Introduction
Few funded research projects in computer science go on for decades. It’s even questionable whether any project should be sustained that long. Nevertheless, the Protégé project at Stanford University began in the 1980s and is still going strong, helping developers to construct reusable ontologies and to build knowledge-based systems. The recognition of our work at the International Semantic Web Conference in October 2014 with the “Ten Years” Award was a great honor. The award has provided an opportunity f o r reflection—both on the Protégé project itself and on the need for computational infrastructure in the AI community.
Protégé has become the most widely used software for building and maintaining ontologies. It is by no means the only solution, and there are known problems with Protégé, but we appreciate that the system is extremely popular. Although his study is now a bit dated, Cardoso (2007) surveyed the Semantic Web community and found that two-thirds of his respondents used Protégé. To date, more than 250,000 people have registered to use the software. Many Fortune 500 companies use Protégé to build their ontologies. Important government projects, such as the development of the National Cancer Institute Thesaurus (Noy et al., 2008; Figure 1) and the World Health Organization’s International Classification of Diseases (ICD-11; Tudorache et al., 2010) depend on the software.
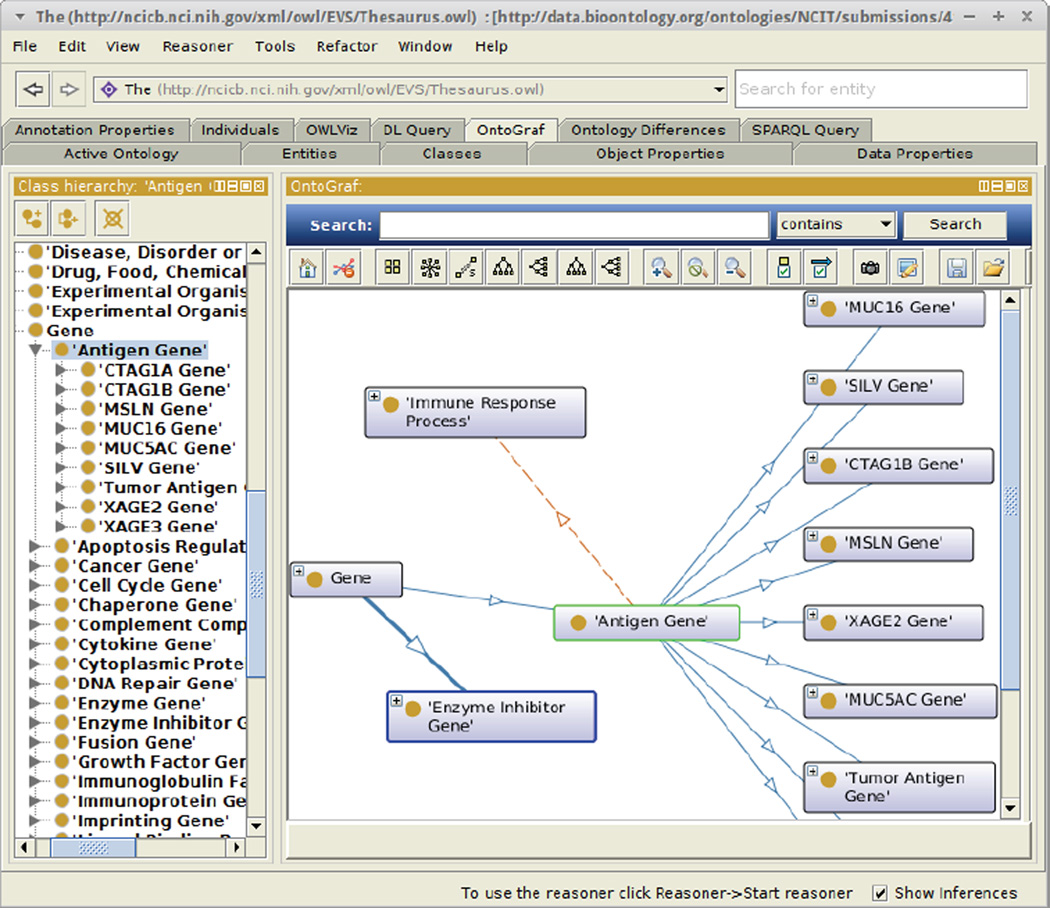
Figure 1.
The Protégé 5 Desktop System. The figure shows the most recent version of the Protégé system used to edit the National Cancer Institute Thesaurus. In the Figure, the user has selected the class Antigen Gene. The visualization displays the relationships among the selected class and other classes in its neighborhood.
Protégé currently exists in a variety of frameworks. A desktop system (Protégé 5) supports many advanced features to enable the construction and management of OWL ontologies (see Figure 1). A Web-based system (WebProtégé) offers distributed access over the Internet using any Web browser and, by design, is much simpler to use for many ontology-engineering tasks (Figure 2). The Web-based version has become extremely popular, and it recently exceeded the desktop-client in its degree of usage. It is extremely handy to be able to point a Web browser to an appropriate server and to begin editing. Like a Google doc, a WebProtégé ontology can be easily shared with a distributed group of users who can engage in collaborative authoring activities from wherever they happen to be logged in. The development environment used by the World Health Organization to manage ICD-11 is based on WebProtégé (Tudorache et al., 2010).
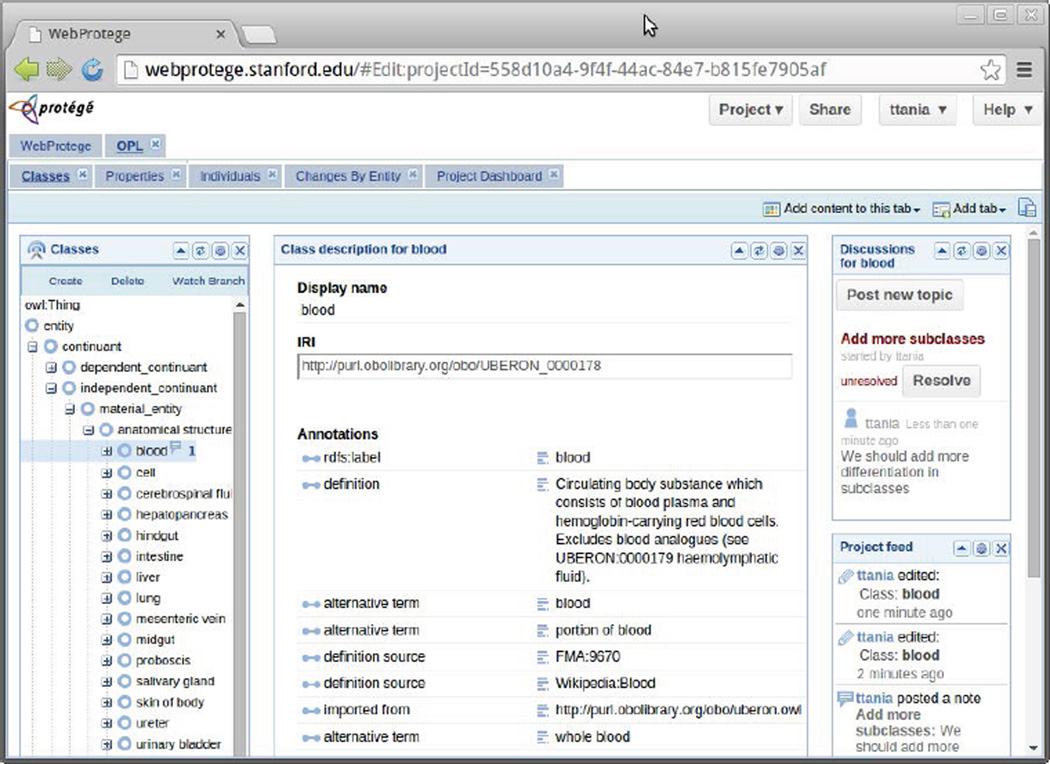
Figure 2.
WebProtégé. The Web-based version of Protégé offers users and their collaborators the opportunity to share and edit ontologies online, much like a Google doc. Here we see the Ontology for Parasite Lifecycle (OPL), an ontology developed by a community of scientists who investigate malaria, toxoplasmosis, and other diseases caused by eukaryotic pathogens (Parikh et al., 2012). In the left-hand pane, the user has selected the class blood. The middle pane shows the properties of the selected class. The right-most pane reveals an audit trail and a threaded discussion regarding the modeling of this entity.
Older versions of the Protégé desktop system have included support for editing ontologies represented in a frame language (namely, in the OKBC framework developed by the DARPA Knowledge Sharing Initiative in the 1990s; Neches et al., 1991). Comparable functionality has not yet been migrated to current versions of Protégé, however. All versions of Protégé may be downloaded from the project’s Web site (http://protege.stanford.edu). They are available under an open-source license.
The paper for which the Protégé team won the “Ten Years” Award (Knublauch et al., 2004) describes the first Protégé system to support the World Wide Web Consortium’s recommended Web Ontology Language (OWL). We had been tracking the emerging OWL specification, and Holger Knublauch, then a post-doctoral fellow in our laboratory, worked with the rest of the team to extend Protégé to create what was, at the time, the only ontology-development platform that could accommodate nearly the complete OWL specification. Protégé’s support for OWL has been enhanced over the years, particularly through a very successful collaboration with Alan Rector’s CO-ODE project at the University of Manchester, and recent versions of Protégé fully support the latest OWL 2.0 specification.
When people think of Protégé, they think of an editor for ontologies, and they think of OWL. Protégé did not start out this way, however. Gennari et al. (2003) traced the history of the first 15 years of the Protégé project, documenting the many shifts in perspective as Protégé progressed from a student project for a Ph.D. dissertation that focused on problems of building knowledge-based systems (Musen, 1989) to a major open-source platform supported by a huge community of diverse users (Musen, 2005).
Adventures in Meta-Land: The Early Years
The Protégé project began at a time before there was an appreciation for the importance of ontologies in providing structure for electronic knowledge bases and long before the rise of ontologies in e-commerce and in information systems generally. The Protégé project was born out of basic research in the intelligent systems community to develop better methods for constructing new knowledge bases.
In the 1980s, researchers at Stanford had had considerable success using a domain-specific knowledge-acquisition tool, known as OPAL (Musen et al., 1987), to build the complex knowledge bases required by a cancer-chemotherapy advisor known as ONCOCIN (Tu et al., 1989). ONCOCIN would take as its input (1) data at a particular point in time about a patient who had cancer and (2) knowledge of the relevant chemotherapy protocol according to which the patient was being treated. The program would generate as output recommendations for laboratory tests to order, drugs to administer, and plans for follow-up care. Encoding such knowledge for ONCOCIN had been an arduous task. With OPAL, it was clear that oncology specialists could use their computers to fill in the blanks of oncology-related forms and to draw chemotherapy flow charts using domain-specific icons much more easily than they could edit the corresponding knowledge bases directly (Figure 3). OPAL thus became an enormously successful tool for building and maintaining cancer-chemotherapy knowledge bases. The problem was that OPAL was something of a hack, as it was impossible to alter the cancer-chemotherapy domain model that supported the user interface without reprogramming the system. If there was a need to build knowledge bases in domains other than cancer chemotherapy, then OPAL was not going to be helpful.
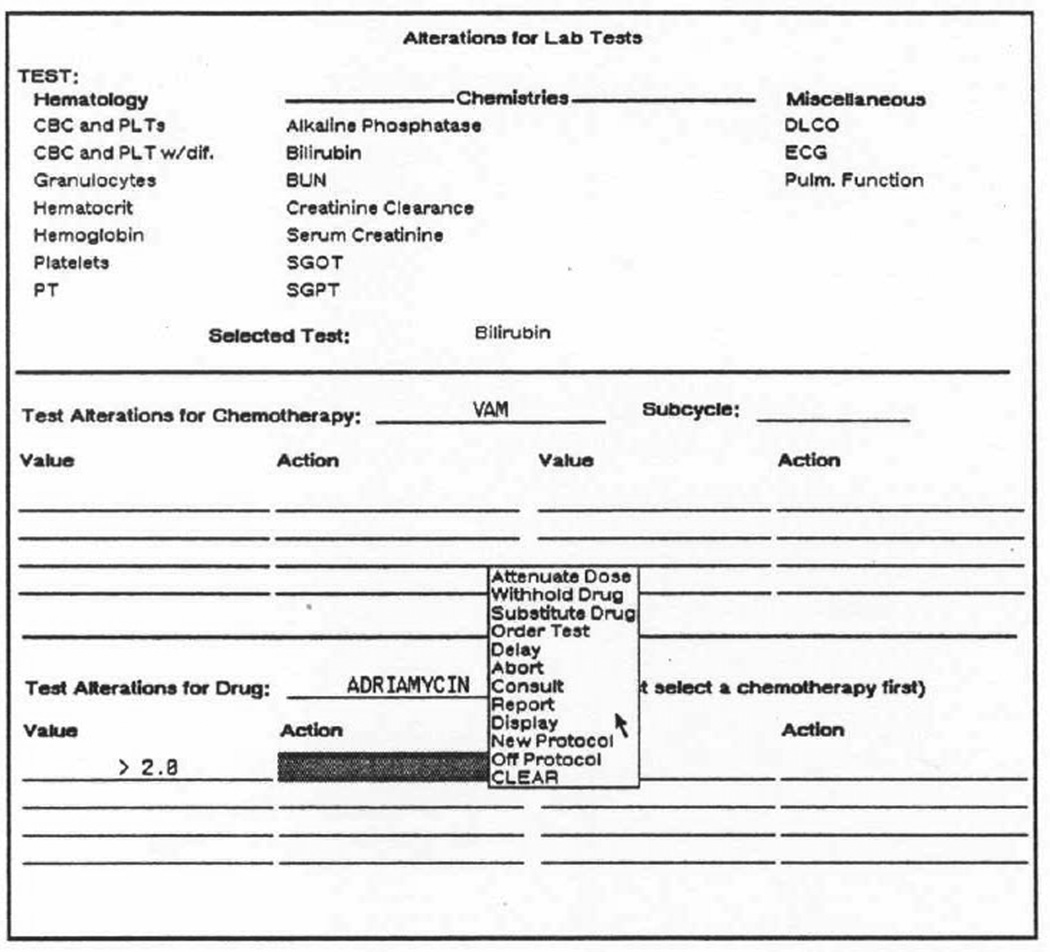
Figure 3.
The OPAL system for entry of cancer-chemotherapy knowledge. In the figure, the user is accessing a domain-specific form to indicate what domain action to take if, when administering a chemotherapy combination known as “VAM,” the patient’s serum bilirubin is measured to be greater than 2.0 mg/dl. OPAL had dozens of such forms, as well as a flowcharting facility used to specify the sequences of chemotherapies and drugs to administer when treating a particular kind of cancer.
The original Protégé system (now bearing the retronym “Protégé-1”) ran on Xerox Lisp machines. The system was designed to allow developers to create and edit the domain models in knowledge-acquisition systems like OPAL (Musen, 1989). A developer could craft a new domain model using Protégé-1, and the system would then generate a domain-specific knowledge-acquisition tool based on that particular model. Protégé-1 addressed the major assumption built into OPAL: the singular domain model of cancer chemotherapy. Protégé-1 still assumed that the run-time system that would interpret the resultant knowledge bases would use a particular problem-solving method (skeletal-plan refinement; Friedland and Iwasaki, 1985), which was good for planning cancer therapy, but not a helpful problem solver for tasks such as diagnosis. If there was a need to build intelligent systems that could reason using alternative problem-solving approaches, then Protégé-1 was not going to be helpful.
Protégé-II, which appeared in the early 1990s, allowed developers to select alternative problem solvers (and even to compose new problem solvers from primitive building blocks) to address particular reasoning requirements (Eriksson et al., 1995). In Protégé-1, developers had been forced to fashion new domain models so that the semantics of the entities in those models were determined by the role that the entities would play in the process of skeletal-plan refinement (Figure 4). Specifically, Protégé-1 had built into it a set of terms and relationships that captured the types of knowledge that our implementation of skeletal-plan refinement required (viz., a hierarchy of skeletal plans that might be instantiated during the planning process, input data that might predicate alternative planning actions, and actions that the planner might take). Developers were expected to describe domain knowledge (e.g., the notion of chemotherapy) by relating it to one of these terms (e.g., a skeletal plan).
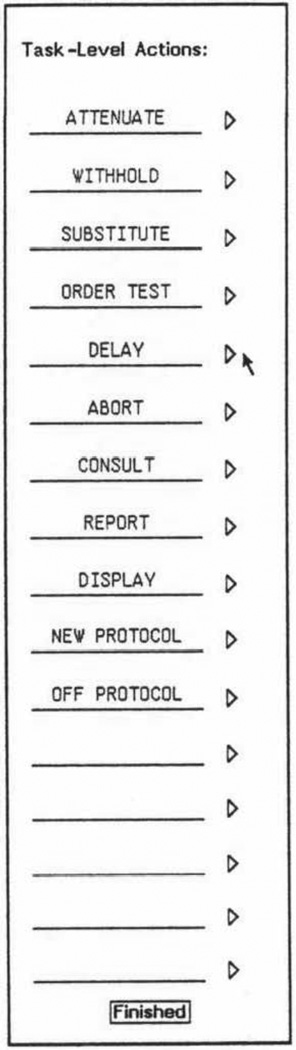
Figure 4.
A Protégé-1 form for entry of classes of task-level actions. Here, the user is entering into a domain-independent form the kinds of actions that a planner should take in the setting of different situations involving the input data provided to the runtime system. The actions correspond to the entries in the menu displayed by the OPAL program in Figure 3. Protégé-1 had dozens of such forms with which a user could specify the domain model for a customizable knowledge-acquisition tool that had much of the functionality of OPAL.
In Protégé-II, the dependency between the entry of domain knowledge and the built-in model of skeletal-plan refinement was eliminated. Developers suddenly were free to define domain entities independently of any given problem solver. The use of ontologies seemed to be the obvious way to create such domain models. The freedom to address the construction of problem-solving systems and the elucidation of domain models as two separate engineering activities meant that the developers using Protégé-II were now operating at a higher plane of abstraction than had been the case with Protégé-1, where modeling was more constrained. Protégé-1, of course, was at a higher plane of abstraction than had been the case with the domain-bound OPAL system.
Protégé-II was seen as an integrated development environment (IDE) for the construction of intelligent systems. It provided an editor, which developers would use to create domain ontologies that Protégé-II would employ to construct an ontology-specific knowledge-acquisition tool that was suitable for the specific class of domain problems described by the ontology (much in the spirit of OPAL), which could elicit the domain knowledge needed to construct an operational knowledge base for particular problems within that class (much in the spirit of an ONCOCIN knowledge base for a particular kind of cancer; Figure 5). Thus, Protégé-II would take any designated domain ontology, and generate semi-automatically an OPAL-like knowledge acquisition tool for entry of the necessary content knowledge needed to instantiate the classes in the ontology for purposes of building an intelligent system (Eriksson et al., 1994). Separately, the developer would fashion a problem solver from a library of components, or else would build a new problem solver from scratch. The developer then would map an explicit representation of the input–output requirements of the completed problem solver to their corresponding classes in the domain ontology—indicating, for example, that administration of chemotherapy was a plan, that administration of a drug within a chemotherapy was a plan, that the result of a particular laboratory test should be treated as input data, and so on (Crubézy and Musen, 2004).
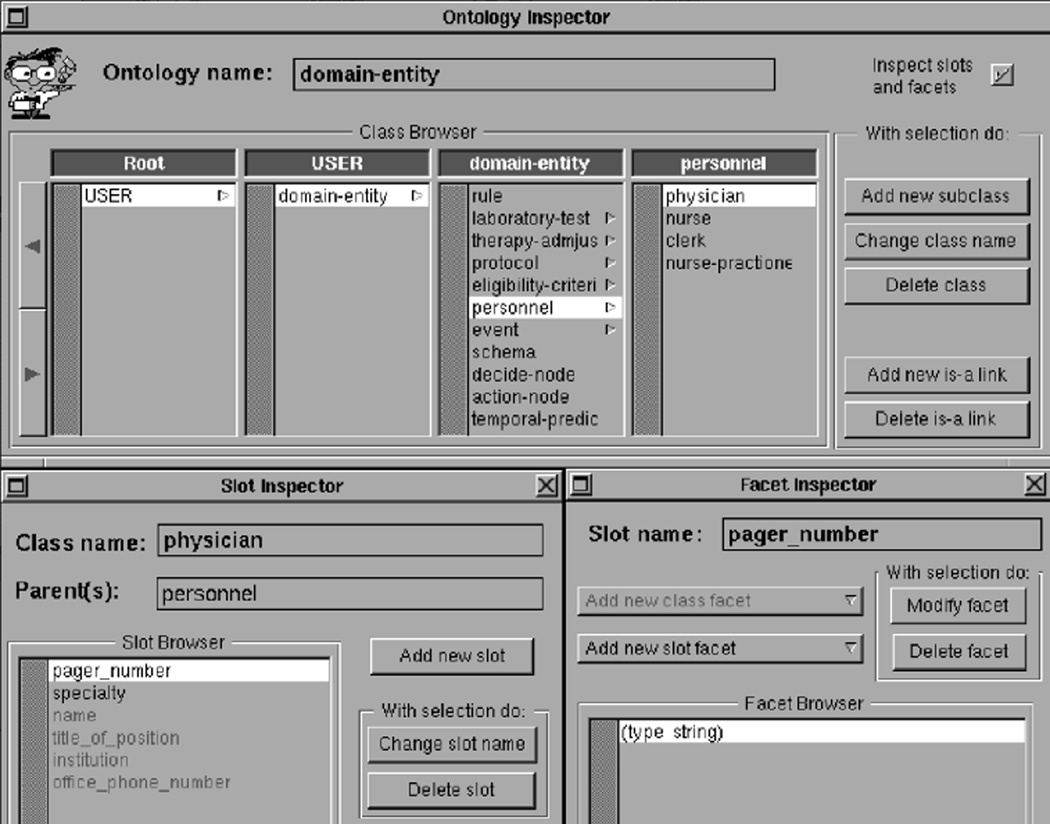
Figure 5.
The Protégé-II ontology editor. Built under the NeXTSTEP operating system, Protégé-II was the first version of Protégé to include a separate ontology editor. Here, the user is specifying that physicians have pager-numbers of data type string.
We used Protégé-II in several projects in our laboratory to build a large number of knowledge bases for intelligent systems (e.g., Musen et al., 1996; Stein et al., 1996). By the end of the 1990s, however, the ontology-editing component of the overall Protégé-II system had begun to take on a life of its own.
The Protégé Ontology Editor—and the New Protégé Community
Twenty years ago, most people in AI felt a bit pretentious using the word ontology in everyday conversation. The world has changed considerably since that time, however. We now recognize ontologies as essential elements of much of AI technology, and we take for granted all the conferences and books and journals that have emerged on the subject (Guarino and Musen, 2015). Whereas at Stanford, the Protégé team continued to build ontologies mainly to structure the frame-based knowledge bases that we were developing in our group, other developers were creating ontologies for a wide range of applications. The idea of the Semantic Web was taking root, and ontologies were being seen as essential for e-commerce and e-science.
Protégé-II, which had been developed under the NeXTSTEP platform, was replaced—just in time —by a version of the system that ran under Windows (Protégé/Win). Within a couple of years, Java became entrenched in the software engineering community, and a new Java-based version of the system, Protégé-2000 (Noy et al., 2001), freed us from reliance on a particular hardware platform while enabling us to create a very flexible plug-in architecture. With the Java-based version of Protégé, we were able to provide support for the development of ontologies in RDF(S) and, soon thereafter, OWL (Knublauch et al., 2004). With subsequent major design changes, Protégé-2000 became simply Protégé 3, followed by Protégé 4, and now Protégé 5 (see Figure 1).
Meanwhile, with interest in ontologies burgeoning throughout AI circles and beyond, the Protégé community grew exponentially. The software—which we were viewing mainly as infrastructure to enable Stanford-based research in intelligent systems—was reaching the stage of becoming a “product.” In 1996, three colleagues and I had some beers while attending a conference in Europe, and we jokingly referred to our gathering as “The First Protégé Users Group Meeting.” Soon, however, the rapidly growing community of Protégé users around the word demanded a real Protégé event. We held a Protégé workshop at Stanford in 1998 for about 50 people. In 2000, some 100 people participated in a Protégé workshop at the University of Manchester. More than 200 people showed up at a similar gathering at the United States National Institutes of Health in 2002. These Protégé conferences were exciting meetings for the entire team: We received extremely valuable feedback on our software, we got to meet the people who regularly corresponded with us on the Protégé-related mailing lists that we had created, and we learned about remarkable applications that members of the community were creating with our technology.
The perception of Protégé was changing radically. The software was no longer viewed mainly as a tool for building knowledge-based systems, but rather as one for building ontologies. The concomitant growth of the Protégé community and the abrupt appearance of scores of Protégé plug-ins that were being contributed by users outside of Stanford proceeded to change the nature of our research activity in a major way. Any alteration of our code base suddenly could affect the work of thousands of other people—and thousands of other people now had their own ideas of where they thought our work should be heading. Overall, the project benefitted tremendously from the expanding user base, and the influx of new ideas and new plug-ins was downright exciting. It’s unusual in academia to feel such a strong connection with the kind of broad community that our work had attracted, and our research group developed a very close, symbiotic relationship with our users.
Although commercial ontology editors with support for OWL began to appear in the marketplace at the end of the last decade, there has been no decline in enthusiasm for Protégé and its plugins. As an academic group, we do not see ourselves in competition with the commercial sector, and “market share” is an irrelevant concept to us and our funders. What is clear, however, is that our users very much value (1) access to Protégé under an open-source license, (2) our team’s commitment to the full implementation of World Wide Web Consortium recommendations, (3) the vast library of Protégé plug-ins that users have developed, and (4) the supportive nature of the extensive Protégé community, whose members, through online media, help one another to troubleshoot specific problems either in using the software or in modeling particular entities. We did not specifically create the Protégé community. Social engineering was never a “deliverable” of any of our federal grants. Nevertheless, the user community that has gravitated toward Protégé is as much a part of the experience of using our tools as is any piece of software. The Protégé project depends critically on the thousands of users who have gathered online to learn from one another and to help us to make Protégé a better system.
What is our Core Mission Anyway?
For the past 30 years, the Protégé project has been funded by several federal agencies to explore and evaluate new ways to build intelligent systems and ontologies. Our most consistent funding has come from the United States National Institutes of Health (particularly the National Library of Medicine and the National Institute for General Medical Sciences). Much of our early Protégé-related funding supported research to develop the infrastructure needed for our “real” goals: the engineering and evaluation of clinical decision-support systems. Protégé at the beginning was viewed merely as a means to an end. More recent grants, however, have supported our original research to study collaborative ontology development (Tudorache et al., 2011; Strohmaier et al., 2013), methods for ontology alignment (Noy and Musen, 2003; Ghazvinian et al., 2009), ontology evaluation through crowdsourcing (Mortensen et al., 2014), ontology visualization (Storey et al., 2001), and methods for the very early stages of ontology conceptualization (Zhang et al., 2015). These academic pursuits are what make our laboratory an exciting place for students and post-docs, and they are what keep the research funding flowing. The new knowledge and techniques that we uncover in our scholarly work contribute new ideas to AI and offer new ways to enhance the technology that comes out of our laboratory.
But there is something of a mismatch. Although our laboratory has received considerable academic recognition for our scholarly accomplishments, this is presumably not why we won the 2014 “Ten Years” Award from the Semantic Web Science Association. The “Ten Years” Award acknowledged the very pragmatic benefits of our work—of creating the first widely used ontology editor for OWL and of building an infrastructure that has been used by more than one quarter of a million people. One of the greatest successes of Protégé is measured in terms of the vast community of users who have benefitted from the technology in a hands-on fashion. And it is this relationship with our user community that makes Protégé more than just another academic project.
It is easy to argue that academic groups really have no business creating software systems of this magnitude. Academic groups lack middle managers who can perform code reviews and who can coordinate software-engineering activities. Such groups lack dedicated user-support personnel and documentation specialists. They also have to survive on external funding, living from one grant or contract to the next. Although the Protégé project generally has been successful in obtaining sponsored research support for its work, there have been some very tense moments during the past 30 years. At one point recently, the project was caught between grants for several months; our future funding had become uncertain due to a reorganization within the National Institutes of Health. Protégé thus became a “poster child” for the general problem of attempting to support long-term research infrastructure using short-term, highly uncertain funding mechanisms (Baker, 2012). Although the National Institutes of Health has been extremely generous to our project, there still are no reliable mechanisms in place to ensure the continuity of research projects such as ours that are delivering infrastructure to a wide community of users in addition to pursuing their own research and development agendas. Our experience makes it obvious that federal funding agencies need better mechanisms for sustaining such projects in a more durable, more coherent manner. Indeed, if the Protégé project does deserve an award, perhaps it is for ensuring its own survival on nothing but “soft money” for the past three decades.
As an academic project, we have been able to learn a great deal from our colleagues around the world. Early work on the KADS project in Europe (Schreiber et al., 2000) had tremendous influence on the Protégé approach to building intelligent systems during the early years. Ontology editors developed by other groups, such as OilEd (Bechhofer et al., 2001), were inspirational to us. Our collaboration with Alan Rector’s group at the University of Manchester helped to ensure that Protégé supported OWL in its complete scope and that the community had excellent documentation for handling OWL within the Protégé framework (Horridge, 2011). At the same time, we began to see many of our ideas picked up in the commercial sector, as companies such as TopQuadrant released their own tools for managing OWL ontologies. As an academic group, we view these commercial tools not so much as “competition” but as confirmation of our work and of many of our design decisions.
The Protégé project has settled down into a delightful ambiguity, where our research grants support our investigation of new methods of ontology engineering and ontology management, while at the same time allowing us to support the thousands of users who rely on our software on a regular basis. Our core mission is therefore a bit blurred, but we believe that we have struck a useful balance, and the feedback that we receive from our users helps us to target our research in beneficial ways, which leads to the kinds of new features in Protégé that simply are not available through current commercial products. Our project is in fact quite fortunate to have both a scholarly component and a built-in technology-transfer activity.
Conclusions
Despite the importance of tools such as Protégé to the AI community, it is unusual for academic groups to derive professional recognition for the development of computational infrastructure. Infrastructure generally is taken for granted, and it almost never leads to the kind of high-impact publications that in scholarly circles are nearly always the coin of the realm. Nevertheless, groups in academia often are in the best position to undertake the development of tools such as Protégé, particularly when the requirements are complex, and when the academics themselves are going to be the first natural users of such software.
We believe that the Protégé project has been successful despite the vagaries of research funding in the United States. Overall, all governments need to put more emphasis on the support of computational infrastructure for AI and to ensure the durability of their investments in this kind of work.
The Protégé project would never have been as successful as it has been without the support of our tremendous user community. Working with this vocal and enthusiastic group of users has been a wonderful experience for the entire Protégé team. It has served as a constant reminder to us that ontologies are no longer arcane laboratory artifacts and that they form the foundation for some of the most interesting work in computer science that is now taking place.
Acknowledgments
The Protégé project is currently supported by NIH grants GM103316 and GM086587 from the National Institute for General Medical Sciences. Previous funding has been provided by the National Library of Medicine, the National Cancer Institute, the Defense Advanced Research Projects Agency, the National Science Foundation, the Engineering and Physical Sciences Research Council of the United Kingdom through a grant to the University of Manchester, the Commission of the European Community through a contract to the University of Amsterdam, and by gifts from industry.
The project would not have been possible without the contributions of the many research staff, post-docs, and students who contributed their talent and effort over the years. These team members have included Yasser Bashir, Andrew Bennett, Sebastian Brandt, Heining Cheng, David Combs, Monica Crubézy, Ol ivier Dameron, Dejing Dou, Nick Drummond, Henrick Eriksson, John Egar, Neil Ernst, Ray Fergerson, Doug Fridsma, John Gennari, Rafael Gonçalves, Bill Grosso, Saeed Hassanpour, Matthew Horridge, Johnson Hou, Luigi Iannone, Simon Jupp, Holger Knublauch, Daniel Lamprecht, Qi Li, Rob Lintern, Dilvan Moreira, Natasha Noy, Csongor Nyulas, Martin O’Connor, John Park, Jan Pöschko, Angel Puerta, Alan Rector, Timothy Redmond, Thomas Rothenfluh, Daniel Rubin, Abraham Sebastian, Yuval Shahar, Ravi Shankar, Philip Shilane, Adam Stein, Margaret-Anne Storey, Marcus Strohmaier, Dmitry Tsarkov, Samson Tu, Tania Tudorache, Jennifer Vendetti, Simon Walk, Eckart Walther, Hao Wang, and Yuhao Zhang.
Biography

Dr. Musen is Professor of Biomedical Informatics at Stanford. He studies intelligent systems, scientific metadata, and decision support. He is the recipient of many honors, including the Donald A. B. Lindberg Award for Innovation in Informatics from the American Medical Informatics Association. Dr. Musen is co-editor-in-chief of the journal Applied Ontology.
References
- Baker M. Databases fight funding cuts: online tools are becoming ever more important to biology, but financial support is unstable. Nature. 2012;489(7414):19. doi: 10.1038/489019a. [DOI] [PubMed] [Google Scholar]
- Bechhofer S, Horrocks I, Goble C, Stevens R. Proceedings of the Joint German/Austrian Conference on Artificial Intelligence. Vol. 2174. Vienna: Springer-Verlag LNAI; 2001. OilEd: a reason-able ontology editor for the Semantic Web; pp. 396–408. [Google Scholar]
- Cardoso J. The Semantic Web vision. Where are we? IEEE Intelligent Systems. 2007;22(5):84–88. [Google Scholar]
- Crubézy M, Musen MA. Ontologies in support of problem solving. In: Staab S, Studer R, editors. Handbook on Ontologies. Berlin: Springer-Verlag; 2004. pp. 321–341. [Google Scholar]
- Eriksson H, Puerta AR, Musen MA. Generation of knowledge-acquisition tools from domain ontologies. International Journal of Human– Computer Studies. 1994;41:425–453. [Google Scholar]
- Eriksson H, Shahar Y, Tu SW, Puerta AR, Musen MA. Task modeling with reusable problem-solving methods. Artificial Intelligence. 1995;79:293–326. doi: 10.1016/0933-3657(95)00006-r. [DOI] [PubMed] [Google Scholar]
- Friedland PE, Iwasaki Y. The concept and implementation of skeletal plans. Journal of Automated Reasoning. 1985;1(2):161–208. [Google Scholar]
- Ghazvinian A, Noy NF, Musen MA. Creating mappings for ontologies in biomedicine: simple methods work. Proceedings of the AMIA Annual Symposium; American Medical Informatics Association; San Francisco, CA. 2009. pp. 198–202. [PMC free article] [PubMed] [Google Scholar]
- Gennari JH, Musen MA, Fergerson RW, Grosso WE, Crubézy M, Eriksson H, Noy NF, Tu SW. The evolution of Protégé: An environment for knowledge-based systems development. International Journal of Human-Computer Studies. 2003;58(1):89–123. [Google Scholar]
- Guarino N, Musen MA. Applied Ontology: The next decade begins. Applied Ontology. 2015;10(1):1–4. [Google Scholar]
- Horridge M. A Practical Guide To Building OWL Ontologies Using Protégé 4 and CO-ODE Tools. 2011 Edition 1.3. http://mowl-power.cs.man.ac.uk/protegeowltutorial/resources/ProtegeOWLTutorialP4_v1_3.pdf. [Google Scholar]
- Knublauch H, Fergerson RW, Noy NF, Musen MA. Third International Conference on the Semantic Web. Hiroshima, Japan: 2004. The Protégé OWL plugin: an open development environment for Semantic Web applications; pp. 229–243. [Google Scholar]
- Mortensen JM, Minty EP, Januszyk M, Sweeney TE, Rector AL, Noy NF, Musen MA. Using the wisdom of the crowds to find critical errors in biomedical ontologies: A study of SNOMED CT. Journal of the American Medical Informatics Association. 2014 doi: 10.1136/amiajnl-2014-002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musen MA. Automated Generation of Model-Based Knowledge-Acquisition Tools. London: Pitman, Artificial Intelligence Research Notes Series; 1989. [Google Scholar]
- Musen MA. Protégé: Community is everything. International Journal of Human–Computer Studies. 2005;62(5):545–552. [Google Scholar]
- Musen MA, Fagan LM, Combs DM, Shortliffe EH. Use of a domain model to drive an interactive knowledge-editing tool. International Journal of Man–Machine Studies. 1987;26:105–121. [Google Scholar]
- Musen MA, Tu SW, Das AK, Shahar Y. EON: A component-based approach to automation of protocol-directed therapy. Journal of the American Medical Informatics Association. 1996;3:367–388. doi: 10.1136/jamia.1996.97084511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neches R, Fikes RE, Finin T, Gruber TR, Patil R, Senator T, Swartout W. Enabling technology for knowledge sharing. AI Magazine. 1991;12(3):36–56. [Google Scholar]
- Noy NF, Musen MA. The PROMPT suite: Interactive tools for ontology merging and mapping. International Journal of Human–Computer Studies. 2003;59(6):983–1024. [Google Scholar]
- Noy NF, Sintek M, Decker S, Crubézy M, Fergerson RW, Musen MA. Creating Semantic Web contents with Protégé-2000. IEEE Intelligent Systems. 2001;16(2):60–71. [Google Scholar]
- Noy NF, de Coronado S, Solbrig H, Fragoso G, Hartel FW, Musen MA. Representing the NCI Thesaurus in OWL DL: Modeling tools help modeling languages. Applied Ontology. 2008;3(3):173–190. doi: 10.3233/AO-2008-0051. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh PP, Zheng J, Logan-Klumpler F, Stoeckert CJ, Jr, Louis C, Topalis P, Protasio AV, Sheth AP, Carrington M, Berriman M, Sahoo SS. The Ontology for Parasite Lifecycle (OPL): towards a consistent vocabulary for lifecycle stages in parasitic organisms. Journal of Biomedical Semantics. 2012;3:5. doi: 10.1186/2041-1480-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Akkermans H, Anjewierden A, de Hoog R, Shadbolt N, Van de Velde W, Wielinga B. Knowledge Engineering and Management: The CommonKADS Methodology. Cambridge, MA: MIT Press; 2000. [Google Scholar]
- Stein A, Musen MA, Shahar Y. Proceedings of the AMIA Fall Symposium. Washington, DC: 1996. Knowledge acquisition for temporal abstraction; pp. 204–208. [PMC free article] [PubMed] [Google Scholar]
- Storey M-A, Musen MA, Silva J, Best C, Ernst N, Fergerson R, Noy N. First International Conference on Knowledge Capture. Victoria, British Columbia, Canada: 2001. Jambalaya: Interactive visualization to enhance ontology authoring and knowledge acquisition in Protégé. In: Workshop on Interactive Tools for Knowledge Capture; pp. 35–43. [Google Scholar]
- Strohmaier M, Walk S, Pöschko J, Lamprecht D, Tudorache T, Nyulas C, Musen MA, Noy NF. How ontologies are made: Studying the hidden social dynamics behind collaborative ontology-engineering projects. Journal of Web Semantics. 2013;20:18–34. doi: 10.1016/j.websem.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SW, Kahn MG, Musen MA, Ferguson JC, Shortliffe EH, Fagan LM. Episodic skeletal-plan refinement based on temporal data. Communications of the Association for Computing Machinery. 1989;32:1439–1455. [Google Scholar]
- Tudorache T, Falconer SM, Nyulas CI, Noy NF, Musen MA. Ninth International Semantic Web Conference. Shanghai, China: 2010. Will Semantic Web technologies work for the development of ICD-11? [Google Scholar]
- Tudorache T, Musen MA. Collaborative development of large-scale biomedical ontologies. In: Ekins S, Hupcey MAZ, Williams AJ, editors. Collaborative Computational Technologies for Biomedical Research. Hoboken, NJ: John Wiley & Sons; 2011. [Google Scholar]
- Zhang Y, Tudorache T, Horridge M, Musen MA. Computer–Human Interaction. Seoul, Korea: 2015. Helping users bootstrap ontologies: an empirical investigation. [Google Scholar]