIK1Ri-8) greater than in ProA-Xa21 control plants (2.8×108) (Figure 4G). These results are consistent with the results of disease lesion length data described above. The lesion lengths are longer and the bacterial numbers are greater when the expression level of XIK1 is lower in the XXIK1Ri lines (Additional file 2: Figure S2 and Additional file 4: Figure S4), suggesting that the resistance of the transgenic lines is in accordance with the expression level of XIK1. Taken together, these results suggest that reduced expression of XIK1 compromises XA21-mediated disease resistance and the LRR-LRK XIK1 plays essential roles in XA21-mediated full innate immunity in rice.
Reduced expression of XIK1 compromises the downstream response of PTI
Previous reports have shown that the genes, OsKS4(LOC_Os04g10060) and Os04g10010(LOC_Os04g10010), function as marker genes involved for the downstream responses associated PTI (Park et al. 2012; Chen et al. 2014a). Thus, we asked whether these two genes in Xa21 plants are also responsive to Xoo. For this purpose, we measured the expression of these two genes in rice plants post the inoculation with Xoo or H2O as a mock treatment. Compared with mock treatment, the expression levels of OsKS4 and Os04g10010 were induced more than two folds at 2 days post Xoo treatment (Figure 5). This result suggests that the XA21-mediated immune signaling was transduced downstream quickly. We then performed qRT-PCR analysis on the expression of the two genes using the leaf tissue samples collected from Kitaake, ProA-Xa21, and XXIK1Ri-1 plants without Xoo inoculation. We found that the expression levels of these two genes were respectively higher in ProA-Xa21 plants than that in Kitaake plants, suggesting that rice plants carrying Xa21 possess a higher level of basal PTI than Kitaake plants. More interestingly, we found that the expression of the two genes, OSKS4 and Os04g10010, were much reduced in XXIK1Ri-1 plants (3.6 ± 0.4 fold and 2.8 ± 0.2 fold, respectivly) compared to ProA-Xa21 plants (8.5 ± 0.1 fold and 4.0 ± 0.7 fold, respectivly) (Figure 6A). We then measured the expression of OSKS4 and Os04g10010 in ProA-Xa21 and XXIK1Ri-1 plants after Xoo inoculation. We found that the induction of both genes were inhibited in the plants silenced for XIK1 (Figure 6B and C). These results suggest that reduced expression of XIK1 compromises XA21-mediated immune response both before and after Xoo inoculation.
Figure 5.
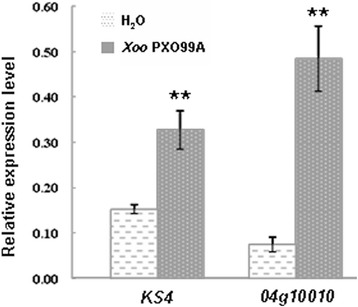
Expression of two PTI marker genes is induced within 2 h after Xoo inoculation. OSKS4 and Os04g10010 are rapidly induced after the infection of Xoo PX099A was used. Expression levels for each gene were normalized to the expression of the ubiquitin5 reference gene. Bars depict average expression level ± SD of three technical replicates. The ** indicate significant differences (P<0.05) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. This experiment was repeated three times with similar results.
Figure 6.
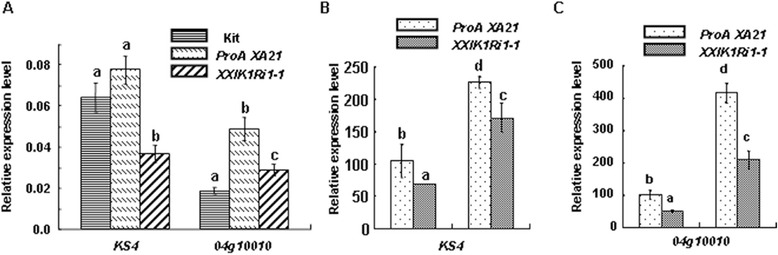
Silencing of XIK1 reduces the basal level of PTI marker genes in ProA-Xa21 plants . The relative expression level of two rice PTI marker genes, OSKS4 and Os04g10010, were determined by qRT-PCR using RNA extracted from rice leaves of six weeks old plants without Xoo inoculation (A) and 2 days post Xoo inoculation (B, C). All data were normalized to the expression of the ubiqutin5 reference gene. The results of one representative experiment were shown. Error bars indicating SD of three technical replicates. Three independent biological experiments were repeated with the similar results. The letters indicate significant differences (P<0.05) for each gene determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
Discussion
Expression of the LRR-RLK gene XIK1 is responsive to Xoo inoculation
Although some resistant proteins have been reported to regulate rice resistance against Xoo, prior studies of RLK family proteins in rice were mainly focused on two RLKs: XA21 and XA3/XA26 (Chen and Ronald 2011). Phosphorylation events play critical roles for these RLKs to initiate the response triggered by PAMPs (Chen et al. 2010; Chen et al. 2014a). There are few reports showing that the expression of these RLKs is induced by pathogens and thus regulates the immune response (Century et al. 1999). Our study reveals that the RLK gene XIK1 was induced by Xoo and that the encoded protein XIK1 positively regulates XA21-mediated immunity (Figures 3 and 4). Previous studies have identified some proteins that play important roles in the regulation of XA21-mediated resistance. For examples, ubiquitin E3 ligase XB3 (Wang et al. 2006) and endoplasmic reticulum chaperone BiP3 were shown to keep the XA21 protein stable (Park et al. 2014). Protein phosphatase 2C XB15 (Park et al. 2008), ATPase XB24 and rice somatic embryogenesis receptor-like kinase 2 (OsSERK2) regulate the phosphorylation of XA21(Chen et al. 2010; Chen et al. 2014a), and the WRKY transcriptional factor XB10 regulates the XA21-mediated immune response through an unknown mechanism (Peng et al. 2008). However, the expressions of these genes were not obviously changed upon the inoculation of the Xoo strain PXO99A. Our finding that the expression of XIK1 was induced by Xoo suggests that some RLKs such as XIK1 regulate PTI via their transcriptional expression (Figure 3). This finding provides new insights in understanding the XA21-mediated PTI signaling and the RLK-mediated innate immunities. We also found that the PTI-related marker genes were down regulated in Xa21 plants silenced for XIK1 (Figures 5 and 6), suggesting that the expression of the genes involved in downstream of XA21-signaling are regulated by XIK1. Additionally, since expression of XIK1 is both tissue- and developmentally-dependent indicates that XIK1 regulates rice development and immunity tissue-specifically and developmentally (Figure 2). This result also provides the explanations why the disease resistance mediated by XA21 is developmentally-dependent.
We also performed a similar Xoo inoculation assay on four RNAi lines silenced for XIK1 in Kitaake rice and found that these RNAi lines were as susceptible as Kitaake plants (data not shown). Thus, our study suggests that XIK1 regulation of rice resistance to Xoo is XA21-dependent.
XIK1 regulates early events in XA21-mediated immunity signaling
LRR-RLKs consist of the largest RLK subfamily in plants with more than 165 members in Arabidopsis (Shiu and Bleecker 2001) and more than 292 members in rice (Shiu et al. 2004). In rice, the well studied LRR-RLK is XA21. Investigation of the molecular mechanisms of XA21-mediated immunity have shown that many proteins are required for XA21 to function (Peng et al. 2008; Park et al. 2010; Wang et al. 2006; Park et al. 2008; Jiang et al. 2013; Chen et al. 2014a). Our study reveals that the LRR-RLK XIK1 positively regulates XA21-mediated immune signaling. As the expression of XIK1 is induced as early as within 2 h post Xoo inoculation and reduced expression of XIK1 compromises the expression of PTI-related genes, we hypothesize that XIK1 regulates the early events of XA21-mediated signaling. Because the XIK1 shares structural similarity with the LRR-RLK XA21, such as the extracellular LRR domain, a transmembrane domain that is required for the plasmembrane-localization of RLK proteins, and the intercellular active kinase domain, we suggest that XIK1 might work as a co-receptor of XA21 for PAMPs recognition and mediate the downstream signaling. As we could not detect the interaction between XIK1 and XA21 by using the intercellular domains of these two RLKs in yeast (data not shown), the extracellular domains of XIK1 and XA21 or the binding of the PAMP might be required for their interaction. To understand the precise mechanism of XIK1 in regulation of XA21-mediated immunity, it is interesting to address these questions in our future work, including whether XIK1 forms a complex with XA21, whether the extra-cellular domains of these two RLKs, or pathogen inoculation are required for their interaction, and whether OsSERK2, a recently identified RLK essential for XA21-mediated immunity is required for XIK1 to function. We also suggest that XIK1 may regulate XA3/XA26, mediated immune response to Xoo, as XA3/XA26 belongs to LRR XII RLK with the similar structure of XA21.
In Arabidopsis, the LRR-LRKs, FLS2 and EFR also mediate PTI by recognizing the PAMPs, flagellin and elongation factor (EF-tu), respectively (Gomez-Gomez and Boller 2000; Zipfel et al. 2006). Previous studies have shown that FLS2- and EFR-mediated immunities share many similar processes as XA21, such as production of reactive oxygen species (ROS), activation of mitogen activated protein kinases (MAPKs), induction of pathogenesis-related genes (PRs) (Gomez-Gomez et al. 1999; Zhang and Zhou 2010; Zipfel et al. 2006) and the requirement of the SERK proteins (i.e. SERK3/BAK1 and SERK4/BKKI for FLS2 and EFR in Arabidopsis, and OsSERK2 for XA21 in rice) for their early signaling (Korner et al. 2013; Yang et al. 2011; Chen et al. 2014a). We hypothesize that the RLKs, encoded by At01g35710 and At04g08850, with high identities of amino acid sequence to XIK1, might also regulate the FLS2- and EFR-mediated early signaling through a similar mechanism as that of XIK1.
LysM-RLKs, another RLK subgroup, have been shown to mediate PTI in rice and Arabidopsis (Miya et al. 2007; Shimizu et al. 2010; Chen and Ronald 2011). The LysM conserved motif of these receptors is reported to mediate plant-pathogen interactions by recognizing chitin molecules derived from fungal pathogens such as Magnaporthe oryzae (Chen and Ronald 2011). Thus, it would also be of great interest to determine whether XIK1 or other proteins with a similar structure of XIK1 regulate LysM-RLK-mediated immunity in the future work.
Conclusion
Our study revealed that the LRR RLK gene XIK1 is pathogen responsive and its expression is induced rapidly upon the inoculation with Xoo. Silencing of XIK1 comprises XA21-mediated resistance to Xoo and XIK1 positively regulates XA21-mediated resistance.
Methods
Plant growth, Xoo inoculation and disease resistance determination
All of the plants were grown in rice fields (Wenjiang, Chengdu, Sichuan, China). Rice Kitaake is used for transcriptional expression pattern analysis. The silencing construct of XIK1 (XIK1RNAi) was introduced into the Xa21 (ProA-Xa21) plants with full resistance to the Xoo strain PXO99A (Hopkins et al. 1992) to obtaine double transgenic plants containing Xa21 and XIK1RNAi. Plants were inoculated with the Xoo strain PXO99A. The inoculation and disease lesion length determination was performed according to the methods described previously (Song et al. 1995). Statistical analysis was performed using a one-way ANOVA followed by post hoc Tukey HSD. The leaves inoculated with Xoo were collected and prepared for transcriptional expression analyses as described below.
RNA extraction and real time RT-PCR analyses
Total RNA was isolated from rice plant tissues using Invitrogen RNA isolation reaction, TRIzol (Invitrogen), following the procedures of the manual. The first strand cDNA was synthesized using the Takara reverse transcription kit (Takara). Quantitative real time PCR (qRT-PCR) was performed on a Bio-Rad CFX96 Real-Time System coupled to a C1000 Thermal Cycler (Bio-Rad). For qRT-PCR reactions, the SsoFastEva Green Super mix was used. The gene expression levels relatively to the ubiquitin5 gene (LOC-06g46770) were analyzed using delta-delta ct method. All of the qRT-PCR primers are shown in Additional file 5: Table S1.
Constructions
For XIK1 RNAi construct, a 361 bp unique cDNA sequence (from 72 nt to 432 nt) of XIK1Ri amplified by primer pair XIK1Ri-1/-2 (shown in Additional file 5: Table S1) and was cloned into _pCR®8™⁄GW/TOPO® (Invitrogen) vector to create the construct pCR8-XIK1Ri. pCR8-XIK1Ri was then inserted into pANDA (Miki and Shimamoto 2004) through LR recombination to create the construct pANDA-XIK1Ri.
Generation of transgenic rice plants
The RNAi construct for XIK1, pANDA-XIK1Ri, was introduced into Xa21 plants through Agrobacterium-mediated transformation according to the method described previously (Chern et al. 2005). Because the ProA-Xa21 transgenic plants are mannose resistant, XIK1Ri transgenic plants in Xa21 genetic background were selected with hygromycin in our study.
Accession numbers
RGAP: LOC_Os02g34790, LOC_Os02g34750
Acknowledgments
The authors thanks for the grants from National Natural Science Foundation of China (NSFC 31171622; 31371705), “Hundred Talents Plan” foundation of Sichuan, Youth foundation of Sichuan (13QNJJ0076) in China and Sichuan Agricultural University “High Talents” start-up foundation to X. W. Chen, the grants Specialized Research Fund for doctoral program of Higher Education (20135103120004) to J. Wang and the grants Specialized Research Fund for doctoral program of Higher Education (20125103120011) to W. T. Li. We also thank Dr. Pamela C Ronald from University of California at Davis for helpful discussion.
Abbreviations
- DPI
Days post inoculation
- XIK1
Xoo-induced kinase 1
Additional files
Identification LRR-RLKs induced by Xoo inoculation in 2 hours. These genes were identified from the array data (http://www.ricearray.org/) of experiment “Comparative transcriptional profiling of rice undergoing infection by X.oryzae pv oryzae or by X.oryzae pv.oryzicola”. We selected the LRR-RLKs which were specially induced by Xoo strain PXO99A in susceptible variety Nipponbare.
Identification of transgenic plants with reduced expression of XIK1. (A) A special fragment was chosen for XIK1 RNAi construct. (B) The relative expression level of XIK1 was determined by qRT-PCR using RNA extracted from rice leaves of RNAi transgenic lines as indicated and ProA-Xa21 plants. All data were normalized to the expression of the ubiqutin5 reference gene. The average expression level from one representative biological experiment was shown. Error bars indicate SD of three technical replicates. Three independent biological experiments were repeated with the similar results obtained.
Determination of the resistance of the T0 and T1 generation of XIK1Ri transgenic plants to Xoo. Six weeks-old plants were inoculated with the Xoo strain PXO99A. The ProA-Xa21 and Kitaake (Kit) were used as resistant and susceptible controls, respectively. From (A) through (C), T0 Transgenic plants carrying XIK1Ri developed long water-soaking lesions. Photograph depicts the representative leaves from the plants at 14 DPI. (D), Lesion length was measured 14 days post inoculation with Xoo strain PXO99A. “Ri(+)” indicates T1 segregants carrying the transgene XIK1Ri whereas “Ri(−)” indicates T1 segregants lacking of XIK1Ri. This experiment was repeated three times with similar results.
Expression level of Xa21 in the plants silenced for XIK1 by qRT-PCR. All data were normalized to the expression of the ubiqutin5 reference gene.
The primers used in this study.
Footnotes
Haitao Hu, Jing Wang and Chan Shi contributed equally to this work.
Competing interests
The authors declare that they have no competing interests
Authors’ contributions
JW and XC conceived this study. HH, CS, CP, WL, MH, CY, JY, JW, BM, YW performed the experiments. HH, CS, WL, JW, SL and XC analyzed data. JW and XC wrote the manuscript. All authors approved the manuscript.
Contributor Information
Haitao Hu, Email: 1175091678@qq.com.
Jing Wang, Email: jingwang406@gmail.com.
Chan Shi, Email: 451623140@qq.com.
Can Yuan, Email: 609669081@qq.com.
Chunfang Peng, Email: 307014162@qq.com.
Junjie Yin, Email: 575929725@qq.com.
Weitao Li, Email: weitao-li@live.cn.
Min He, Email: cqhemin_1983@163.com.
Jichun Wang, Email: wang_jichun88@163.com.
Bintian Ma, Email: btma02@vip.sina.com.
Yuping Wang, Email: wypscwj@126.com.
Shigui Li, Email: lishigui_sc@263.net.
Xuewei Chen, Email: xwchen88@163.com.
References
- Century KS, Lagman RA, Adkisson M, Morlan J, Tobias R, Schwartz K, Smith A, Love J, Ronald PC, Whalen MC. Short communication: developmental control of Xa21-mediated disease resistance in rice. Plant J. 1999;20(2):231–236. doi: 10.1046/j.1365-313x.1999.00589.x. [DOI] [PubMed] [Google Scholar]
- Chen X, Ronald PC. Innate immunity in rice. Trends Plant Sci. 2011;16(8):451–459. doi: 10.1016/j.tplants.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chern M, Canlas PE, Ruan D, Jiang C, Ronald PC. An ATPase promotes autophosphorylation of the pattern recognition receptor XA21 and inhibits XA21-mediated immunity. Proc Natl Acad Sci U S A. 2010;107(17):8029–8034. doi: 10.1073/pnas.0912311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zuo S, Schwessinger B, Chern M, Canlas PE, Ruan D, Zhou X, Wang J, Daudi A, Petzold CJ, Heazlewood JH, Ronald PC (2014a) An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol Plant, doi:10.1093/mp/ssu003 [DOI] [PMC free article] [PubMed]
- Chen X, Zuo S, Schwessinger B, Chern M, Canlas PE, Ruan D, Zhou X, Wang J, Daudi A, Petzold CJ, Heazlewood JH, Ronald PC. An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol Plant. 2014;7(5):874–892. doi: 10.1093/mp/ssu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Canlsa PE, Fitzgerald H, Ronald PC. NRR, a negative regulator of disease resistance in rice that interacts with Arabidopsis NPR1 and rice NH1. The Plant Journal. 2005;43(5):623–635. doi: 10.1111/j.1365-313X.2005.02485.x. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119(2):397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89(4):575–585. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Gish LA, Clark SE. The RLK/Pelle family of kinases. Plant J. 2011;66(1):117–127. doi: 10.1111/j.1365-313X.2011.04518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5(6):1003–1011. doi: 10.1016/S1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999;18(3):277–284. doi: 10.1046/j.1365-313X.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- Greeff C, Roux M, Mundy J, Petersen M. Receptor-like kinase complexes in plant innate immunity. Front Plant Sci. 2012;3:209. doi: 10.3389/fpls.2012.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins CM, White FF, Choi SH, Guo A, Leach JE. Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 1992;5(6):451–459. doi: 10.1094/MPMI-5-451. [DOI] [PubMed] [Google Scholar]
- Hwang SG, Kim DS, Jang CS. Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and Arabidopsis. Genetica. 2011;139(8):1023–1032. doi: 10.1007/s10709-011-9604-y. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Chen X, Ding X, Wang Y, Chen Q, Song WY. The XA21 binding protein XB25 is required for maintaining XA21-mediated disease resistance. Plant J. 2013;73(5):814–823. doi: 10.1111/tpj.12076. [DOI] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000;14(1):108–117. [PMC free article] [PubMed] [Google Scholar]
- Kolade OO, Bamford VA, Ancillo Anton G, Jones JD, Vera P, Hemmings AM. In vitro characterization of the cysteine-rich capping domains in a plant leucine rich repeat protein. Biochim Biophys Acta. 2006;1764(6):1043–1053. doi: 10.1016/j.bbapap.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Korner CJ, Klauser D, Niehl A, Dominguez-Ferreras A, Chinchilla D, Boller T, Heinlein M, Hann DR. The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol Plant Microbe Interact. 2013;26(11):1271–1280. doi: 10.1094/MPMI-06-13-0179-R. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90(5):929–938. doi: 10.1016/S0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Mazzola M, Leach JE, Nelson R, White FF. Analysis of the interaction between Xanthomonas oryzae pv. oryzae and the rice cultivars IR24 and IR-BB21. Phytopathology. 1994;84(8):392–397. doi: 10.1094/Phyto-84-392. [DOI] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45(4):490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104(49):19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu JH, Lee HS, Kao TH. Characterization of a pollen-expressed receptor-like kinase gene of Petunia inflata and the activity of its encoded kinase. Plant Cell. 1994;6(5):709–721. doi: 10.1105/tpc.6.5.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Peng Y, Chen X, Dardick C, Ruan D, Bart R, Canlas P, Ronald PC. Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol. 2008;6(9):e231. doi: 10.1371/journal.pbio.0060231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Bart R, Chern M, Canlas PE, Bai W, Ronald PC. Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS One. 2010;5(2):e9262. doi: 10.1371/journal.pone.0009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M, Valent B, Wang GL (2012) The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24(11): 4748–4762, doi:10.1105/tpc.112.105429 [DOI] [PMC free article] [PubMed]
- Park CJ, Song MY, Kim CY, Jeon JS, Ronald PC, Park CJ, Song MY, Kim CY, Jeon JS, Ronald PC. Rice BiP3 regulates immunity mediated by the PRRs XA3 and XA21 but not immunity mediated by the NB-LRR protein, Pi5. Biochem Biophys Res Commun. 2014;448(1):70–75. doi: 10.1016/j.bbrc.2014.04.093. [DOI] [PubMed] [Google Scholar]
- Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant. 2008;1(3):446–458. doi: 10.1093/mp/ssn024. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, Shibuya N. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64(2):204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE (113):re22, doi:10.1126/stke.2001.113.re22 [DOI] [PubMed]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16(5):1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai W, Zhu L, Fauquet C, Ronald PC. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270(5243):1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, et al. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8(4):735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Song WY, Ruan DL, Sideris S, Ronald PC. The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol Plant Microbe Interact. 1996;9(9):850–855. doi: 10.1094/MPMI-9-0850. [DOI] [PubMed] [Google Scholar]
- Wang YS, Pi LY, Chen X, Chakrabarty PK, Jiang J, De Leon AL, et al. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell. 2006;18(12):3635–3646. doi: 10.1105/tpc.106.046730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Cao Y, Xu C, Li X, Wang S. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor Appl Genet. 2006;113(7):1347–1355. doi: 10.1007/s00122-006-0388-x. [DOI] [PubMed] [Google Scholar]
- Yang DH, Hettenhausen C, Baldwin IT, Wu J. The multifaceted function of BAK1/SERK3: plant immunity to pathogens and responses to insect herbivores. Plant Signal Behav. 2011;6(9):1322–1324. doi: 10.4161/psb.6.9.16438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhou JM. Plant immunity triggered by microbial molecular signatures. Mol Plant. 2010;3(5):783–793. doi: 10.1093/mp/ssq035. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125(4):749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]


