Abstract
The evolution of bacterial resistance to conventional antimicrobials is a widely documented phenomenon with gravely important consequences for public health. However, bacteria also produce a vast repertoire of natural antimicrobials, presumably in order to kill competing species. Bacteriocins are a common class of protein-based antimicrobials that have been shown to have an important role in the ecology and evolution of bacterial communities. Relative to the evolution of antibiotic resistance, little is known about how novel resistance to these toxic compounds evolves. In this study, we present results illustrating that, although resistance is able to evolve, it remains critically dependent on the environmental context. Resistance to bacteriocins, in particular the pyocin S2, evolves readily when iron is present but less so when iron is limiting, because the receptor for this pyocin is also required for iron uptake during iron limitation. This suggests that although resistance to bacteriocins can easily evolve, environmental conditions will determine how and when resistance occurs.
Introduction
Bacteria are prolific producers of toxic compounds that inhibit the growth of potential competitors. Many of these naturally occurring compounds have been developed into drugs that combat bacterial pathogens (Fleming, 1929). However, as resistance to the ever-shrinking pool of effective antibiotics increases (Davies et al., 2013), there has been a renewed effort to seek out novel compounds that inhibit bacterial growth. One potential source of new antimicrobials are bacteriocins—highly potent antimicrobial peptides produced by bacteria that kill closely related species—and it has recently been suggested that bacteriocins may represent a viable alternative to traditional antibiotics (Cotter et al., 2013). However, little is known about how resistance to bacteriocins might evolve and the consequences this will have on the resistant bacteria (Perron et al., 2006, 2015).
Although few studies have explicitly examined how resistance to bacteriocins evolves, several studies examining the ecology of bacteriocin production have found high levels of both bacteriocin production and resistance in environmental isolates (Hawlena et al., 2010a, b, 2012; Kohler et al., 2010a). Furthermore, many studies identifying the molecular targets of different types of bacteriocins have shown that resistance to certain types of bacteriocins is possible through changes to cell surface receptors or intracellular targets (del Castillo et al., 2001; Kramer et al., 2006). Moreover, although resistance is possible, many bacteria remain sensitive (Hawlena et al., 2010a; Kohler et al., 2010b), which may indicate that resistance is either hard to evolve or there are costs associated with resistance mutations, as is commonly observed for antibiotics (Andersson and Levin, 1999).
In this study, we were interested in investigating how resistance to bacteriocins produced by the human pathogen, Pseudomonas aeruginosa, evolves. P. aeruginosa produces a number of different bacteriocins (termed pyocins) (Michel-Briand and Baysse, 2002), and previous studies have shown the pyocin S2 can have an important role in shaping the ecology and evolution of this organism (Inglis et al., 2009, 2011, 2012). The pyocin S2, is a soluble (S-type) pyocin that kills cells by breaking down DNA owing to its C-terminal endonuclease domain. Like many other bacteriocins, S2 is translocated into the cell via important and highly conserved cell surface receptors. In this case, S2 binds to FpvA, which is also responsible for the uptake of iron-bound siderophores, an essential source of iron for bacterial growth (Denayer et al., 2007). As both the pyocin S2 and iron-scavenging siderophores share the same receptor, we posited that changes in environmental iron concentrations, which can be caused naturally by fluctuations in pH (Robin et al., 2008; Lujan et al., 2015), might greatly affect the likelihood of resistance evolution. For example, when iron is present in the environment, changes to receptor expression will have little effect on growth; in contrast, when iron concentrations are low and the uptake of siderophores is required for growth, resistance may be less likely and more costly to evolve. In order to test these predictions, we co-cultured an S2-producing strain of P. aeruginosa (PAO1) and an S2-sensitive strain (O:9) for 70 generations under two different environmental conditions: iron rich and iron limited. We found that resistance evolved less often under iron limitation and resistant mutants were less fit than their counterparts that evolved in iron-rich conditions, suggesting that environmental constraints may have a greater role in bacteriocin resistance as they commonly target essential receptors involved in nutrient acquisition. Surprisingly, the evolution of bacteriocin resistance was not linked with any specific genetic changes to the FpvA receptor but may instead be associated with mutations in other genes that are known to affect its regulation (Beare et al., 2003; Schalk et al., 2009).
Materials and methods
Strains
P. aeruginosa strain PAO1, was used as the bacteriocin producer and serotype O:9 as the bacteriocin-sensitive competitor. PAO1 is a known producer of pyocin S2, whereas serotype O:9 is sensitive to S2 pyocins (Denayer et al., 2007). Bacteriocin production, sensitivity and insensitivity were confirmed using a simple plate assay where the production of the relevant bacteriocin is determined by overlaying bacteria mixed in semi-solid agar on plates that have been spotted with bacteria of another strain, as described by Fyfe et al. (1984). If the strain inoculated on the plate produces bacteriocin that kills the strain mixed with semi-solid agar, a halo-shaped zone of clearing can be observed in the bacterial lawn after incubating at 37 °C for 18 h. The absence of a clear halo indicates that either the overlaid strain is insensitive to the bacteriocin producer or the inoculated strain does not produce any bacteriocin.
Transfer regime
Overnight cultures for both strains were grown with shaking at 0.65 g and 37 °C for 18 h and then diluted to an optical density (OD) at 600 nm (OD600 nm) of 1.8 to ensure similar numbers of bacteria per millilitre. Thirty-ml glass universals containing 6 ml of Casamino acids medium (CAA; 5 g Casamino acids, 1.18 g K2HPO4·3H2O, 0.25 g MgSO4·7H2O, per litre) were inoculated with a total of 104 overnight culture cells of 90% serotype O:9 and 10% PAO1. To create an iron-limited environment, half of the tubes (12 in total) were also supplemented with 20 mm sodium bicarbonate and 100 μg ml−1 apo-transferrin (Sigma, Dorset, UK), a natural iron-chelator (Meyer et al., 1996). Inoculated tubes were subsequently grown for 96 h with shaking at 0.65 g in a 37 °C incubator. After 96 h, individual cultures were serially diluted and plated from a range of dilutions (10−4 to 10−1) on rifampicin 312.5 μg ml−1 agar plates (incubated for 18 h at 37 °C) to select for serotype O:9 (Inglis et al., 2009). Overnight cultures of O:9 were initiated from these plates by picking 100 individual, bacterial colonies, or if <100 colonies were present at a 10−1 dilution, the entire contents of the plate was transferred. To limit co-evolution between O:9 and PAO1, new overnights of PAO1 were initiated from frozen stocks at each transfer. These overnight cultures were then used to inoculate new tubes of CAA medium with a total of 104 cells of 90% serotype O:9 and 10% PAO1, and this selection procedure was repeated for a further nine transfers. Overnight cultures of serotype O:9 were also frozen down at the beginning of each transfer, and colony-forming units for O:9 were recorded at the end of each transfer.
Measuring resistance
Overnight cultures of ancestral and evolved O:9 were started from frozen stocks and incubated as described above. In all, 1 μl of each overnight culture was subsequently added to six independent wells containing 99 μl of CAA (iron-rich medium) combined with one part filtered PAO1 supernatant in a 96-well microtitre plate. The microtitre plates were then incubated for 24 h with shaking at 37 °C. Growth in the CAA and PAO1 supernatant mix was measured using OD600 nm. To control for the effect of other extracellular metabolites apart from pyocin S2 that may cause growth inhibition, 1 μl of each overnight culture was subsequently added to six independent wells containing 99 μl of CAA (iron-rich medium) combined with one part filtered PAO1150-2 supernatant in a 96-well microtitre plate. PAO1150-2 is a transposon knockout mutant of psy2 that no longer produces pyocin S2 and so should have no growth-inhibiting effect on ancestral and evolved O:9 clones. Microtitre plates were incubated for 24 h with shaking at 37 °C, and growth was measured using OD600 nm.
Growth in iron-limited conditions
In order to determine whether differences in cell density could explain our observed results, six replicate cultures of O:9 were grown in CAA and CAA supplemented with 20 mm sodium bicarbonate and 100 μg ml−1 apo-transferrin. Cultures were incubated for 24 h with shaking at 37 °C and subsequently serially diluted and plated to CAA agar plates so colony-forming units could be counted.
We were also interested to see whether evolved cultures of O:9 showed any reduction in their ability to grow under iron limitation. We, therefore, started overnight cultures of ancestral and evolved O:9 from frozen stocks and incubated as described above. In all, 1 μl of each overnight culture was subsequently added to six independent wells containing 99 μl of CAA supplemented with 20 mm sodium bicarbonate and 100 μg ml−1 apo-transferrin. Microtitre plates were incubated for 24 h with shaking at 37 °C, and growth was measured using OD600 nm.
Sequencing fpvA
We amplified and sequenced the fpvA gene of six randomly isolated bacterial colonies, which evolved under iron-limited conditions, and six randomly isolated bacterial colonies, which evolved under iron-rich conditions. PCR primers were designed to cover the full length of the gene. PCR products were aligned and analysed using MEGA6 (Tamura et al., 2013).
Analyses
In order to determine whether growth in the presence or absence of iron affected resistance evolution, a one-way analysis of variance was performed on growth data, using the evolutionary history of each strain (that is, whether it evolved in iron or iron-limited conditions) as a discrete explanatory variable. One-sample T-tests were used to compare growth between evolved populations and the non-evolved ancestral strain. All statistical analyses were carried out in R v. 3. 1. 2.
Results
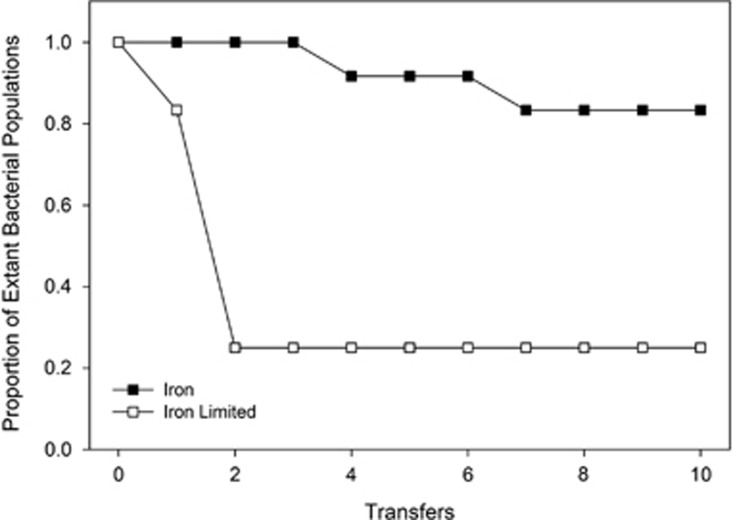
In this experiment, bacteria sensitive to the pyocin S2 (serotype O:9) were co-cultured with pyocin-producing bacteria PAO1, under two different environmental conditions: freely available iron and iron limitation. The presence and density of the sensitive strain O:9 was recorded at each transfer. Under iron-limited growth conditions, 9 of the 12 replicate populations went extinct during the transfer regime, whereas when iron was present only 2 of the 12 replicate populations went extinct (P<0.0123 in a two-tailed Fisher's Exact Test; Figure 1). Given that the ancestral O:9 shows very limited growth in the presence of PAO1 (Figure 2a), we interpret this as greater evolution of resistance in populations evolved under iron-rich conditions.
Figure 1.
Bacterial extinction during experimental evolution. Bacteriocin-sensitive bacteria (O:9) were grown in the presence of a bacteriocin producer (PAO1) in either iron-limited or -rich media. Bacteriocin-sensitive bacteria (O:9) were isolated after each round of growth and passaged to fresh media containing new cultures of the bacteriocin producer (PAO1). Significantly more bacteriocin-sensitive populations went extinct under iron-limited conditions compared with when iron was present in the environment after circa 70 bacterial generations (that is, 10 transfers).
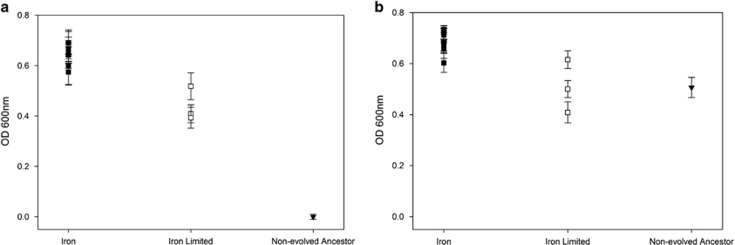
Figure 2.
(a) Resistance to the pyocin S2. The surviving bacterial populations of O:9 that evolved in both iron-rich and -limited conditions, as well as the ancestral O:9 strain, were grown in the supernatant of PAO1 in order to test for resistance to the pyocin S2. All evolved populations displayed some form of resistance compared with the non-evolved ancestor, with bacterial populations evolved under iron limitation displaying lower levels of growth compared with populations grown in iron-rich conditions. Error bars represent the s.e.m. of four replicates from each population. (b) Evolved resistance is specific to pyocin S2. To rule out the effect of other growth-inhibiting compounds produced by the S2 pyocin producer PAO1, evolved bacterial populations and the non-evolved ancestor were grown in the supernatant of a S2 pyocin knockout mutant (PAO1150-2). Under these conditions, the non-evolved ancestor was able to grow, indicating that evolved resistance is indeed due to pyocin S2. Error bars represent the s.e.m. of four replicates from each population.
In order to confirm that the remaining populations were resistant to the pyocin S2, we grew each of the populations and the non-evolved O:9 ancestor in a mixture of PAO1 supernatant and fresh media. We found that all remaining populations had evolved greatly increased resistance to the supernatant, relative to their ancestor (t9=55.92, P<0.001 for those evolved in iron-rich conditions and t2=11.19, P<0.008 for those evolved in iron-limited conditions). Moreover, bacterial populations evolved under iron-rich conditions had higher levels of growth compared with ones evolved in iron-limited conditions (Figure 2a; F1,11=51.3, P<0.001), which indicates that either lower levels of resistance evolved under iron-limited conditions or that evolved resistance was more costly.
To further investigate the specificity and extent of resistance evolution, we also grew the pyocin-resistant bacteria in the supernatant of a pyocin S2 knockout mutant (PAO1150-2) at the same time as the above experiment. Unlike growth in the presence of pyocin S2-containing supernatant, ancestral O:9 had similar growth to bacterial populations evolved in iron-limited conditions (t2=0.03, P>0.97), showing that there was no obvious evolution of resistance to potential toxins that might affect O:9 in the supernatant, in addition to pyocin S2. The O:9 populations evolved in iron-rich conditions displayed slightly higher growth than both the ancestral strain (t9=12.59, P<0.001) and strains evolved in iron-limited media (F1,11=22.3, P<0.001) (Figure 2b), which is consistent with a combination of increased adaptation to laboratory conditions and lower costs associated with resistance to pyocin S2, relative to populations evolved under iron-limited conditions. Importantly, there was also no difference in the relative densities of strains evolved under iron-limited and -rich conditions (F1,22=0.318, P>0.57) when grown in the presence and absence of pyocin S2 in the supernatant, suggesting that there was no difference in the extent of resistance evolution between treatments but rather that costs of resistance were greater when evolved under iron-limited conditions.
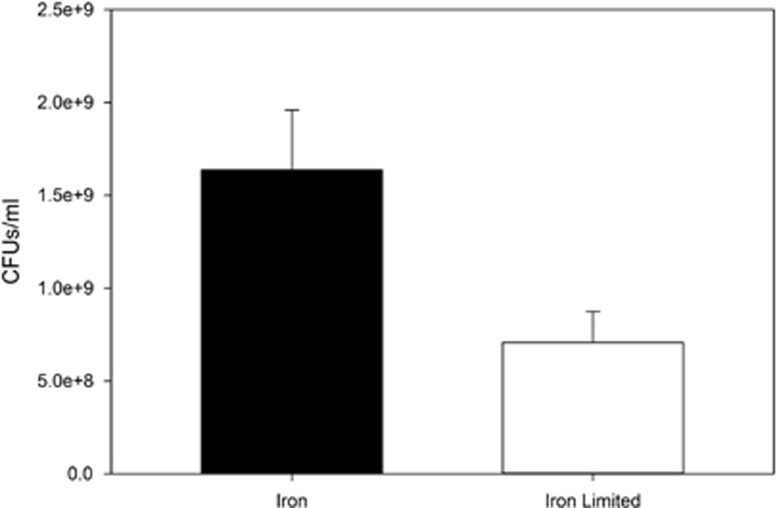
Differences in resistance evolution between iron-rich and -limited environments could also be affected (over and above the expression of siderophore receptors) if population size, and hence mutation supply rate, is reduced in iron-limited conditions. We found that when O:9 is grown by itself (that is, not in the presence of PAO1) it reaches a twofold greater density in iron-rich compared with iron-limited conditions (t11=2.56, P<0.029; Figure 3). It is quite possible that this difference may have contributed to resistance evolution, hence we carried out additional experiments to determine whether resistance evolution resulted in poor growth in iron-limited media, further constraining pyocin resistance in this environment as hypothesised.
Figure 3.
Differential growth in iron and iron-limited conditions. P. aeruginosa serotype O:9 was grown in both iron-rich and -limited conditions in order to see how population size varies between these two conditions. O:9 reaches higher final densities when iron is present compared with iron-limited conditions, which may influence resistance evolution between both treatments. Error bars represent the s.e.m. of six replicates grown in each condition.
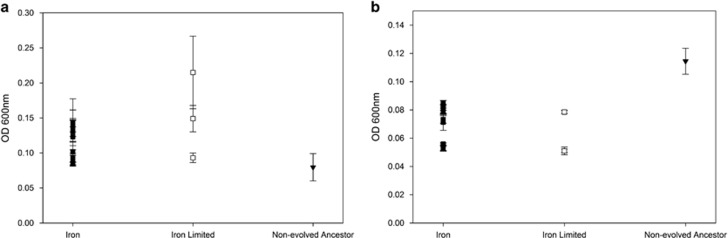
To see whether resistant bacterial populations might have evolved changes in their FpvA-mediated siderophore uptake, we first tested their ability to grow under iron-limited conditions where functional FpvA receptors are required for iron uptake. We found that when resistant populations were grown in iron-rich conditions there was no difference in growth between the evolved populations (F1,11=2.52, P>0.14), and although growth was slightly higher in populations evolved in iron-rich conditions compared with the ancestral strain (t9=5.27, P<0.001), there was no difference between the ancestral strain and those evolved in iron-limited conditions (t2=2.06, P>0.18) (Figure 4a). However, when resistant populations that evolved in both iron-rich and -limited conditions were grown in iron-limited media they had greatly impaired growth compared with the ancestral strain (t9=10.93, P<0.001 and t2=5.93, P<0.028, respectively) and grew equally poorly compared with each other (F1,11=1.58, P>0.23) (Figure 4b).
Figure 4.
(a) Growth in iron-rich conditions. The surviving bacterial populations of O:9 that evolved in both iron-rich and -limited conditions, as well as the ancestral O:9 strain, were grown in iron-rich conditions. No significant difference in growth was observed between both evolved populations or between populations evolved under iron-limited conditions and the ancestral strain. There was a slight difference in growth between populations evolved in iron-rich conditions and the ancestral strain. Error bars represent the s.e.m. of four replicates from each population. (b) Growth in iron-limited conditions. Interestingly, when these evolved populations were grown in iron-limited conditions, they show reduced growth compared with the non-evolved ancestor, presumably owing to changes in the iron uptake system. Error bars represent the s.e.m. of four replicates from each population.
As resistant populations grew so poorly in iron-limited conditions, we wanted to further investigate whether any genetic changes had occurred to FpvA, which might explain the populations' reduced growth. However, sequences of resistant clones from the populations evolved in both iron-rich and -limited environments showed no specific mutations associated with resistance when compared with each other and the ancestor. This suggests that changes in other genes or promoter regions involved in the regulation or expression of FpvA may instead underlie the resistant phenotype.
Discussion
In this study we found that resistance to the bacteriocin S2 is able to evolve in both iron-rich and -limited environments. However, under conditions where iron is freely available resistance evolved more frequently and resistant mutants were fitter (in terms of growth) compared with their counterparts evolved under iron limitation. A contributing factor to this observed difference in resistance evolution between environments could be the dual function of the receptor responsible for iron uptake (Denayer et al., 2007). Under iron-limited conditions, P. aeruginosa produces iron-chelating siderophores, such as pyoverdine (Meyer, 2000). The receptor FpvA, involved in translocating the iron-bound pyoverdine complex into the bacterial cell, is also co-expressed during iron limitation in order to facilitate iron uptake. However, FpvA also has a crucial role in translocating pyocin S2 (Denayer et al., 2007), which may explain why under iron-limited conditions the effect of pyocin S2 is increased (Ohkawa et al., 1980).
An obvious mechanism for resistance would, therefore, be the loss of function in the FpvA receptor or reduction in its expression, which would prevent pyocin from entering the cell. Under conditions where iron is easily accessible, this may be a likely form of resistance. However, when iron is scarce and FpvA is required for pyoverdine uptake, cells would suffer a large fitness cost in terms of growth rate. Correspondingly, we find that when bacteria have evolved bacteriocin resistance in iron-rich conditions, they grow poorly under iron limitation (Figure 4b), showing that iron uptake has indeed been impaired. Similarly, bacteria that have evolved bacteriocin resistance in iron-limited conditions also display equally poor growth under iron limitation (Figure 4b). This is somewhat surprising as mutations that confer resistance without affecting iron uptake would be strongly selected for in this environment, suggesting that resistance may only be able to evolve at the expense of pyoverdine-mediated iron acquisition.
Another important factor that may have also influenced the evolution of resistance between iron-rich and -limited environments is mutation supply rate (Arjan et al., 1999). When iron is present, bacterial populations are more than twice as numerous as when iron is limiting (Figure 3a), making bacteriocin resistance mutations more likely to occur. This may also explain why resistant mutants derived from iron-rich environments grow better than resistant mutants evolved in iron-limited conditions (Figures 2a and b), as other mutations that increase bacterial fitness either directly or through compensating for costly resistant mutations are more likely to fix with increased mutation supply (Perron et al., 2007).
Iron-uptake receptors are common targets for many bacteriocins, for example, FepA, CirA and FhuA are all involved in iron acquisition and act as receptors for a number of different colicins (Cascales et al., 2007). Similarly, BtuB, which essential for growth in Escherichia coli and is responsible for translocating vitamin B12 across the outer membrane, is also a frequent target receptor for most other colicins (Cascales et al., 2007). The targeting of essential and important receptors is, in general, a good strategy for reducing the chance of resistance evolving or at least making resistance extremely costly in many natural environments, as seen in our experiments and in other bacteriocin-resistant strains (for example, colicin-resistant E. coli have reduced fitness compared with their ancestral, sensitive strains (Kerr et al., 2002)). Analogously, bacteriophages commonly target costly receptors, and changes that confer resistance to bacteriophage have also been shown to exert fitness costs on bacterial hosts (Smith et al., 1987; Buckling and Brockhurst, 2012). For example, the loss pili in Pseudomonas syringae not only provides resistance to the bacteriophage θ6 but also reduces the fitness of P. syringae on host plants and subsequent bacterial disease (Romantschuk and Bamford, 1985; Romantschuk et al., 1993).
However, targeting individual receptors in many cases may only allow bacteria to kill specific competitors (in fact, many bacteriocins kill only closely related strains and can even act as selfish genetic elements promoting their own transmission in a bacterial population (Inglis et al., 2013)). This specificity reduces their scope as general antibiotics, but instead they could be used effectively for targeting particular strains of interest (for example, multidrug resistant or highly virulent bacteria). Furthermore, by better understanding how resistance to bacteriocin evolves and taking into account environmental conditions that influence resistance evolution, it may be possible to effectively reduce its occurrence.
Acknowledgments
We thank Joan Strassmann and David Queller for their helpful comments. This work was funded by the AXA Research Fund, BBSRC, ERC and NERC. AB is also supported by the Royal Society.
The authors declare no conflict of interest.
References
- Andersson DI, Levin BR. (1999). The biological cost of antibiotic resistance. Curr Opin Microbiol 2: 489–493. [DOI] [PubMed] [Google Scholar]
- Arjan JA, Visser M, Zeyl CW, Gerrish PJ, Blanchard JL, Lenski RE. (1999). Diminishing returns from mutation supply rate in asexual populations. Science 283: 404–406. [DOI] [PubMed] [Google Scholar]
- Beare PA, For RJ, Martin LW, Lamont IL. (2003). Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol Microbiol 47: 195–207. [DOI] [PubMed] [Google Scholar]
- Buckling A, Brockhurst M. (2012). Bacteria-virus coevolution. Adv Exp Med Biol 751: 347–370. [DOI] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K et al. (2007). Colicin biology. Microbiol Mol Biol Rev 71: 158–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Ross RP, Hill C. (2013). Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol 11: 95–105. [DOI] [PubMed] [Google Scholar]
- Davies SC, Fowler T, Watson J, Livermore DM, Walker D. (2013). Annual Report of the Chief Medical Officer: infection and the rise of antimicrobial resistance. Lancet 381: 1606–1609. [DOI] [PubMed] [Google Scholar]
- del Castillo FJ, del Castillo I, Moreno F. (2001). Construction and characterization of mutations at codon 751 of the Escherichia coli gyrB gene that confer resistance to the antimicrobial peptide microcin B17 and alter the activity of DNA gyrase. J Bacteriol 183: 2137–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayer S, Matthijs S, Cornelis P. (2007). Pyocin S2 (Sa) kills Pseudomonas aeruginosa strains via the FpvA type I ferripyoverdine receptor. J Bacteriol 189: 7663–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. (1929). On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br J Exp Pathol 10: 226–236. [Google Scholar]
- Fyfe JAM, Harris G, Govan JRW. (1984). Revised pyocin typing method for Pseudomonas aeruginosa. J Clin Microbiol 20: 47–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawlena H, Bashey F, Lively CM. (2010. a). The evolution of spite: population structure and bacteriocin-mediated antagonism in two natural populations of xenorhabdus bacteria. Evolution 64: 3198–3204. [DOI] [PubMed] [Google Scholar]
- Hawlena H, Bashey F, Mendes-Soares H, Lively CM. (2010. b). Spiteful Interactions in a natural population of the bacterium Xenorhabdus bovienii. Am Nat 175: 374–381. [DOI] [PubMed] [Google Scholar]
- Hawlena H, Bashey F, Lively CM. (2012). Bacteriocin-mediated interactions within and between coexisting species. Ecol Evol 2: 2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis RF, Gardner A, Cornelis P, Buckling A. (2009). Spite and virulence in the bacterium Pseudomonas aeruginosa. Proc Natl Acad Sci USA 106: 5703–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis RF, Roberts PG, Gardner A, Buckling A. (2011). Spite and the scale of competition in Pseudomonas aeruginosa. Am Nat 178: 276–285. [DOI] [PubMed] [Google Scholar]
- Inglis RF, Brown SP, Buckling A. (2012). Spite versus cheats: competition among social strategies shapes virulence in Pseudomonas aeruginosa. Evolution 66: 3472–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis RF, Bayramoglu B, Gillor O, Ackermann M. (2013). The role of bacteriocins as selfish genetic elements. Biol Lett 9: 20121173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJM. (2002). Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418: 171–174. [DOI] [PubMed] [Google Scholar]
- Kohler T, Donner V, van Delden C. (2010. a). Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J Bacteriol 192: 1921–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T, Perron GG, Buckling A, van Delden C. (2010. b). Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog 6: e1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer NE, van Hijum SA, Knol J, Kok J, Kuipers OP. (2006). Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob Agents Chemother 50: 1753–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan AM, Gomez P, Buckling A. (2015). Siderophore cooperation of the bacterium Pseudomonas fluorescens in soil. Biol Lett 11: 20140934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. (1996). Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun 64: 518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JM. (2000). Proverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch Microbiol 174: 135–142. [DOI] [PubMed] [Google Scholar]
- Michel-Briand Y, Baysse C. (2002). The pyocins of Pseudomonas aeruginosa. Biochimie 84: 499–510. [DOI] [PubMed] [Google Scholar]
- Ohkawa I, Shiga S, Kageyama M. (1980). Effect of iron concentration in the growth medium on the sensitivity of Pseudomonas aeruginosa to pyocin S2. J Biochem 87: 323–331. [DOI] [PubMed] [Google Scholar]
- Perron GG, Zasloff M, Bell G. (2006). Experimental evolution of resistance to an antimicrobial peptide. Proc Biol Sci 273: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron GG, Gonzalez A, Buckling A. (2007). Source-sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proc Biol Sci 274: 2351–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron GG, Inglis RF, Pennings PS, Cobey S. (2015). Fighting microbial drug resistance: a primer on the role of evolutionary biology in public health. Evol Appl 8: 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin A, Vansuyt G, Hinsinger P, Meyer JM, Briat JF, Lemanceau P. (2008). Iron dynamics in the rhizosphere: consequences for plant health and nutrition. Adv Agron 99: 183–225. [Google Scholar]
- Romantschuk M, Bamford DH. (1985). Function of pili in bacteriophage phi 6 penetration. J Gen Virol 66(Pt 11): 2461–2469. [DOI] [PubMed] [Google Scholar]
- Romantschuk M, Nurmiaholassila EL, Roine E, Suoniemi A. (1993). Pilus-mediated adsorption of Pseudomonas syringae to the surface of host and nonhost plant-leaves. J Gen Microbiol 139: 2251–2260. [Google Scholar]
- Schalk IJ, Lamont IL, Cobessi D. (2009). Structure-function relationships in the bifunctional ferrisiderophore FpvA receptor from Pseudomonas aeruginosa. Biometals 22: 671–678. [DOI] [PubMed] [Google Scholar]
- Smith HW, Huggins MB, Shaw KM. (1987). The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J Gen Microbiol 133: 1111–1126. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]