Abstract
Glycans and other saccharide moieties attached to proteins and lipids, or present on the surface of a cell, are actively involved in numerous physiological or pathological processes. Their structural flexibility (that is based on the formation of various kinds of linkages between saccharides) is making glycans superb “identity cards”. In fact, glycans can form more “words” or “codes” (i.e., unique sequences) from the same number of “letters” (building blocks) than DNA or proteins. Glycans are physicochemically similar and it is not a trivial task to identify their sequence, or - even more challenging - to link a given glycan to a particular physiological or pathological process. Lectins can recognise differences in glycan compositions even in their bound state and therefore are most useful tools in the task to decipher the “glycocode”. Thus, lectin-based biosensors working in a label-free mode can effectively complement the current weaponry of analytical tools in glycomics. This review gives an introduction into the area of glycomics and then focuses on the design, analytical performance, and practical utility of lectin-based electrochemical label-free biosensors for the detection of isolated glycoproteins or intact cells.
Keywords: biosensors, glycomics, electrochemical impedance spectroscopy, electrochemistry, label-free detection, lectins
Glycomics
There is ever growing interest to switch from studying DNA/proteins, since genomics/proteomics cannot answer many fundamental questions of the cell physiology and pathology. Glycomics is able to reveal finely tuned reading mechanisms in the cell orchestra based on graded affinity, avidity and multivalency of glycans (covalently attached sugar chains to proteins and lipids) [1]. For many decades it was believed the role of the glycans (sugars) was mainly nutritional, but the story becomes more and more complex as the structure of glycans itself [2]. Glycans are better equipped to be an information coding tool compared to DNA and proteins due to a number of possible “words” (unique sequences) formed from the same number of “letters” (i.e. building units). Theoretical number of all possible hexamers (i.e. consisting of 6 building units) for glycans (1.44×1015) is few orders of magnitude larger compared to peptides (6.4 ×106) or DNA (4,096) [3,4]. The size of the cellular glycome is estimated to be in excess of 100,000–500,000 glycan modified biomolecules (proteins and lipids) with a number of unique glycans to be 3,000-7,000 [4,5,6]. This variation can explain human complexity in light of a paradoxically small genome [7,8].
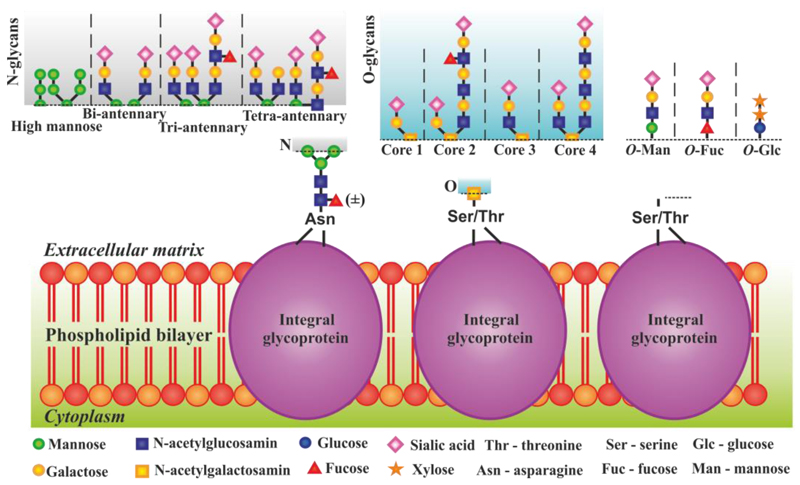
The main reason for glycomics being behind advances in genomics and proteomics is an enormous complexicity of glycans on one side with similar physico-chemical properties of glycans on the other side [9]. Glycan are attached to the protein backbone either via N- or O-linkage (N-glycans or O-glycans) with various branching points [10,11] (Figure 1). For example a subtle change in the structure of sialic acids (e.g. addition of one oxygen) or a position sialic acid residue is attached to the backbone of glycan chain have dramatic consequences upon decoding [12]. Glycan sequence cannot be directly read from the genome, but is formed by an array (up to 500) of enzymes [7,13,14,15,16,17]. As a result, a cell can produce glycans distinct from those of neighbouring cells [17] or two molecules of the same protein from the same tissue may have different glycans and different roles [7,18].
Figure 1.
Graphical representation of glycan complexity showing variability of sugar building blocks, multiple branching and attachment points.
It is predicted 50-90% of human proteins are glycosylated [6,19,20] and cells are surrounded by a protective layer of glycans - ‘glycocalyx’ [4]. This “sugary” coat is working as a busy information centre regulating interaction between cells and within cells (elongation; inflammation; fertilisation; cell growth/signalling; host-pathogen interaction; cell-cell interaction; immune response and cancer) [21,22,23,24,25,26,27,28]. Moreover, glycans can serve as a “quality control” indicator during protein synthesis with production of a misfolded protein in the absence of correct glycosylation [29]. Glycans stabilise tertiary structure of proteins, enhance protein resistance towards action of proteases and limit unwanted nonspecific lateral protein-protein interactions [23,29]. There are different blood groups and use of inappropriate blood can lose rather than safe lives, but less known is the fact changes in glycan composition are behind this phenomenon [5]. Even removal of “old” blood cells from the blood is triggered by the changed glycan density on the surface of blood cells [30,31]. Moreover, host organism has to keep out beneficial intestinal bacteria at some distance (again based on glycan recognition) from the intestine to avoid harmful immune response [32].
Changed glycan identity has a dramatic effect in the pathological processes such as progression of various diseases (e.g. Chagas disease, sleeping sickness, lyme disease, chronic inflammation, HIV/influenza invasion and various forms of cancer) [33,34,35,36,37,38,39,40]. Binding of viruses to beneficial intestinal flora with enhancement of viral stability for subsequent viral transmission is mediated by glycans, as well [41]. Thus, better understanding of glycan mediated pathogenesis can help to develop more efficient tools for disease treatment. There are actually some strategies developed for “neutralisation” of various forms of HIV viruses [42,43,44], more efficient vaccines against autoimmune diseases [45,46,47] and better diagnosis and therapy of various diseases [48]. Moreover, many previously established and even commercially successful strategies how to treat diseases are currently revisited in light of glycan recognition in order to lower side effects, enhance serum half-life or to decrease cellular toxicity [6,7,49].
A new emerging and powerful glycomics tools such as biochips/arrays, high throughput mass spectroscopy, liquid spectroscopy and nuclear magnetic resonance are behind huge progress in the field [50,51,52,53,54,55,56,57]. Biochips/arrays are clearly benefiting from more mature and successful technology of DNA biochips [58,59,60,61,62, 63,64,65,66,67]. The advantage of using biochips is a minute consumption of biorecognition elements printed at high density with high throughput of assays. Moreover, biochip technology allows quantitative and systematic identification of glycan interactions. Biochips are usually based on a fluorescent detection with a need to fluorescently label a sample or a biorecognition element [68,69]. This can cause unwanted variability in labelling and biorecognition [50] and thus other formats of analysis working in a label-free mode of detection are vital.
Lectins
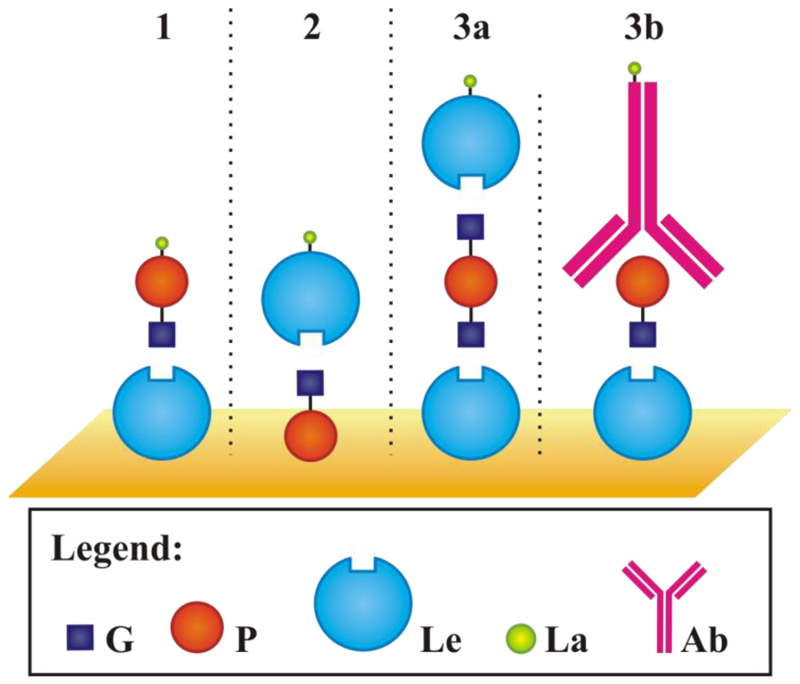
Lectins (lat. legere = to choose) are proteins able to recognize and reversibly bind to free or bound mono- and oligosaccharides [70,71]. They cause agglutination and precipitation of saccharides-containing biomolecules and even whole cells (erythrocytes), hence the archaic term agglutinins. However, they are not catalytic active and do not participate in the immune response of higher organisms. Lectins are usually found in viruses, bacteria and fungi, plants and animals, too. They are therefore a relatively heterogenous group of oligomeric proteins belonging to distinct families with similar sequences, these days used for the identification of new ones. They are natural glycocode decipherers [4], with a wide range of applications in sensitive, rapid and high-throughput assays. Lectins are mostly isolated from natural sources using different methods and nowadays they are also produced using recombinant DNA technology [72]. For biosensing purposes three distinct configurations with lectins involved can be applied i.e. lectins directly immobilised on a solid support with subsequent detection of glycan (Figure 2-1), an inverse configuration with attachment of a glycan followed by lectin biorecognition (Figure 2-2) and a sandwich format with a glycan sandwiched between surface-bound lectin and a second biorecognition pair (lectin - Figure 2-3a or antibody Figure 2-3b).
Figure 2.
Typical lectin-based biorecognition configurations with a direct detection (1), an indirect detection (2) and two forms of a sandwiched assay protocol (3a and 3b). G – glycan, P – protein, Le – lectin, La – label, Ab – antibody.
Label-free biosensors
According to IUPAC a biosensor is defined as a self-contained integrated device, which is capable of providing specific quantitative or semi-quantitative analytical information using a biological recognition element (biochemical receptor) which is retained in direct spatial contact with a transduction element [73,74]. In a biochip/microarray format of assays the part with an immobilised biorecognition element is not an integral part of a reader device and such devices are thus not biosensors. Label-free mode of operation is an essential tool for studying glycomics, since labelling can introduce unwanted variability in a biorecognition event [50]. In this chapter we will deal only with biosensors working in a label-free mode of action and construction/application of lectin biochips/arrays is thus not considered.
Electrochemical biosensors
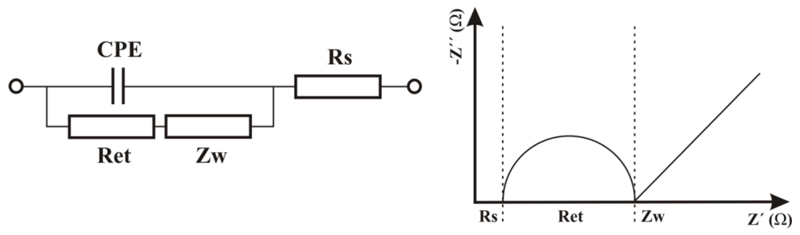
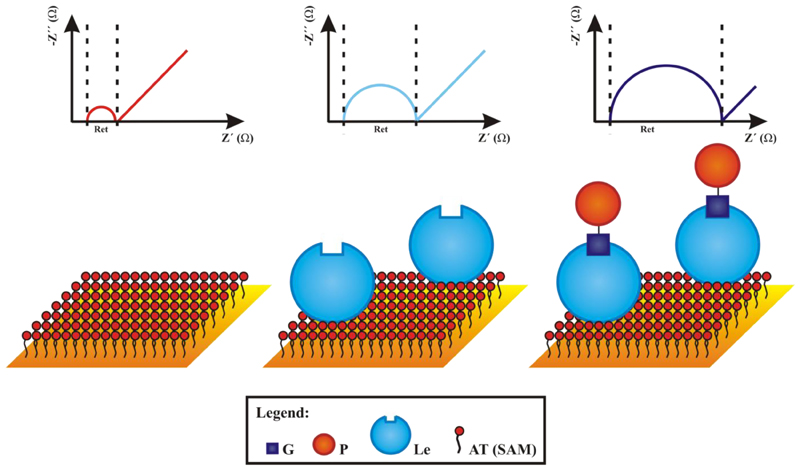
Electrochemical detection is quite often utilised in combination with other techniques such as separation, enzymatic digestion of glycans in the field of glycomics for some time [75]. A true potential of the technique is in biosensor construction due to advances compared to other detection means such as operational simplicity, extraordinary sensitivity, low cost and rapid, real-time detection [76]. The most frequently used label-free electrochemical technique is EIS (electrochemical impedance spectroscopy). This technique is based on an electric perturbation (small alternating current amplitude) of a thin layer on the conductive surface with ability to provide characteristics (impedance, resistance and capacitance) of this interface utilisable in sensing by employment of an equivalent circuit for data evaluation (Figure 3 left). EIS results are typically transformed into a Nyquist plot, which can provide information about electron transfer resistance Ret in a direct way (Figure 3 right). When a biorecognition took place, the double layer is modified and a subtle change in double layer characteristics can be used for detection (i.e. Ret as shown in Figure 4). EIS investigation is most frequently performed in the presence of a redox probe, a mixture of ferricyanide and ferrocyanide, with change in the charge transfer resistivity Ret of the interface used for detection. EIS is extensively used as a non-destructive technique for reliable analysis of surface conditions such as binding and desorption processes at electrode surfaces [77,78,79,80]. Subsequently, EIS allows complex biorecognition events to be probed in a simple, sensitive and label-free manner and is being increasingly popular to develop electrochemical lectin-based biosensors for glycan determination.
Figure 3.
A scheme of a typical equivalent circuit applied for EIS data evaluation (on left) with a Nyquist plot representation used for Ret reading (on right). CPE – capacitance, Ret – electron transfer resistance, Rs – resistance of the solution and Zw – a Warburg element.
Figure 4.
Schematic representation of a change in Ret with increased loading of biomolecules on a surface modified by SAM. For meaning of other symbols see Figure 2, AT - alkanethiol.
Although the topic of this review has been partly covered in recent excellent reviews [6,55,81,82], the main aim of this paper is to catch current/recent trends in the emerging field of electrochemical label-free detection of glycans employing lectins allowing to work in concentration ranges down to fM – aM level. In the context of this review paper, by a label-free biorecognition is considered any approach in which an initial interaction between lectin and a glycan is performed without any label present on lectin or a glycan structure, while in the subsequent step needed to generate the analytical signal lectin or an antibody might be labelled (see Figure 7 and 8).
Figure 7.
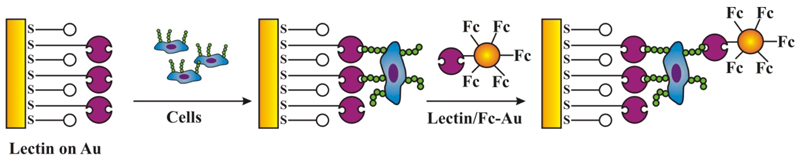
Step by step modification of gold electrode by lectin linked to SAM with an incubation of the biosensor with cells and final signal generation by formation of sandwich with lectin bound to electrochemical probe ferrocene (Fc) loaded gold nanoparticle.
Figure 8.
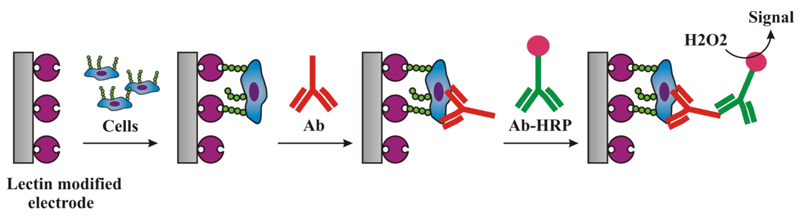
Step by step modification of an electrode by lectin with an incubation of the biosensor with cells and final signal generation by formation of sandwich with primary antibody and secondary antibody labelled with peroxidase.
Detection of glycans on glycoproteins/liposaccharides
Initial efforts to detect glycan determinants of the glycoproteins by electrochemical means were launched by the group of Prof. Joshi [83,84]. In the first report a competitive form of assay was chosen to detect various saccharidic analytes including the cancer-associated T antigen. Arachis hypogaea agglutinin (peanut agglutinin, PNA) was covalently immobilised on the surface of a mixed SAM (self-assembled monolayer) with an activated –COOH group as an attachment point. A detection principle was based on a competition between analyte and nanoparticle-labelled analyte. Both types of analytes were mixed together and incubated with a lectin on the transducer surface. After binding, CdS quantum dot nanoparticles were electrochemically stripped and cadmium released was detected using square wave voltammetry. Finally, the concentration of the analyte was inversely proportional to the electrochemical signal obtained [83]. Even though a label was used in a detection scheme, the biorecognition of the analyte by the lectin was of label-free form of interaction. In the following study the group focused on a truly label-free detection scheme based on electrochemical impedance spectrometry (EIS) employed for the first time in 2007 [84]. A sialic acid binding Sambucus nigra agglutinin (SNA) and a galactose binding PNA were covalently immobilised on printed circuit board electrodes. The assays were used for detection of artificial (glycan attached to the gold nanoparticle) and natural glycoconjugates (glycoprotein asialofetuin and its sialylated form fetuin). The assays were really quick with a response time of 80 s and with sensitivity of glycoprotein detection down to 10 pg mL-1 (e.g. 150 fM), while using a cost-effective electrode material [84].
A group of Prof. Oliveira put a substantial effort to use lectin modified surfaces with EIS detection for discrimination between human samples from patients infected by dengue fever (DF) or dengue hemorrhagic fever (DHF) and those of healthy individuals (H). Dengue fever is a mosquito-borne virus disease with a range of effects from an acute febrile illness called dengue fever (DF) to more severe, life threatening illnesses, called dengue hemorrhagic fever (DHF). The latter has a higher mortality rate. The dengue virus non-structural glycoprotein, NS1, may present different molecular mass depending on glycosylation. In the initial study two lectins concanavalin A (Con A) and a lectin isolated from Cratylia mollis seeds (CramoLL) were immobilised on gold nanoparticles with polyvinyl butyral and deposited on the gold electrode. Ovalbumin was chosen as a glycoprotein standard to probe analytical utility of both immobilised lectins with detection limit in the low nM range [85]. Real samples were subsequently analysed on a Con A modified surface with EIS analysis. The results showed differences in the Ret between healthy serum samples and DF samples with a potential to detect infection [86]. In the following study human serum samples DF, DHF and H were analysed with EIS on the electrode with immobilised Con A [87]. A 3-D graph representation revealed a clear clustering of serum samples with their distinct separation in the 3-D space as a result of different glycan pattern. Thus, this data presentation may allow the distinction among sub-classes of an immunological response as in the case of samples with Dengue virus contamination [87]. Besides Con A, another lectin CramoLL was tested to prove its utility as a sensing element to distinguish between serums of patients contaminated with dengue serotypes [88]. In order to enhance loading of the lectin on the surface, Fe3O4 nanoparticles were employed within polyvinyl butyral matrix. The biosensor exhibited a wide linear range to different concentrations of serum samples with high reproducibility of assays (RSD in the range 2.85-3.77%). Moreover the selectivity of detection was excellent, as well, with only 5.2% of the specific signal for the control sample. Authors suggested the platform of detection can be useful as a reusable and sensitive biosensor device in dengue diagnosis [88]. The most recent contribution from the group for detection of dengue serotypes was based on lectin isolated from Bauhinia monandra (BmoLL) [89]. The lectin was immobilised via electrostatic docking on gold nanoparticle-polyaniline nanocomposite deposited on gold electrode. More complex picture of the interaction of samples with different serotypes of a disease with the lectin was shown in a 3-D graph based on three circuit elements from EIS data fitting. These results again showed clear distinction between serotypes by separate occupation of space within a 3-D graph. As in the previous case, negative sample showed only a subtle change in the Ret (0.46 kΩ), while change of Ret for serum samples varied from 5.58 to 26.7 kΩ. This proves that not only different serotypes can be revealed, but the biosensor is able clearly distinguish between healthy and infected serum samples [89]. CramoLL lectin immobilised through electrostatic interaction on a gold nanoparticles modified surface was successfully used in analysis of liposaccharides isolated from different bacterial strains [90]. The results showed differences in the biosensor response depending on the source liposaccharides were isolated from. The biosensor showed high reproducibility of assays expressed as RSD of measurements ranging from 4.89 to 5.78% [90].
In the last report presented here EIS was used for evaluation of capacitance changes rather than changes in Ret after binding took place [91]. The surface of the silicon chip with an array of gold electrodes was interfaced with nanoporous alumina membrane with high density of nanowells on the top of each particular electrode. Two lectins SNA and MAA (Maackia amurensis agglutinin) were immobilised on gold electrodes modified by SAM in a covalent fashion by amine coupling. The biosensor performance was tested with a standard glycoprotein fetuin or asialofetuin and protein isolated from a cultured human pancreatic cancer cell line BXPC-3. The results obtained with both glycoforms of fetuin were in a good agreement with glycan composition and affinity of lectins for a particular glycan. The biosensor offered high reliability of assays and a good agreement with enzyme-linked lectin assays (ELLA). The detection limit of a biosensor for its analyte was 5 orders of magnitude lower compared to ELLA (i.e. 20 fM vs. 4.6 nM). An assay time for the biosensor of 15 min was much shorter compared to 4 h needed for ELLA. Moreover, a minute amount of sample (10 μl) was sufficient for the analysis by the biosensor [91].
A systematic investigation EIS can offer in a label-free detection of glycans started in our group recently with the focus on careful control of an immobilisation protocol and a blocking procedure. Results obtained so far are promising with detection limit down to 1 fM level with a linearity range spanning 7 orders of magnitude [92]. Moreover, integration of gold nanoparticles within EIS lectin-based biosensing device offered detection limit down to 1 aM level (unpublished results).
EIS has been used to probe lectin specificity on glycan patterned surfaces with the aim to investigate lectin affinity depending on the glycan surface density [93] or depending on glycan neighbouring functional groups [94]. These studies are important for further and more comprehensive understanding of the true potential lectins have in preparing biosensors or lectin-based diagnostic devices. A wide range of “traditional” immobilisation protocols for attachment of glycan to the surface is available including covalent binding on activated/modified gold and glass, adsorption on nitrocellulose or polystyrene-coated glass, adsorption of hydrophobic glycans (linked to aliphatic hydrocarbon) on hydrophobic surfaces, immobilisation of biotinylated glycans on streptavidin-modified surfaces, etc. [95,96,97,98]. In order to fulfil a requirement to control both orientation and density of glycan on surfaces new and reliable patterning protocols are required [99,100]. Recent efforts in the application of glycan patterned surfaces addressed these issues by application of thiolated glycans forming a mixed self-assembled monolayer in the presence of a diluting thiol on gold surfaces. This protocol allowed to control density of glycans effectively on the surface, exposure of glycan from a modified layer and an overall charge of the surface [94]. Copper(I)-catalysed azide-alkyne cycloaddition better known as a “click” chemistry was successfully applied to control density of glycans on the surface by grafting glycan bearing azido terminating arm on alkyne modified boron-doped diamond surface [93].
Detection of glycans on cell surfaces
A group of Prof. Ju has been mostly involved in the use of lectins in profiling of glycan determinant on viable cells of various cancer diseases such as tumour cell line BGC-823 [101], a human chronic myelogenous leukaemia line K562 [102] and human gastric carcinoma cells BGC-823 [103,104,105]. These results showed the prospect of using lectins in glycoprofiling in combination with electrochemical detection platform. Since these assays were based on attachment of cells on a modified surface and only then labelled lectins were used for glycan recognition the assay format does not fulfil definition for the biosensor or for a label-free detection and thus are not discussed here in details.
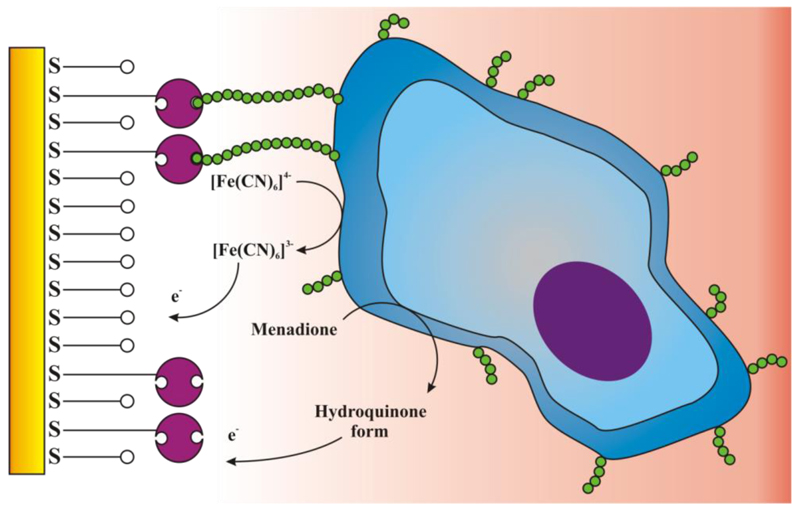
A group of Prof. Mikkelsen pioneered application of lectins in sensing of viable cells based on glycan-lectin recognition [106,107]. Respiration of attached viable cells was chosen for a signal generation with a mixture of two redox probes – ferricyanide and menadione (vitamin K) present in the assay buffer (Figure 5). The role of a hydrophobic redox probe menadione was to reach respiratory enzymes present either in the cytoplasm or within cytoplasmatic membrane and to transfer electrons to the hydrophilic redox probe ferricyanide working as a shuttle to transfer electrons to the external electrode [108].
Figure 5.
Graphical representation of a label-free approach of lectin-based cell biorecognition utilising intracellular enzymatic machinery for electrochemical signal generation.
Ten different lectins were immobilised on the surface of various membranes by a glutaraldehyde-assisted covalent coupling and physically attached to the platinum electrode. Six microbial species were incubated with the lectin modified membranes for 2 min and ferrocyanide released (a product of ferricyanide reduction by microbes) was detected using chronocoulometry. Principal component analysis was used for data treatment and all six microbial species were successfully distinguished. Moreover results obtained by this novel lectin-based biosensor device were in a general agreement with classical agglutination test [106]. The initial study was later extended to distinguish four different E. coli subspecies with the same electrochemical approach and implementation of principal component analysis. Moreover, authors integrated the device with a screen-printed array for cost-effective and parallel assays. The total time of analysis was 40 min and the method was successfully validated with optical density assays for monitoring of cell growth during shake-flask fermentation [107].
A group of Prof. Ju besides development of various label-based detection tools for analysis of viable cells was active in label-free ones, as well. In the first paper four lectins were covalently immobilised via amine coupling on the electrode surface modified by carbon nanohorns to enhance accessibility of glycans from the cell surface [109]. Carbon nanohorn modified surface was more efficient in binding of unprocessed mammalian tumour cells K562 when compared to the surface modified by single- and multi-walled carbon nanotubes. The highest sensitivity of cell detection was observed on the surface modified by wheat germ agglutinin, what suggest for high expression of (GlcNAc)2 and/or sialic acid on the cell surface. Results were in an agreement with those obtained by a flow cytometry, confirming the validity of the proposed method. Subsequently EIS-based method was applied for detection of 30-azido-30-deoxythymidine, a model drug employed in inhibition of nucleotide-sugar transport affecting glycosylation of cell surfaces. Physiological differentiation of the cells induced by addition of sodium butyrate was also monitored by this approach. Results really showed such dynamic changes of the cell glycosylation can be effectively tracked by the method [109]. In the most recent study from the group Con A was immobilised on an electrode modified by multi-walled carbon nanotubes and the tumour cells were detected with a linear response from 1×104 to 1×107 cells mL-1 [110] (Figure 6). Ability of lectin biosensors to detect subtle changes in the glycan density on the surface of the cells was proved by using an inhibitor of a specific mannosidase II, an enzyme responsible for selective removal of mannose units from the surface. The study showed increased Ret resistance using EIS, suggesting increased amount of mannose units on the cell surface. EIS results were highly reproducible with RSD of assays from 4.8 to 6.2% depending on the concentration of cells in the sample [110].
Figure 6.
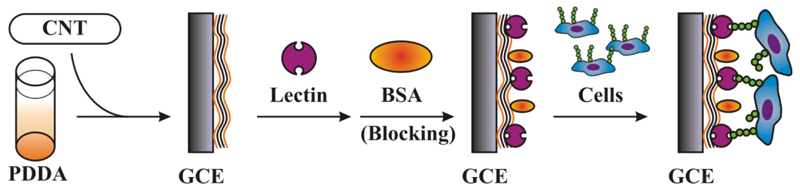
Step by step modification of a glassy carbon electrode (GCE), by a dispersion of CNTs in poly(diallyldimethylammonium chloride) (PDDA), lectin immobilisation and blocking of the surface by bovine serum albumin (BSA) with a final incubation of the biosensor with cells.
Sulfate-reducing bacteria are anaerobic microorganisms producing sulphide, a highly corrosive and toxic chemical, what can cause serious problem for industries, economies and ecological systems. Therefore, a rapid and easy method for their detection based on EIS with Con A covalently immobilised on 11-mercaptoundecanoic acid SAM layer on gold electrode was developed [111]. A remarkable sensitivity of the biosensor is underlined by a linear relationship between Ret and concentration of bacterial cells in the range from 1.8×100 to 1.8×107 cfu mL-1 (cfu = colony forming unit). Moreover, the electrochemical lectin biosensor was successfully validated by a classical biochemical method of cell counting taking 15 days to complete [111].
Nine different lectins were used for probing 3 different microbial strains on screen-printed gold electrodes [112]. The Ret using EIS varied linearly with the logarithmic value of E. coli concentration from 5.0×103 to 5.0×107 cfu mL-1 using Con A. Different response profiles found for different lectins were treated by principal component analysis, which allowed classification and distinction of bacteria. Moreover, electrochemical monitoring of β-galactosidase activity within the surface attached bacteria helped to distinguish between E. coli and Staphylococcus aureus, which showed a similar affinity for Con A. EIS-based method of bacteria detection showed advantages compared to conventional plate counting method such as a short assay time of 1 h. Moreover, the method required only a small amount of test solutions and cheap reagents [112].
Two lectins Con A and Ricinus communis agglutinin (RCA) electrostatically adsorbed on a positively charged film prepared by layer-by-layer approach were employed for detection of gram-negative bacteria, yeasts and mammalian cells with a high sensitivity [113]. The biosensor revealed different binding pattern on two lectin modified surfaces with a detection limit down to 1×103 cfu mL-1 for microbes, while a detection limit down to 1×104 cfu mL-1 was achieved for human cells. A linear dependence of bacterial concentration on Ret spanned from 2 to 4 orders of magnitude depending on a particular cell type [113].
Due to large surface area and presence of a high density of glycan determinants on the cell surface a sandwich assay for detection of cells is possible. In this case the cell of interest is captured by the lectin followed by introduction of a second labelled reagent (i.e. the same lectin or an antibody) to the system for electrochemical signal generation. This novel sandwich platform of detection was introduced in 2010 and applied to discriminate between glycan determinants present on the cell surface of normal and cancer cells derived from human lung, liver and prostate [114]. The method was based on binding of cells on the surface with either covalently linked SNA or Con A. After cell binding took place lectin attached to gold nanoparticle loaded with a redox probe interacted with the cells forming a sandwich configuration. Electrochemical signal in the form of a differential pulse voltammetry was generated by a redox probe. The results showed mannose was present at high levels in both normal and cancer cells, while enhanced amount of sialic acid was expressed only on cancer cells. The results of the method were in a good agreement with the results obtained from fluorescent microscopy investigation. A typical limit of detection was in the range from 7×103 to 1.1×105 cells mL-1 depending on the cell type. An extremely high glycan density on the surface of cancer cells QGY-7703 resulted in a remarkable detection limit down to 5 cells mL-1. When the biosensor was calibrated, calculation of average number of glycan molecules on a single cell surface was possible i.e. 4.5 × 109 sialic acid molecules for A549 cells [114]. A similar sandwich concept was employed for detection of K562 leukemic cells [115]. In that case Con A was covalently immobilised on a SAM layer on gold via amine coupling and cells bound to the lectin modified surface were electrochemically “visualised” by the use of Con A attached to gold nanoparticle loaded with ferrocene redox probe (Figure 7). Cyclic voltammetry was used to detect redox probe and the current was proportional to the concentration of cells in the range from 1.0×102 to 1.0×107 cells mL-1. Moreover, the biosensor was able to quantify an absolute number of cells bound to the biosensor surface and the number of mannose units on the surface of a single cell (i.e. 4.7×109 mannose determinants), a density agreeing with an enzymatic method of analysis [115].
A sandwich assay protocol described above was slightly modified by the group of Prof. Zhu with application of a primary antibody (against P-glycoprotein) and a secondary antibody labelled with horseradish peroxidase [116] (Figure 8). The main function of the enzyme was to oxidise a redox probe for a signal generation. Such biosensor was able to detect HeLa cells in the concentration ranging from 8.0×102 to 2.0×107 cells mL-1 with a limit of detection of 500 cells mL-1. Moreover, the biosensor provided information about the number of mannose moieties (4×1010 molecules) and number of P-glycoproteins (8.5×106 molecules) on a single HeLa carcinoma cell [116].
Conclusions
A short introduction about emerging field of glycomics with the focus on the impact of changed glycan “identity” in the cell physiology or pathology was given. Challenges ahead in the field and current array of tools available to crack the “glycocode” were briefly described. In more details electrochemical lectin based biosensors working in a label-free mode of operation were characterised showing practical utility in analysis of real samples or for identification of various cells including harmful ones. EIS has been successfully applied as a transducer for label-free lectin biosensors for detection of glycans. In almost all cases discussed here an equivalent circuit showed in Figure 3 was applied with Ret element employed as an output signal. Only in one study a fixed frequency was utilised as a reading signal [84] and in another study change of the capacitance of the lectin-based interface was read for biosensor calibration and application [91]. Most likely the main reason for an occasional use of capacitance compared to Ret as a transducing signal is a requirement to have a high capacitance biorecognition layer, which should not be permeable to ions from the solution, what is not that easy to achieve [79,117,118]. Even though a single element of Ret was sufficient as a biosensor output signal for most studies Prof. Oliveira introduced a 3-D map with additional EIS elements involved [87,89]. This approach allowed a lectin biosensor to distinguish between two serotypes with similar Ret and this might be a way to enhance overall robustness of the method. Advantages of the use of EIS in combination with biosensing were already discussed (i.e. high sensitivity, wide linear range, label-free mode of detection and a short analysis time). Few disadvantages discussed in the literature i.e. relatively high cost of the method compared to other electrochemical techniques and a complex data analysis are not that problematic nowadays [119]. For example the cost of EIS module is a fraction of a purchasing cost for Surface Plasmon Resonance instrument and data evaluation can be visually checked during a fitting procedure. Moreover the software usually allows to show data in other than Nyquist plots, what can effectively identify problems during measurement. A potential limitation of using EIS in the array format might be time of 5 min needed to apply a whole range of frequencies during analysis, what can be overcome by a measurement done at a fixed frequency. This approach can have an additional benefit – i.e. a possibility to detect biorecognition in a real time and both association and dissociation phase of binding event can be read [120]. Other value-added benefits for lectin biosensors can be estimated from a clever and sophisticated use of nanomaterials including carbon nanotubes and graphene in combination with EIS or field-effect sensing [121].
Table 1.
Listing of lectins together with their systematic name, source and specificity for a particular glycan [122,123,124,125,126,127,128,129]
| Lectin | Systematic name | Source | Specificity |
|---|---|---|---|
| AAA | Anguilla anguilla | A | α-L-Fuc |
| AAL | Aleuria aurantia | F | α-L-Fuc |
| ABL | Agaricus bisporus | F | Gal β(1-3)GalNAc, GlcNAc |
| AOL | Aspergillus oryzae | MO | α-L-Fuc |
| APA (abrin) | Abrus precatorius | P | Gal β(1-3)GalNAc |
| BPA | Bauhinia purpurea | P | Gal β(1-3)GalNAc |
| CFL | Cratylia floribunda | P | α-D-Man, α-D-Glc |
| CFT agglutinin | Codium fragile subsp. tomentosoides | P | GalNAc |
| Con A | Canavalia ensiformis | P | α-D-Man, α-D-Glc |
| Con Br | Canavalia brasiliensis | P | α-D-Man, α-D-Glc |
| DBA | Dolichos biflorus | P | Gal β(1-3)GalNAc |
| DGL | Dioclea grandiflora | P | α-D-Man, α-D-Glc |
| DSA (DSL) | Datura stramonium | P | GlcNAc β (1-4)GlcNAc |
| DVL | Dioclea violacea | P | α-D-Man, α-D-Glc |
| ECA | Erythrina cristagalli | P | Gal β(1-4)GalNAc |
| EUE | Euonymus europaeus | P | Gal α (1-3)Gal |
| Favin | Vicia faba | P | α-D-Man, α-D-Glc |
| GNA | Galanthus nivalis | P | α-D-Man |
| GSA-I | Griffonia (Bandeiraea) simplicifolia | P | Gal α (1-3)Gal |
| GSA-II | Griffonia (Bandeiraea) simplicifolia | P | GlcNAc |
| HAA | Helix aspersa | A | α-GalNAc |
| HBA | Hevea brasiliensis | P | GlcNAc |
| HHL | Hippeastrum hybrid | P | D-Man α (1-3) (1-6) bound |
| HPA | Helix pomatia | A | α-GalNAc, α-GclNAc, α-Gal |
| CHA | Cymbidium hybrid | P | α-D-Man |
| Jacalin (AIL) | Artocarpus integrifolia | P | Galβ(1-3)GalNAc |
| LBA | Phaseolus lunatus | P | D-GalNAc, GlcNAc β (1-4) bound |
| LCA (LCH) | Lens culinaris | P | α-D-Man, α-D-Glc |
| LEA | Lycopersicon esculentum | P | [GlcNAc β (1-4)]2-4 |
| LFA (LMA) | Limax flavus | A | Neu5Ac α (2-3), (2-6), (2-8) bound |
| LOA | Listera ovata | P | D-Man α (1-3) bound |
| LPA | Limulus polyphermus | A | Neu5Ac |
| LTA (Lotus lectin) | Lotus tetragonolobus | P | α-L-Fuc |
| MAA (MAL, MAH) | Maackia amurensis | P | Neu5Ac α (2-3) bound |
| MHA | Myrianthus holstii | P | GlcNAc |
| MPA | Maclura pomifera | P | Galβ(1-3)GalNAc |
| NPA | Narcissus pseudonarcissus | P | α-D-Man |
| PHA-E (E4-PHA) | Phaseolus vulgaris | P | Man oligosaccharides |
| PHA-L (L4-PHA) | Phaseolus vulgaris | P | branched β (1-6) GlcNAc |
| PNA | Arachis hypogaea | P | Galβ(1-3)GalNAc |
| PSA | Pisum sativum | P | α-D-Man |
| PSL | Psathyrella velutina | F | β-D-GlcNAc |
| PWM | Phytolacca americana | P | [GlcNAc]3 |
| RCA-I | Ricinus communis | P | β-D-Gal |
| RCA-II | Ricinus communis | P | Gal β (1-4) GalNAc |
| RPA | Robinia pseudoacacia | P | β-D-GalNAc |
| SBA | Glycine max | P | α, β-Gal, α, β-GalNAc |
| SJA | Sophora japonica | P | Galβ(1-3)GalNAc |
| SNA-I | Sambucus nigra | P | Neu5Ac α (2-6)Gal |
| SNA-II | Sambucus nigra | P | Gal, GalNAc |
| SOH | Soja hispida | P | β-D-GalNAc |
| STA | Solanum tuberosum | P | [β-D-GlcNAc]2-5 |
| TML | Tritrichomonas mobiliensis | MO | Neu5Ac α(2-3), (2-6) bound |
| UEA-I | Ulex europaeus | P | α-L-Fuc |
| UEA-II | Ulex europaeus | P | GlcNAc β (1-4)GlcNAc |
| VML | Vatairea macrocarpa | P | α-D-GalNAc |
| VVA (VVL) | Vicia villosa | P | α-D-GalNAc |
| WBA | Psophocarpus tetragonolobus | P | β-D-GalNAc |
| WFA | Wisteria floribunda | P | α, β-GalNac |
| WGA | Triticum vulgaris | P | β-D-GlcNAc, Neu5Ac |
P – Plant, A – Animal, F – Fungi, MO – Microorganism, Glc – Glucose, GlcNAc – N-Acetylglucosamine, Man – Mannose, Gal – Galactose, GalNAc – N-Acetylgalactosamine, Neu5Ac – N-Acetylneuraminic acid (NANA, sialic acid), Fuc – Fucose
Acknowledgement
The financial support from Slovak scientific grant agency VEGA 2/0127/10 and from the Slovak research and development agency APVV 0282-11 is acknowledged. This contribution/publication was the result of the project implementation: Centre for materials, layers and systems for applications and chemical processes under extreme conditions—stage II, supported by the Research and Development Operational Program funded by the ERDF. The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013) / ERC Grant Agreement n. 311532.
References
- 1.Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nature Methods. 2005;2:817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- 2.Varki A, Cummings R, Esko J, et al. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 1999. [PubMed] [Google Scholar]
- 3.Gabius H-J, Siebert H-C, André S, Jiménez-Barbero J, Rüdiger H. Chemical biology of the sugar code. ChemBioChem. 2004;5:740–764. doi: 10.1002/cbic.200300753. [DOI] [PubMed] [Google Scholar]
- 4.Gabius H-J, André S, Jiménez-Barbero J, Romero A, Solís D. From lectin structure to functional glycomics: principles of the sugar code. Trends Biochem Sci. 2011;36:298–313. doi: 10.1016/j.tibs.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Cummings RD. The repertoire of glycan determinants in the human glycome. Mol BioSystems. 2009;5:1087–1104. doi: 10.1039/B907931A. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham S, Gerlach JQ, Kane M, Joshi L. Glyco-biosensors: recent advances and applications for the detection of free and bound carbohydrates. Analyst. 2010;135:2471–2480. doi: 10.1039/C0AN00276C. [DOI] [PubMed] [Google Scholar]
- 7.Schmaltz RM, Hanson SR, Wong C-H. Enzymes in the synthesis of glycoconjugates. Chem Rev. 2011;111:4259–4307. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]
- 8.van Kasteren SI, Kramer HB, Jensen HH, Campbell SJ, Kirkpatrick J, Oldham NJ, Anthony DC, Davis BG. Expanding the diversity of chemical protein modification allows post-translational mimicry. Nature. 2007;446:1105–1109. doi: 10.1038/nature05757. http://www.nature.com/nature/journal/v446/n7139/full/nature05757.html_-_a1 [DOI] [PubMed] [Google Scholar]
- 9.Bertozzi CR, Kiessling LL. Chemical glycobiology. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 10.Mislovičová D, Katrlík J, Paulovičová E, Gemeiner P, Tkac J. Comparison of three distinct ELLA protocols for determination of apparent affinity constants between Con A and glycoproteins. Colloids Surf B: Biointerf. 2012;94:163–169. doi: 10.1016/j.colsurfb.2012.01.036. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee DK. N-glycans in cell survival and death: Cross-talk between glycosyltransferases. Biochim Biophys Acta Gen Subj. 2012 doi: 10.1016/j.bbagen.2012.01.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariño K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nature Chem Biol. 2010;6:713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- 13.Harvey DJ, Merry AH, Royle L, Campbell MP, Dwek RA, Rudd PM. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics. 2009;9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]
- 14.Lepenies B, Seeberger PH. The promise of glycomics, glycan arrays and carbohydrate-based vaccines. Immunopharm Immunotox. 2010;32:196–207. doi: 10.3109/08923970903292663. [DOI] [PubMed] [Google Scholar]
- 15.Horlacher T, Seeberger PH. Carbohydrate arrays as tools for research and diagnostics. Chem Soc Rev. 2008;37:1414–1422. doi: 10.1039/B708016F. [DOI] [PubMed] [Google Scholar]
- 16.Laurent N, Voglmeir J, Flitsch SL. Glycoarrays - tools for determining protein–carbohydrate interactions and glycoenzyme specificity. Chem Commun. 2008:4400–4412. doi: 10.1039/B806983M. [DOI] [PubMed] [Google Scholar]
- 17.Rillahan CD, Paulson JC. Glycan Microarrays for Decoding the Glycome. Annu Rev Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakus JF, Mahal LK. New technologies for glycomic analysis: toward a systematic understanding of the glycome. Annu Rev Anal Chem. 2011;4:367–392. doi: 10.1146/annurev-anchem-061010-113951. [DOI] [PubMed] [Google Scholar]
- 19.Wu C, Wong C. Chemistry and glycobiology. Chem Commun. 2011;47:6201–6207. doi: 10.1039/C0CC04359A. [DOI] [PubMed] [Google Scholar]
- 20.Voglmeir J, Sardzík R, Weissenborn MJ, Flitsch SL. Enzymatic glycosylations on arrays. OMICS. 2010;14:437–444. doi: 10.1089/omi.2010.0035. [DOI] [PubMed] [Google Scholar]
- 21.Ghazarian H, Idoni B, Oppenheimer SB. A glycobiology review: carbohydrates, lectins and implications in cancer therapeutics. Acta Histochem. 2010;113:236–247. doi: 10.1016/j.acthis.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang P, Chiu PCN, Lee C, Chang L, Panico M, Morris HR, et al. Human sperm binding is mediated by the sialyl-Lewisx oligosaccharide on the zona pellucida. Science. 2011;333:1761–1764. doi: 10.1126/science.1207438. [DOI] [PubMed] [Google Scholar]
- 23.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 24.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 25.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nature Rev Cancer. 2011;11:678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakaidani Y, Nomura T, Matsuura A, Ito M, Suzuki E, Murakami K, et al. O-Linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell–matrix interactions. Nature Commun. 2011;2 doi: 10.1038/ncomms1591. Art No.: 583. [DOI] [PubMed] [Google Scholar]
- 27.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta Gen Subj. 2012 doi: 10.1016/j.bbagen.2011.12.001. in press. [DOI] [PubMed] [Google Scholar]
- 28.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gamblin DP, Scanlan EM, Davis BG. Glycoprotein synthesis: an update. Chem Rev. 2009;109:131–163. doi: 10.1021/cr078291i. [DOI] [PubMed] [Google Scholar]
- 30.Bratosin D, Mazurier J, Debray H, Lecocq M, Boilly B, et al. Flow cytofluorimetric analysis of young and senescent human erythrocytes probed with lectins. Evidence that sialic acids control their life span. Glycoconj J. 1995;12:258–267. doi: 10.1007/BF00731328. [DOI] [PubMed] [Google Scholar]
- 31.Marikovsky Y, Marikovsky M. Clearance of senescent erythrocytes: wheat germ agglutinin distribution on young and old human erythrocytes. Glycoconj J. 2002;19:1–4. doi: 10.1023/A:1022513327982. [DOI] [PubMed] [Google Scholar]
- 32.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schauer R, Kamerling JP. The chemistry and biology of Trypanosomal trans-sialidases: virulence factors in chagas disease and sleeping sickness. ChemBioChem. 2011;12:22462264. doi: 10.1002/cbic.201100421. [DOI] [PubMed] [Google Scholar]
- 34.Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, et al. Shotgun glycomics: a microarray strategy for functional glycomics. Nature Methods. 2011;8:85–90. doi: 10.1038/nmeth.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnamoorthy L, Bess JW, Jr, Preston AB, Mahal LK, et al. HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nature Chem Biol. 2009;5:244–250. doi: 10.1038/nchembio.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirabayashi J. Glycome 'fingerprints' provide definitive clues to HIV origins. Nature Chem Biol. 2009;5:198–199. doi: 10.1038/nchembio0409-198. [DOI] [PubMed] [Google Scholar]
- 37.Katrlík J, Švitel J, Gemeiner P, Kožár T, Tkac J. Glycan and lectin microarrays for glycomics and medicinal applications. Med Res Rev. 2010;30:394–418. doi: 10.1002/med.20195. and references cited therein. [DOI] [PubMed] [Google Scholar]
- 38.Dube DH, Bertozzi CR. Glycans in cancer and inflammation - potential for therapeutics and diagnostics. Nature Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 39.Soundararajan V, Zheng S, Patel N, Warnock K, Raman R, Wilson IA, Raguram S, Sasisekharan V, Sasisekharan R. Networks link antigenic and receptor-binding sites of influenza hemagglutinin: Mechanistic insight into fitter strain propagation. Sci Rep. 2011;1(Article number: 200) doi: 10.1038/srep00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012 doi: 10.1038/nature10831. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doores KJ, Fulton Z, Hong V, Patel MK, Scanlan CN, Wormald MR, et al. A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity. Proc Natl Acad Sci USA. 2010;107:17107–17112. doi: 10.1073/pnas.1002717107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P, Wang S, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anthony RM, Kobayashi T, Wermeling F, Ravetch JV. Intravenous gammaglobulin suppresses inflammation through a novel TH2 pathway. Nature. 2011;475:110–113. doi: 10.1038/nature10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG anti-Inflammatory activity with a recombinant IgG Fc. Science. 2008;320:373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 48.Garcia I, Marradi M, Penades S. Glyconanoparticles: multifunctional nanomaterial for biomedical applications. Nanomedicine. 2010;5:777–792. doi: 10.2217/nnm.10.48. [DOI] [PubMed] [Google Scholar]
- 49.Van Bueren JJL, Rispens T, Verploegen S, Van Der Palen-Merkus T, Stapel S, Workman LJ, et al. Anti-galactose-α-1,3-galactose IgE from allergic patients does not bind α-galactosylated glycans on intact therapeutic antibody Fc domains. Nature Biotechnol. 2011;29:574–576. doi: 10.1038/nbt.1912. [DOI] [PubMed] [Google Scholar]
- 50.Gemeiner P, Mislovičová D, Tkáč J, Švitel J, Pätoprstý V, et al. Lectinomics: II. A highway to biomedical/clinical diagnostics. Biotechnol Adv. 2009;27:1–15. doi: 10.1016/j.biotechadv.2008.07.003. references cited therein. [DOI] [PubMed] [Google Scholar]
- 51.El-Boubbou K, Huang X. Glyco-nanomaterials: translating insights from the “sugar-code” to biomedical applications. Curr Med Chem 2011. 2011;18:2060–2078. doi: 10.2174/092986711795656144. [DOI] [PubMed] [Google Scholar]
- 52.Feizi T, Chai W. Oligosaccharide microarrays to decipher the glyco code. Nature Rev Mol Cell Biol. 2004;5:582–588. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- 53.Nilsson CL. Lectins: proteins that interpret the sugar code. Anal Chem. 2003;75:348A–353A. doi: 10.1021/ac031373w. [DOI] [PubMed] [Google Scholar]
- 54.Turnbull JE, Field RA. Emerging glycomics technologies. Nature Chem Biol. 2007;3:74–77. doi: 10.1038/nchembio0207-74. [DOI] [PubMed] [Google Scholar]
- 55.Gerlach JQ, Cunningham S, Kane M, Joshi L. Glycobiomimics and glycobiosensors. Biochem Soc Trans. 2010;38:1333–1336. doi: 10.1042/BST0381333. [DOI] [PubMed] [Google Scholar]
- 56.Alley WR, Jr, Vasseur JA, Goetz JA, Svoboda M, Mann BF, Matei DE, Menning N, Hussein A, Mechref Y, Novotny MV. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Prot Res. 2012;11:2282–2300. doi: 10.1021/pr201070k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kolarich D, Lepenies B, Seeberger PH. Glycomics, glycoproteomics and the immune system. Curr Opin Chem Biol. 2012;16:214–220. doi: 10.1016/j.cbpa.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 58.Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nature Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- 59.Love KR, Seeberger PH. Carbohydrate arrays as tools for glycomics. Angew Chem - Int Ed. 2002;41:3583–3586. doi: 10.1002/1521-3773(20021004)41:19<3583::AID-ANIE3583>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 60.Mellet CO, Fernandez JMG. Carbohydrate microarrays. ChemBioChem. 2002;3:819–822. doi: 10.1002/1439-7633(20020902)3:9<819::AID-CBIC819>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 61.Park S, Shin I. Fabrication of carbohydrate chips for studying protein–carbohydrate interactions. Angew Chem - Int Ed. 2002;41:3180–3182. doi: 10.1002/1521-3773(20020902)41:17<3180::AID-ANIE3180>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 62.Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nature Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 63.Houseman BT, Mrksich M. Carbohydrate arrays for the evaluation of protein binding and enzymatic modification. Chem Biol. 2002;9:443–454. doi: 10.1016/S1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]
- 64.Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, et al. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiol. 2005;15:31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 65.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, et al. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nature Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 66.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycopatterns. ChemBioChem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 67.Zheng T, Peelen D, Smith LM. Lectin arrays for profiling cell surface carbohydrate expression. J Am Chem Soc. 2005;127:9982–9983. doi: 10.1021/ja0505550. [DOI] [PubMed] [Google Scholar]
- 68.Nagl S, Schaeferling M, Wolfbeis OS. Fluorescence analysis in microarray technology. Microchim Acta. 2005;151:1–21. doi: 10.1007/s00604-005-0393-9. [DOI] [Google Scholar]
- 69.Borisov SM, Wolfbeis OS. Optical biosensors. Chem Rev. 2008;108:423–461. doi: 10.1021/cr068105t. [DOI] [PubMed] [Google Scholar]
- 70.Sharon N, Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiol. 2004;14:53R–62R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 71.Lis H, Sharon N. Lectins: carbohydrate-specific proteins that mediate cellular recognition. Chem Rev. 1998;98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 72.Bučko M, Mislovičová D, Nahálka J, Vikartovská A, Šefčovičová J, Katrlík J, Tkáč J, Gemeiner P, Lacík I, Štefuca V, Polakovič M, et al. Immobilization in biotechnology and biorecognition: from macro- to nanoscale systems. Chem Papers. 2012;66:983–998. doi: 10.2478/s11696-012-0226-3. [DOI] [Google Scholar]
- 73.Thévenot DR, Toth K, Durst RA, Wilson GS. Electrochemical biosensors: recommended definitions and classification. Biosens Bioelectron. 2001;16:121–131. doi: 10.1016/S0956-5663(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 74.Labuda J, Oliveira Brett AM, Evtugyn G, Fojta M, Mascini M, Ozsoz M, Palchetti I, Paleček E, Wang J. Electrochemical nucleic acid-based biosensors: concepts, terms, and methodology (IUPAC Technical Report) Pure Appl Chem. 2010;82:1161–1187. doi: 10.1351/PAC-REP-09-08-1. [DOI] [Google Scholar]
- 75.Jelinek R, Kolusheva S. Carbohydrate biosensors. Chem Rev. 2004;104:5987–6015. doi: 10.1021/cr0300284. [DOI] [PubMed] [Google Scholar]
- 76.Wang J. Electrochemical biosensing based on noble metal nanoparticles. Microchim Acta. 2012 doi: 10.1007/s00604-011-0758-1. [DOI] [Google Scholar]
- 77.Katz E, Willner I. Probing biomolecular interactions at conductive and semiconductive surfaces by impedance spectroscopy: routes to impedimetric immunosensors, DNA-sensors, and enzyme biosensors. Electroanal. 2003;15:913–947. doi: 10.1002/elan.200390114. [DOI] [Google Scholar]
- 78.Pejcic B, De Marco R. Impedance spectroscopy: over 35 years of electrochemical sensor optimization. Electrochim Acta. 2006;51:6217–6229. doi: 10.1016/j.electacta.2006.04.025. [DOI] [Google Scholar]
- 79.Daniels JS, Pourmand N. Label-free impedance biosensors: opportunities and challenges. Electroanal. 2007;19:1239–1257. doi: 10.1002/elan.200603855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lisdat F, Schäfer D. The use of electrochemical impedance spectroscopy for biosensing. Anal Bioanal Chem. 2008;391:1555–1567. doi: 10.1007/s00216-008-1970-7. [DOI] [PubMed] [Google Scholar]
- 81.Sánchez-Pomales G, Zangmeister RA. Recent advances in electrochemical glycobiosensing. Int J Electrochem. 2011;2011 doi: 10.4061/2011/825790. Article ID 825790, 11 pages. [DOI] [Google Scholar]
- 82.Zeng X, Andrade CAS, Oliveira MDL, Sun X-L. Carbohydrate–protein interactions and their biosensing applications. Anal Bioanal Chem. 2012;402:3161–3176. doi: 10.1007/s00216-011-5594-y. [DOI] [PubMed] [Google Scholar]
- 83.Dai Z, Kawde A-N, Xiang Y, La Belle JT, Gerlach J, Bhavanandan VP, Joshi L, Wang J. Nanoparticle-based sensing of glycan-lectin interactions. J Am Chem Soc. 2006;128:10018–10019. doi: 10.1021/ja063565p. [DOI] [PubMed] [Google Scholar]
- 84.La Belle JT, Gerlach JQ, Svarovsky S, Joshi L. Label-free impedimetric detection of glycan-lectin interactions. Anal Chem. 2007;79:6959–6964. doi: 10.1021/ac070651e. [DOI] [PubMed] [Google Scholar]
- 85.Oliveira MDL, Correia MTS, Coelho LCBB, Diniz FB. Electrochemical evaluation of lectin-sugar interaction on gold electrode modified with colloidal gold and polyvinyl butyral. Colloids Surf B: Biointerf. 2008;66:13–19. doi: 10.1016/j.colsurfb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 86.Oliveira MDL, Correia MTS, Diniz FB. A novel approach to classify serum glycoproteins from patients infected by dengue using electrochemical impedance spectroscopy analysis. Synth Met. 2009;159:2162–2164. doi: 10.1016/j.synthmet.2009.09.022. [DOI] [Google Scholar]
- 87.Oliveira MDL, Correia MTS, Diniz FB. Concanavalin A and polyvinyl butyral use as a potential dengue electrochemical biosensor. Biosens Bioelectron. 2009;25:728–732. doi: 10.1016/j.bios.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 88.Oliveira MDL, Nogueira ML, Correia MTS, Coelho LCBB, Andrade CAS. Detection of dengue virus serotypes on the surface of gold electrode based on Cratylia mollis lectin affinity. Sens Actuat B: Chem. 2011;155:789–795. doi: 10.1016/j.snb.2011.01.049. [DOI] [Google Scholar]
- 89.Andrade CAS, Oliveira MDL, de Melo CP, Coelho LCBB, Correia MTS, Nogueira ML, Singh PR, Zeng X. Diagnosis of dengue infection using a modified gold electrode with hybrid organic-inorganic nanocomposite and Bauhinia monandra lectin. J Colloid Interf Sci. 2011;362:517–523. doi: 10.1016/j.jcis.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 90.Oliveira MDL, Andrade CAS, Correia MTS, Coelho LCBB, Singh PR, Zeng X. Impedimetric biosensor based on self-assembled hybrid cystein-gold nanoparticles and CramoLL lectin for bacterial lipopolysaccharide recognition. J Colloid Interf Sci. 2011;362:194–201. doi: 10.1016/j.jcis.2011.06.042. [DOI] [PubMed] [Google Scholar]
- 91.Nagaraj VJ, Aithal S, Eaton S, Bothara M, Wiktor P, Prasad S. NanoMonitor: a miniature electronic biosensor for glycan biomarker detection. Nanomedicine. 2010;5:369–378. doi: 10.2217/nnm.10.11. [DOI] [PubMed] [Google Scholar]
- 92.Bertók T, Gemeiner P, Mikula M, Gemeiner P, Tkac J. An ultrasensitive electrochemical label-free detection of a glycoprotein by a lectin-based biosensor device. 2012 submitted. [Google Scholar]
- 93.Szunerits S, Niedziǒlka-Jönsson J, Boukherroub R, Woisel P, Baumann J-S, Siriwardena A. Label-free detection of lectins on carbohydrate-modified boron-doped diamond surfaces. Anal Chem. 2010;82:8203–8210. doi: 10.1021/ac1016387. [DOI] [PubMed] [Google Scholar]
- 94.Loaiza OA, Lamas-Ardisana PJ, Jubete E, Ochoteco E, Loinaz I, Cabañero G, García I, Penadés S. Nanostructured disposable impedimetric sensors as tools for specific biomolecular interactions: sensitive recognition of concanavalin A. Anal Chem. 2011;83:2987–2995. doi: 10.1021/ac103108m. [DOI] [PubMed] [Google Scholar]
- 95.Feizi T, Chai W. Oligosaccharide microarrays to decipher the glyco code. Nat Rev Mol Cell Biol. 2004;5:582–588. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- 96.Feizi T, Fazio F, Chai W, Wong CH. Carbohydrate microarrays - A new set of technologies at the frontiers of glycomics. Curr Opin Struct Biol. 2003;13:637–645. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 97.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 98.Culf AS, Cuperlovic-Culf M, Ouellette RJ. Carbohydrate microarrays: Survey of fabrication techniques. OMICS. 2006;10:289–310. doi: 10.1089/omi.2006.10.289. [DOI] [PubMed] [Google Scholar]
- 99.Ratner DM, Adams EW, Su J, O’Keefe BR, Mrksich M, Seeberger PH. Probing protein-carbohydrate interactions with microarrays of synthetic oligosaccharides. Chem Bio Chem. 2004;5:379–382. doi: 10.1002/cbic.200300804. [DOI] [PubMed] [Google Scholar]
- 100.Jadhav SA. Self-assembled monolayers (SAMs) of carboxylic acids: an overview. Central Eur J Chem. 2011;9:369–378. doi: 10.2478/s11532-011-0024-8. [DOI] [Google Scholar]
- 101.Ding L, Cheng W, Wang X, Ding S, Ju H. Carbohydrate monolayer strategy for electrochemical assay of cell surface carbohydrate. J Am Chem Soc. 2008;130:7224–7225. doi: 10.1021/ja801468b. [DOI] [PubMed] [Google Scholar]
- 102.Ding L, Ji Q, Qian R, Cheng W, Huangxian J. Lectin-based nanoprobes functionalized with enzyme for highly sensitive electrochemical monitoring of dynamic carbohydrate expression on living cells. Anal Chem. 2010;82:1292–1298. doi: 10.1021/ac902285q. [DOI] [PubMed] [Google Scholar]
- 103.Cheng W, Ding L, Lei J, Ding S, Ju H. Effective cell capture with tetrapeptide-functionalized carbon nanotubes and dual signal amplification for cytosensing and evaluation of cell surface carbohydrate. Anal Chem. 2008;80:3867–3872. doi: 10.1021/ac800199t. [DOI] [PubMed] [Google Scholar]
- 104.Xue Y, Ding L, Lei J, Yan F, Ju H. In situ electrochemical imaging of membrane glycan expression on micropatterned adherent single cells. Anal Chem. 2010;82:7112–7118. doi: 10.1021/ac101688p. [DOI] [PubMed] [Google Scholar]
- 105.Ding L, Qian R, Xue Y, Cheng W, Ju H. In situ scanometric assay of cell surface carbohydrate by glyconanoparticle-aggregation-regulated silver enhancement. Anal Chem. 2010;82:5804–5809. doi: 10.1021/ac100866e. [DOI] [PubMed] [Google Scholar]
- 106.Ertl P, Mikkelsen SR. Electrochemical biosensor array for the identification of microorganisms based on lectin - lipopolysaccharide recognition. Anal Chem. 2001;73:4241–4248. doi: 10.1021/ac010324l. [DOI] [PubMed] [Google Scholar]
- 107.Ertl P, Wagner M, Corton E, Mikkelsen SR. Rapid identification of viable Escherichia coli subspecies with an electrochemical screen-printed biosensor array. Biosens Bioelectron. 2003;18:907–916. doi: 10.1016/S0956-5663(02)00206-3. [DOI] [PubMed] [Google Scholar]
- 108.Heiskanen A, Yakovleva J, Spégel C, Taboryski R, Koudelka-Hep M, Emnéus J, Ruzgas T. Amperometric monitoring of redox activity in living yeast cells: comparison of menadione and menadione sodium bisulfite as electron transfer mediators. Electrochem Commun. 2004;6:219–224. doi: 10.1016/j.elecom.2003.12.003. [DOI] [Google Scholar]
- 109.Ding L, Cheng W, Wang X, Xue Y, Lei J, Yin Y, Ju H. A label-free strategy for facile electrochemical analysis of dynamic glycan expression on living cells. Chem Commun. 2009;46:7161–7163. doi: 10.1039/b918008g. [DOI] [PubMed] [Google Scholar]
- 110.Xue Y, Bao L, Xiao X, Ding L, Lei J, Ju H. Noncovalent functionalization of carbon nanotubes with lectin for label-free dynamic monitoring of cell-surface glycan expression. Anal Biochem. 2011;410:92–97. doi: 10.1016/j.ab.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 111.Wan Y, Zhang D, Hou B. Monitoring microbial populations of sulfate-reducing bacteria using an impedimetric immunosensor based on agglutination assay. Talanta. 2009;80:218–223. doi: 10.1016/j.talanta.2009.06.057. [DOI] [PubMed] [Google Scholar]
- 112.Gamella M, Campuzano S, Parrado C, Reviejo AJ, Pingarrón JM. Microorganisms recognition and quantification by lectin adsorptive affinity impedance. Talanta. 2009;78:1303–1309. doi: 10.1016/j.talanta.2009.01.059. [DOI] [PubMed] [Google Scholar]
- 113.Xi F, Gao J, Wang J, Wang Z. Discrimination and detection of bacteria with a label-free impedimetric biosensor based on self-assembled lectin monolayer. J Electroanal Chem. 2011;656:252–257. doi: 10.1016/j.jelechem.2010.10.025. [DOI] [Google Scholar]
- 114.Zhang X, Teng Y, Fu Y, Xu L, Zhang S, He B, Wang C, Zhang W. Lectin-based biosensor strategy for electrochemical assay of glycan expression on living cancer cells. Anal Chem. 2010;82:9455–9460. doi: 10.1021/ac102132p. [DOI] [PubMed] [Google Scholar]
- 115.Ding C, Qian S, Wang Z, Qu B. Electrochemical cytosensor based on gold nanoparticles for the determination of carbohydrate on cell surface. Anal Biochem. 2011;414:84–87. doi: 10.1016/j.ab.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 116.Zhang J-J, Cheng F-F, Zheng T-T, Zhu J-J. Design and implementation of electrochemical cytosensor for evaluation of cell surface carbohydrate and glycoprotein. Anal Chem. 2010;82:3547–3555. doi: 10.1021/ac9026127. [DOI] [PubMed] [Google Scholar]
- 117.Berggren C, Bjarnason B, Johansson G. Capacitive biosensors. Electroanal. 2001;13:173–180. doi: 10.1002/1521-4109(200103)13:3<173::AID-ELAN173>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 118.Tkac J, Davis JJ. Label-free Field Effect Protein Sensing. In: Davis JJ, editor. Engineering the bioelectronic interface: Applications to analyte biosensing and protein detection. Royal Society of Chemistry, Cambridge: 2009. pp. 193–224. [DOI] [Google Scholar]
- 119.Lasia A. Electrochemical impedance spectroscopy and its applications. In: Conway BE, Bockris J, White RE, editors. Modern aspects of electrochemistry. Kluwer Academic/Plenum Publishers; New York: 1999. pp. 143–248. [Google Scholar]
- 120.Dijksma M, Kamp B, Hoogvliet JC, van Bennekom WP. Development of an electrochemical immunosensor for direct detection of interferon-γ at the attomolar level. Anal Chem. 2001;73:901–907. doi: 10.1021/ac001051h. http://pubs.acs.org/doi/abs/10.1021/ac001051h_-_ac001051hAF1 [DOI] [PubMed] [Google Scholar]
- 121.Vedala H, Chen Y, Cecioni S, Imberty A, Vidal S, Star A. Nanoelectronic detection of lectin-carbohydrate interactions using carbon nanotubes. Nano Lett. 2011;11:170–175. doi: 10.1021/nl103286k. [DOI] [PubMed] [Google Scholar]
- 122.Mislovičová D, Gemeiner P, Kozarova A, Kožár T. Lectinomics I. Relevance of exogenous plant lectins in biomedical diagnostics. Biologia. 2009;64:1–19. doi: 10.2478/s11756-009-0029-3. [DOI] [Google Scholar]
- 123.Kaku H, Peumans WJ, Goldstein IJ. Isolation and characterization of a second lectin (SNA-II) present in elderberry (Sambucus nigra L.) bark. Arch Biochem Biophys. 2010;277:255–262. doi: 10.1016/0003-9861(90)90576-K. [DOI] [PubMed] [Google Scholar]
- 124.Rahaie M, Kazemi SS. Lectin-based Biosensors: As Powerful Tools in Bioanalytical Applications. Biotechnol. 2010;9:428–443. doi: 10.3923/biotech.2010.428.443. [DOI] [Google Scholar]
- 125.Cunningham S, Gerlach JQ, Kane M, Joshi L. Glyco-biosensors: Recent advances and applications for the detection of free and bound carbohydrates. Analyst. 2010;135:2471–2480. doi: 10.1039/C0AN00276C. [DOI] [PubMed] [Google Scholar]
- 126.Bertók T, Šefčovičová J, Gemeiner P, Tkáč J. Lectinomics: A Tool in Clinical Diagnostics. Chem Listy. 2012;106:20–26. [Google Scholar]
- 127. http://www.pdb.org
- 128. http://www.sigmaaldrich.com
- 129. http://www.sigmaaldrich.com