Abstract
Systemic sclerosis (SSc) is an autoimmune disease seriously affecting patient´s quality of life. The heterogeneity of the disease also means that identification and subsequent validation of biomarkers of the disease is quite challenging. A fully validated single biomarker for diagnosis, prognosis, disease activity and assessment of response to therapy is not yet available. The main aim of this study was to apply an alternative assay protocol to the immunoassay-based analysis of this disease by employment of sialic acid recognizing lectin Sambucus nigra agglutinin (SNA) to glycoprofile serum samples. To our best knowledge this is the first study describing direct lectin-based glycoprofiling of serum SSc samples. Three different analytical methods for glycoprofiling of serum samples relying on application of lectins are compared here from a bioanalytical point of view including traditional ELISA-like lectin-based method (ELLA), novel fluorescent lectin microarrays and ultrasensitive impedimetric lectin biosensors. Results obtained by all three bioanalytical methods consistently showed differences in the level of sialic acid present on glycoproteins, when serum from healthy people was compared to the one from patients having SSc. Thus, analysis of sialic acid content in human serum could be of a diagnostic value for future detection of SSc, but further work is needed to enhance selectivity of assays for example by glycoprofiling of a fraction of human serum enriched in antibodies for individual diagnostics.
Keywords: Lectin, sialic acid, electrochemical impedance spectroscopy, lectin microarray, enzyme-linked lectin assay, systemic sclerosis
1. Introduction
Systemic sclerosis (SSc) is a chronic and progressive autoimmune disease of a connective tissue and can affect virtually every organ in the body [1–4]. Total survival time of patients is shortened with a mortality rate of 15% after 10 years due to heart failure and renal insufficiency. Initial classification of patients was done using American College of Rheumatology preliminary criteria developed in 1980 [5], but over time it was obvious that such classification was not well-suited for identification of early stage of the disease in SSc patients. Analysis of serum autoantibodies might identify the early stage of the disease, but the level of autoantibodies in individual patients can vary significantly [6]. The heterogeneity of the disease also means that identification and subsequent validation of biomarkers of the disease is quite challenging and the situation might be even more complicated since SSc patients might have other autoimmune diseases [7].
Despite significant progress in searching for prospective disease biomarkers by an increase in the number of published SSc biomarker studies, a fully validated single biomarker for diagnosis, prognosis, disease activity and assessment of response to therapy is not yet available [8]. Different techniques such as protein microarrays [9], peptide aptamer microarrays [10], optical biosensor [11] and ELISA [12] proved to be promising approaches for finding/analysis of protein(autoantibody)-based biomarkers of SSc. Recent studies suggest that microRNAs [13] and glycans might play an important role in the disease as judged from presence of anti-glycan antibodies [14] or other glycan-recognizing proteins (Siglecs) [15] in samples from SSc patients. Other autoimmune diseases are emerging at an increasing speed due to combination of genetic and environmental factors. At the same time all these factors together result in the dysregulation of synthesis of glycans stimulating immune system [16]. An increasing number of studies prove that protein glycosylation plays an important role in regulation of immune responses and in the development of autoimmune diseases, as well [17].
Recent advances in material chemistry, especially with introduction of various nanomaterials, together with launching of novel transducing protocols and biorecognition elements are behind a huge development in ultrasensitive and multiplexed analysis of a wide range of disease biomarkers [18–23]. It is well-known that presence of labels can negatively affect binding [24, 25] and thus label-free methods of analysis are preferential option for diagnostic purposes. One of the most sensitive biosensing detection platform is electrochemical impedance spectroscopy (EIS), offering robust, extremely sensitive method for monitoring of biorecognition events in a label-free mode of operation [19, 26–28]. This method is based on the application of small amplitude alternating voltage (perturbation) to the interfacial layer on the electrode, measuring the double layer capacitance (CDL) and charge transfer resistance (RCT), respectively [29–31]. Moreover, recently we developed reliable EIS assay for detection of glycoproteins directly in human serum with low level of non-specific binding [28].
In this study, we present a novel approach for analysis of a systemic sclerosis disease by detection of glycan moieties on proteins in diluted sera without any further pre-treatment using SNA lectin recognizing terminal sialic acid of a glycan moiety on glycoproteins. Moreover, in this study three different bioanalytical approaches for glycoprofiling of human sera are described, including traditional ELISA-like method enzyme-linked lectin assay (ELLA), novel multiplexed lectin microarray format of analysis and ultrasensitive EIS-based biosensing with immobilized lectins. Since glycosylation is the most common posttranslational modification of proteins (≥50% of all eukaryotic proteins and ≥70% of all therapeutic proteins) [19, 32] a huge scientific effort is devoted to better understanding of their role in cell processes or to apply such knowledge to design better therapeutics or diagnostic tools [33–40]. Thus, glycoprofiling of samples from SSc patients can be an alternative way for future early stage diagnostics of the SSc disease and other diseases including various types of cancer associated with aberrant glycosylation.
2. Experimental section
2.1. Materials
11-Mercaptoundecanoic acid (MUA), potassium hexacyanoferrate(III), potassium hexacyanoferrate(II) trihydrate, sodium chloride, potassium chloride, potassium phosphate monobasic, potassium phosphate dibasic, 1,3-propanesultone, N-hydroxysuccinimide (NHS), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), sodium sulfate, N,N-dimethylethylene diamine, dicyclohexylcarbodiimide (DCC), ethanolamine hydrochloride, dichloromethane, tetrahydrofuran (THF), Tween 20, Ricinus communis agglutinin (RCA recognizing galactose, caution: it is a biological toxin), concanavalin A (Con A recognizing mannose and glucose), anti-BSA (bovine serum albumin) antibody, polyvinylalcohol (PVA) and protein A from Staphylococcus aureus were purchased from Sigma Aldrich (USA). (R)-Lipoic acid was purchased from TCI Europe. Sambucus nigra agglutinin (SNA recognizing sialic acid) lectin from Sambucus nigra was purchased from EYLabs (USA). Ethanol for UV/Vis spectroscopy (ultrapure) was purchased from Slavus (Slovakia). Phosphate buffer saline tablets (PBST) were from Merck (Slovakia). Eight biotinylated lectins Aleuria aurantia lectin (AAL), Lens culinaris agglutinin (LCA), Maackia amurensis lectin (MAL), Phaseolus vulgaris agglutinin (PHAE), Ricinus communis agglutinin and Sambucus nigra agglutinin, concanavalin A and wheat-germ agglutinin (WGA); and avidin-peroxidase (AV-HRP) were purchased from Vector Laboratories (USA). CF555-streptavidin fluorescent label was purchased from Biotium (USA). Sulfobetaine ((R)-3-((2-(5-(1,2-dithiolan-3-yl)-pentanamido)ethyl)dimethylammonio)-propane-1-sulfonate (DPS) was synthesized according to a previously published protocol [28].
2.2. Apparatus
Electrochemical investigation was performed on a potentiostat PGSTAT 128N (Ecochemie, Netherlands) run under Nova Software 1.10 (Ecochemie, Netherlands) in a three electrode cell system, using modified gold electrode, auxiliary Pt and reference Ag/AgCl electrode.
Lectin microarray was prepared by spotting samples using SpotBot3 Microarray Protein edition (Arrayit, USA) on epoxide coated slides Nexterion E (Schott, Germany), which were scanned after experiment using InnoScan710 scanner (Innopsys, France) at a wavelength of 532 nm. The slide image was evaluated using the software Mapix 5.5.0 by evaluation of the intensity of fluorescence and intensity of all independent array spots on the array.
Enzyme-linked lectin assay (ELLA) was performed on MaxiSorp plates (Nunc A/S, Roskilde, Denmark) with the absorbance read at 490 nm with a microplate reader Synergy HT (BioTek).
A peak force tapping mode atomic force microscopy (ScanAsyst, Bruker, USA) in air was carried out on a Bioscope Catalyst instrument and Olympus IX71 microscope in conjunction with NanoScope 8.15 software at a scan rate 0.5 line/s (512 samples/line with 512 lines) with the peak force setpoint of 2 nN. Microarray epoxy-coated slides were modified as described above and scanned using SCANASYST-AIR silicon tip on nitride lever (Bruker, USA, with f0=50-90 kHz and k=0.4 N/m), sharpened to a tip radius of 2 nm. The software provides value of root mean square roughness (Rq) reflecting roughness of the surface.
2.3. Electrode pretreatment, biosensor preparation and EIS analysis
Polycrystalline gold electrode surface was treated as described previously, namely using electrochemical reductive desorption, mechanical polishing, chemical treatment and once again electrochemical polishing and gold oxide stripping procedure [41]. Shortly, in the first step previously bound thiol molecules were desorbed from the surface using cyclic voltammetry (CV) in 100 mM NaOH under anaerobic conditions (100 scans run from -500 mV to -1,500 mV at a scan rate of 100 mV/s), then the electrodes were mechanically polished using polishing pad and alumina slurry (particle size 1 and 0.3 μm, each for 5 min) and left in hot piranha solution for 20 min (H2O2 and H2SO4 at 1+3 ratio, caution: handle with a special care). After that, CV was run again in 100 mM H2SO4 (25 scans run from -200 mV to +1,500 mV at a scan rate of 100 mV/s) for the electrochemical polishing and gold oxide stripping (10 scans run from +750 mV to +200 mV at a scan rate of 100 mV/s), respectively. Immediately after a gold oxide stripping procedure was completed, the electrodes were washed with DW and ultrapure ethanol, left to dry in a dustless environment and subsequently used for self-assembled layer (SAM) formation from 1 mM thiol stock solution.
Electrode was modified by a mixed SAM from 1+1 ratio of 1 mM of sulfobetaine derivative (DPS) and 11-mercaptoundecanoic acid (MUA) by a simple passive incubation overnight. Then, SNA lectin was immobilized using standard amine coupling with carboxylic groups of MUA being activated by a 1+1 mixture of 0.2 M EDC and 0.05 M NHS for 15 min. SNA was covalently immobilized on the activated SAM layer from a 40 μL stock solution (1 mg/mL i.e. 6.7 μM) by 1 h incubation. The surface after lectin immobilization was not blocked by small molecules i.e. ethanolamine since betaine-modified surface was reasonably resistant to protein adsorption in a wide range of protein concentrations [28]. Since concentration of proteins incubated with the biosensor was much lower compared to lectin concentration applied during immobilization we decided to skip blocking by small molecules. All EIS measurements were performed in an electrolyte containing 5 mM potassium hexacyanoferrate(III), 5 mM potassium hexacyanoferrate(II) and 0.1 M KCl. The EIS analysis was run at 50 different frequencies (ranging from 0.1 Hz to 100 kHz) under Nova Software 1.10 (Ecochemie, Netherlands). Data acquired were evaluated by the same software using a Nyquist plot with a Randles-Erschler equivalent circuit R(C[RW]) employed. It was observed that after EDC/NHS activation charge transfer resistance (RCT) decreased due to presence of lower density of negative charge in an agreement with a previous study [42] and after immobilization of lectin RCT increased again. Relative change of RCT after incubation with an analyte to RCT value of a reference surface (biosensor surface after the lectin immobilization and stabilization in sterile 100 mM KCl solution) was used for biosensing purposes. Each analyte/sample was measured at least in triplicate with an independent biosensor device, and results are shown with a standard deviation (±SD) calculated in MS Excel. Human serum samples were diluted in a sterile and filtered 10 mM PBS buffer, pH 7.4. All stock solutions (lectin, standard glycoproteins and human sera) were stored at −20 °C in aliquots.
2.4. Microarray analysis
Lectin microarray experiments were performed with a phosphate buffer solution (PBS) as a printing buffer, PBST (a phosphate buffer solution with addition of 0.05% Tween 20) as a washing buffer and PBST containing 1 M ethanolamine applied as a blocking buffer. Shortly, at least four different concentrations of anti-BSA or diluted serum samples, respectively, were spotted using spotter on epoxide coated slides using a previously optimized protocol. Spotting temperature was set to 10°C and humidity to 60%. Subsequently, the slide was placed into a humidity chamber for 1 h at the room temperature with humidity of 80–90%, blocked using a blocking buffer at room temperature for 1 h and with slow shaking, rinsed under a mild stream of a printing buffer in a Petri dish and drained. Then, 70 μl of 50 μg/mL biotinylated SNA in a binding buffer was applied to the slide surface and incubated for 1 h. After lectin incubation, the slide was incubated with the Biotium CF555-streptavidin solution (5 mg/mL in a printing buffer) for 15 min and finally after washing the slide was scanned with a reader.
2.5. Enzyme-linked lectin binding assay
MaxiSorp plates were incubated with solution of protein A (4 μg/ml) in PBS buffer for 4 h. All following washing and incubation steps were performed with PBS buffer with Tween 20 (0.05%) The plate was washed 3 times with 200 μl of a washing buffer and the surface was subsequently blocked with 0.5% solution of PVA (1 h). After washing, individual SSc samples (2,000 x diluted) were added into the wells. Plate was incubated at 4 °C overnight and to washed empty wells a solution of biotinylated SNA (1 μg/ml) was added and incubated for 1 h and washed afterwards. Then, AV-HRP (0.25 mg/ml) was added into the each well and incubated for 1 h. After washing, AV-HRP bound to lectins was determined with a substrate o-phenylenediamine (OPD) dissolved in a citrate-phosphate buffer pH 4.6 in the presence of hydrogen peroxide (incubation for 15 min in the dark). Finally, after addition of 3.6 M H2SO4 the absorbance at 490 nm was measured.
2.6. Total serum protein concentration according to Lowry
The amount of total serum proteins in human serum was analyzed according to Lowry [43]. Shortly, 500 μL of a 100x diluted sample was mixed with 2.5 mL of a reagent (1% CuSO4.5H2O + 2% potassium sodium tartrate + 0.2 M NaOH + 4% Na2CO3) and left to react at room temperature for 10 min. This step was followed by an incubation with 0.25 mL of Folin-Ciocalteus phenol reagent (1+1 mixture with DW), left to react at room temperature for another 30 min, and right after this step, absorbance at 500 nm was measured.
2.7. Characterization of human samples
Ten female patients (n=10, mean age (range)=38.2 (24-53) years) fulfilling the criteria of LeRoy for SSc [44] and ten control females (n=10, mean age (range)=38.3 (31-51) years) were included in the study. SSc patients were recruited from the National Institute of Rheumatic Diseases in Piešt’any, Slovakia. Control subjects were recruited by the laboratory staff of the Institute of Experimental Endocrinology, Slovak Academy of Sciences Bratislava, Slovakia. The mean IL-1 beta (3.0 vs. 4.3 pg/mL), IL-6 (0.6 vs. 0.0 pg/mL), TNF protein (17.5 vs. 13.7 pg/mL) concentrations did not differ between SSc patients and controls. The disease activity of SSc patients, assessed using EULAR Systemic Sclerosis Activity Score [45], was “inactive”. All the SSc patients were positive for the following antibodies: anti-nuclear, anti-centromeric, and anti-Scl-70. None of the patients had been treated with glucocorticoids during the past five years. The following drugs were used by SSc patients: metalcaptase, pentoxifylline, naftidrofuryl, xantinol, nitroglycerin or other vasodilatation drugs, ginkgo biloba extract, non-steroidal anti-inflammatory drugs, calcium channel blockers, alendronate, azathioprine. The last dose of the medicaments was administered 24 h prior to the investigation. All subjects gave informed written consent and the study was approved by the Ethics Committee of the National Institute of Rheumatic Diseases, Piešt’any, Slovakia. All buffer components were dissolved in deionized water (DW, G = 0.033 μS).
3. Results and discussion
3.1. Selection of lectins
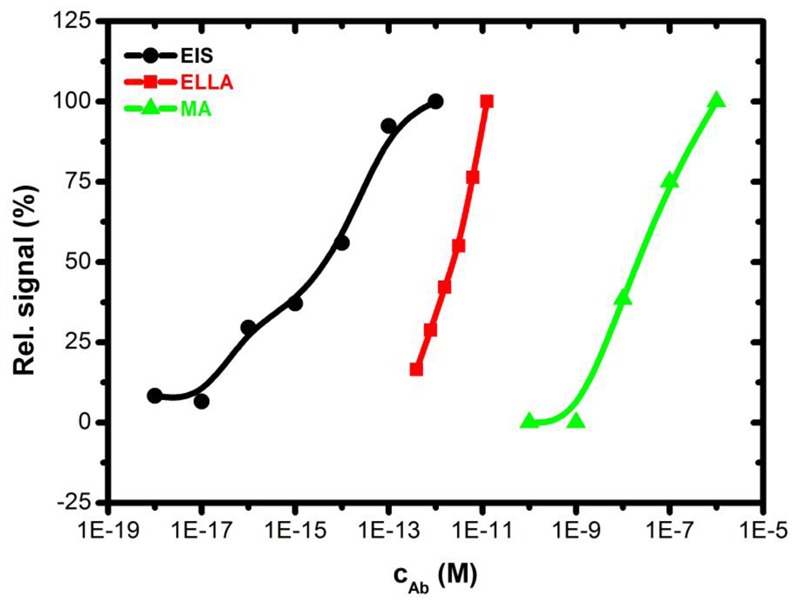
The first step in this study was selection of proper lectins most suitable for the glycoprofiling analysis. For that purpose, a lectin microarray procedure (MA) was employed offering much higher throughput of analysis compared to the other two methods investigated, i.e. ELLA and impedimetric lectin biosensors. Lectins involved in the study were selected to keep in mind an ability to bind to biantennary complex glycan types often present on human IgG (and other proteins) in human sera at relatively high concentrations and known to be aberrantly glycosylated during a progress of specific (mostly autoimmune) diseases [46]. Antibody against BSA was selected as a glycoprotein to perform calibration of lectin microarray technique with eight different lectins. In Fig. S1 a calibration plot for analysis of interaction of anti-BSA antibody with three lectins Con A, RCA and SNA due to their high binding preference is shown. Lectins like Helix pomatia lectin and Maackia amurensis lectin showing only low binding capacity and Phaseolus vulgaris lectin showing a binding pattern similar to Con A. Preliminary glycoprofiling of serum samples from patients and healthy individuals also showed that from eight lectins Con A, RCA, AAL, LCA, MAL, PHAE, WGA and SNA, especially SNA lectin is suitable to discriminate these two types of serum samples (data not shown). Thus, a calibration curve for anti-BSA for all three bioanalytical methods based on SNA lectin was constructed (Fig. 1). From these results it is clear that the most sensitive is the EIS method (working concentration range 10 aM – 1 pM), followed by ELLA method (390 fM – 12 pM) and MA (10 nM – 1 μM) (Fig. 1). Detection limit for EIS-based biosensor is well below 100 aM concentration range, what is the lowest limit of detection of any lectin-based label-free biosensor device published so far, when further signal amplification strategy was not carried out (i.e. integration of gold nanoparticles or a sandwich configuration) [19]. Moreover, EIS provides linear range spanning 4-5 orders of magnitude while the other two methods offer quite narrow linear range.
Figure 1.
Calibration curve for anti-BSA using various bioanalytical devices (EIS – electrochemical impedance spectroscopy-based lectin biosensor, ELLA – enzyme linked lectin assays, MA – lectin microarray) with SNA lectin immobilized. Relative response for each bioanalytical method is plotted, when 100% means a signal of 22.2% for rel. ∆RCT for EIS, 0.862 arbitrary units of absorbance at 490 nm for ELLA and 99.1 arbitrary units of fluorescence for MA. Ab is an anti-BSA antibody.
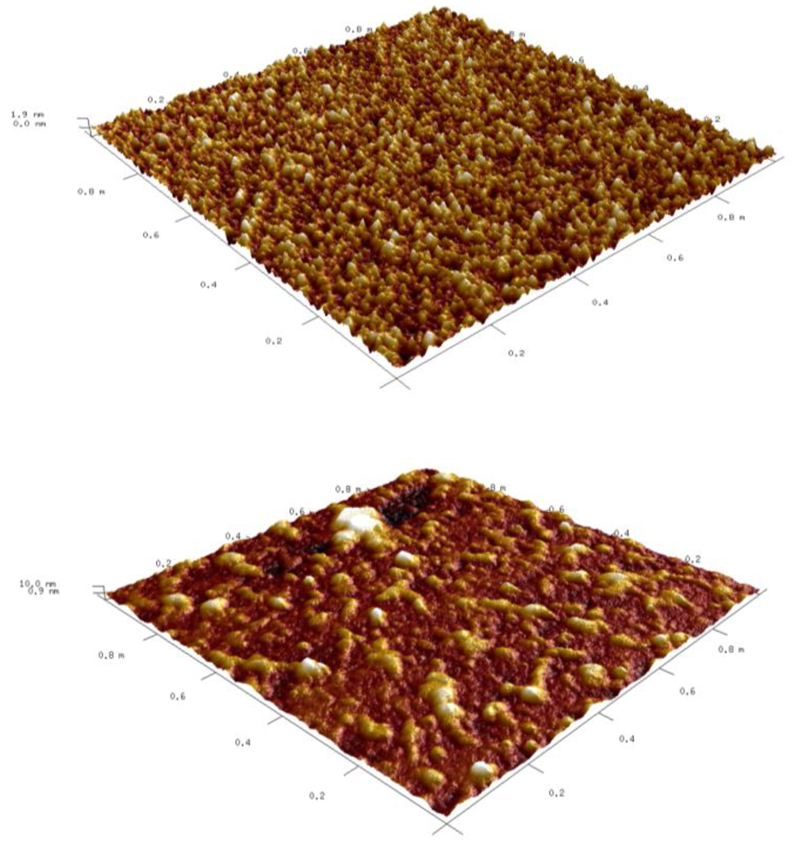
An interaction between a lectin and a glycoprotein on the microarray slide was also investigated using atomic force microscopy working in a peak force tapping mode. As shown in Fig. 2, it was possible to clearly distinguish between an unmodified surface (epoxy slides) and surface with adsorbed glycoprotein, lectin and avidin-peroxidase being spotted on the same slide. The surface roughness factor Rq for a bare microarray slide was 0.53 nm, with Rmax being 5.5 nm, and an increase in these values after completing whole spotting and incubation procedure to a final Rq of 2.4 nm and Rmax of 27 nm was observed. These results together with a height profile (Fig. S2) provide evidence about isolated protein features on the surface of a microarray slide.
Figure 2.
AFM images (1x1 μm) of unmodified microarray slide (upper image with Rq=0.5 nm) and the same slide after the sample spotting and incubation with a lectin (bottom image with Rq=2.4 nm). Isolated islets of the proteins (INV, Con A and streptavidin bound to it) may be observed.
The INV is a glycoprotein with dimensions 10.6 x 11.9 x 13.8 nm (pdb code: 3KF5), Con A with a size of 6.3 x 8.7 x 8.9 nm (pdb code: 3CNA) and streptavidin with dimensions 8.2 x 8.1 x 4.7 nm (pdb code: 4GJS) [47]. A total height of the peak showed in Fig. S2 of 24 nm thus correlates very well with the real size of all proteins present on a lectin microarray surface after completion of spotting and incubation steps, supporting conclusion about effective attachment of all protein components. In our previous study it was shown that AFM could visualize monomers, dimers and tetramers of Con A lectin [28].
3.2. Glycoprofiling of serum samples
According to the above described protocols, the three bioanalytical methods were used for the glycoprofiling of diluted human sera – a lectin microarray technique with fluorescently labeled lectins, an enzyme-linked lectin assay (ELLA) with horseradish peroxidase-labeled lectins and a label-free EIS lectin-based biosensor device. Since IgGs are present in human sera in very high concentration, it can be anticipated that lectin-based devices are sensitive mainly to changes in glycan composition on IgGs. Each of these methods provided a signal, which was expressed as a value relative to concentration of total serum proteins. From a group of ten healthy people and ten people having SSc, a mean value obtained in this way was calculated.
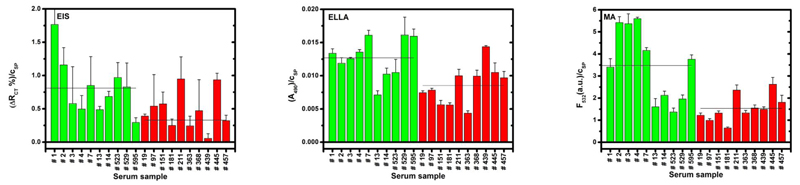
All three methods applied in SNA glycoprofiling of human sera from healthy individuals and those suffering from SSc showed the same binding pattern. A signal obtained from all three analytical methods is stronger for sera from healthy individuals compared to SSc patients (Fig. 3). Quantitative analysis showed that output response ratio for control sample compared to sample from people having SSc was 1.39 (EIS), 1.51 (ELLA) and 2.27 (MA). The variation in the response ratio between control and SSc samples is a result of different immobilization protocol for attachment of SNA lectin during construction of three bioanalytical devices. Recent study from the Joshi´s group pointed out to the fact that recognition of glycoprotein by lectins is to high extent influenced by lectin and glycoprotein orientation [48]. The obvious result of the study is the fact that samples from healthy individuals contains more sialic acid compared to samples from SSc patients.
Figure 3.
Analysis of the level of sialic acid present in serum proteins from healthy control (green bars) and patients with SSc (red bars) using SNA lectin. Mean value measured by all three bioanalytical methods (EIS – electrochemical impedance spectroscopy-based lectin biosensor, ELLA – enzyme-linked lectin assays and MA – lectin microarray) for both types of samples is shown as a thin line. Each sample was measured at least in triplicate with a mean value and RSD shown and the signal is related to the total serum protein concentration (cSP) obtained according to a Lowry procedure, which was expressed in g/L. P-value for all three methods was p≤0.05 (obtained from a standard Student´s test).
Although lectin microarrays are behind recent discoveries of the role of glycans in physiological/pathological processes, revealing new prospective cancer biomarkers and mapping post-translational regulation of the human glycome [49–51], to our best knowledge this is the first time lectin microarrays were applied in glycoprofiling of samples of human sera with SSc. The same is true for impedimetric lectin biosensors, which have not been applied in analysis of this type of human samples [19, 26]. ELLA has not been applied in glycoprofiling of SSc sera and thus glycoprofiling of SSc sera is a completely novel approach for analysis of this type of samples with a potential to be applied in diagnostics of this disease after further development in the future.
Lectin microarray could detect glycoproteins down to low nM level, ELLA down to pM level (Fig. 1) and EIS-based lectin biosensor down to fM-aM level [28, 32, 52]. From all these three methods of glycoprofiling lectin microarray is the best, followed by ELLA and EIS-based lectin biosensors in terms of multiplexing, while in case sensitivity of assays is the main requirement than this order is completely reversed (Tab. 1). There is a room for improvement of multiplexing performance of EIS method to some degree, since in a recent study it was shown that EIS assay can be performed on a chip having 6 electrodes [53] and there are some companies offering electrochemical assays on 96 electrodes within wells on the ELISA plate. Thus, moderate multiplexing of EIS assay format is feasible.
Table 1.
Key parameters for three lectin-based glycoprofiling methods
| Glycoprofiling method | Sensitivity | Multiplexing | Dynamic range | Assay reproducibility | Label-free |
|---|---|---|---|---|---|
| Lectin microarray | + | +++ | ++ | ++ |  |
| ELLA | ++ | ++ | + | +++ |  |
| EIS-lectin biosensor | +++ | + | +++ | +* |  |
- low assay reproducibility of the EIS-lectin biosensor is due to the fact that for every single analysis of samples a new biosensor was used and that sample assay reproducibility rather means reproducibility of the biosensor construction, including the gold electrodes surface regeneration
Even though lectin glycoprofiling can provide information about changes in glycan composition when serum from healthy individuals and patients with SSc are compared, to be able to predict presence of this disease in a particular individual, further development is needed. Presence of any kind of inflammation in both groups, i.e. healthy individuals and patients having SSc, can influence total plasma glycome and also glycan composition on antibodies. In a recent study plasma from 107 patients was analyzed [54]. Results showed that inflammation after cardiac surgery influenced total plasma glycome in all examined patients in a similar way, while changes in glycan composition on antibodies were individual-dependent [54]. To make glycoprofiling more selective it would be possible to separate antibodies from serum and to glycoprofile only this protein fraction. Alternatively glycan biochips, glycan biosensors or antigen microarrays could be applied to selectively fish out specific antibodies considered as SSc biomarkers with their subsequent glycoprofiling by lectins.
It is now apparent that analysis of a single biomarker is not sufficient for reliable detection and monitoring of various diseases and rather analysis of panel of protein biomarkers is needed [55]. Although multiplexed analysis of up to seven different protein profiles are needed to predict various types of cancer with high specificity and sensitivity [56], a recent study suggest that in multiplexed analysis of diseases besides protein profiles also DNA/RNA profiles of selected biomarkers should be performed [57]. We believe that in future multiplexed analysis of various disease states will require not only analysis of proteomic and genomic profiles of selected biomarkers, but that analysis of glycosylation changes will be equally important. This is especially true for monitoring of disease states of diseases such as SSc and other ones exhibiting large variations in protein profiles with a huge heterogeneity of the disease. This study can be the first step in the direction to include glycoprofiling into multiplexed assay formats for analysis of disease biomarkers. Moreover,EIS covering wide dynamic concentration range fulfill requirement to measure both high and low concentration of biomarkers in the same sample in presence of highly abundant serum proteins.
4. Conclusions
A glycoprofiling of serum from SSc patients by three different lectin-based biodevices was performed. This is the first study showing potential of glycoprofiling to distinguish serum samples from healthy individuals or those having SSc. To our best knowledge, none of the three glycoprofiling protocols showed here, i.e. lectin EIS-based biosensors, ELLA and lectin microarray, have been applied for analysis of sera from SSc patients. From 8 different lectins tested in a lectin microarray format, especially SNA lectin is suitable for in-depth analysis of subtle changes in the glycan composition of serum samples. A direct comparison of all three glycan detection methods revealed that especially lectin EIS-based biosensor devices are the best in discriminating glycan changes between sera from healthy individuals and those having SSc. At the same time the biosensor device proved to be the most sensitive device offering large concentration window for glycan analysis. The main problems of lectin biosensors, i.e. low throughput of analysis and reproducibility of their preparation, can be addressed by using array of disposable and low-cost electrodes prepared either using a screen-printed process or using a thin film deposition.
Supplementary Material
Acknowledgement
The financial support from the Slovak scientific grant agency VEGA 2/0162/14 and from the Slovak research and development agency APVV 0282-11 is acknowledged. The research leading to these results has received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement No. 311532 and this work has received funding from the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement No. 317420. This publication is the result of the project implementation: Applied research in the field of industrial biocatalysis, ITMS code: 26240220079 supported by the Research & Development Operational Programme funded by the ERDF. Research leading to these results was supported by BASF Slovakia. Authors thank to Danka Žišková for supporting experiments.
References
- [1].Barnes J, Mayes MD. Epidemiology of systemic sclerosis: Incidence, prevalence, survival, risk factors, malignancy, and environmental triggers. Curr Opin Rheumatol. 2012;24:165–170. doi: 10.1097/BOR.0b013e32834ff2e8. [DOI] [PubMed] [Google Scholar]
- [2].Bhattacharyya S, Wei J, Varga J. Understanding fibrosis in systemic sclerosis: Shifting paradigms, emerging opportunities. Nat Rev Rheumatol. 2012;8:42–54. doi: 10.1038/nrrheum.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Imrich R, Lukac J, Rovensky J, Radikova Z, Penesova A, Kvetnansky R, Huckova M, Vigas M, Macho L, Koska J. Lower adrenocortical and adrenomedullary responses to hypoglycemia in premenopausal women with systemic sclerosis. J Rheumatol. 2006;33:2235–2241. [PubMed] [Google Scholar]
- [4].Radstake TR, Gorlova O, Rueda B, Martin J-E, Alizadeh BZ, Palomino-Morales R, Coenen MJ, Vonk MC, Voskuyl AE, Schuerwegh AJ. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Gen. 2010;42:426–429. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Masi AT. Preliminary criteria for the classification of systemic sclerosis (scleroderma) Arthritis Rheum. 1980;23:581–590. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- [6].Nihtyanova SI, Denton CP. Autoantibodies as predictive tools in systemic sclerosis. Nat Rev Rheumatol. 2010;6:112–116. doi: 10.1038/nrrheum.2009.238. [DOI] [PubMed] [Google Scholar]
- [7].Elhai M, Avouac J, Kahan A, Allanore Y. Systemic sclerosis at the crossroad of polyautoimmunity. Autoimmun Rev. 2013;12:1052–1057. doi: 10.1016/j.autrev.2013.05.002. [DOI] [PubMed] [Google Scholar]
- [8].Castelino FV, Varga J. Current status of systemic sclerosis biomarkers: Applications for diagnosis, management and drug development. Exp Rev Clin Immunol. 2013;9:1077–1090. doi: 10.1586/1744666X.2013.848792. [DOI] [PubMed] [Google Scholar]
- [9].Abel L, Kutschki S, Turewicz M, Eisenacher M, Stoutjesdijk J, Meyer HE, Woitalla D, May C. Autoimmune profiling with protein microarrays in clinical applications. BBA-Proteins Proteom. 2014;1844:977–987. doi: 10.1016/j.bbapap.2014.02.023. [DOI] [PubMed] [Google Scholar]
- [10].Straw S, Ferrigno PK, Song Q, Tomlinson D, Del Galdo F. Proof of concept study to identify candidate biomarkers of fibrosis using high throughput peptide aptamer microarray and validate by enzyme linked immunosorbant assay. J Biomed Sci Eng. 2013;6:32. [Google Scholar]
- [11].Cuccioloni M, Moroncini G, Mozzicafreddo M, Pozniak K, Nacci G. Biosensor-based binding assay for platelet-derived growth factor receptor-α autoantibodies in human serum. J Anal Bioanal Tech. 2013;7:38–72. [Google Scholar]
- [12].Gugliesi F, Bawadekar M, De Andrea M, Dell’Oste V, Caneparo V, Tincani A, Gariglio M, Landolfo S. Nuclear DNA sensor IFI16 as circulating protein in autoimmune diseases is a signal of damage that impairs endothelial cells through high-affinity membrane binding. PLoS One. 2013;8:e63045. doi: 10.1371/journal.pone.0063045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhu H, Luo H, Zuo X. Micrornas: Their involvement in fibrosis pathogenesis and use as diagnostic biomarkers in scleroderma. Exp Mol Med. 2013;45:e41. doi: 10.1038/emm.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nishijima C, Sato S, Takehara K. Anti-agalactosyl IgG antibodies in sera from patients with systemic sclerosis. J Rheumatol. 2001;28:1847–1851. [PubMed] [Google Scholar]
- [15].York MR, Nagai T, Mangini AJ, Lemaire R, van Seventer JM, Lafyatis R. A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists. Arthritis Rheum. 2007;56:1010–1020. doi: 10.1002/art.22382. [DOI] [PubMed] [Google Scholar]
- [16].Lauc G, Huffman JE, Pučić JE, Zgaga L, Adamczyk B, Mužinić A, Novokmet M, Polašek O, Gornik O, Krištić J, Keser T, et al. Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet. 2013;9:e1003225. doi: 10.1371/journal.pgen.1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. 2008;9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- [18].Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA. Emerging principles for the therapeutic exploitation of glycosylation. Science. 2014;343:1235681. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]
- [19].Kluková L’, Bertók T, Kasák P, Tkac J. Nanoscale controlled architecture for development of ultrasensitive lectin biosensors applicable in glycomics. Anal Methods. 2014;6:4922–4931. doi: 10.1039/C4AY00495G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Luo X, Davis JJ. Electrical biosensors and the label free detection of protein disease biomarkers. Chem Soc Rev. 2013;42:5944–5962. doi: 10.1039/c3cs60077g. [DOI] [PubMed] [Google Scholar]
- [21].Mu B, Zhang J, McNicholas TP, Reuel NF, Kruss S, Strano MS. Recent advances in molecular recognition based on nanoengineered platforms. Acc Chem Res. 2014;47:979–988. doi: 10.1021/ar400162w. [DOI] [PubMed] [Google Scholar]
- [22].Xu JJ, Zhao WW, Song S, Fan C, Chen HY. Functional nanoprobes for ultrasensitive detection of biomolecules: An update. Chem Soc Rev. 2014;43:1601–1611. doi: 10.1039/c3cs60277j. [DOI] [PubMed] [Google Scholar]
- [23].Shen J, Li Y, Gu h, Xia F, Zuo X. Recent development of sandwich assay based on the nanobiotechnologies for proteins, nucleic acids, small molecules, and ions. Chem Rev. 2014;114:7631–7677. doi: 10.1021/cr300248x. [DOI] [PubMed] [Google Scholar]
- [24].Gemeiner P, Mislovičová D, Tkáč J, Švitel J, Pätoprstý V, Hrabárová E, Kogan G, Kožár T. Lectinomics: II. A highway to biomedical/clinical diagnostics. Biotechnol Adv. 2009;27:1–15. doi: 10.1016/j.biotechadv.2008.07.003. [DOI] [PubMed] [Google Scholar]
- [25].Katrlík J, Švitel J, Gemeiner P, Kožár T, Tkac J. Glycan and lectin microarrays for glycomics and medicinal applications. Med Res Rev. 2010;30:394–418. doi: 10.1002/med.20195. [DOI] [PubMed] [Google Scholar]
- [26].Bertók T, Katrlík J, Gemeiner P, Tkac J. Electrochemical lectin based biosensors as a label-free tool in glycomics. Microchim Acta. 2013;180:1–13. doi: 10.1007/s00604-012-0876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hushegyi A, Tkac J. Are glycan biosensors an alternative to glycan microarrays? Anal Methods. 2014;6:6610–6620. doi: 10.1039/C4AY00692E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bertok T, Klukova L, Sediva A, Kasák P, Semak V, Micusik M, Omastova M, Chovanová L, Vlček M, Imrich R, Vikartovska A, et al. Ultrasensitive impedimetric lectin biosensors with efficient antifouling properties applied in glycoprofiling of human serum samples. Anal Chem. 2013;85:7324–7332. doi: 10.1021/ac401281t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fernandes FC, Santos A, Martins DC, Goes MS, Bueno PR. Comparing label free electrochemical impedimetric and capacitive biosensing architectures. Biosens Bioelectron. 2014;57C:96–102. doi: 10.1016/j.bios.2014.01.044. [DOI] [PubMed] [Google Scholar]
- [30].Santos A, Carvalho FC, Roque-Barreira M-C, Bueno PR. Impedance-derived electrochemical capacitance spectroscopy for the evaluation of lectin–glycoprotein binding affinity. Biosens Bioelectron. 2014;62:102–105. doi: 10.1016/j.bios.2014.06.034. [DOI] [PubMed] [Google Scholar]
- [31].Carvalho FC, Martins DC, Santos A, Roque-Barreira M-C, Bueno PR. Analysing affinity between ArtinM lectin and myeloid leukemia cells by impedimetric and piezoelectric label free sensorial methods. Biosensors. 2014;4:358–369. doi: 10.3390/bios4040358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bertok T, Sediva A, Vikartovska A, Tkac J. Comparison of the 2D and 3D nanostructured lectin-based biosensors for in situ detection of sialic acid on glycoproteins. Int J Electrochem Sci. 2014;9:890–900. [PMC free article] [PubMed] [Google Scholar]
- [33].Alley WR, Mann BF, Novotny MV. High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem Rev. 2013;113:2668–2732. doi: 10.1021/cr3003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Arya SK, Bhansali S. Lung cancer and its early detection using biomarker-based biosensors. Chem Rev. 2011;111:6783–6809. doi: 10.1021/cr100420s. [DOI] [PubMed] [Google Scholar]
- [35].Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gilgunn S, Conroy PJ, Saldova R, Rudd PM, O'Kennedy RJ. Aberrant PSA glycosylation-a sweet predictor of prostate cancer. Nat Rev Urol. 2013;10:99–107. doi: 10.1038/nrurol.2012.258. [DOI] [PubMed] [Google Scholar]
- [37].Kim J-H, Resende R, Wennekes T, Chen H-M, Bance N, Buchini S, Watts AG, Pilling P, Streltsov VA, Petric M, Liggins R, et al. Mechanism-based covalent neuraminidase inhibitors with broad-spectrum influenza antiviral activity. Science. 2013;340:71–75. doi: 10.1126/science.1232552. [DOI] [PubMed] [Google Scholar]
- [38].Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P-S, Wang S-K, Stanfield RL, Julien J-P, Ramos A, Crispin M, Depetris R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Song R, Oren DA, Franco D, Seaman MS, Ho DD. Strategic addition of an n-linked glycan to a monoclonal antibody improves its HIV-1 neutralizing activity. Nat Biotechnol. 2013;31:1047–1052. doi: 10.1038/nbt.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tkac J, Davis JJ. An optimised electrode pre-treatment for SAM formation on polycrystalline gold. J Electroanal Chem. 2008;621:117–120. [Google Scholar]
- [42].Liu L, Deng D, Xing Y, Li S, Yuan B, Chen J, Xia N. Activity analysis of the carbodiimide-mediated amine coupling reaction on self-assembled monolayers by cyclic voltammetry. Electrochim Acta. 2013;89:616–622. [Google Scholar]
- [43].Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- [44].LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger T, Jr, Rowell N, Wollheim F. Scleroderma (systemic sclerosis): Classification, subsets and pathogenesis. J Rheumatol. 1988;15:202. [PubMed] [Google Scholar]
- [45].Valentini G, Cerinic M, Bombardieri S. Consensus statement: Core variables in the assessment of the patient with systemic sclerosis. Scleroderma Care Res. 2005;2 [Google Scholar]
- [46].Zauner G, Selman MHJ, Bondt A, Rombouts Y, Blank D, Deelder AM, Wuhrer M. Glycoproteomic analysis of antibodies. Mol Cel Proteom. 2013;12:856–865. doi: 10.1074/mcp.R112.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. http://www.rcsb.org/pdb/home/home.do, http://www.rcsb.org/pdb/home/home.do.
- [48].Gerlach JQ, Kilcoyne M, Joshi L. Microarray evaluation of the effects of lectin and glycoprotein orientation and data filtering on glycoform discrimination. Anal Methods. 2014;6:440–449. [Google Scholar]
- [49].Agrawal P, Kurcon T, Pilobello KT, Rakus JF, Koppolu S, Liu Z, Batista BS, Eng WS, Hsu K-L, Liang Y. Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode. P Natl Acad Sci. 2014;111:4338–4343. doi: 10.1073/pnas.1321524111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ribeiro JP, Mahal LK. Dot by dot: Analyzing the glycome using lectin microarrays. Curr Opin Chem Biol. 2013;17:827–831. doi: 10.1016/j.cbpa.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hirabayashi J, Yamada M, Kuno A, Tateno H. Lectin microarrays: Concept, principle and applications. Chem Soc Rev. 2013;42:4443–4458. doi: 10.1039/c3cs35419a. [DOI] [PubMed] [Google Scholar]
- [52].Bertok T, Sediva A, Katrlik J, Gemeiner P, Mikula M, Nosko M, Tkac J. Label-free detection of glycoproteins by the lectin biosensor down to attomolar level using gold nanoparticles. Talanta. 2013;108:11–18. doi: 10.1016/j.talanta.2013.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Luo X, Xu Q, James T, Davis JJ. Redox and label-free array detection of protein markers in human serum. Anal Chem. 2014;86:5553–5558. doi: 10.1021/ac5010037. [DOI] [PubMed] [Google Scholar]
- [54].Novokmet M, Lukic E, Vuckovic F, Ethuric Z, Keser T, Rajsl K, Remondini D, Castellani G, Gasparovic H, Gornik O, Lauc G. Changes in IgG and total plasma protein glycomes in acute systemic inflammation. Sci Rep. 2014;4:4347. doi: 10.1038/srep04347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rusling JF, Kumar CV, Gutkind JS, Patel V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst. 2010;135:2496–2511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wilson MS, Nie W. Multiplex measurement of seven tumor markers using an electrochemical protein chip. Anal Chem. 2006;78:6476–6483. doi: 10.1021/ac060843u. [DOI] [PubMed] [Google Scholar]
- [57].Wei F, Liao W, Xu Z, Yang Y, Wong DT, Ho CM. Bio/abiotic interface constructed from nanoscale DNA dendrimer and conducting polymer for ultrasensitive biomolecular diagnosis. Small. 2009;5:1784–1790. doi: 10.1002/smll.200900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.