Abstract
T follicular helper (Tfh) cells are a CD4 T cell subset that is important for supporting plasma cell and germinal center (GC) responses1,2. The initial induction of Tfh cell properties occurs within the first few days following activation by antigen recognition on dendritic cells (DCs), though how DCs promote this cell-fate decision is not fully understood1,2. Moreover, although Tfh cells are uniquely defined by expression of the follicle-homing receptor CXCR51,2, the guidance receptor promoting the earlier localization of activated T cells at the B cell follicle–T zone interface has been unclear3–5. Here we show that the G-protein coupled receptor EBI2 (GPR183) and its ligand 7α,25-dihydroxycholesterol (7α,25-OHC) mediate positioning of activated CD4 T cells at the follicle–T zone interface. In this location they interact with activated DCs and are exposed to Tfh cell-promoting ICOS ligand. IL2 is a cytokine that has multiple influences on T cell fate, including negative regulation of Tfh cell differentiation6–10. We demonstrate that activated DCs in the outer T zone further augment Tfh cell differentiation by producing membrane and soluble forms of CD25, the IL2 receptor α chain, and quenching T cell-derived IL2. Mice lacking EBI2 in T cells or CD25 in DCs have reduced Tfh cells and mount defective T cell-dependent plasma cell and GC responses. These findings demonstrate that distinct niches within the lymphoid organ T zone support distinct cell fate decisions, and they establish a function for DC-derived CD25 in controlling IL2 availability and T cell differentiation.
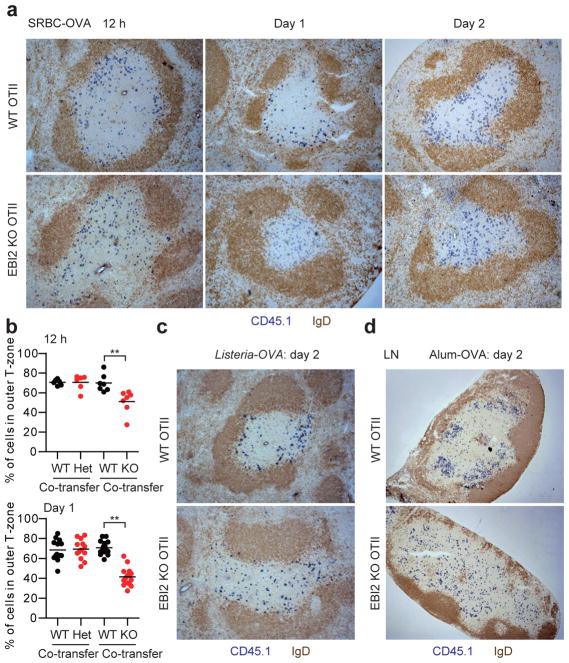
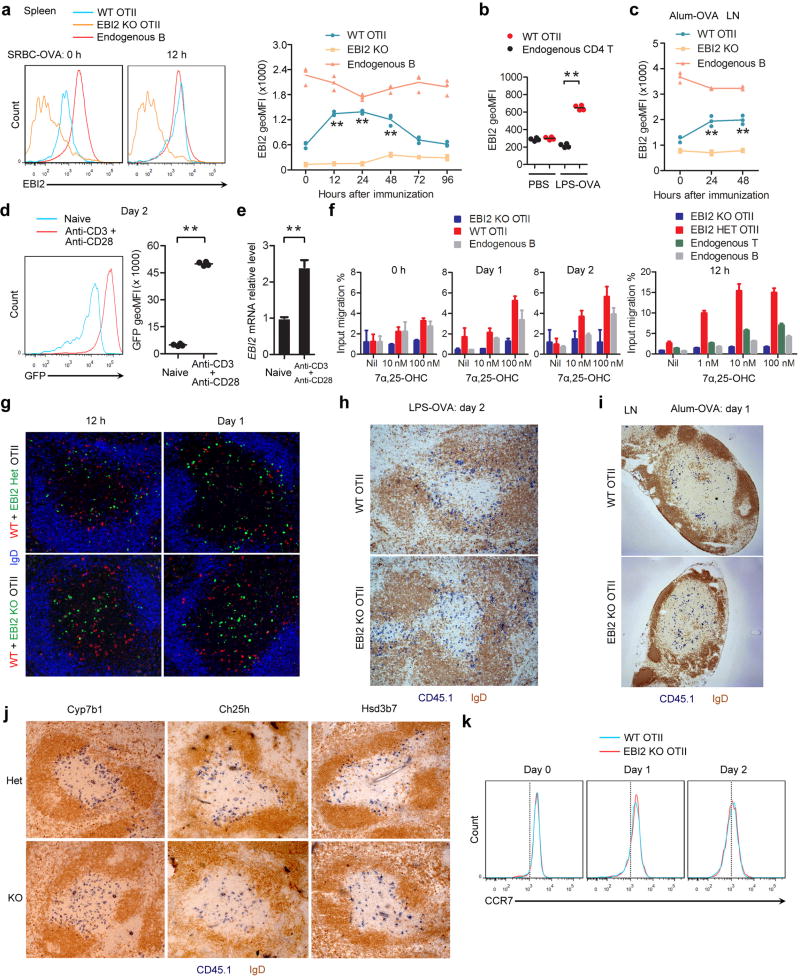
EBI2 is expressed by CD4 T cells11–14, but whether it has a role in positioning T cells during the early stages of activation has been unclear. Using an ovalbumin (OVA) specific TCR transgenic (OTII) system involving transfer of OTII T cells to wild-type (WT) hosts, we found that EBI2 was upregulated on cognate splenic T cells within 12 hours of immunization with a particulate form of OVA (sheep red blood cell (SRBC) conjugated), and it remained high at day 2 (Extended Data Fig. 1a). Similar EBI2 induction occurred following immunization with OVA in LPS, on lymph node (LN) T cells after immunization with OVA in alum, and in vitro following T cell activation by anti-CD3 and -CD28 (Extended Data Fig. 1b–e). Migration to 7α,25-OHC was augmented at these time points (Extended Data Fig. 1f). Analysis of spleen sections showed that transferred WT T cells accumulated in the outer T zone at 12 hours and day 1 of the SRBC-OVA response and the cells remained enriched in this location at day 2 (Fig. 1a). EBI2 knockout (KO) T cells, by contrast, failed to accumulate in the outer T zone at either time point and instead remained dispersed throughout the T zone (Fig. 1a). Quantitative analysis using a mixed transfer system confirmed that the activated EBI2 KO cells had less access than control cells to the outer T zone (Fig. 1b and Extended Data Fig. 1g). Similar findings were made at day 2 after immunization with OVA-expressing Listeria monocytogenes (Fig. 1c) and with OVA in LPS (Extended Data Fig. 1h). WT OTII T cells also moved to the B–T zone interface in LNs following immunization with alum-OVA, but EBI2-deficient T cells failed to relocalize (Fig. 1d and Extended Data Fig. 1i). Activated T cell positioning in the outer T zone was directed by 7α,25-OHC as it was dependent on the enzymes needed for its synthesis (Cyp7b1 and Ch25h) and catabolism (Hsd3b7) (Extended Data Fig. 1j).
Figure 1. EBI2 promotes positioning of newly activated CD4 T cells in the outer T zone.
(a) Immunohistochemical analysis of spleens for transferred WT or EBI2 KO OTII CD45.1+ T cells (blue) and endogenous B cells (IgD, brown) at 12 h, 1 day and 2 days after SRBC-OVA immunization. (b) Fraction of WT and EBI2 het or KO OTII T cells in the outer ¼ of the splenic T zone at 12 h and 1 day after SRBC-OVA. Sections were stained as in Extended Data Fig. 1g. See Methods for details. (c, d) As for a except mice were immunized with Listeria-OVA (c) or alum-OVA and inguinal LNs were analyzed (d). **, p<0.01 by student’s t-test. Data are representative of three (a, b) or two (c–e) experiments with at least three (a) or two (b–e) mice per group.
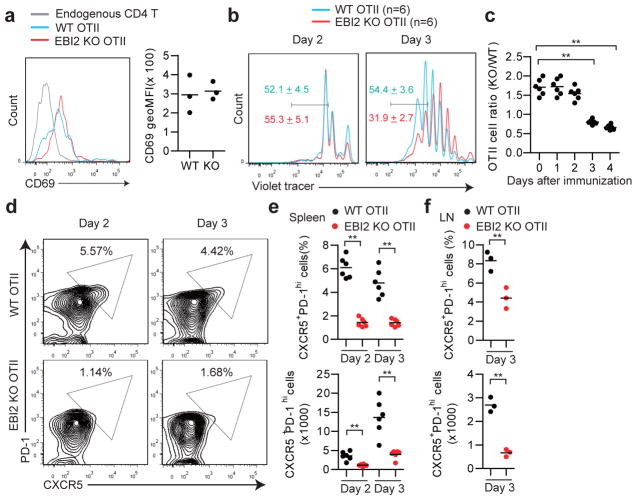
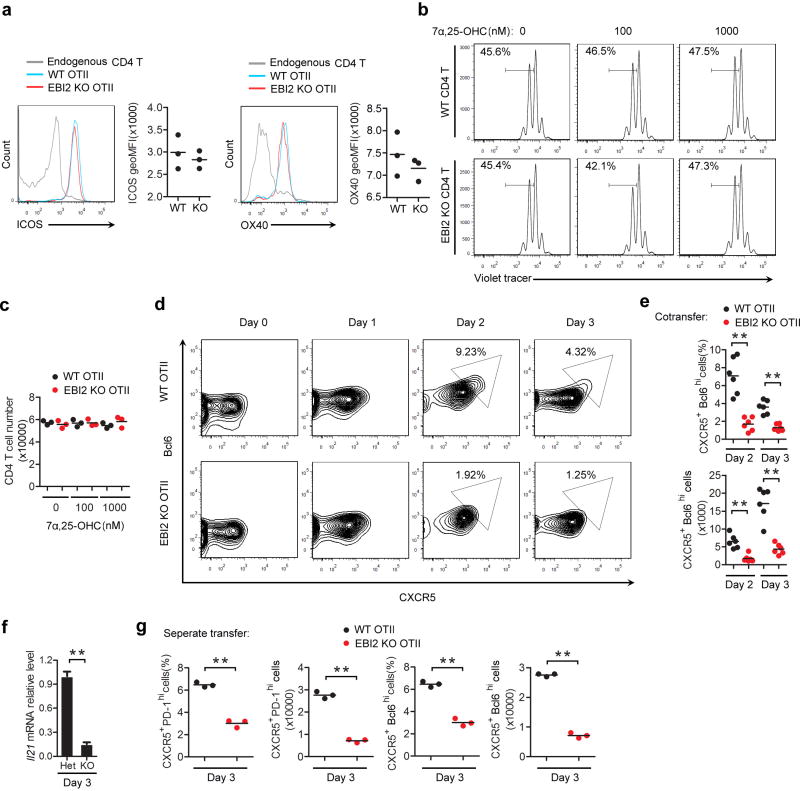
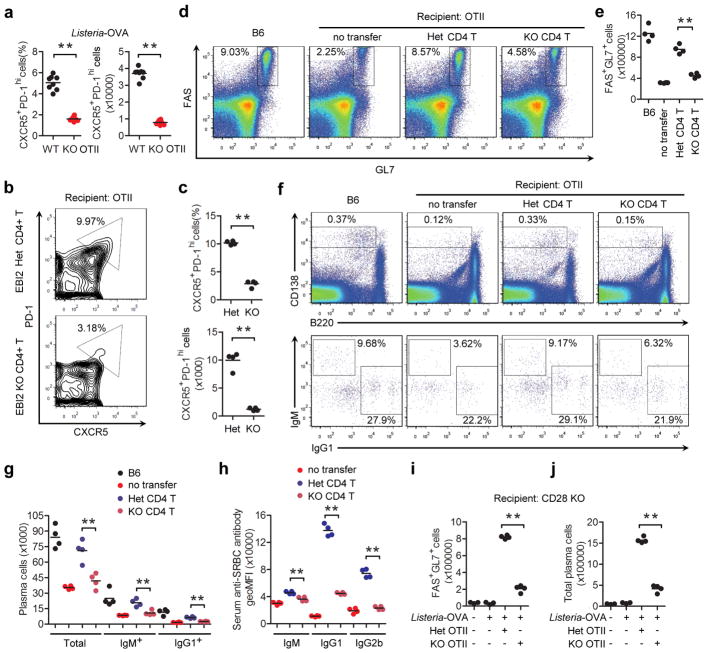
Flow cytometric analysis for the early activation marker, CD69, showed that co-transferred EBI2 KO and WT T cells were comparably activated at day 2 of the SRBC-OVA response (Fig. 2a) indicating similar initial exposure to cognate MHC class II-peptide complexes. Upregulation of the costimulatory molecules ICOS and OX40 also occurred to an equivalent extent (Extended Data Fig. 2a). Proliferation began by day 2 and at this time point the WT and EBI2 KO cells responded similarly (Fig. 2b, c). However, by day 3, the EBI2-deficient cells were undergoing less proliferation and their numbers increased more slowly (Fig. 2b, c). This was not due to a direct effect of 7α,25-OHC on T cell proliferation (Extended Data Fig. 2b, c). Tracking of differentiation markers on the in vivo activated T cells revealed that EBI2 KO cells were compromised in their induction of a Tfh cell phenotype, as assessed by CXCR5, PD-1 (Fig. 2d, e), Bcl6, and Il21 expression (Extended Data Fig. 2d–f). EBI2-deficient OTII T cells also differentiated less efficiently into Tfh cells in LNs (Fig. 2f). We also observed reduced Tfh cell responses to Listeria-OVA, reduced polyclonal EBI2 KO Tfh cell responses to SRBCs, and reduced GC and plasma cell responses to these antigens (Supplemental Information and Extended Data Fig. 3a–j).
Figure 2. Defective differentiation of EBI2-deficient T cells to follicular helpers.
(a) CD69 expression on WT OTII, EBI2 KO OTII and endogenous CD4 T cells in transfer recipient spleens 2 days after SRBC-OVA immunization. Histograms show representative FACS and graphs show summary geoMFI data for 3 mice of each type. (b) Proliferation of co-transferred WT and EBI2 KO OTII T cells monitored by violet tracer dye dilution at days 2 and 3 after immunization. Numbers indicate mean % (±SD) of cells in the indicated gate (n=6). (c) Summary of data from b shown as a ratio of KO/WT OTII T cell number at the indicated days. (d) Flow cytometric analysis of co-transferred WT and EBI2 KO OTII T cells for PD-1 and CXCR5 at days 2 and 3 following immunization. Numbers indicate frequency of cells in gated region. (e) Summary of data of the type in d. Upper plot shows frequency and lower plot number of CXCR5+PD-1hi CD4+ OTII T cells. (f) Frequency and number of CXCR5+PD-1hi CD4+ OTII T cells in peripheral LNs of mice of the type in d, immunized with alum-OVA. *, p<0.05 and **, p<0.01 by ANOVA (d, f) or student’s t-test (g). Data are representative of three (a–f) or two (g) experiments with at least three mice per group.
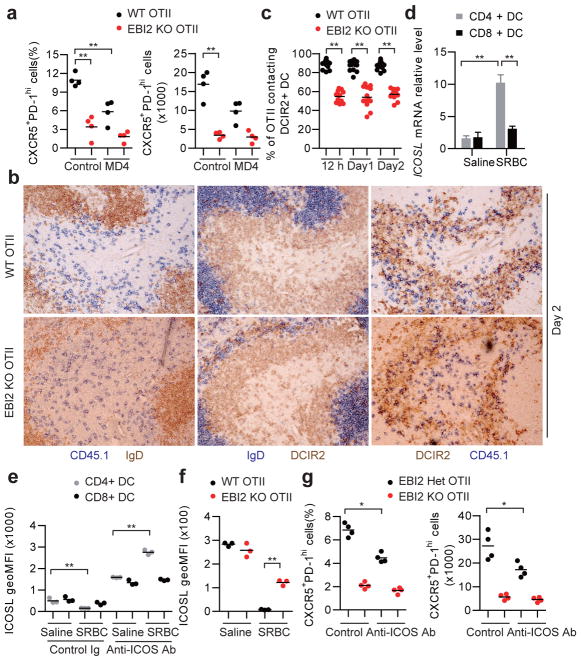
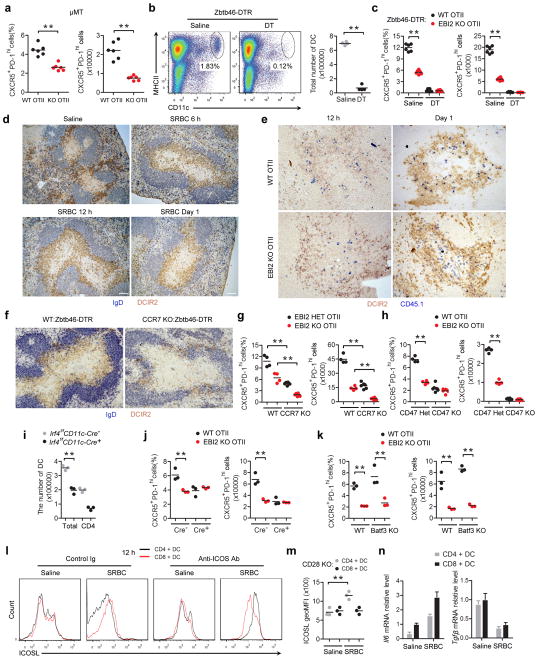
Tfh cell differentiation is promoted by interaction with both DCs and B cells, and time course studies indicate that DCs are critical early while B cells play a later role1,2. Consistent with these requirements, WT OTII T cells showed a partial reduction in Tfh cell differentiation at day 3 of the response in MD4 Ig-transgenic mice that lack cognate B cells capable of OVA antigen presentation to OTII T cells (Fig. 3a). Importantly, however, EBI2 KO OTII T cells formed less Tfh cells than WT OTII T cells in the MD4 recipients, indicating that T cell EBI2 expression augmented Tfh cell development at early time points in a B cell independent manner. Similar observations were made in B cell-deficient μMT mice (Extended Data Fig. 4a). These findings led us to test whether EBI2 was required in T cells for some type of interaction with DCs. Ablation of DCs using Zbtb46-DTR mice15 caused a complete block in Tfh cell generation (Extended Data Fig. 4b, c), consistent with previous studies using other DC ablation approaches16. Splenic CD4 and DCIR2 co-expressing DCs relocalize from bridging channels to the outer T zone within 6 hours of immunization with SRBCs17,18, and the cells remain in this region for at least 2 days (Fig. 3b and Extended Data Fig. 4d). At 12 hour, day 1 and day 2 time points, almost the entire population of activated WT OTII T cells colocalized with the activated DCIR2+ DCs (Fig. 3b, c and Extended Data Fig. 4e). By contrast, activated EBI2 KO T cells were broadly distributed and only partially overlapped with the DCIR2+ DCs (Fig. 3b, c and Extended Data Fig. 4e). Using mice with deficiencies in DCs we established that CD4+ but not CD8+ DCs were important for Tfh cell induction by SRBC-OVA (Supplementary Information and Extended Data Fig. 4f–k).
Figure 3. T cell EBI2 is required for CD4+ DC-mediated augmentation of Tfh cell induction.
(a) Frequency and number of CXCR5+PD-1hi WT and EBI2 KO OTII T cells in control or MD4 recipient spleens at day 3 after immunization with SRBC-OVA. (b) Immunohistochemical analysis of consecutive sections from WT transfer recipient spleens at day 2 after immunization, stained for : left panels, WT or EBI2 KO OTII CD45.1+ T cells (blue) and B cells (IgD, brown); center panels, DCIR2+ DCs (brown) and B cells (IgD, blue); right panels, OTII CD45.1+ T cells (blue) and DCIR2+ DCs (brown) (c) Frequency of WT or EBI2 KO OTII T cells contacting DCIR2+ DCs determined in sections of the type in b and Extended Data Fig. 4e, at the indicated times after immunization. (d) Icosl mRNA abundance in splenic CD4+ and CD8+ CD11c+ DCs from mice immunized 12 h earlier with saline or SRBCs, shown relative to the saline control. (e) Summary data of ICOSL surface levels for DCs of the type in d from mice also treated with control or ICOS blocking antibody. (f) ICOSL surface levels on CD4+ splenic DCs from mice that had received WT or EBI2 KO OTII T cells, 12 h after immunization with SRBC-OVA. Recipient mice were CD28 KO. See Supplementary Information for details. (g) Frequency and number CXCR5+PD-1hi WT and EBI2 KO OTII T cells in spleens from mice treated with ICOS blocking antibody, analyzed at day 3 after immunization. *, p<0.05 and **, p<0.01 by ANOVA (a–d, e) or student’s t-test (f, g). Data are representative of three (a–c) or two (d–g) experiments with at least three (a, c–g) or two (b) mice per group (error bars (d), s.e.m.).
ICOS signaling is important for Tfh cell differentiation1,2 and splenic DCs upregulated ICOSL mRNA following activation by SRBCs, with the extent of upregulation being greater in CD4+ than CD8+ DCs (Fig. 3d). However, flow cytometric analysis showed 12 hour activated CD4+ DCs had low surface ICOSL staining (Fig. 3e and Extended Data Fig. 4l). Since ICOSL undergoes rapid ectodomain shedding following ICOS engagement19,20, we considered the possibility that surface levels were reduced due to interactions with ICOS-high activated T cells. Consistent with this idea, when SRBC-immunized mice were also treated with an ICOS blocking antibody to prevent ICOS-induced ICOSL shedding19,20, CD4+ but not CD8+ DCs showed increased ICOSL surface abundance compared to unimmunized mice (Fig. 3e and Extended Data Fig. 4l). We took advantage of the sensitivity of ICOSL to ICOS-induced shedding as a method to measure the amount of interaction between DCs and cognate T cells. Twelve hours following SRBC-OVA immunization, ICOSL levels were higher on CD4+ DCs in mice harboring EBI2-deficient OTII T cells than WT OTII T cells (Fig. 3f). These data suggest that the reduced Tfh differentiation of EBI2 KO OTII T cells occurs at least in part due to lower ICOS engagement with ICOSL on CD4+ DCs. However, in mice treated with an ICOS blocking antibody, though Tfh differentiation of control OTII T cells was reduced, EBI2-deficient OTII T cells were still more defective (Fig. 3g), indicating an ICOS independent influence of EBI2 in augmenting Tfh cell fate. An assessment of mRNA levels of other factors established to have an effect on Tfh differentiation (IL6, TGFβ) showed that they were similarly expressed in CD4+ and CD8+ DCs and they were therefore not considered likely factors accounting for the EBI2-dependence of Tfh cell differentiation (Extended Data Fig. 4n).
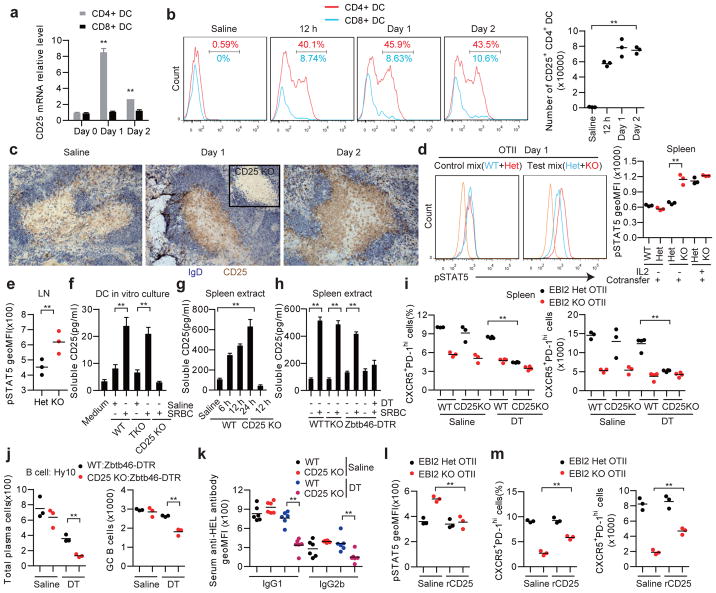
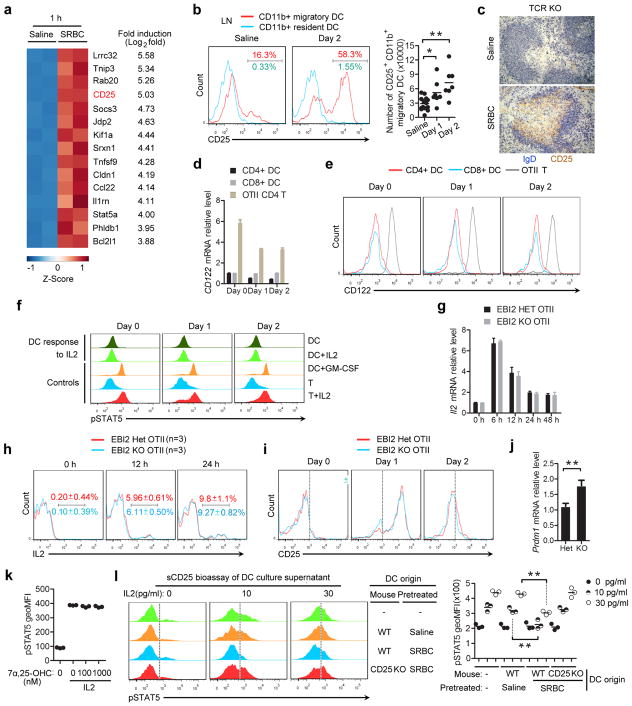
To search for surface or secreted DC-derived factors that might augment Tfh cell differentiation we performed RNAseq analysis on CD4+ DCs from the spleens of saline or SRBC immunized mice. This analysis revealed CD25, the high affinity IL2 receptor α-chain, as one of the most strongly induced genes in SRBC-activated DCs (Extended Data Fig. 5a). CD25 mRNA induction occurred rapidly after immunization and remained elevated for at least 2 days (Fig. 4a) and many CD4+ DCs were surface positive for CD25 over this time frame (Fig. 4b). CD8+ DCs showed little induction of CD25 mRNA or protein under these immunization conditions (Fig. 4a, b). Analysis of LN DCs following OVA plus alum immunization revealed upregulation of CD25 on migratory CD11b+ DCs at day 1 and 2 (Extended Data Fig. 5b). Staining of spleen sections identified CD25+ cells in the unstimulated T zone that are likely CD25hi regulatory T cells, but also showed broad induction of CD25 in the outer T zone within 12 hours of SRBC immunization in a pattern resembling the DCIR2+ DC distribution (Fig. 4c). Similar appearance of CD25 staining in the T zone was seen in T cell-deficient mice, providing evidence that expression by activated DCs was being detected (Extended Data Fig. 5c). CD25 needs to associate with CD122 (IL2Rβ) and IL2Rγ to transmit signals in response to IL210. Despite expression of CD25, activated CD4+ DCs showed minimal CD122 mRNA and protein expression and they did not respond to IL2 as assessed by intracellular pSTAT5 staining (Extended Data Fig. 5d–f). These data led us to consider the possibility that activated DCs express CD25 to alter IL2 availability in the outer T zone.
Figure 4. DC CD25 expression reduces IL2 signaling in activated CD4 T cells, favoring their differentiation to follicular helpers.
(a, b) CD25 transcript (a) and surface (b) levels on CD4+ and CD8+ splenic DCs from mice immunized with saline or with SRBCs 12 h, 1 or 2 days earlier. Transcript levels are plotted relative to the day 0 mean for each DC type. (c) Immunohistochemical analysis of spleens from WT mice immunized with saline or SRBCs, stained to detect IgD (blue) and CD25 (brown). Inset shows 12 h SRBC immunized CD25 KO. (d) Flow cytometry of pSTAT5 in WT, EBI2 Het or EBI2 KO OTII T cells in mice that received mixtures of CD45.2 and CD45.1/2 marked cells, stained ex vivo 1 day after SRBC-OVA immunization. Left panels show example histogram plots of gated OTII T cells and right graph shows summary geoMFI data for 3 mice of each type in one experiment, including mice injected with IL2 as a positive control. Orange histogram indicates endogenous CD4+ T cells. (e) Summary of pSTAT5 levels in control (Het) and EBI2 KO OTII T cells in LNs at day 3 after alum-OVA immunization. (f) Soluble CD25 detected by ELISA in culture supernatants of splenic CD4+ DCs from WT, TCR KO or CD25 KO mice immunized 1 day earlier with saline or SRBCs, or medium alone. (g, h) Soluble CD25 detected by ELISA in spleen extracts taken from WT, CD25 KO, TCR KO, or Zbtb46-DTR treated with DT, immunized as indicated (g) or 12 h (h) prior to analysis. (i) Frequency and number of CXCR5+PD-1hi control (EBI2 Het) and EBI2 KO OTII T cells in spleens from WT:Zbtb46-DTR or CD25 KO:Zbtb46-DTR mixed BM chimeras pretreated with saline or DT, at day 3 after SRBC-OVA. (j) HEL-binding plasma cell and GC B cell numbers in spleens from WT:Zbtb46-DTR (control) or CD25 KO:Zbtb46-DTR chimeras treated with saline or DT and transferred with Hy10 B cells, at day 5 after immunization with HEL-SRBC. (k) Serum IgG1 and IgG2b anti-HEL antibody in mice of the type in j analyzed by FACS of HEL-conjugated mouse RBCs. (l, m) Summary geoMFI of pSTAT5 levels (l) and CXCR5+PD-1hi cell frequencies and numbers of control (Het) and EBI2 KO OTII T cells in WT mice at day 1 (l) and day 3 (m) after SRBC-OVA immunization and treatment with saline or recombinant CD25 (rCD25). TKO, TCRβδ KO. *, p<0.05 and **, p<0.01 by ANOVA (a, b, f–h, k) or student’s t-test (d, e, i, j, l). Data are representative of three (a, b, d) or two (c, e–m) experiments with at least three (a, b, d–f, h–m) or two (c, g) mice per group (error bars (a, f–h), s.e.m.).
IL2 has pleiotropic roles in directing T cell fate and this includes a negative influence on Tfh cell differentiation6–10. Despite equivalent IL2 production and receptor expression (Supplementary Information and Extended Data Fig. 5g–i), EBI2 KO T cells showed more IL2R signaling than WT T cells at day 1 after immunization as evidenced by higher pSTAT5 levels (Fig. 4d), suggesting that the EBI2 KO T cells were being exposed to more IL2. The elevated induction of pSTAT5 in EBI2 KO T cells was also seen in LNs following immunization with alum-OVA (Fig. 4e). Blimp1, encoded by Prdm1, is induced by IL2 and negatively regulates expression of Bcl6, a factor essential for Tfh cell development2. In agreement with higher IL2 exposure, the EBI2 KO T cells showed greater Prdm1 expression at day 3 (Extended Data Fig. 5j).
The above findings led us to test whether DCs antagonize IL2 availability to activated T cells in the outer T zone. Consistent with this possibility, when in vivo activated DCs were incubated briefly in vitro they were found to release soluble CD25 (sCD25) into the culture supernatant (Fig. 4f). DC production of sCD25 was not dependent on interaction with T cells (Fig. 4f). Analysis of spleen tissue extracts showed elevated sCD25 production at 6 hours after SRBC immunization and this was maintained at 24 hours (Fig. 4g) and occurred in a T cell-independent and DC-dependent manner (Fig. 4h). To determine whether the sCD25 functioned as an IL2 antagonist, supernatants from cultures of activated CD4+ DCs were tested in a bioassay for their ability to inhibit IL2R signaling. Regulatory T cells were used as the reporter cells in this bioassay since they were more sensitive to low dose IL2 than Tfh cells (unpubl. obs.). Supernatants from cultured SRBC-activated WT but not CD25 KO CD4+ DCs were able to antagonize IL2R signaling in T cells (Extended Data Fig. 5l).
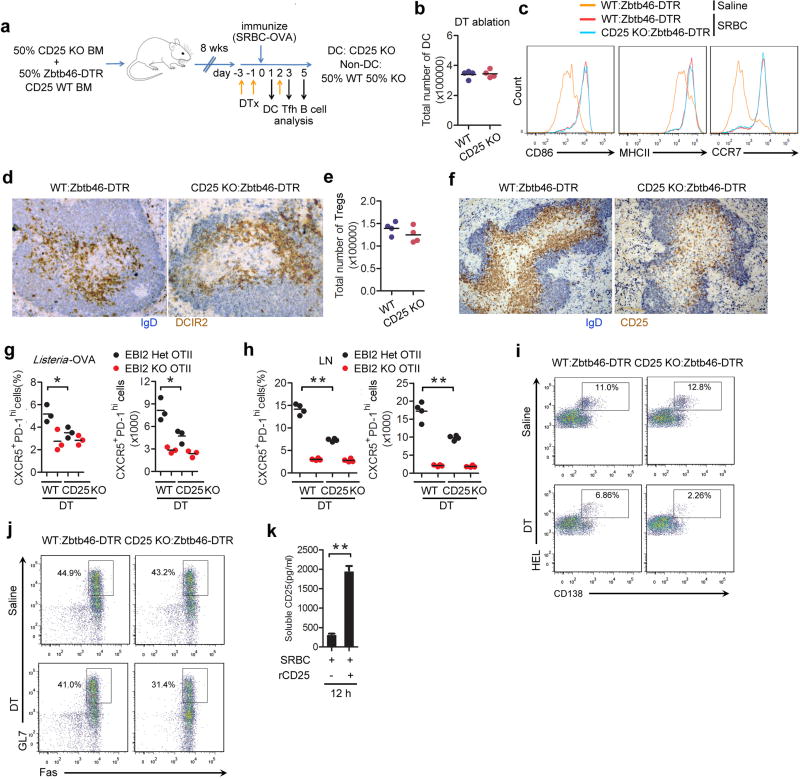
To determine whether DCs were regulating T cell differentiation in vivo by production of CD25 we generated BM chimeric mice that lacked CD25 in DCs (Supplementary Information and Extended Data Figs. 6a–e). In these recipients, control (EBI2 Het) OTII T cells were compromised in their ability to take on a Tfh cell fate whereas EBI2 KO T cells were only mildly affected (Fig. 4i). Staining of tissue sections from the DT-treated BM chimeras established that the high CD25 abundance in the outer T zone was dependent on CD25 expression by DCs (Extended Data Fig. 6f). Splenic Tfh cell responses to Listeria-OVA and LN Tfh cell responses to alum-OVA were also diminished in mice lacking CD25 on DCs (Extended Data Fig. 6g, h). Moreover, mice lacking CD25 in DCs and harboring transgenic B cells specific for hen egg lysozyme (HEL) mounted reduced plasma cell and GC responses to HEL-conjugated SRBCs (Fig. 4j and Extended Data Fig. 6i, j) and this was associated with reduced serum anti-HEL IgG1 and IgG2b antibody levels (Fig. 4k). To further test whether reduced exposure of EBI2 KO T cells to sCD25 could account for the defective Tfh cell induction, we treated mice with recombinant sCD25 at day 0 and 1 of the OTII T cell response to SRBC-OVA. This treatment was sufficient to elevate sCD25 levels in tissue extracts (Extended Data Fig. 6k), to antagonize pSTAT5 over-induction in the EBI2 KO T cells (Fig. 4l) and to partially rescue Tfh cell differentiation (Fig. 4m). A full restoration of the Tfh response was not expected given that EBI2-deficient cells are also compromised in accessing ICOSL and possibly other Tfh cell-promoting signals from cells in the outer T zone.
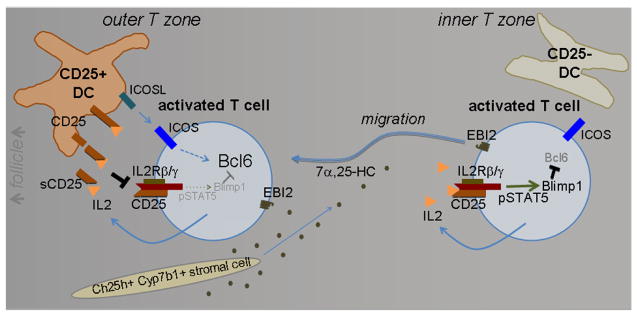
This work establishes a role for EBI2 and 7α,25-OHC in positioning of activated T cells at the follicle-T zone interface, promoting contact with Tfh cell-priming ICOSLhi CD25+ DCs (Supplementary Information and Extended Data Fig. 7). Given that interactions with both DCs and B cells are important for full Tfh cell differentiation1,2, we suggest that T cell EBI2 upregulation initially acts to favor interaction with Tfh cell-promoting DCs and subsequently with activated B cells. While EBI2 upregulation is important for Tfh cell induction, the receptor is downregulated at week 2 of the response and this may facilitate Tfh cell retention in GCs14. Soluble CD25 was first detected in human serum ~30 years ago and it has since been reported in human and mouse serum in a large number of studies and it has been correlated with various disease conditions21–23. While there has been evidence that sCD25 can antagonize certain IL2 functions10,22,24 the significance of sCD25 in vivo has been unclear. Moreover, the function of CD25 in myeloid cells has been mysterious10,25,26, with some in vitro studies suggesting it suppresses27 and others that it augments28,29 T cell responses. We show that DC production of CD25 plays an important role in quenching IL2 in the outer T zone and it thereby cooperates with other factors, including ICOSL, to facilitate Tfh cell differentiation (Extended Data Fig. 7). While our findings show DCs produce sCD25, we do not exclude the possibility that membrane associated CD25 on DCs also has a regulatory role. Strengths of IL2 signaling influence Treg cell activity, Th17, Th1 and Th2 cell development, and CD8 T cell proliferation and differentiation10,30. We suggest that CD25-mediated IL2-quenching by DCs will be a general mechanism acting to guide a range of IL2-sensitive cell activation and differentiation processes.
Methods
Mice and bone marrow chimeras
Wild-type C57BL/6NCr and C57BL/6-cBrd/cBrd/Cr (B6-Ly5.2) mice of 7–9 weeks of age were purchased from the National Cancer Institute (Frederick, MD, USA). Ebi2−/− (containing a GFP reporter in place of the Ebi2 coding exon), Cyp7b1−/−, Ch25h−/−, Hsd3b7−/−, Ccr7−/−, HEL-specific MD4 Ig-transgenic, HEL-specific Hy10 mice, and OVA-specific OTII TCR-transgenic mice have been described (11,17,31 and references therein). B cell-deficient μMT mice were kindly provided by Tony Defranco and Christopher Allen (University of California, San Francisco, CA, USA). Cd28−/− mice were kindly provided by K. Mark Ansel (University of California, San Francisco). TCRβδ−/−, Cd47−/−, Batf3−/− and Zbtb46-diphtheria toxin receptor (DTR) mice were from Jackson laboratories (Jax, Bar Harbor, ME, USA). Irf4fl/fl CD11c-Cre+ mice were from Jax and kindly provided by Shomyseh Sanjabi (Gladstone Institutes, San Francisco). Cd25−/− mice were from Jax and were kindly provided by Markus Muschen (University of California, San Francisco). Bone marrow (BM) chimeras were generated as described11 and analyzed after 6–12 weeks. Mixed BM chimeras were made by mixing equal amounts of the two types of BM prior to transfer. The sample sizes were guided by previous studies in our laboratory. No animals were excluded from analysis, and sample size estimates were not used. The mouse genotype was not blinded from the investigator. Mice of a given genotype were randomly assigned to groups. However, littermate mice were evenly distributed into control or treatment groups and mice of both groups were co-caged whenever possible. In experiments involving transfers of OTII T cells, since the TCR transgene is on the Y chromosome, male mice were used as donors and recipients. In other experiments, similar numbers of male and female mice were used. All mice were adult and were studied between 7 and 20 weeks of age. Animals were housed in a specific-pathogen free environment in the Laboratory Animal Research Center at the University of California, San Francisco, and all experiments conformed to ethical principles and guidelines approved by the Institutional Animal Care and Use Committee.
Adoptive transfer, immunizations, DC ablation and treatments
For analysis of CD4+ T cell position, activation or Tfh cell differentiation in spleens, 1–5 × 106 WT and/or EBI2 KO OTII cells were adoptively transferred into mice. One day after cell transfer, recipients were immunized intraperitoneally (i.p.) with 2×108 SRBCs (Colorado Serum Company, Denver, CO) conjugated with OVA (Sigma-Aldrich, St. Louis, MO, USA) as described32 with minor modifications detailed below, with 25 μg OVA plus 25 μg LPS (E. coli 0111:B4, Sigma-Aldrich), intravenously (i.v.) with 2×109 heat-killed Listeria-OVA as described33, or subcutaneously with 25 μg OVA in 200 μl Alum (InvivoGen, San Diego, CA, USA). For conjugation of OVA with SRBCs, 1 ml of SRBCs were washed with PBS three times, incubated with 4 ml of 30 mg/ml ice cold OVA in PBS and crosslinked with 1 ml of 100 mg/ml EDCI (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide, Sigma-Aldrich) for 1 hour on ice with occasional mixing, followed by washing four times in PBS to remove the free OVA and confirmation of the conjugation by flow cytometry. For HEL-specific antibody responses, 1×105 Hy10 B cells34 were adoptively transferred into desired recipients. One day after cell transfer, recipients were i.p. immunized with SRBCs conjugated with a low affinity mutant of HEL termed HEL2× (ref 35) as described17. To visualize cell proliferation, cells were labeled with CellTrace™ violet tracer (Molecular Probes, invitrogen, Carlsbad, CA, USA) according to manufacturer instructions. For DC ablation, Zbtb46-DTR full or mixed chimeras were injected i.p. with 20 ng DT (Sigma-Aldrich) per gram of body weight 3 days before cell transfer and received 4 ng DT per gram of body weight on the third day after the initial DT injection and in some cases again 3 days later. To block ICOS, mice were injected i.v. with anti-mouse ICOS antibody (rat IgG2b, clone 7E.17G9, BioXCell, West Lebanon, NH, USA) or rat IgG2b isotype control one day before and 2 days after cell transfer (0.5 mg/mouse/injection). To block IL2, two doses of recombinant CD25 protein (25 μg/mouse/dose, Sino Biological Inc., Beijing, China) were injected into mice at the same time with immunization or one day after immunization.
Flow cytometry and cell sorting
All antibody conjugates were from Biolegend (San Diego, CA, USA) or BD Biosciences (San Jose, CA, USA). EBI2 surface staining, T cell staining, GC B cell staining and intracellular Ig staining for plasma cells were performed as described17,31. EBI2 surface staining was performed with a goat polyclonal antibody against the N-terminus (clone A20, Santa Cruz Biotechnology, Dallas, TX, USA) as described17. Tfh cell staining was performed with antibodies including biotin-conjugated anti-CXCR5 (BD Biosciences), PE-Cy7-conjugated anti-PD-1 (clone RMP1-30, Biolegend) and Alexa 647-conjugated anti-Bcl6 (clone k112-91, BD Biosciences) as described36. Staining of CD25 on splenic or LN DC was performed using Alexa 647-conjugated anti-CD25 (clone PC61, Biolegend), PE-Cy7-conjugated anti-CD11c (clone N418, Tonbo Biosciences), FITC-conjugated anti-I-Ab (clone AF6-120.1, BD Biosciences), PE-conjugated anti-DCIR2 (clone 33D1, eBioscience) or PE-conjugated anti-CD11b (clone M1/70, Biolegend), Pacific Blue-conjugated anti-CD8a (clone 53-6.7, Biolegend), and biotin-conjugated anti-CD103 (clone 2E7, Biolegend). To assess pSTAT5 levels directly ex vivo, spleens were immediately mashed using cell strainers into Cytofix/Cytoperm buffer (BD Biosciences). For peripheral LN analyses, the brachial, axillary, and inguinal LN were pooled. After fixation for 30 minutes at 37°C, the cells were washed, resuspended in Perm Buffer III (BD Biosciences) and incubated on ice for 30 minutes. After an additional wash, cells were stained for surface and intracellular antigens, including pSTAT5 (pY694, BD Biosciences), for 45 minutes at room temperature. Where indicated, 4 μg of human IL2 was injected intravenously into mice 2 hours prior to analysis as a positive control for pSTAT5. Data were collected on an LSRII and a FACSVerse (BD Biosciences) and were analyzed with FlowJo software (TreeStar). DCs, OTII CD4+ T cells or Tfh cells were sorted using a FACSAria III (BD) as described31.
Transwell migration assay
Splenocytes were allowed to transmigrate for 4 hours across 5 μm transwell filters (Corning Costar, Corning, NY, USA) towards medium or 7α,25-OHC (Avanti Polar Lipids, Alabaster, AL, USA) and enumerated by flow cytometry as described17.
Immunohistochemistry and immunofluorescence microscopy
Cryosections of 7 μm were fixed and stained immunohistochemically as described17,31 with: FITC-conjugated anti-IgD (clone 11-26c.2a, BD Biosciences), biotin-conjugated anti-CD45.1 (clone A20, Biolegend, San Diego, CA, USA), biotin-conjugated anti-DCIR2 (clone 33D1, Biolegend) or biotin-conjugated anti-CD25 (clone PC61.5, BD Biosciences) followed by HRP-conjugated anti-FITC, AP-conjugated anti-FITC, and/or AP-conjugated SA (Jackson Immunoresearch, Newmarket, Suffolk, UK). For staining of DCIR2 and CD25, a tyramide amplification kit was used (TSA Biotin System; Perkin Elmer; Waltham, MA, USA). For immunofluorescence, staining was performed with biotin conjugated anti-CD45.1, rabbit anti-GFP (Molecular Probes, Eugene, OR, USA), and goat anti-mouse IgD (GAM/IGD(FC)/7S, Cedarlane Laboratories), followed by AMCA-conjugated donkey anti-goat IgG (Jackson Immunoresearch), Alexa 488-conjugated donkey anti-rabbit IgG and Alexa 647-conjugated streptavidin (Invitrogen). Images were captured with a Zeiss AxioOberver Z1 inverted microscope.
Image Quantification
Immunofluorescence images were analyzed using IMARIS (ver 7.3.0). White-pulp cords containing circular T zones were used to quantify outer T-zone positioning of co-transferred WT (red) and EBI2 HET or KO cells (green). OTII cells were defined using the Spots function in IMARIS and coordinates for each OTII cell were exported into R. The center and average radius of the T zone was measured using the Measurement Points function in IMARIS. The “outer T-zone” was defined as the area further than 3/4ths of the average radius from the center of the T zone. The distance of each OTII cell from the center of the T zone and the proportion of cells in the outer T-zone was calculated using R. An average of 70 cells were present for each co-transferred group per T-zone (average of 140 total). IHC images were analyzed manually using the Cell Counter Plugin in ImageJ (ver 1.49). OTII cells that were in contact with DCIR2+ DC were distinguished from lone OTII cells using separate counters.
Soluble CD25 ELISA and bioassay
To test the production of sCD25 by DCs in vitro, mice were first i.p. immunized with SRBCs. At the time of analysis, spleen CD4+ DCs were enriched by depletion of T, B and NK cells with a cocktail of biotin-conjugated antibodies and isolated by positive selection using biotin-conjugated anti-DCIR2 to purities of over 90% (Miltenyi Biotec). The purified DCs were cultured in vitro for 8 hours and the presence of soluble CD25 in culture supernatants was detected using a CD25 ELISA kit (R&D Systems Inc, Minneapolis, MN, USA). To detect sCD25 in spleen tissue, each spleen was mashed into 1 ml of medium through a 70μm cell strainer and centrifuged at 300 g for 10 min and 3,000 g for 15 min at 4°C. The cell-free supernatant was subjected to the ELISA assay. To test for antagonism of IL2 mediated signaling, DC culture supernatants were mixed with different dosages of recombinant mouse IL2 (Biolegend) for 2 hours and added to splenocytes at 37°C for 30 minutes. pSTAT5 levels in CD25+CD4+ T cells were analyzed as described above.
Detection of SRBC or HEL-specific antibody responses
To assay for anti-SRBC or anti-HEL IgM and IgG from mouse serum, 50 μl of SRBCs or HEL-conjugated mouse RBCs (5 × 107 cells/ml in PBS) was incubated with 2 μl of serum for 1 hour at room temperature. After washing, the RBCs were incubated with fluorescent antibodies against mouse IgM, IgG1 and IgG2b for flow cytometric analysis.
Quantitative RT-PCR
Total RNA from sorted cells was isolated and reverse-transcribed, and quantitative PCR was performed as described31. Data were analyzed using the comparative CT (2−ΔΔCt) method using Hprt as the reference.
RNA-Seq Analysis
Spleens were taken one hour after saline or SRBC immunization. CD4+ DCs were pre-enriched using MACS manual cell separation columns with anti-CD11c microbeads (Miltenyi Biotec) and further sorted based on surface markers of CD11c+I-Ab+CD4+CD8-. Cells were sorted twice on a FACSAira III to purities of over 99%. Sorted DCs (106) were snap frozen and then RNA was extracted with the QIAGEN RNeasy Kit. RNA quality was checked with the Agilent 2100 Bioanalyzer (RNA integrity number >9 for all samples). Barcoded sequencing libraries were generated with 100 ng of RNA with the Ovation RNA-Seq System V2 and Encore Rapid Library System. Sequencing was performed on an Illumina HiSeq 2500 (UCSF Human Genetics Core) with 100-bp paired-end reads. Sequences were reported as FASTQ files, which were aligned to the mm9 mouse genome with STAR (Spliced Transcript Alignment to a Reference). Generation of Log2FC values and further analyses were performed with a Bioconductor package on RStudio. The RNAseq data have been deposited in the Gene Expression Omnibus (NCBI) data repository under accession code GEO: GSE71165.
Statistical analysis
Prism software (GraphPad) was used for all statistical analysis. Statistical comparisons were performed using an ANOVA or a two-tailed Student’s t-test. P values were considered significant when less than 0.05.
Extended Data
Extended Data Figure 1. EBI2 and 7α,25-OHC promote positioning of newly activated CD4 T cells in the outer T zone.
(a) Flow cytometric analysis of EBI2 expression on splenic OTII T cells and endogenous B cells in transfer recipients at 0 and 12 h after SRBC-OVA immunization. EBI2 KO cells were used as a staining control. Left histograms show example FACS data and right panel shows summary data across the indicated time points as geometric mean fluorescence intensity (geoMFI). (b) EBI2 expression on OTII and endogenous T cells in transfer recipients 2 days after saline or LPS-OVA immunization. Left histograms show example flow cytometric data and right panel shows summary geoMFI data for 4 mice. (c) Summary geoMFI time course data of EBI2 expression on LN OTII T cells in transfer recipients at the indicated times after alum-OVA immunization. (d) GFP expression in EBI2GFP/+ CD4 T cells that were unstimulated (naïve) or treated with anti-CD3 plus anti-CD28 for 2 days. Left histograms show example flow cytometric data and right panel shows summary geoMFI data for 3 mice. (e) Ebi2 mRNA abundance in cells of the type in d, determined by RT-qPCR and shown relative to the naïve cells. (f) Migration of OTII T cells and endogenous cells to the indicated amounts of 7α,25-OHC in transwell assays. Cells were from unimmunized (0 h) or immunized (day 1, 2) transfer recipient mice in one experiment (left panel) or from 12 hour immunized transfer recipients in a second experiment (right panel). Data are shown as % of input cells of each type that migrated. (g) Immunofluorescence analysis of spleen showing the distribution of co-transferred WT CD45.1+ (red) and EBI2 het or KO (GFP+, green) OTII T cells and endogenous B cells (IgD, blue) at 12 h and 1 day after immunization. (h, i) Immunohistochemical analysis of WT spleens (h) and inguinal LNs (i) showing the distribution of transferred control (WT) or EBI2 deficient (KO) OTII CD45.1+ T cells (blue) and endogenous B cells (IgD, brown) at day 2 after LPS-OVA immunization (h) or day 1 after alum-OVA immunization (i). (j) Immunohistochemical analysis of Cyp7b1, Ch25h or Hsd3b7 control (het, upper panels), or KO (lower panels) spleens showing the distribution of transferred WT OTII T cells (CD45.1, blue) and endogenous B cells (IgD, brown) at day 2 after SRBC-OVA immunization. (k) CCR7 expression on WT and EBI2 KO OTII T cells in transfer recipient spleens at the indicated days after SRBC-OVA immunization. **, p<0.01 by ANOVA (a, c) or student’s t-test (b, d, f). Data are representative of two (a–i, k) or three (j) independent experiments with at least three (a–c, k) or two (d–j) mice per group (error bars (e), s.e.m.).
Extended Data Figure 2. Defective differentiation of EBI2-deficient T cells to follicular helpers.
(a) ICOS and OX40 expression on WT, EBI2-deficient (KO) OTII and endogenous CD4 T cells in transfer recipient spleens two days after SRBC-OVA immunization. (b, c) In vitro proliferation of WT and EBI2 KO T cells in response to anti-CD3 plus anti-CD28 in the presence of the indicated amounts of 7α,25-OHC, shown as violet tracer dye dilution profiles (b) and total CD4 T cell numbers (c) at day 3 of culture. Numbers in b indicate frequency of cells that have undergone 2 or more divisions. (d) Flow cytometric analysis of co-transferred WT and EBI2 KO OTII T cells for CXCR5 and intracellular Bcl6 expression at the indicated days following SRBC-OVA immunization. Numbers indicate frequency of cells in gated region. (e) Summary of data of the type in d. Upper plot shows frequency and lower plot number of CXCR5+Bcl6hi OTII T cells. (f) Il21 mRNA abundance in CXCR5+ PD-1hi control (Het) or EBI2 KO OTII T cells sorted from recipient spleens at day 3 after immunization with SRBC-OVA, determined by RT-QPCR and shown relative to the Het control. (f) Frequency and number of CXCR5+ PD-1hi (left panels) or CXCR5+ Bcl6hi (right panels) WT and EBI2 KO OTII T cells in mice that received the cells as separate transfers, at day 3 following SRBC-OVA immunization. **, p<0.01 by ANOVA (e) or student’s t-test (f, g). Data are representative of three (a, d, e, g) or two (b, c, f) independent experiments with at least three mice per group (error bars (f), s.e.m.).
Extended Data Figure 3. EBI2-deficient T cells support reduced plasma cell and GC response.
(a) Frequency and number of CXCR5+PD-1hi CD4+ control (WT) and EBI2 KO OTII T cells in spleens of day 3 Listeria-OVA immunized transfer recipients. (b) PD-1 and CXCR5 flow cytometric analysis of control (Het) and EBI2 KO polyclonal CD4 T cells co-transferred to OTII recipients, 8 days after immunization with unconjugated SRBCs. (c) Summary of data of the type in b shown as CXCR5+PD-1hi cell frequency and number. (d) Flow cytometric analysis for the GC markers FAS and GL7 on endogenous B cells in OTII TCR transgenic mice that received no cells, control (HET) CD4 T cells or EBI2 KO CD4 T cells, or in WT B6 control mice, 12 days after immunization with unconjugated SRBCs. (e) Summary of data from d shown as number of FAS+GL7+ GC B cells. (f) Flow cytometric analysis for CD138hi B220lo plasma cells (upper plots) and intracellular IgM and IgG1 staining of these cells (lower plots) in mice of the type in d. (g) Summary of data from f shown as number of cells. (h) Serum anti-SRBC antibody levels in mice of the type in d, determined by flow cytometric analysis of SRBCs stained with immune sera, plotted as geoMFI. (i, j) Number of Fas+GL7+ GC cells and CD138hiB220int plasma cells in Listeria-OVA immunized CD28 KO mice that had received control (het) or EBI2 KO OTII cells, analyzed at day 5. **, p<0.01 by ANOVA (g, h) or student’s t-test (a, e, c, i, j). Data are representative of two independent experiments with at least three mice per group.
Extended Data Figure 4. T cell EBI2 is required for CD4+ DC-mediated augmentation of Tfh cell induction.
(a) Frequency and number of CXCR5+PD-1hi OTII T cells in μMT recipients determined by flow cytometric analysis. (b) Flow cytometric analysis for CD11c and MHC class II on splenocytes from Zbtb46-DTR mice treated with saline or DT for one day. Graph shows summary data for DC number in 4 mice of each type. (c) Frequency and number CXCR5+PD-1hi WT and EBI2 KO OTII T cells in spleens from Zbtb46-DTR BM chimeras treated with saline or diphtheria toxin (DT), at day 3 after immunization with SRBC-OVA. (d) Immunohistochemical analysis of spleen sections from WT mice without immunization (saline) or SRBC immunized for the indicated times, stained to detect IgD+ B cells (blue) and DCIR2+ DCs (brown). (e) Immunohistochemical analysis of spleen sections from recipients of WT or EBI2 KO OTII T cells at 12 h and 1 day after immunization SRBC-OVA immunization, stained for OTII CD45.1+ T cells (blue) and DCIR2+ DCs (brown). (f) Immunohistochemical analysis of spleen sections from WT:Zbtb46-DTR or CCR7 KO:Zbtb46-DTR mixed BM chimeras treated with DT, at day 2 after immunization. (g) Frequency and number of CXCR5+PD-1hi WT and EBI2 KO OTII T cells in spleens from WT:Zbtb46-DTR (control) or CCR7 KO:Zbtb46-DTR mixed BM chimeras treated with DT, at day 3 after immunization. (h) Frequency and number of CXCR5+PD-1hi control (het) and EBI2 KO co-transferred OTII T cells in spleens of CD47 KO recipients at day 3 after SRBC-OVA immunization. (i) Number of total and CD4+ DC in spleens from Irf4f/f CD11c-Cre− or + mice. (j) As for g but in Irf4f/f CD11c-Cre−or + recipient mice. (k) As for g but in Batf3 KO recipient mice. (l) ICOSL surface levels for DCs from mice immunized 12 h earlier with saline or SRBCs and also treated with control or ICOS blocking antibody. (m) ICOSL surface levels for CD4+ or CD8+ DCs from CD28 KO mice immunized 12 h earlier with saline or SRBCs. (n) Il6 and Tgfβ mRNA abundance in sorted CD4+ and CD8+ splenic DCs from mice treated with saline or SRBC six hours earlier, determined by RT-QPCR, shown relative to the control CD8+ DC. **, p<0.01 by ANOVA (g, k) or student’s t-test (a–c, i, j, m). Data are representative of three (a–e) or two (f–n) independent experiments with at least three (a–c, g–n) or two (d–f) mice per group (error bars (n), s.e.m.).
Extended Data Figure 5. DCs produce membrane and soluble CD25 and inhibit IL2R signaling.
(a) Heat map of RNAseq data from sorted CD4+ splenic DC showing the top 15 most induced genes at 1 h following SRBC versus saline immunization. (b) CD25 surface levels in CD11b+ migratory and resident DCs from LNs of mice immunized with saline or alum-OVA 2 days earlier. Graph on right shows summary data for total number of migratory CD25+ DCs. (c) Immunohistochemical analysis of spleen sections from TCRβδ KO mice immunized one day earlier with saline or SRBC, stained to detect IgD (blue) and CD25 (brown). (d, e) CD122 (Il2rb) mRNA determined by RT-QPCR (d) and surface staining (e) on the indicated cell types isolated from spleens of WT OTII T cell recipients at day 0, 1 and 2 after SRBC-OVA immunization. Transcript data are plotted relative to the signal in CD4+ DCs at day 0. (f) Intracellular flow cytometric analysis of pSTAT5 in CD4+ DCs or, as a positive control CD4+ T cells, that were untreated or incubated with IL2 (200 pg/ml) or, as a further positive control, GM-CSF (100 pg/ml). (g) Il2 mRNA in control (Het) and EBI2 KO OTII T cells isolated from recipient mice at the indicated times after SRBC-OVA immunization. (h) Intracellular flow cytometry for IL2 in cells of the type in f at 0, 12 and 24 h. Percentages show mean (±SEM) for 3 mice at each time point. (i) Flow cytometric analysis of CD25 expression on co-transferred WT and EBI2 KO OTII T cells in WT recipients at the indicated days after SRBC-OVA immunization. (j) Prdm1 (encoding Blimp1) transcript levels in sorted CXCR5+PD-1hi control (het) and EBI2 KO OTII T cells from SRBC-OVA immunized mice at day 3, plotted relative to the mean level in the Het group. (k) Summary of pSTAT5 staining data for OTII T cells from mice immunized one day earlier with SRBC-OVA, incubated with the indicated amounts of 7α,25-OHC plus IL2 (200 pg/ml) for one hour. (l) Flow cytometry of pSTAT5 in CD25+ (regulatory) T cells exposed to the indicated amounts of IL2 that had been pre-mixed with supernatants (s/n) from 8 h cultures of splenic CD4+ DCs from WT or CD25 KO mice immunized with saline or SRBCs 1 day before. Graph on right shows summary data from one experiment. **, p<0.01 by ANOVA (b, l) or student’s t-test (j). Data are representative of one (a) or two (b–l) independent experiments with at least two (a) or three (b–l) mice per group (error bars (g, j), s.e.m.).
Extended Data Figure 6. DC CD25 expression reduces IL2 signaling in activated CD4 T cells, favoring their differentiation to follicular helpers.
(a) Diagram of CD25 KO:Zbtb46-DTR BM chimera generation and time line of experiment. DTx, DT treatment. (b–d) Numbers (b), surface marker expression (c) and outer T zone positioning (d) of CD4+ DCIR2+ DCs in WT:Zbtb46-DTR and CD25 KO:Zbtb46-DTR mixed BM chimeras pretreated with DT, at day 1 after saline or SRBC immunization. (e) Number of Foxp3+ CD25+ regulatory T cells in mice of the type in k except that the mice were immunized for three days. (f) Immunohistochemical analysis of spleen sections from mice of the type in a, stained to detect IgD (blue) and CD25 (brown). (g, h) Frequency and number of CXCR5+PD-1hi control (EBI2 Het) and EBI2 KO OTII T cells in spleens (g) or LNs (h) from WT:Zbtb46-DTR or CD25 KO:Zbtb46-DTR mixed BM chimeras pretreated with saline or DT, at day 3 after immunization with Listeria-OVA (g) or alum-OVA (h). (i, j) Flow cytometric analysis for HEL-binding CD138+ plasma cells (i) and HEL-binding GL7+ Fas+ GC B cells (j) in spleens from WT:Zbtb46-DTR (control) or CD25 KO:Zbtb46-DTR mixed BM chimeras that had received Hy10 B cells and been treated with DT, at day 5 after immunization with HEL-SRBC. (k) Soluble CD25 detected by ELISA in spleen extracts taken from 12 h SRBC immunized mice, at day 1 after saline or recombinant CD25 treatment. **, p<0.01 by student’s t-test (h, k). Data are representative of two independent experiments with at least three (b, c, g–k) or two (d, f) mice per group (error bars (k), s.e.m.).
Extended Data Figure 7. Model of how EBI2-dependent positioning of activated T cells in association with CD25+ DCs in the outer T zone favors Tfh cell differentiation.
Initially, cognate T cells throughout the T zone are activated by antigen recognition and promptly start upregulating EBI2 and making IL2. EBI2 guides cells to the 7α,25-OHC high outer T zone and in this location they interact with activated DCs producing membrane and shed CD25 that binds and quenches IL2. This limits IL2R signaling on the T cell via pSTAT5 and allows induction of Bcl6 by other inputs such as ICOSL. T cells that lack EBI2 or remain in the inner T zone for other reasons are exposed to autocrine IL2 and this induces Blimp1, a repressor of Bcl62, disfavoring the Tfh cell fate.
Supplementary Material
Acknowledgments
We thank Chris Allen, Mark Ansel, Tony Defranco, Markus Muschen, and Shomyseh Sanjabi for mice, Jinping An for expert help with the mouse colony, Ying Xu for help with quantitative PCR, and Abul Abbas and Michael Barnes for comments on the manuscript. J.G.C. is an investigator of the Howard Hughes Medical Institute. E. L. is supported by the UCSF Biomedical Sciences (BMS) Graduate program and the National Science Foundation (grant no. 1144247). This work was supported in part by NIH grant AI40098.
Footnotes
Author contributions. J.L. designed and performed experiments, interpreted the results and prepared the manuscript. E.L. performed a number of experiments including staining and quantitation of cell distribution in sections and helped prepare the manuscript. T.Y. performed experiments identifying the defects in EBI2 KO T cells. J.G.C. designed experiments, supervised research and wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Ramiscal RR, Vinuesa CG. T-cell subsets in the germinal center. Immunol Rev. 2013;252:146–155. doi: 10.1111/imr.12031. [DOI] [PubMed] [Google Scholar]
- 2.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garside P, Ingulli E, Merica RR, Johnson JG, Noelle RJ, Jenkins MK. Visualization of specific B and T lymphocyte interactions in the lymph node. Science. 1998;281:96–99. doi: 10.1126/science.281.5373.96. [DOI] [PubMed] [Google Scholar]
- 4.Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 5.Lee SK, Rigby RJ, Zotos D, Tsai LM, Kawamoto S, Marshall JL, Ramiscal RR, Chan TD, Gatto D, Brink R, Yu D, Fagarasan S, Tarlinton DM, Cunningham AF, Vinuesa CG. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J Exp Med. 2011;208:1377–1388. doi: 10.1084/jem.20102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, Watowich SS, Qi H, Dong C, Wang D. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol Chem. 2012;287:11234–11239. doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira JP, Kelly LM, Xu Y, Cyster JG. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 2009;460:1122–1126. doi: 10.1038/nature08226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannedouche S, Zhang J, Yi T, Shen W, Nguyen D, Pereira JP, Guerini D, Baumgarten BU, Roggo S, Wen B, Knochenmuss R, Noel S, Gessier F, Kelly LM, Vanek M, Laurent S, Preuss I, Miault C, Christen I, Karuna R, Li W, Koo DI, Suply T, Schmedt C, Peters EC, Falchetto R, Katopodis A, Spanka C, Roy MO, Detheux M, Chen YA, Schultz PG, Cho CY, Seuwen K, Cyster JG, Sailer AW. Oxysterols direct immune cell migration via EBI2. Nature. 2011;475:524–527. doi: 10.1038/nature10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, Yang XV, Wu J, Kuei C, Mani NS, Zhang L, Yu J, Sutton SW, Qin N, Banie H, Karlsson L, Sun S, Lovenberg TW. Oxysterols direct B-cell migration through EBI2. Nature. 2011;475:519–523. doi: 10.1038/nature10226. [DOI] [PubMed] [Google Scholar]
- 14.Suan D, Nguyen A, Moran I, Bourne K, Hermes JR, Arshi M, Hampton HR, Tomura M, Miwa Y, Kelleher AD, Kaplan W, Deenick EK, Tangye SG, Brink R, Chtanova T, Phan TG. T follicular helper cells have distinct modes of migration and molecular signatures in naive and memory immune responses. Immunity. 2015;42:704–718. doi: 10.1016/j.immuni.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao KH, Niec RE, Nussenzweig MC. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209:1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goenka R, Barnett LG, Silver JS, O’Neill PJ, Hunter CA, Cancro MP, Laufer TM. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J Immunol. 2011;187:1091–1095. doi: 10.4049/jimmunol.1100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi T, Cyster JG. EBI2-mediated bridging channel positioning supports splenic dendritic cell homeostasis and particulate antigen capture. Elife. 2013;2:e00757. doi: 10.7554/eLife.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi T, Li J, Chen H, Wu J, An J, Xu Y, Hu Y, Lowell CA, Cyster JG. Splenic Dendritic Cells Survey Red Blood Cells for Missing Self-CD47 to Trigger Adaptive Immune Responses. Immunity. 2015;43:764–775. doi: 10.1016/j.immuni.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logue EC, Bakkour S, Murphy MM, Nolla H, Sha WC. ICOS-induced B7h shedding on B cells is inhibited by TLR7/8 and TLR9. J Immunol. 2006;177:2356–2364. doi: 10.4049/jimmunol.177.4.2356. [DOI] [PubMed] [Google Scholar]
- 20.Marczynska J, Ozga A, Wlodarczyk A, Majchrzak-Gorecka M, Kulig P, Banas M, Michalczyk-Wetula D, Majewski P, Hutloff A, Schwarz J, Chalaris A, Scheller J, Rose-John S, Cichy J. The role of metalloproteinase ADAM17 in regulating ICOS ligand-mediated humoral immune responses. J Immunol. 2014;193:2753–2763. doi: 10.4049/jimmunol.1302893. [DOI] [PubMed] [Google Scholar]
- 21.Rubin LA, Kurman CC, Fritz ME, Biddison WE, Boutin B, Yarchoan R, Nelson DL. Soluble interleukin 2 receptors are released from activated human lymphoid cells in vitro. J Immunol. 1985;135:3172–3177. [PubMed] [Google Scholar]
- 22.Maier LM, Anderson DE, Severson CA, Baecher-Allan C, Healy B, Liu DV, Wittrup KD, De Jager PL, Hafler DA. Soluble IL-2RA levels in multiple sclerosis subjects and the effect of soluble IL-2RA on immune responses. J Immunol. 2009;182:1541–1547. doi: 10.4049/jimmunol.182.3.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chistiakov DA, Chistiakova EI, Voronova NV, Turakulov RI, Savost’anov KV. A variant of the Il2ra / Cd25 gene predisposing to graves’ disease is associated with increased levels of soluble interleukin-2 receptor. Scand J Immunol. 2011;74:496–501. doi: 10.1111/j.1365-3083.2011.02608.x. [DOI] [PubMed] [Google Scholar]
- 24.Russell SE, Moore AC, Fallon PG, Walsh PT. Soluble IL-2Ralpha (sCD25) exacerbates autoimmunity and enhances the development of Th17 responses in mice. PLoS One. 2012;7:e47748. doi: 10.1371/journal.pone.0047748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kronin V, Vremec D, Shortman K. Does the IL-2 receptor alpha chain induced on dendritic cells have a biological function? Int Immunol. 1998;10:237–240. doi: 10.1093/intimm/10.2.237. [DOI] [PubMed] [Google Scholar]
- 26.Liang D, Zuo A, Shao H, Born WK, O’Brien RL, Kaplan HJ, Sun D. Role of CD25+ dendritic cells in the generation of Th17 autoreactive T cells in autoimmune experimental uveitis. J Immunol. 2012;188:5785–5791. doi: 10.4049/jimmunol.1200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popov A, Driesen J, Abdullah Z, Wickenhauser C, Beyer M, Debey-Pascher S, Saric T, Kummer S, Takikawa O, Domann E, Chakraborty T, Kronke M, Utermohlen O, Schultze JL. Infection of myeloid dendritic cells with Listeria monocytogenes leads to the suppression of T cell function by multiple inhibitory mechanisms. J Immunol. 2008;181:4976–4988. doi: 10.4049/jimmunol.181.7.4976. [DOI] [PubMed] [Google Scholar]
- 28.Fukao T, Koyasu S. Expression of functional IL-2 receptors on mature splenic dendritic cells. Eur J Immunol. 2000;30:1453–1457. doi: 10.1002/(SICI)1521-4141(200005)30:5<1453::AID-IMMU1453>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 29.Wuest SC, Edwan JH, Martin JF, Han S, Perry JS, Cartagena CM, Matsuura E, Maric D, Waldmann TA, Bielekova B. A role for interleukin-2 trans-presentation in dendritic cell-mediated T cell activation in humans, as revealed by daclizumab therapy. Nat Med. 2011;17:604–609. doi: 10.1038/nm.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klatzmann D, Abbas AK. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat Rev Immunol. 2015;15:283–294. doi: 10.1038/nri3823. [DOI] [PubMed] [Google Scholar]
- 31.Yi T, Wang X, Kelly LM, An J, Xu Y, Sailer AW, Gustafsson JA, Russell DW, Cyster JG. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity. 2012;37:535–548. doi: 10.1016/j.immuni.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlsson F, Getahun A, Rutemark C, Heyman B. Impaired antibody responses but normal proliferation of specific CD4+ T cells in mice lacking complement receptors 1 and 2. Scand J Immunol. 2009;70:77–84. doi: 10.1111/j.1365-3083.2009.02274.x. [DOI] [PubMed] [Google Scholar]
- 33.Muraille E, Giannino R, Guirnalda P, Leiner I, Jung S, Pamer EG, Lauvau G. Distinct in vivo dendritic cell activation by live versus killed Listeria monocytogenes. Eur J Immunol. 2005;35:1463–1471. doi: 10.1002/eji.200526024. [DOI] [PubMed] [Google Scholar]
- 34.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 35.Paus D, Phan TG, Chan TD, Gardam S, Basten A, Brink R. Antigen recognition strength regulates the choice between extrafollicular plasma cell and germinal center B cell differentiation. J Exp Med. 2006;203:1081–1091. doi: 10.1084/jem.20060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meli AP, King IL. Identification of mouse T follicular helper cells by flow cytometry. Methods Mol Biol. 2015;1291:3–11. doi: 10.1007/978-1-4939-2498-1_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.