Abstract
Abnormal signaling of the protein kinase Akt has been shown to contribute to human diseases such as diabetes and cancer, but Akt has proven to be a challenging target for drugging. Using iterative in situ click chemistry we recently developed multiple protein catalyzed capture (PCC) agents that allosterically modulate Akt enzymatic activity in a protein based assay. Here we utilize similar PCCs to exploit endogenous protein degradation pathways. We use the modularity of the anti-Akt PCCs to prepare Proteolysis Targeting Chimeric molecules (PROTACs) that are shown to promote the rapid degradation of Akt in live cancer cells. These novel PROTACs demonstrate that the epitope targeting selectivity of PCCs can be coupled with non-traditional drugging moieties to inhibit challenging targets.
INTRODUCTION
Protein-catalyzed capture agents (PCCs) are a class of ligands that are built using in situ click chemistry (Lewis et al., 2002) to allow a protein of interest to select its own high-affinity binders (Agnew et al., 2009). These synthetic peptides have some similarities to monoclonal antibodies, but are a fraction of the size and can exhibit a high level of stability (Farrow et al., 2013; Pfeilsticker et al., 2013). A recent advance of this technology permits the targeted development of a PCC against a specific epitope of a given protein (Das et al., 2015; Farrow et al., 2015; Nag et al., 2013). Unlike the case for small molecule ligands, the generalized PCC epitope targeting strategy does not rely on the presence of a hydrophobic binding pocket. This opens up several non-traditional approaches towards altering enzymatic activity, including the targeting of sites that can allosterically influence that activity (Millward et al., 2011) or by disrupting protein-activator associations (Deyle et al., 2015). A second possibility is simply to use the synthetic flexibility of the PCC as a selective targeting moiety for labeling the target with a molecular signal, such as a degradation signal, as is used for the case of Proteolysis Targeting Chimeric molecules (PROTACs). Here we explore the use of an epitope targeted PCC developed against an allosteric site of Akt2 as an in-cell allosteric activator and PROTAC.
Akt is a serine/threonine protein kinase with three closely related isoforms (Akt1-3) and is involved in cellular processes such as glucose metabolism, apoptosis, and cell proliferation (Engelman, 2009; Manning and Cantley, 2007). Aberrant Akt signaling is implicated in diabetes and in many cancers, making it an attractive drug and diagnostic target (Lawlor and Alessi, 2001). We previously reported the development of a PCC targeting the C-terminal hydrophobic motif (HM) of Akt2 that includes the Ser474 residue (Nag et al., 2013). Phosphorylation of Akt at Ser474 leads to allosteric activation of Akt and increases the kinase activity 10 fold (Yang et al., 2002). We thus hypothesized that targeting the Ser474 site could lead to compounds that influence Akt kinase activity. We increased the interaction footprint of the PCC with Akt2 by expanding it into two distinct triligands through in situ click chemistry screens. One of the triligands, tri_a, was shown to allosterically activate Akt enzymatic activity in in vitro kinase assays (Nag et al., 2013).
Although specific peptides (typically macrocycles) have been designed for cell entry (Chu et al., 2015), the tri_a PCC is a branched structure consisting of linear branches, and so does not naturally enter into live cells. We recently reported on a strategy for the delivery of PCCs into cells. The target of the PCC was a cell-penetrating enzyme (Botulinum Neurotoxin Serotype A), and so the PCC was carried into cells as a type of Trojan horse cargo (Farrow, et al., 2015). A second more common approach (Hassane et al., 2009) is to append a cell penetrating peptide (CPP) to the PCC, and that is the route we choose here. CPP-labeled tri_a was found to penetrate into live cells. The influence of that compound on in-cell kinase activity and on cellular proliferation was then explored in two cancer cell lines. We next modified the tri_a to present a HIF-1α degradation signal, and explored the capacity of this compound to promote in-cell Akt degradation.
RESULTS AND DISCUSSION
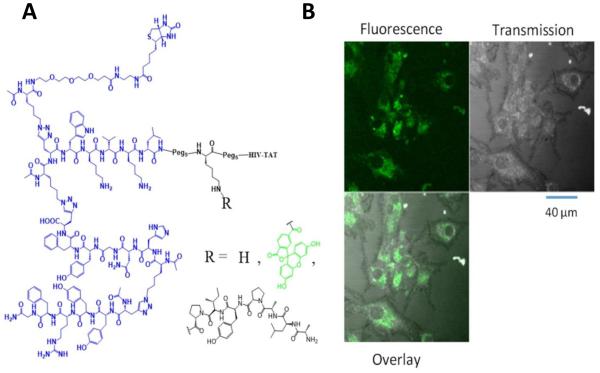
We conjugated the PCC agent to the HIV TAT peptide, which is a CPP that efficiently penetrates cell membranes via endocytosis, and allows CPP-bound molecules to enter cells (Heitz et al., 2009). Figure 1A shows the structure of TAT-conjugated tri_a, where the TAT sequence is separated from the capture agent by two PEG spacers placed on either side of a protected-lysine residue. This permits further functionalization as desired during the solid phase peptide synthesis via the ε-amino group (adding a dye, signaling peptide, etc.). To validate cellular uptake, we treated U87 cells with fluorescein-labeled tri_a (CPP-tri_a-FL, Figure 1B) and acquired simultaneous fluorescence and transmission images. U87 cells are particularly useful for imaging since they grow in a uniform monolayer (Camphausen et al., 2005). We found that the CPP-tri_a-FL is able to efficiently penetrate the cell membrane and enter the cells. No fluorescence signals were detected outside the cells, and the uniform distribution of the fluorescence through the cell cytoplasm suggests that the PCCs were released into the cells (rather than completely trapped within endosomes).
Figure 1. Akt activating PCC, tri_a, and exploiting the modularity of this PCC.
(A) Structure of Akt-activating N-terminal triligand, tri_a. The lysine residue between the PEG spacers can be further functionalized with fluorescein or the degradation-inducing Hif-1α peptide. Complete structures are shown in Figure S1. R = H is referred to as CPP-tri_a, R = fluorescein is CPP-tri_a-FL, and R = ALAPYIP (Hif-1α degradation peptide) is CPP-tri_a-PR. (B) Live-cell confocal images of the fluorescein-labeled capture agent CPP-tri_a-FL delivered into U87 cells.
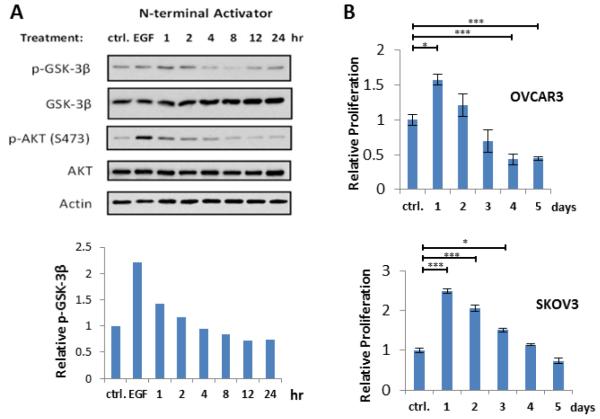
We next performed functional assays to confirm that the in vitro effects translate into live cells. The enzymatic activity of Akt can be monitored by measuring the phosphorylated level of its direct substrate, glycogen synthase kinase 3 beta (GSK-3β) (Engelman, 2009). SKOV3 ovarian cancer cells were dosed with TAT-coupled tri_a (CPP-tri_a, Figure 1A, S1) for various times. Following cell lysis, the relative levels of various proteins were measured via western blotting (Figure 2A). Both untreated cells and EGF-stimulated cells were used as controls. The level of phospho(p)-GSK-3β was seen to increase after 1 hour of treatment, decrease over the next few hours, and finally increase once more – perhaps alluding to a feedback mechanism in the cell. The initial increase in the p-GSK-3β level indicates that the capture agent is binding Akt and stimulating its enzymatic activity in cells. The p-Akt level initially increases and then steadily decreases over time. Notably, EGF-stimulated cells display a large increase in p-Akt, but the corresponding level of p-GSK-3β in EGF-stimulated cells is still lower than the initial levels in cells treated with CPP-tri_a.
Figure 2. tri_a activates Akt in cells.
(A) Western blots of SKOV3 cells treated with 50 μM tri_a Akt-activating capture agent. Densitometry was performed on the p-GSK-3β blot and the relative intensities were plotted on the bar graph below the blotting assays. (B) XTT cell proliferation assay results from OVCAR3 and SKOV3 cells treated with 50 μM tri_a for times varying from 1-5 days. A student’s t test was used to determine significant differences where * = p-value ≤ 0.05, and *** = p-value ≤ 0.01. See Figure S2 for parallel inhibition experiments.
We next performed an XTT cell proliferation assay to determine if Akt activation by CPP-tri_a stimulates cell proliferation (Xu et al., 2012). Dehydrogenase enzymes in live cells reduce XTT tetrazolium salt to a vividly colored formazan dye that can be used to quantify the number of viable cells (Berridge et al., 2005; Scudiero et al., 1988). We found that CPP-tri_a affected both OVCAR3 and SKOV3 cell lines (Figure 2B). Interestingly, for both cell lines, a sharp increase in proliferation after 24 hours is followed by a steady decrease over a few days – once more suggesting a possible feedback mechanism. Eventually, after several days of treatment, cellular proliferation falls back to baseline levels for SKVO3 cells while OVCAR3 cells are inhibited relative to the untreated control. An initial increase in cell number after treatment with CPP-tri_a is consistent with previous studies showing that Akt activation promotes cell cycle progression during the G1 and G2 phases of the cell cycle (Kandel et al., 2002; Ramaswamy et al., 1999). Additionally, activated Akt is known to inhibit the pro-apoptotic Bad protein (Datta et al., 1997; Peso et al., 1997), as well as the FoxO and Myc family of proteins (Arden, 2004; Zhu et al., 2008). Such enhanced cell proliferation can be used in beneficial ways. Many pathological disorders are associated with aberrant cell death signaling, including various neurodegenerative diseases (Mattson and Sherman, 2003). Akt activation was recently shown to prevent neuronal cell death (Jo et al., 2012). Thus, an Akt activator might prove useful as a tool to probe the Akt signaling pathway and to help develop potential therapeutics for disease-associated apoptosis abnormalities. In any case, these results further corroborate the in vitro data that tri_a directly activates Akt.
PROTACs represent an example of using cellular signals to control levels of specific proteins (Buckley and Crews, 2014; Sakamoto et al., 2001). In particular, PROTACs utilize the quality-control machinery of the cell by artificially labeling proteins for proteasomal degradation. We sought to turn CPP-tri_a into a PROTAC (CPP-tri_a-PR, Figure 3, S1) by encoding a peptide ligand for the E3 ubiquitin ligase von Hippel Lindau protein (VHL) directly into the CPP-labeled PCC agents (Hines et al., 2013). The peptide represents the VHL binding site from hypoxia-inducible factor (HIF-1α) protein.
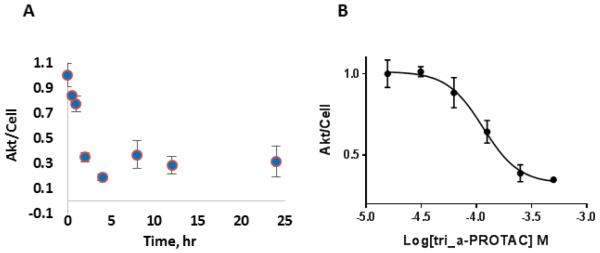
Figure 3. Tri_a-PROTAC knocks down Akt in cells.
(A) In-cell ELISA measurement of Akt in OVCAR3 cells treated with tri_a-PROTAC containing the degradation-inducing Hif-1α sequence for various times. (B) In-cell ELISA measurement of Akt after 4 hour treatment at variable tri_a-PROTAC doses. Results show dose-dependent degradation of Akt with an EC50 value of 128±19 μM. The data are normalized to the untreated control and relative levels of Akt/cell are plotted. Figure S3 contains additional degradation data with CPP-tri_1p-PR.
When OVCAR3 cells were treated with CPP-tri_a-PR over a variable time period up to 24 hours Akt levels diminished. After 30 minutes of treatment, a relative decrease in Akt level was measurable, with a continuing decrease that reached a nadir after 4 hours (Figure 3A). Similarly, an inhibitory capture agent, CPP-tri_i (see Fig S2 for inhibition data), was also modified into a PROTAC (CPP-tri_i-PR, Figure S3) and also effectively removed Akt from cells (supplemental information), with a slightly slower time constant. We also observed a dose-dependent decrease in Akt upon treatment with the CPP-tri_a-PR, and determined an EC50 degradation constant of 128±19 μM (Figure 3B).
SIGNIFICANCE
Previous PROTACs have been developed by adding a degradation signal to a modified protein ligand (Rodriguez-Gonzalez et al., 2008) or previously discovered inhibitors (Gustafson et al., 2015; Long et al., 2012). Here we report that an epitope-targeted PCC can be modified into a PROTAC to direct synthetic degradation to specific proteins in cells. We also show an Akt activator and inhibitor can both serve as PROTACs (Figure 3 and Supplement), indicating that ubiquitination and degradation of this protein can be induced regardless of the activation state of the protein. Targeting proteins for enzymatic destruction is an attractive alternative to developing traditional small molecule inhibitors, especially for proteins (such as Akt) that are difficult to inhibit with traditional methods (Neklesa and Crews, 2012).
The principle concept reported here is the use of an epitope targeted PCC as simply a vehicle for directing PROTAC-initiated degradation to a specific target. A limitation of this approach, as reported, is that the PCCs are not intrinsically cell penetrant, and so were modified with a CPP for the in-cell assays. We have recently reported on epitope targeted macrocyclic PCCs (Das et al., 2015), and such peptide architectures have been modified for both cell permeability (Chu et al., 2015; Hewitt et al., 2015; Walensky and Bird, 2014) and other desired pharmacokinetic properties (Bock et al., 2013). Other promising proteolysis tags include members of the IMiD family, such as thalidomide, which is a drug with a rich history (Bartlett et al., 2004). In 2010 it was discovered that thalidomide targets the E3 ubiquitin ligase member cereblon (Ito et al., 2010). Related examples of cereblon-targeted degradation include the use of pomalidomide (Lu et al., 2015) and phthalimide (Winter et al., 2015) for PROTAC development.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
All peptides were synthesized via solid phase peptide synthesis as described previously (Nag et al., 2013)
Cell culture
All cell lines were purchased from American Type Culture collection. U87 (ATCC number HTB14), OVCAR3 (ATCC number HTB161), SKOV3 (ATCC number HTB77) cells were cultured under conditions specified by the provider.
Fluorescence microscopy
Cells were seeded onto chambered coverglass slides (Sigma) and allowed to attach overnight. The following day, cells were serum starved and treated with fluorescent capture agent. Live cells were then imaged using a Zeiss LSM 5 Exciter microscope in the Caltech Biological Imaging Center.
Immunoblotting
Western blots were performed according to standard protocols. Briefly, cells were lysed with cell lysis buffer (Cell Signaling Technology) containing protease and phosphatase inhibitors (Cell Signaling Technology). Cell lysates were quantified with a Bradford protein assay (Thermo Scientific) and prepared for gel electrophoresis in Laemmli sample buffer and reducing agent. 20 μg of cell lysate were added to precast polyacrylamide gels (Bio-Rad) and proteins were separated by electrophoresis followed by transfer to PVDF membrane. Membranes were then blocked and probed with primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies. The bands were visualized chemiluminescent substrate (Thermo Scientific). The following antibodies were used according to manufacturer protcol: p-GSK3β (Cell Signaling, 9323), GSK3 β (Cell Signaling, 12456), AKT (Cell Signaling, 4691), p-AKT (S473) (Cell Signaling, 4060), Actin (Cell Signaling, 8456), HRP-linked Anti-rabbit IgG (Cell Signaling, 7074).
Cell Proliferation Assay
XTT assay kit was purchased from Cell Signaling Technology (#9095) and used according to manufacturer protocol. Briefly, 1×104 cells/well were seeded in a 96-well plate. The following day, cells were serum starved and treated with PCC agent. Following treatment, XTT was added to the media and after 1 hour the absorbance at 450 nm was measured.
In-cell ELISA assay
In-cell ELISA kits were purchased from Thermo Scientific (#62215) and used according to manufacturer protocol. Briefly, 1×104 cells were seeded in 384-well plates and allowed to attach overnight. The following day, cells were serum starved and treated with capture agent. Following capture agent treatment, cells were fixed, permeablized, blocked, and then stained with primary and HRP-conjugated secondary antibody and developed with colorimetric peroxidase substrate. The absorbance was measured at 450 nm to quantify the protein. Cells were then incubated with Janus Green whole-cell stain and the absorbance was measured at 615 nm to quantify relative number of cells per well. The Akt signal was then normalized to the relative cell number for each well to determine the Akt/cell.
Supplementary Material
Highlights.
An Akt-targeted PCC allosterically activates Akt kinase activity in cells
The anti-Akt PCC was functionalized with a degradation tag to create a novel PROTAC
Combining PCCs with degradation tags is a new strategy to create PROTACs
In Brief.
A Protein Catalyzed Capture Agent (PCC) was developed against the C-terminal hydrophobic motif of Akt, a traditionally “undruggable” site, and shown to be an allosteric activator. Functionalization of the PCC with a degradation tag led to a new anti-Akt PROTAC.
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute grant 1U54CA199090-01 (JRH PI) and by the Institute for Collaborative Biotechnologies (W911NF-09-0001) from the U.S. Army Research Office. JRH is a founder and board member of Indi Molecular. Indi Molecular is seeking to commercialize the PCC agent technology.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information includes structures, characterization and synthetic procedures used to develop CPP-tri_a and CPP-tri_a-PR. Additionally, procedures and results for inhibitory capture agent CPP-tri_i and CPP-tri_i-PR are also included.
AUTHOR CONTRIBUTIONS
R.K.H., J.O.V., S.D. and A.N. designed the compounds. R.K.H., J.O.V., G.T., and K.T. synthesized the compounds. R.K.H and J.O.V. designed and conducted the experiments. R.K.H., J.O.V., A.S., and J.R.H analyzed the data. R.K.H, J.O.V., and J.R.H wrote the manuscript.
REFERENCES
- Agnew HD, Rohde RD, Millward SW, Nag A, Yeo W-S, Hein JE, Pitram SM, Tariq AA, Burns VM, Krom RJ, et al. Iterative In Situ Click Chemistry Creates Antibody-like Protein-Capture Agents. Angew. Chem. Int. Ed. 2009;48:4944–4948. doi: 10.1002/anie.200900488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden KC. FoxO: Linking New Signaling Pathways. Mol. Cell. 2004;14:416–418. doi: 10.1016/s1097-2765(04)00213-8. [DOI] [PubMed] [Google Scholar]
- Bartlett JB, Dredge K, Dalgleish AG. The evolution of thalidomide and its IMiD derivatives as anticancer agents. Nat. Rev. Cancer. 2004;4:314–322. doi: 10.1038/nrc1323. [DOI] [PubMed] [Google Scholar]
- Berridge MV, Herst PM, Tan AS. Tetrazolium dyes as tools in cell biology: New insights into their cellular reduction. In: El-Gewely MR, editor. Biotechnology Annual Review. Elsevier: 2005. pp. 127–152. [DOI] [PubMed] [Google Scholar]
- Bock JE, Gavenonis J, Kritzer JA. Getting in Shape: Controlling Peptide Bioactivity and Bioavailability Using Conformational Constraints. ACS Chem. Biol. 2013;8:488–499. doi: 10.1021/cb300515u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DL, Crews CM. Small-Molecule Control of Intracellular Protein Levels through Modulation of the Ubiquitin Proteasome System. Angew. Chem. Int. Ed. 2014;53:2312–2330. doi: 10.1002/anie.201307761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camphausen K, Purow B, Sproull M, Scott T, Ozawa T, Deen DF, Tofilon PJ. Influence of in vivo growth on human glioma cell line gene expression: Convergent profiles under orthotopic conditions. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8287–8292. doi: 10.1073/pnas.0502887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Q, Moellering RE, Hilinski GJ, Kim Y-W, Grossmann TN, Yeh JT-H, Verdine GL. Towards understanding cell penetration by stapled peptides. MedChemComm. 2015;6:111–119. [Google Scholar]
- Das S, Nag A, Liang J, Bunck DN, Umeda A, Farrow B, Coppock MB, Sarkes DA, Finch AS, Agnew HD, et al. A General Synthetic Approach for Designing Epitope Targeted Macrocyclic Peptide Ligands. Angew. Chem. Int. Ed. 2015;54:13219–13224. doi: 10.1002/anie.201505243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt Phosphorylation of BAD Couples Survival Signals to the Cell-Intrinsic Death Machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- Deyle KM, Farrow B, Qiao Hee Y, Work J, Wong M, Lai B, Umeda A, Millward SW, Nag A, Das S, et al. A protein-targeting strategy used to develop a selective inhibitor of the E17K point mutation in the PH domain of Akt1. Nat. Chem. 2015;7:455–462. doi: 10.1038/nchem.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat. Rev. Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Farrow B, Hong SA, Romero EC, Lai B, Coppock MB, Deyle KM, Finch AS, Stratis-Cullum DN, Agnew HD, Yang S, et al. A Chemically Synthesized Capture Agent Enables the Selective, Sensitive, and Robust Electrochemical Detection of Anthrax Protective Antigen. ACS Nano. 2013;7:9452–9460. doi: 10.1021/nn404296k. [DOI] [PubMed] [Google Scholar]
- Farrow B, Wong M, Malette J, Lai B, Deyle KM, Das S, Nag A, Agnew HD, Heath JR. Epitope Targeting of Tertiary Protein Structure Enables Target-Guided Synthesis of a Potent In-Cell Inhibitor of Botulinum Neurotoxin. Angew. Chem. Int. Ed. 2015;54:7114–7119. doi: 10.1002/anie.201502451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson JL, Neklesa TK, Cox CS, Roth AG, Buckley DL, Tae HS, Sundberg TB, Stagg DB, Hines J, McDonnell DP, et al. Small-Molecule-Mediated Degradation of the Androgen Receptor Through Hydrophobic Tagging. Angew. Chem. Int. Ed. 2015;54:9659–9662. doi: 10.1002/anie.201503720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassane FS, Saleh AF, Abes R, Gait MJ, Lebleu B. Cell penetrating peptides: overview and applications to the delivery of oligonucleotides. Cell. Mol. Life Sci. 2009;67:715–726. doi: 10.1007/s00018-009-0186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br. J. Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt WM, Leung SSF, Pye CR, Ponkey AR, Bednarek M, Jacobson MP, Lokey RS. Cell-Permeable Cyclic Peptides from Synthetic Libraries Inspired by Natural Products. J. Am. Chem. Soc. 2015;137:715–721. doi: 10.1021/ja508766b. [DOI] [PubMed] [Google Scholar]
- Hines J, Gough JD, Corson TW, Crews CM. Posttranslational protein knockdown coupled to receptor tyrosine kinase activation with phosphoPROTACs. Proc. Natl. Acad. Sci. 2013;110:8942–8947. doi: 10.1073/pnas.1217206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a Primary Target of Thalidomide Teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Jo H, Mondal S, Tan D, Nagata E, Takizawa S, Sharma AK, Hou Q, Shanmugasundaram K, Prasad A, Tung JK, et al. Small molecule-induced cytosolic activation of protein kinase Akt rescues ischemia-elicited neuronal death. Proc. Natl. Acad. Sci. 2012;109:10581–10586. doi: 10.1073/pnas.1202810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ES, Skeen J, Majewski N, Di Cristofano A, Pandolfi PP, Feliciano CS, Gartel A, Hay N. Activation of Akt/Protein Kinase B Overcomes a G2/M Cell Cycle Checkpoint Induced by DNA Damage. Mol. Cell. Biol. 2002;22:7831–7841. doi: 10.1128/MCB.22.22.7831-7841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor MA, Alessi DR. PKB/Akt a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 2001;114:2903–2910. doi: 10.1242/jcs.114.16.2903. [DOI] [PubMed] [Google Scholar]
- Lewis WG, Green LG, Grynszpan F, Radić Z, Carlier PR, Taylor P, Finn MG, Sharpless KB. Click Chemistry In Situ: Acetylcholinesterase as a Reaction Vessel for the Selective Assembly of a Femtomolar Inhibitor from an Array of Building Blocks. Angew. Chem. 2002;114:1095–1099. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Long MJC, Gollapalli DR, Hedstrom L. Inhibitor Mediated Protein Degradation. Chem. Biol. 2012;19:629–637. doi: 10.1016/j.chembiol.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, Hines J, Winkler JD, Crew AP, Coleman K, et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol. 2015;22:1–9. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB Signaling: Navigating Downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Sherman M. Perturbed signal transduction in neurodegenerative disorders involving aberrant protein aggregation. NeuroMolecular Med. 2003;4:109–131. doi: 10.1385/NMM:4:1-2:109. [DOI] [PubMed] [Google Scholar]
- Millward SW, Henning RK, Kwong GA, Pitram S, Agnew HD, Deyle KM, Nag A, Hein J, Lee SS, Lim J, et al. Iterative in Situ Click Chemistry Assembles a Branched Capture Agent and Allosteric Inhibitor for Akt1. J. Am. Chem. Soc. 2011;133:18280–18288. doi: 10.1021/ja2064389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Das S, Yu MB, Deyle KM, Millward SW, Heath JR. A Chemical Epitope-Targeting Strategy for Protein Capture Agents: The Serine 474 Epitope of the Kinase Akt2. Angew. Chem. Int. Ed. 2013;52:13975–13979. doi: 10.1002/anie.201305882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa TK, Crews CM. Chemical biology: Greasy tags for protein removal. Nature. 2012;487:308–309. doi: 10.1038/487308a. [DOI] [PubMed] [Google Scholar]
- Peso L. del, González-García M, Page C, Herrera R, Nuñez G. Interleukin-3-Induced Phosphorylation of BAD Through the Protein Kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- Pfeilsticker JA, Umeda A, Farrow B, Hsueh CL, Deyle KM, Kim JT, Lai BT, Heath JR. A Cocktail of Thermally Stable, Chemically Synthesized Capture Agents for the Efficient Detection of Anti-Gp41 Antibodies from Human Sera. PLoS ONE. 2013;8:e76224. doi: 10.1371/journal.pone.0076224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM, Sellers WR. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gonzalez A, Cyrus K, Salcius M, Kim K, Crews CM, Deshaies RJ, Sakamoto KM. Targeting steroid hormone receptors for ubiquitination and degradation in breast and prostate cancer. Oncogene. 2008;27:7201–7211. doi: 10.1038/onc.2008.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudiero DA, Shoemaker RH, Paull KD, Monks A, Tierney S, Nofziger TH, Currens MJ, Seniff D, Boyd MR. Evaluation of a Soluble Tetrazolium/Formazan Assay for Cell Growth and Drug Sensitivity in Culture Using Human and Other Tumor Cell Lines. Cancer Res. 1988;48:4827–4833. [PubMed] [Google Scholar]
- Walensky LD, Bird GH. Hydrocarbon-Stapled Peptides: Principles, Practice, and Progress. J. Med. Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Lao Y, Zhang Y, Gillespie DA. Akt: A Double-Edged Sword in Cell Proliferation and Genome Stability. J. Oncol. 2012;2012:e951724. doi: 10.1155/2012/951724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D. Crystal structure of an activated Akt/Protein Kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat. Struct. Mol. Biol. 2002;9:940–944. doi: 10.1038/nsb870. [DOI] [PubMed] [Google Scholar]
- Zhu J, Blenis J, Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc. Natl. Acad. Sci. 2008;105:6584–6589. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.