Abstract
Wilms’ Tumor Gene 1 (WT1) is an attractive target antigen for cancer immunotherapy because it is overexpressed in many hematologic malignancies and solid tumors but has limited, low-level expression in normal adult tissues. Multiple HLA class I and class II restricted epitopes have been identified in WT1, and multiple investigators are pursuing the treatment of cancer patients with WT1 based vaccines and adoptively transferred WT1 reactive T cells. Here we isolated an HLA-A*0201 restricted WT1 reactive T cell receptor (TCR) by stimulating PBL of healthy donors with the peptide WT1:126-134 in vitro. This TCR mediated peptide recognition down to a concentration of ~0.1 ng/ml when pulsed onto T2 cells as well as recognition of HLA-A*0201+ target cells transfected with full-length WT1 cDNA. However, it did not mediate consistent recognition of many HLA-A*0201+ tumor cell lines or freshly isolated leukemia cells that endogeneously expressed WT1. We dissected this pattern of recognition further and observed that WT1:126-134 was more efficiently processed by immunoproteasomes compared to standard proteasomes. However, pretreatment of WT1+ tumor cell lines with Interferon gamma (IFNγ) did not appreciably enhance recognition by our TCR. In addition, we highly overexpressed WT1 in several leukemia cell lines by electroporation with full-length WT1 cDNA. Some of these lines were still not recognized by our TCR suggesting possible antigen processing defects in some leukemias. These results suggest WT1:126-134 may not be a suitable target for T cell based tumor immunotherapies.
Introduction
The adoptive transfer of melanoma reactive tumor infiltrating T lymphocytes (TIL) can mediate cancer regression in approximately 50% of patients with metastatic melanoma (1). In addition, the adoptive transfer of normal peripheral lymphocytes genetically modified by the insertion of tumor reactive T cell receptors (TCRs) or chimeric antigen receptors (CARs) can mediate in vivo tumor regression in multiple histologies (2-8). However choosing a tumor specific antigenic target is critical because adoptively transferred T cells reactive with epitopes presented on normal tissues even at very low levels can induce severe toxicities (5, 9, 10).
Wilms’ Tumor Gene 1 (WT1) encodes a zinc finger transcription factor critical for cell growth and differentiation (11). WT1 is highly expressed in the majority of acute myeloid leukemias (AML) and acute lymphoid leukemias (ALL) and has been reported to be expressed in a variety of solid cancers including tumors of the lung, breast, digestive organs, brain, head and neck, thyroid, and female genital tract (12). Although expression of WT1 is critical during embryogenesis, its expression in normal adult tissues appears to be limited primarily to renal glomerular podocytes and CD34+ hematopoetic stem cells (13, 14).
Multiple HLA class I and class II restricted T cell epitopes in WT1 have been studied (15-21), and many of these have been associated with specific recognition by reactive T cells of a few WT1+ tumors, most frequently leukemias. However, only a few of these investigations reported broad recognition of large panels of WT1+ tumor cells expressing the relevant HLA molecule. Based on the identification of these epitopes, multiple clinical trials have been conducted in which patients with WT1+ tumors were vaccinated with peptides or dendritic cells (DCs) electroporated with WT1 mRNA (12, 22). Although some antitumor responses were reported in these trials, the majority of patients did not benefit clinically. As an alternate approach, Chapuis et al. reported the findings of a clinical trial in which patients with high-risk leukemias were treated with adoptively transferred allogeneic WT1 reactive T cell clones. The authors reported long-term persistence of the clones in the peripheral blood of patients. Transient responses were observed in 2 of 11 patients, and stable disease was noted in 3 others (23). More recently, this group isolated a high avidity HLA-A*0201 restricted TCR specifically reactive with WT1:126-134 and is currently conducting a clinical trial in which patients with high risk or relapsed AML, MDS, or CML are being treated with adoptively transferred T cells genetically modified to express this TCR (Clinicaltrials.gov ID# NCT01640301).
Despite low level expression of WT1 in some normal adult tissues including kidney podocytes and CD34+ hematopoetic stem cells, no toxicities associated with targeting WT1 on normal cells have been reported in the above-referenced trials. Some reports have suggested that WT1 reactive T cells can distinguish between tumor cells that overexpress WT1 and normal tissues with lower levels of expression. In particular, Gao et al. reported that WT1 reactive T cells specifically lysed leukemia cell lines and inhibited colony formation by transformed CD34+ progenitor cells from patients with CML, whereas colony formation by normal CD34+ progenitor cells was unaffected (24). In addition, using a murine model, Asai et al. reported that although WT1-specific T cells lysed kidney podocytes in vitro, they did not induce renal damage when adoptively transferred in vivo (25).
Based on these studies suggesting WT1 may be a promising tumor associated antigen, we isolated a high avidity HLA-A*0201 restricted WT1 reactive TCR for potential use in a gene therapy clinical trial. This high avidity TCR mediated recognition of sub-nanomolar levels of peptide pulsed on T2 cells as well as HLA-A*0201+ target cells transfected with full length WT1 cDNA. However, this TCR did not mediate consistent recognition of multiple HLA-A*0201+ WT1+ tumor cell lines or fresh leukemias, raising doubts concerning this epitope as a suitable immunotherapeutic target.
Materials and Methods
Cell lines, primary human cell cultures, and peptides
A comprehensive list of cell lines used as target cells in the experiments described here is provided in Supplementary Table 1. Cell lines were either purchased from ATCC or were kindly provided by Dr. Terry Fry (NIH/NCI). As part of the studies presented here, all cell lines were evaluated by FACS for expression of HLA-A*0201 and by quantitative RT-PCR for expression of WT1. COS-7 and 293 cells stably expressing HLA-A*0201 (COS-A2 and 293-A2 respectively) were generated previously by retroviral transduction with a vector encoding this protein. COS-A2 and 293-A2 cells transiently expressing full-length WT1, MART-1, or GFP were generated by transfection with mammalian expression vectors (either pCDNA3.1 or pCMV6-Entry) encoding these proteins using Lipofectamine 2000 (Invitrogen). 293 cells expressing standard proteasomal subunits or inducible subunits 1, 2, and 5 of the immunoproteasome were kindly provided by Dr. Benoit Van den Eynde (26). These cells transiently expressing HLA-A*0201 and full-length WT1 or MART-1 were generated by transfection with mammalian expression vectors (either pCDNA3.1 or pCMV6-Entry) encoding these proteins using Lipofectamine 2000 (Invitrogen). For some experiments, tumor cell lines were electroporated with full-length WT1, MART-1, or GFP cDNAs using an Amaxa® system in Nucleofector® kit V solution (Maxcyte) to overexpress these proteins. For some experiments, tumor cell lines were treated for 2 days with 200 IU/ml IFNγ prior to use as target cells.
Primary human leukemia cells from 4 patients were kindly provided by Dr. Terry Fry. These samples were bone marrow aspirates from patients enrolled in a WT1 vaccine trial collected prior to treatment (Clinicaltrials.gov identifier NCT00923910). 3 were from patients with ALL and 1 was from a patient with AML. All samples contained >60% leukemic cells, and all were previously confirmed to express HLA-A*0201 and WT1 as a requirement for inclusion in the vaccine trial.
Peptides were purchased from commercial sources (Peptide 2.0, Peproteomics, and Pennisula Laboratories), and molecular weights were verified by laser desorption mass spectrometry.
Isolation of WT1 reactive T cells from PBL of healthy donors
CD8+ T lymphocytes from 5 healthy HLA-A*0201+ donors were stimulated with the WT1:126-134 peptide (RMFPNAPYL) using a previously reported protocol (23). Briefly, short-term dendritic cell (DC) cultures were generated from PBMCs by stimulating cells that adhered to tissue culture treated flasks with 800 IU/ml GM-CSF (Leukine®/sargramostin; Sanofi-Aventis U.S. LLC) and 1000 IU/ml IL-4 (R&D Systems) in Cellgenix DC media (Cellgenix) supplemented with 1% heat inactivated human AB serum. The next day, the following maturation cytokines were added to the DC cultures: 10 ng/ml TNFα (R&D Systems), 10 ng/ml IL-1β (R&D Systems), 10 ng/ml IL-6 (R&D Systems), and 1 μg/ml PGE2 (Sigma). The following day, the DCs were harvested, pulsed with 2.5 μg/ml peptide for 2-4 hours at 37 C, irradiated (4000 rads), and resuspended in RPMI 1640 supplemented with 10% heat inactivated human AB serum, 2 mM L-glutamine, 50 units/ml penicillin, and 50 μg/mL streptomycin (CTL media). On the same day, autologous CD8+ T lymphocytes were isolated from whole PBMCs using anti-CD8 coated paramagnetic beads (Miltenyi Biotech). CD8+ T cells were stimulated with peptide-pulsed DCs in 24 well plates (~5e6 CD8+ T cells and ~1e6 DCs in 2 ml per well) in CTL media containing 30 ng/ml IL-21 (Peprotech). Two days later, the following cytokines were added to the cultures: 5 ng/ml IL-7 (Peprotech), 12.5 IU/ml IL-2 (Chiron), and 1 ng/ml IL-15 (Peprotech). Cultures in individual wells were maintained separately and were fed and split as necessary with media containing IL-2, IL-7, and IL-15. Two and four weeks after the initial stimulation, cultures were restimulated with autologous peptide-pulsed (2.5 μg/ml) irradiated (4000 rads) PBMCs in 12 well plates (~2e6 responder cells and ~10e6 peptid-pulsed PBMCs in 4 ml per well) in CTL media containing 30 ng/ml IL-21 (Peprotech). Two days after each restimulation, IL-2, IL-7, and IL-15 were added, and cultures in individual wells were maintained separately and were fed and split as necessary with media containing IL-2, IL-7, and IL-15. ~10 days after each stimulation, cultures were evaluated for recognition of the WT1 peptide and/or target cells expressing HLA-A*0201 and the full length WT1 protein in IFNγ secretion assays (described in more detail later in the Material and Methods section).
After two in vitro stimulations, one culture that demonstrated specific recognition of T2 cells loaded with the WT1 peptide and 293-A2 cells transfected with full-length WT1 cDNA was cloned by limiting dilution. Briefly, 10 96-well U bottom plates were set up in which each well contained 2 T cells, 1e5 irradiated allogeneic PBMCs (as feeder cells), 30 ng/ml OKT3 (Miltenyi Biotech), and 600 IU/ml IL-2 in 200 μl CTL media. Growth-positive wells were selected ~2 weeks and were further expanded in the presence of allogeneic feeder cells, OKT3, and IL2. The resulting limiting dilution cultures were evaluated for recognition of the WT1 peptide and target cells expressing HLA-A*0201 and the full length WT1 protein by means of specific IFNγ secretion.
TCR cloning and construction of a retroviral vector encoding TCR α and β chains
For 10 individual limiting dilution T cell cultures that demonstrated recognition of WT1 peptide and 293-A2 cells transfected with full-length WT1 cDNA, total RNA was isolated using RNeasy mini kits (Qiagen). TCR α and β chains were then identified using 5’-rapid amplification of cDNA ends (RACE)-PCR. 5’- RACE-ready first-strand cDNA was prepared from total RNA using a SMARTer-RACE cDNA amplification kit (Clontech). 3’-truncated TCR α and β chain fragments were then amplified by PCR (Advantage 2 PCR kit; Clontech) using several primers within α and β chain constant regions. Gel-purified PCR products were then sequenced (Macrogen). DNA sequences were analyzed for the presence of specific human TCR α and β chain sequences using the International Immunogenetics Information Systems Web site. A retroviral vector encoding codon-optimized TCR α and β chains from the dominant TCR clonotype was constructed using an MSGV1 backbone by Gene Oracle. In this vector, the variable and complementarity determining regions of the human TCR α and β chains were linked to murine constant regions to improve expression on the surfaces of human T cells (27). The vector encoded both the α and β chains separated by a picornavirus ribosomal skip element. Retroviral supernatants were generated by cotransfecting 293 cells that stably expressed MMLV gag and pol proteins with the MSGV1 TCR vector and a vector encoding the RD114 feline endogenous virus retroviral envelope protein using Lipofectamine 2000 (Invitrogen). Supernatants were collected 2 and 3 days post-transfection and were diluted 1:1 with fresh DMEM containing 10% FCS.
Genetic modification of T lymphocytes using retroviral transduction
MSGV1 retroviral particles encoding TCR α and β chains produced as described above were used to transduce PBMCs from 3 HLA-A*0201+ donors twice on days 2 and 3 after stimulation with 50 ng/ml OKT3 and 300 IU/mL rhIL-2 in media comprised of 50% AIMV/50% CM (50/50 media). Nontissue culture-treated plates were coated overnight with Retronectin (Takara Bio) at 4 C (10 μg/ml; 2 ml/6-well plate well). Retronectin-coated plates were subsequently blocked with PBS containing 2% bovine serum albumin for 30 min at room temperature and were then washed with PBS. After aspirating the wash buffer, the diluted retroviral supernatants described above were added to the Retronectin-coated plates (4 ml/6-well plate well). The plates were then centrifuged at 2,000×g for 2 h at 32 C. During the centrifugation, 2-day stimulated PBMCs were harvested and resuspended at 5e5 cells/ml in 50/50 media containing 300 IU/mL rhIL-2. After spin-loading the plates, approximately half of the retroviral supernatant was aspirated, and PBMCs were added to the wells (4 ml or 2e6 cells/6-well plate well). The cells were then centrifuged at 1,000×g for 10 min and then incubated overnight at 37 C. Transductions were repeated on day 3 post-OKT3 stimulation using the same protocol. Four to 24 hours after the second transduction, T cells were harvested and resuspended in fresh medium containing 300 IU/mL rhIL-2 and allowed to expand in vitro. One to 4 weeks after transduction, TCR expression was evaluated by FACS, and T cell function was evaluated by measuring IFNγ secretion and/or 41BB expression in response to appropriate target cells.
Evaluation of WT1 expression in tumors by qRT-PCR
RNA from tumor cell lines was isolated using RNeasy mini kits (Qiagen) and was reverse transcribed into cDNA using “High-capacity RNA-to-cDNA” kits (Applied Biosystems). Copies of WT1 RNA transcripts were quantified in triplicate samples normalized to copies of GAPDH RNA transcripts using a TaqMan® Gene Expression Assay (HS01103751; Applied Biosystems).
IFNγ secretion, 41BB expression, and cytolysis assays
Recognition of WT1 by human T cell populations was evaluated based on specific IFNγ secretion or 41BB expression in response to T2 cells preincubated with peptide, HLA-A2+ WT1+ tumor cell lines, and 293 or COS-7 cells expressing HLA-A*0201 and WT1 using overnight coculture assays. In some experiments, tumor cells were pretreated with IFNγ (200 U/mL) 2 days prior to the assay or were electroporated with WT1 cDNA 1 day before the assay. Responder T cells (1e5) were coincubated with stimulator cells (0.5-1e5; 200 μl per 96 well plate well) ~20 h at 37 C, and the concentration of human IFNγ in coculture supernatants was measured by ELISA using commercially available reagents (Thermo Scientific). In some experiments, 41BB expression on responding T cells was measured by FACS as described below.
For some IFNγ treated tumor cell lines, cytolysis by T cells expressing a WT1 reactive TCR was measured using a commercially available LDH Cytotoxicity Assay (Pierce Biotechnology) according to the manufacturer's instructions. Responder T cells (1e5) were coincubated with stimulator cells at a 10:1 effector to target ratio (E:T) (0.1e5; 100 μl per 96 well plate well) ~4 h at 37 C, and the concentration of LDH in coculture supernatants was measured. The percent specific cytotoxicity was calculated as follows:
FACS analyses
Tumor cell lines were characterized for expression of HLA-A2 using a FITC-conjugated monoclonal antibody against HLA-A2 (One Lambda). T cells that were transduced with the retroviral vector encoding human variable / murine constant region TCR α and β chains were characterized for expression of CD4, CD8, and TCR using a monoclonal antibody specifically reactive with the constant region of the murine β chain (eBiosciences) and antibodies against human CD4 or CD8 (BD Biosciences). For some experiments, 41BB expression on T cells was evaluated by FACS ~24 hours after stimulation with target cells using a monoclonal anti-CD137 antibody (Miltenyi). FACS analyses were performed on a FACSCalibur®, Fortessa®, or Canto® flow cytometer (BD Biosciences), and data were analyzed using FlowJo software (Tree Star).
Results
Generation of WT1 reactive T cell clones from PBL of healthy HLA-A*0201+ donors
To generate WT1 reactive T lymphocytes, we followed a previously reported protocol in which CD8+ PBL from healthy donors were stimulated in vitro with autologous DCs pulsed with the WT1:126-134 peptide (23). Cultures were maintained individually and were restimulated twice with peptide pulsed autologous PBMCs. From each of 5 healthy donors, we generated bulk T cell populations that specifically recognized the WT1 peptide loaded onto T2 cells (Supplementary Table 2). However, only two cultures from healthy donor 4 and one culture from healthy donor 5 specifically recognized 293-A2 cells that had been transiently transfected with full-length WT1 cDNA suggesting these cultures could recognize processed antigen (Supplementary Table 2).
The bulk culture from healthy donor 4 with the most pronounced recognition of 293-A2 cells transfected with WT1 (culture #6 in Supplementary Table 2) underwent limiting dilution cloning, and 130 growth-positive wells were screened for recognition of peptide and transfectants. 108 of those demonstrated specific peptide and transfectant reactivities, and data from 10 of those are presented in Table 1.
Table 1.
IFNγ secretion (pg/ml) by T cell clones from healthy donor H4a
| Clone ID | T2 + HBV peptideb | T2 + WT1 peptideb | 293-A2-GFPc | 293-A2-WT1c |
|---|---|---|---|---|
| P1A10 | 56 | >2000d | 42 | >2000 |
| P1G11e | 67 | >2000 | 53 | 177 |
| P2H11 | 48 | >2000 | 59 | >2000 |
| P5A7 | 24 | >2000 | 40 | 370 |
| P5D2 | 35 | >2000 | 56 | >2000 |
| P5D4 | 34 | >2000 | 23 | >2000 |
| P6H3 | 40 | >2000 | 0 | >2000 |
| P8A6 | 20 | >2000 | 17 | >2000 |
| P8B5 | 35 | >2000 | 3 | >2000 |
| P9G11 | 29 | >2000 | 29 | >2000 |
CD8+ PBL from healthy donor H4 were stimulated in vitro 2 times with the WT1:126-134 peptide as described in the Material and Methods section. Bulk culture #6 was cloned by limiting dilution, and 130 growth-positive wells were screened for recognition of peptide and transfectants. 108 of those demonstrated specific peptide and transfectant reactivities, and data from 10 of those are presented here. IFNγ in coculture supernatants was measured by ELISA.
T2 cells were pre-loaded with 1 μg/ml of a control peptide from HBV with high binding affinity for HLA-A*0201 or WT1:126-134 ~1 hour prior to the coculture.
293 cells stably expressing high levels of HLA-A*0201 by means of retroviral transduction and antibiotic selection were transiently transfected with GFP cDNA as a negative control or full-length WT1 cDNA ~24 hours prior to the coculture.
underlined values indicate IFNγ secretion >100 pg/ml and >2X background with any negative control target cell.
TCR sequencing of cDNA from all clones except P1G11 indicated the presence of a single α chain (TRAV12-1*01; CDR3: CVVNTPPNTDKLIF) and a single β chain (TRBV7-2*01; CDR3: CASTPFTSGSGWDEQFF). Clone P1G11 contained a distinct TCR α chain (TRAV6*02; CDR3: CAFSGCARQLTF) and β chain (TRBV10-3*01; CDR3: CAISESMASGDNNEQFF).
Identification, cloning, and function of a WT1-reactive TCR in human T cells
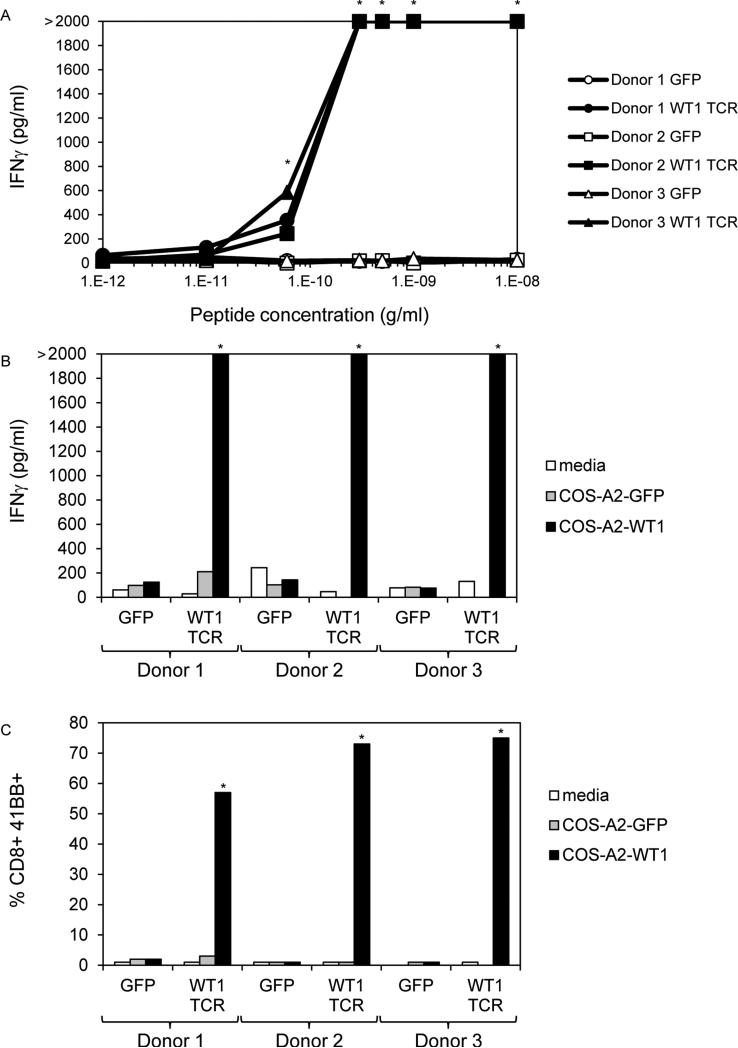
For the 10 limiting dilution cultures presented in Table 1, we attempted to identify TCRs by 5’ RACE using α and β chain constant region primers. TCR sequencing of cDNA from all clones except P1G11 indicated the presence of the same single α chain (TRAV12-1*01; CDR3: CVVNTPPNTDKLIF) and the same single β chain (TRBV7-2*01; CDR3: CASTPFTSGSGWDEQFF). Clone P1G11 contained a distinct TCR α chain (TRAV6*02; CDR3: CAFSGCARQLTF) and β chain (TRBV10-3*01; CDR3: CAISESMASGDNNEQFF). We constructed a retroviral vector encoding the dominant α and β chains separated by a picornavirus ribosomal skip element. In this vector, the α and β chain variable and complementarity determining regions were grafted onto murine constant regions to allow for more efficient pairing and more stable expression on the surface of T cells genetically modified to express this TCR (27). This retroviral vector was used to transduce PBL from 3 HLA-A*0201+ allogeneic donors. By evaluating the expression of the murine TCR β chain on the surfaces of transduced cells by FACS, both CD8+ and CD4+ T cells expressed the TCR, and the overall transduction efficiencies in PBL from Donors 1, 2, and 3, were 81%, 89%, and 94% respectively. The transduced T cells were then tested for recognition of titrated amounts of peptide pulsed on T2 cells as evaluated by IFNγ secretion. The WT1 TCR mediated recognition of a peptide concentration as low as ~0.1 ng/ml (Figure 1A). We also evaluated recognition of COS-A2 cells transiently transfected with full-length cDNA, and the WT1 TCR consistently mediated recognition of these target cells indicating it could recognize a peptide that had been processed and presented in the context of HLA-A*0201 (Figure 1B). In addition to measuring IFNγ secretion, we measured the ability of the transduced T cells to upregulate the costimulatory molecule CD137 (41BB) in response to transiently transfected COS-A2 cells (Figure 1C). After overnight stimulation with COS-A2 cells expressing WT1, the expression of 41BB on CD8+ T cells from Donors 1, 2, and 3, were 73%, 75%, and 57% respectively indicating the majority of the α and β chains of the transduced TCRs paired correctly and were functional.
Figure 1.
Recognition of WT1 peptide and transfectants by PBL retrovirally transduced to express a WT1 reactive TCR. PBL from 3 HLA-A*0201+ donors were transduced with retroviral particles encoding GFP as a negative control or with the WT1 reactive TCR (TRAV12-1*01; CDR3: CVVNTPPNTDKLIF; TRBV7-2*01; CDR3: CASTPFTSGSGWDEQFF). A. Transduced T cells were cocultured overnight with T2 cells pre-pulsed with titrated concentrations of peptide, and IFNγ in coculture supernatants was measured by ELISA. B and C. COS-7 cells stably expressing high levels of HLA-A*0201 by means of retroviral transduction and antibiotic selection were transiently transfected with GFP cDNA as a negative control or full-length WT1 cDNA. The next day, transduced T cells were cocultured overnight with these transfected cells. IFNγ in coculture supernatants was measured by ELISA (B), and 41BB expression on CD8+ T cells was measured by FACS (C). * indicates >100 pg/ml IFNγ secretion or >5% CD8+ 41BB+ cells by WT1 TCR transduced T cells, >2X background with any negative control target cell, and >2X background of GFP transduced T cells with the same target cell. Bars that reach 2000 pg/ml indicate off-scale IFNγ levels > 2000 pg/ml.
Lack of consistent recognition of WT1+ tumor cell lines and primary leukemias by T cells expressing the WT1-reactive TCR
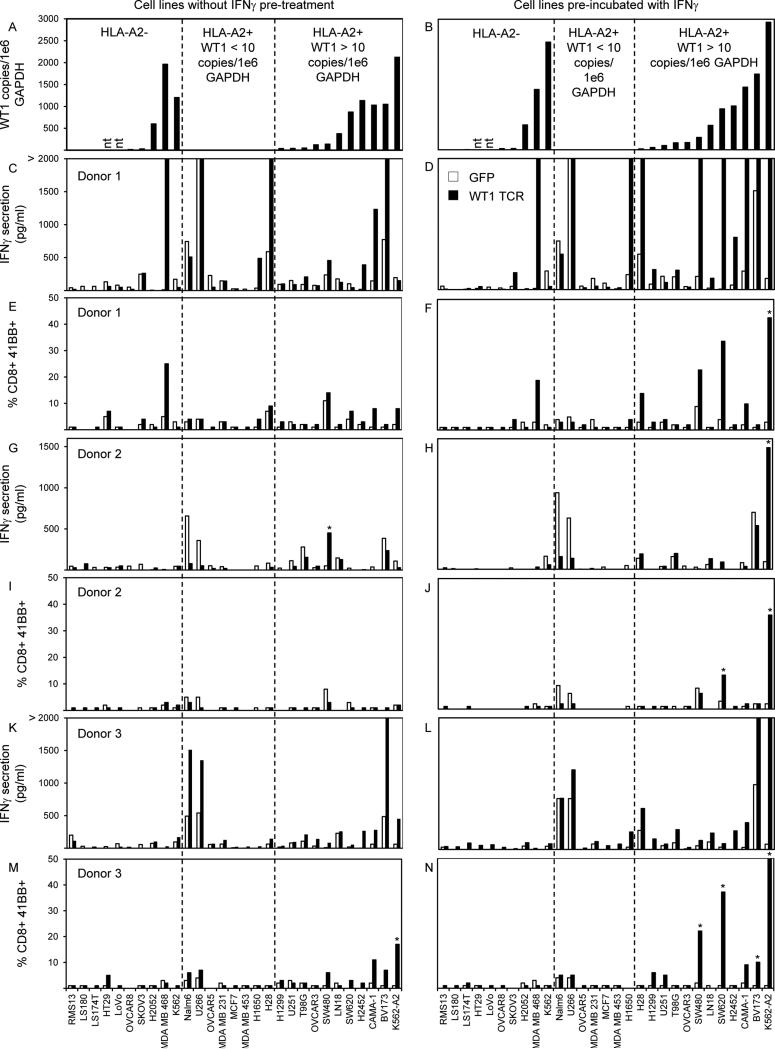
To determine if the WT1 TCR we isolated could mediate recognition of naturally occurring WT1 peptide, we evaluated recognition of multiple HLA-A*0201+ WT1+ tumor cell lines in comparison to HLA-A*0201- and/or WT1- cell lines (Figure 2). WT1 expression in tumor cell lines was quantified as the number of WT1 RNA copies relative to the housekeeping gene GAPDH measured by quantitative RT-PCR (Figure 2A), and HLA-A2 expression was measured by FACS (data not shown). Recognition of target cell lines was evaluated by means of IFNγ secretion and 41BB expression after overnight coculture with WT1 TCR transduced T cells (Figure 2C, E, G, I, K, and M). Recognition of HLA-A*0201+ WT1+ cell lines was considered positive and specific if IFNγ secretion was >100 pg/ml or >5% CD8+ cells expressed 41BB cells and if these levels were >2X background compared to any negative control target cell and >2X background of GFP transduced T cells cultured with the same target cell. Using these criteria, there appeared to be some specific recognition of the SW480 colon cancer cell line by T cells from donor 2 based on IFNγ secretion and of the K562 leukemia cell line genetically modified to express HLA-A*0201 by 41BB expression in donor 3. However, these reactivities were not consistent among the 3 donors, and there was no consistent recognition of multiple HLA-A*0201+ WT1+ tumor cells in any of the 3 donors. T cells from donor 1 secreted high background levels of IFNγ in response to the HLA-A*0201- tumor cell line MDA MB 468 and to the HLA-A*0201+ WT1- tumor cell lines U266 and HT28 (Figure 2C). The cause of this background was unclear, but it was not likely due to class I MHC alloreactivity because MDA MB 468 and U266 do not share any class I MHC molecules. Class II MHC alloreactivity in this patient could not be discounted. Nonetheless, the specificity of the TCR for WT1 in donor 1 was clearly demonstrated in Figure 1 B and C in which COS-A2 cells expressing WT1 were specifically recognized in comparison to COS-A2 cells expressing GFP because the only difference between these 2 target cells was the expression of WT1.
Figure 2.
WT1 RNA expression in tumor cell lines and recognition by PBL retrovirally transduced to express a WT1 reactive TCR. Quantitative RT-PCRs for WT1 and GAPDH were conducted on RNAs isolated from tumor cell lines with (B) and without (A) 48 hour pre-treatment with 200 U/ml IFNγ. WT1 RNA copies were divided by GAPDH copies and expressed as WT1 copy number per 1e6 GAPDH copy number. PBL from 3 HLA-A*0201+ donors were transduced with retroviral particles encoding GFP (empty bars) as a negative control or with the WT1 reactive TCR (TRAV12-1*01; CDR3: CVVNTPPNTDKLIF; TRBV7-2*01; CDR3: CASTPFTSGSGWDEQFF) (black bars). Transduced T cells were cocultured overnight with tumor cell lines pre-treated with (D, F, H, J, L, N) or without (C, E, G, I, K, M) IFNγ. IFNγ in coculture supernatants was measured by ELISA (C, D, G, H, K, L), and 41BB expression on CD8+ T cells was measured by FACS (E, F, I, J, M, N). * indicates >100 pg/ml IFNγ secretion or >5% CD8+ 41BB+ cells by WT1 TCR transduced T cells, >2X background with any negative control target cell, and >2X background of GFP transduced T cells with the same target cell. Bars that reach 2000 pg/ml indicate off-scale IFNγ levels > 2000 pg/ml.
It has previously been reported that standard proteasomes process antigens differently than immunoproteasomes and that some epitopes are preferentially processed and presented more efficiently by cells expressing one or the other (26). For example, the HLA-A*0201 restricted epitope from the MART-1 melanoma antigen (MART-1:27-35) was reported to be more efficiently processed by cells expressing standard proteasomes, whereas the HLA-B40 restricted epitope from the cancer testis antigen MAGE-A3 (MAGE-A3:114-122) has previously been reported to be more efficiently processed by cells expressing immunoproteasomes (28). To determine if the presence of standard proteasomes or immunoproteasomes might influence the processing and presentation of the WT1:126-134 epitope, we evaluated recognition of 293 cells expressing either standard proteasomes or immunoproteasomes after transient transfection with HLA-A*0201 and full-length WT1 cDNA by T cells expressing the WT1 TCR (Table 2). As a control, we also evaluated recognition of MART-1:27-35 by a MART-1 reactive TIL (DMF5). As previously reported, the DMF5 cells recognized MART-1 more efficiently in 293 cells expressing standard proteasomes in comparison to those containing immunoproteasomes (Table 2). In contrast, T cells from all 3 donors transduced with the WT1 TCR specifically recognized WT1 more efficiently in 293 cells expressing immunoproteasomes in comparison to those containing standard proteasomes.
Table 2.
Recognition of WT1 peptide and transfectants by PBL retrovirally transduced to express a WT1 reactive TCRa
| Donor 1 |
Donor 2 |
Donor 3 |
|||||
|---|---|---|---|---|---|---|---|
| DMF5b | GFP | WT1 TCR | GFP | WT1 TCR | GFP | WT1 TCR | |
| media | 28 | 30 | 29 | 28 | 28 | 30 | 30 |
| T2 + MART peptidec | 9980g | 37 | 37 | 34 | 29 | 52 | 40 |
| T2 + WT1 peptidec | 56 | 35 | >20000 | 32 | >20000 | 45 | >20000 |
| COS-A2 + MARTd | 4710 | 31 | 37 | 36 | 33 | 83 | 43 |
| COS-A2 + WT1d | 27 | 31 | 11449 | 34 | 8482 | 77 | 7254 |
| 293 SP + A2/MARTe | 872 | 29 | 28 | 28 | 28 | 29 | 29 |
| 293 SP + A2/WT1e | 28 | 29 | 52 | 28 | 66 | 31 | 61 |
| 293 IP + A2/MARTf | 79 | 29 | 29 | 33 | 30 | 34 | 41 |
| 293 IP + A2/WT1f | 28 | 29 | 1963 | 32 | 3606 | 36 | 1947 |
PBL from 3 HLA-A*0201+ donors were transduced with retroviral particles encoding GFP as a negative control or with the WT1 reactive TCR (TRAV12-1*01; CDR3: CVVNTPPNTDKLIF; TRBV7-2*01; CDR3: CASTPFTSGSGWDEQFF). Transduced T cells were cocultured overnight with the indicated target cells, and IFNγ in coculture supernatants was measured by ELISA.
DMF5 is a MART-1 reactive T cell population included as a positive control for the transfections.
T2 cells were pre-loaded with 1 μg/ml of a control peptide from HBV with high binding affinity for HLA-A*0201 or WT1:126-134 ~1 hour prior to the coculture.
COS-7 cells stably expressing high levels of HLA-A*0201 by means of retroviral transduction and antibiotic selection were transiently transfected with GFP cDNA as a negative control or full-length WT1 cDNA ~24 hours prior to the coculture.
293 cells expressing components of the standard proteasome were transiently co-transfected with HLA-A*0201 cDNA and either full-length MART-1 or WT1 cDNA ~24 hours prior to the coculture.
293 cells expressing inducible subunits 1, 2, and 5 of the immunoproteasome were transiently co-transfected with HLA-A*0201 cDNA and either full-length MART-1 or WT1 cDNA ~24 hours prior to the coculture.
underlined values indicate IFNγ secretion >100 pg/ml and >2X background with any negative control target cell.
It has previously been reported that standard proteasomes can convert to immunoproteasomes in cell lines treated with IFNγ (29). To determine if the presence of immunoproteasomes in HLA-A*0201+ WT1+ cell lines might enhance recognition by T cells expressing the WT1 TCR, we pretreated tumor cells with IFNγ prior to using them as targets in coculture assays (Figure 2B, D, F, H, J, L, and N). WT1 expression in IFNγ treated tumor cell lines was quantified as the number of WT1 RNA copies relative to the house keeping gene GAPDH measured by quantitative RT-PCR, and HLA-A2 expression was measured by FACS (Figure 2B). No significant differences in WT1 expression were observed in IFNγ treated cells compared to untreated cells (Figure 2A vs. 2B). However, expression of HLA-A2 on the surfaces of most tumor cells was enhanced by pre-treatment with IFNγ (data not shown). Recognition of IFNγ treated target cell lines was evaluated by means of IFNγ secretion and 41BB expression after overnight coculture with WT1 TCR transduced T cells (Figure 2D, F, H, J, L, and N). Recognition of HLA-A*0201+ WT1+ cell lines was considered positive and specific if IFNγ secretion was >100 pg/ml or >5% CD8+ cells expressed 41BB cells and if these levels were >2X background with any negative control target cell and >2X background of GFP transduced T cells with the same target cell. Using these criteria, there appeared to be some specific recognition of the K562 leukemia cell line genetically modified to express HLA-A*0201 assessed by either IFNγ secretion or 41BB expression from all 3 donors. In addition, there appeared to be some irregular reactivities against SW480, SW620, and BV173 by T cells from donors 2 and 3. However, these reactivities were not consistent among the 3 donors, and there was no consistent recognition of multiple HLA-A*0201+ WT1+ tumor cells in any of the 3 donors.
We have previously identified T cell populations that specifically lysed tumor cell lines in vitro that did not secrete IFNγ in response to the same target cells (30). Therefore, in addition to evaluating IFNγ secretion and 41BB expression, we measured the cytolytic capacity of T cells expressing the WT1 reactive TCR using a commercially available LDH cytotoxicity assay. T cells from all three donors specifically lysed T2 cells pulsed with the WT1 peptide and COS-7 cells expressing HLA-A*0201 transfected with full length WT1 cDNA. However, none of these T cell populations specifically lysed any of the HLA-A*0201+ WT1+ tumor cell lines tested (Supplementary Figure 1).
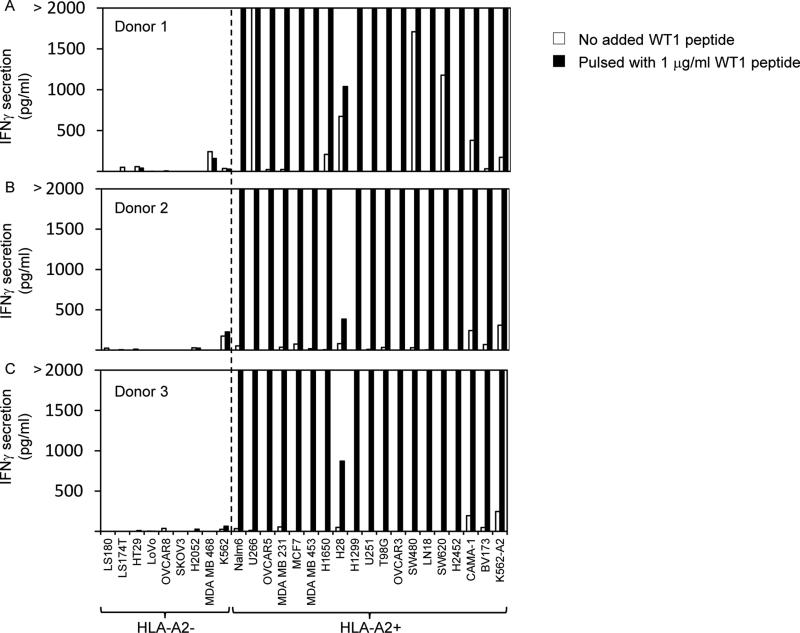
HLA-A2 expression on tumor cell lines was evaluated by FACS; however, to determine if the levels of HLA-A*0201 on the cell surfaces were sufficient to enable recognition by T cells expressing the WT1 TCR, we pulsed the tumor cell lines with the WT1:126-134 peptide and evaluated recognition (Figure 3). All of the HLA-A*0201+ tumor cell lines pre-pulsed with the WT1:126-134 peptide were specifically recognized by at least 2 of the 3 donors, whereas none of the HLA-A*0201- tumor cell lines pre-pulsed with this peptide were specifically recognized.
Figure 3.
Recognition of WT1 peptide-pulsed tumor cell lines by PBL retrovirally transduced to express a WT1 reactive TCR. PBL from 3 HLA-A*0201+ donors were transduced with retroviral particles encoding GFP as a negative control or with the WT1 reactive TCR (TRAV12-1*01; CDR3: CVVNTPPNTDKLIF; TRBV7-2*01; CDR3: CASTPFTSGSGWDEQFF). A-C. Transduced T cells were cocultured overnight with tumor cell lines pre-pulsed with 1 μg/ml of WT1 peptide or in the absence of exogenously pulsed peptide, and IFNγ in coculture supernatants was measured by ELISA.
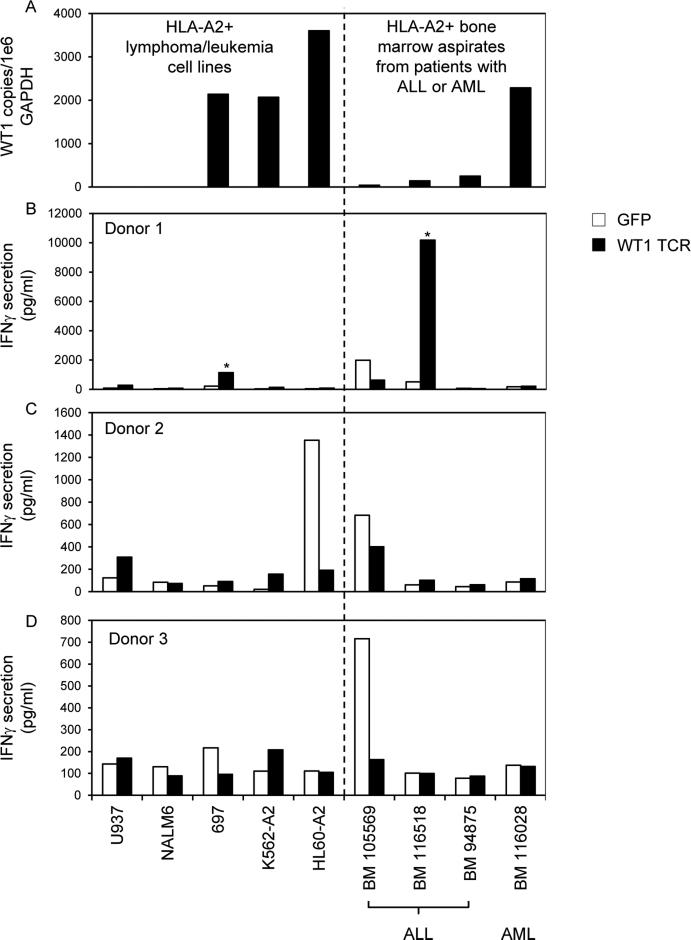
To determine if the WT1 TCR we isolated could mediate recognition of primary leukemia cells, we evaluated recognition of bone marrow aspirates from 3 patients with ALL and 1 patient with AML (Figure 4). WT1 expression in tumor cell lines and primary leukemias was quantified as the number of WT1 RNA copies relative to the housekeeping gene GAPDH measured by quantitative RT-PCR, and HLA-A2 expression was measured by FACS (Figure 4A). Only the bone marrow aspirate from the patient with AML contained >1000 copies of WT1 RNA per 1e6 GAPDH. Recognition of target cells was evaluated by means of IFNγ secretion after overnight coculture with WT1 TCR transduced T cells (Figure 4B, C, and D). Recognition of HLA-A*0201+ WT1+ cells was considered positive and specific if IFNγ secretion was >100 pg/ml and if these levels were >2X background with any negative control target cell and >2X background of GFP transduced T cells with the same target cell. Using these criteria, there appeared to be some specific recognition of the 697 leukemic cell line and the bone marrow 116518 primary leukemia cells by T cells from donor 1. However, these reactivities were not consistent among the 3 donors, and there was no consistent recognition of multiple primary HLA-A*0201+ WT1+ tumor cells in any of the 3 donors (Figure 4).
Figure 4.
WT1 RNA expression in tumor cell lines and bone marrow aspirates from HLA-A*0201+ patients with leukemia and recognition by PBL retrovirally transduced to express a WT1 reactive TCR. (A) Quantitative RT-PCRs for WT1 and GAPDH were conducted on RNAs isolated from tumor cell lines and bone marrow aspirates. WT1 RNA copies were divided by GAPDH copies and expressed as WT1 copy number per 1e6 GAPDH copy number. (B-D) PBL from 3 HLA-A*0201+ donors were transduced with retroviral particles encoding GFP as a negative control or with the WT1 reactive TCR (TRAV12-1*01; CDR3: CVVNTPPNTDKLIF; TRBV7-2*01; CDR3: CASTPFTSGSGWDEQFF). Transduced T cells were cocultured overnight with tumor cell lines and bone marrow aspirates. IFNγ in coculture supernatants was measured by ELISA. * indicates >100 pg/ml IFNγ secretion by WT1 TCR transduced T cells, >2X background with any negative control target cell, and >2X background of GFP transduced T cells with the same target cell.
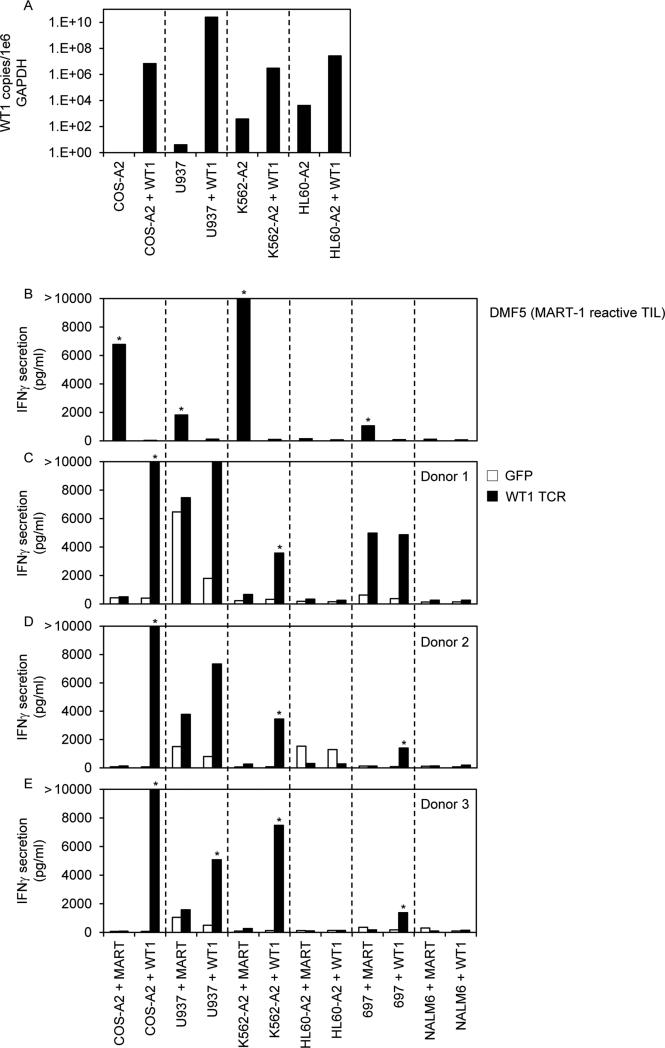
Finally, to determine if the lack of recognition was associated with the level of WT1 expression in tumors, we highly overexpressed WT1 cDNA in some leukemia cell lines by means of electroporation (Figure 5). As evaluated by qRT-PCR, the electroporated cell lines expressed 1e4 to 1e9 more WT1 RNA copies than the parental cell lines (Figure 5A). We evaluated recognition of these electroporated cell lines by T cells expressing our WT1 reactive TCR and observed specific and consistent recognition of COS-A2 and K562-A2 cells electroporated with WT1 cDNA from all 3 donors. There was also some recognition of the HLA-A*0201+ U937 and 697 cell lines electroporated with WT1 cDNA, but this was inconsistent. In contrast, the HLA-A*0201+ HL60-A2 and NALM6 cell lines electroporated with WT1 cDNA were not recognized by T cells from any donor despite the presence of nearly equal copies of WT1 mRNA and the housekeeping gene GAPDH. These cell lines do not naturally express the melanoma antigen MART-1. Therefore, as controls for evaluating the ability of these lines to process and present antigen, we also electroporated them with MART-1 cDNA and evaluated recognition by the MART-1 reactive TIL, DMF5. COS-A2, K562-A2, U937, and 697 cells electroporated with MART-1 cDNA were specifically recognized by DMF5; however, the HL60-A2 and NALM6 cell lines were not (Figure 5B) further suggesting possible antigen processing defects in these cells.
Figure 5.
WT1 RNA expression in tumor cell lines electroporated with WT1 cDNA and recognition by PBL retrovirally transduced to express a WT1 reactive TCR. (A) Quantitative RT-PCRs for WT1 and GAPDH were conducted on RNAs isolated from tumor cell lines electroporated with WT1 cDNA or mock-electroporated. WT1 RNA copies were divided by GAPDH copies and expressed as WT1 copy number per 1e6 GAPDH copy number. (B-E) PBL from 3 HLA-A*0201+ donors were transduced with retroviral particles encoding GFP as a negative control or with the WT1 reactive TCR (TRAV12-1*01; CDR3: CVVNTPPNTDKLIF; TRBV7-2*01; CDR3: CASTPFTSGSGWDEQFF). Transduced T cells (C-E) and a MART-1 reactive T cell line (DMF5) (A) were cocultured overnight with tumor cell lines previously electroporated with MART-1 or WT1 cDNA. IFNγ in coculture supernatants was measured by ELISA. * indicates >100 pg/ml IFNγ secretion by WT1 TCR transduced T cells, >2X background with any negative control target cell, and >2X background of GFP transduced T cells with the same target cell. Bars that reach 2000 pg/ml indicate off-scale IFNγ levels > 2000 pg/ml.
Discussion
Clinicaltrials.gov currently lists 15 immunotherapy trials that target WT1 either by means of vaccination or adoptive T cell transfer for patients with cancers that express WT1 and are actively recruiting. Also, in a National Cancer Institute pilot project aimed at prioritizing cancer antigens, WT1 was identified as the highest priority antigen (31). Therefore, we attempted to isolate a high avidity TCR specifically reactive with WT1 for use in a gene therapy protocol. In the report of a recent clinical trial, patients with high-risk leukemias were treated with adoptively transferred allogeneic T cell clones specifically reactive with the HLA-A*0201 restricted peptide WT1:126-134 pulsed onto TAP-deficient B-LCL cells (23). We attempted to use identical methods to raise T cells reactive with this same epitope. We stimulated PBL from five HLA-A*0201+ healthy donors in vitro with WT1:126-134 pulsed mature DCs in the presence of IL21 and subsequently IL2 and IL7. From all five donors we generated T cell populations that specifically recognized the peptide (Supplementary Table 2). However, only one T cell clone from one healthy donor also consistently recognized HLA-A*0201+ target cells transfected with full-length WT1 cDNA. From this T cell clone, we isolated a WT1 reactive TCR and grafted the α and β variable chain regions of this TCR onto murine constant regions to improve expression on the surfaces of T cells genetically modified to express this receptor (27). We generated a retroviral construct encoding this TCR and used it to transduce PBL from 3 different HLA-A*0201+ donors. These genetically modified T cells consistently recognized peptide pulsed T2 cells and HLA-A*0201+ target cells transfected with full-length WT1 cDNA. However, they did not reproducibly recognize HLA-A*0201+ WT1+ tumor cells.
There are many potential explanations for the lack of tumor recognition by the WT1:126-134 reactive TCR described here. When we genetically introduced WT1 into target cells either by means of transfection or electroporation, WT1 mRNA copies were 1e4 to 1e9 fold higher in the genetically modified cells compared to the parental cell lines (Figure 5). It is possible that our TCR did not have sufficient avidity to mediate recognition of the low levels of WT1 endogeneously expressed in tumors compared to cells genetically modified to express full-length WT1 cDNA. The TCR we isolated specifically recognized the WT1:126-134 peptide at a concentration of ~0.1 ng/ml when pulsed onto T2 cells. Certainly other investigators have described TCRs with higher functional avidities; however, this degree of peptide recognition has often been associated with potent recognition of tumor cells. As examples, tumor reactive TCRs with similar or lower peptide avidities have been described for surviving (32), NY-ESO-1 (33), and TRP2 (34). One group identified an HLA-A*0201 restricted WT1:126-134 reactive TCR that did not appear to mediate peptide recognition below 1 nM, and yet T cells expressing that TCR were reported to lyse WT1+ K562-A2 cells but not WT1- C1R-A2 cells. Interestingly, in that manuscript, the authors made several modifications to their TCR that improved binding to HLA-A*0201/WT1:26-134 tetramers, but these modifications did not improve the functional avidity of the TCR (35). These comparisons suggest that the lack of consistent tumor recognition by our TCR may not solely be a reflection of its peptide avidity.
To investigate the lack of consistent tumor recognition further, we considered the possibility that WT1:126-134 might be preferentially processed either by standard proteasomes or immunoproteasomes as has been observed for other tumor associated antigens (28). We evaluated recognition of WT1+ target cells expressing either of these proteasomes and observed the WT1:126-134 peptide was preferentially processed and presented by immunoproteasomes. This may in part explain why cell lines derived from solid tumors were not efficiently recognized as they generally express standard proteasomal subunits (28, 29). However, hematologic malignancies have been reported to express significantly higher levels of immunoproteasomal subunits (36), so this finding of preferential processing may not explain the lack of recognition of many leukemic cell lines. Subsequently, we evaluated recognition of tumor cells treated with IFNγ which has been reported to induce the expression of immunoproteasomes and upregulate HLA class I expression (37). However, our WT1 reactive TCR did not mediate consistent recognition of IFNγ treated tumors. We also highly overexpressed WT1 cDNA in some leukemia cell lines by means of electroporation to determine if the lack of recognition was simply associated with the level of WT1 expression in the tumors. Three of four electroporated cell lines expressing as much or more WT1 mRNA copies than the housekeeping gene GAPDH were not consistently recognized by our TCR suggesting possible antigen processing defects in these cells. Finally, we evaluated recognition of primary leukemia cells from three patients with ALL and one with AML in the event that all of our previous findings were related to the use of cultured cell lines that might not be relevant to our ultimate clinical goal of treating cancers in patients. However, our TCR did not consistently mediate recognition of these fresh leukemias either.
Although we speculate that the HLA-A*0201 restricted WT1:126-134 peptide may not be a suitable target epitope for cancer immunotherapy, other investigators have reported preclinical studies suggesting the opposite. One group used a WT1:126-134 reactive TCR they isolated from an HLA-A*0201- donor to genetically modify previously non-reactive T cells (38). Another team used a TCR-like single-chain variable fragment (scFv) antibody specific for the WT1:126-134/HLA-A*0201 complex (39). Both groups demonstrated lysis of the HLA-A*0201+ WT1+ human leukemia cell line BV173 cell line in vitro in comparison to one HLA-A*0201- tumor cell line, Kyo-1 or SKOV3 respectively. The WT1:126-134 reactive TCR we described here induced significant IFNγ secretion in response to the BV173 cell line in two of the three donors evaluated in comparison to the SKOV3 cell line (Figure 2, donors 1 and 3). However, in comparison to multiple other HLA-A*0201- and WT- tumor cell lines, we could not conclude that the observed IFNγ secretion was specific. In the previously reported preclinical studies, the TCR like antibody or T cells expressing the WT1:126-134 reactive TCR inhibited the growth of BV173 in immunodeficient mice compared to mock transduced T cells or lower affinity antibodies. However, it is unclear what effect the TCR or antibody would have had on a WT1- tumor.
In addition to preclinical studies, several investigators have targeted the HLA-A*0201 restricted WT1:126-134 epitope in human clinical trials, some of which are still ongoing. In the previously mentioned clinical trial in which patients with high-risk leukemias were treated with adoptively transferred allogeneic WT1:126-134 reactive T cell clones (23),11 patients were treated, and the authors reported evidence of antileukemic activity in two of them. One patient with advanced progressive disease had a transient response, and a second patient with minimal residual disease at the time of treatment had a prolonged remission. In a recent review article (40), the authors suggested the lack of clinical efficacy in that trial may have been attributed to their inability to consistently generate high avidity WT1 reactive T cell clones. Therefore, they screened >1000 clones from >70 HLA-A*0201+ normal donors, and isolated the TCR with the highest avidity for the HLA-A*0201/WT1:126-134 complex for use in gene therapy trials. They are currently conducting a clinical trial in which patients with high risk or relapsed AML, MDS, or CML are being treated with adoptively transferred T cells genetically modified to express this TCR (Clinicaltrials.gov ID# NCT01640301). They reported preliminary clinical trial results at the 2014 meeting of The American Society of Hematology (abstract 3939) and at the 2015 meeting of The American Association for Cancer Research (abstract SY31-03), but the clinical efficacy of this TCR has yet to be fully elucidated. Another group demonstrated they could consistently isolate T cells reactive with this peptide from multiple donors and that those T cells specifically lysed numerous HLA-A*0202+ WT1+ leukemias and solid tumors in vitro (41). This group is currently conducting two clinical trials in which patients are being treated with allogeneic T cells that have been pre-sensitized with WT1 peptides (Clinicaltrials.gov ID numbers NCT00620633 and NCT01758328) but findings have not yet been reported.
Several other MHC class I and class II restricted WT1 epitopes have been identified (15-21). It may be possible to target some of these other epitopes effectively using specific TCRs. However, there are concerns about toxicities that may be induced by targeting WT1 expressed in normal tissues with high avidity TCRs. Low levels of expression of WT1 have been found in some normal adult tissues including kidney podocytes (25), CD34+ hematopoetic stem cells (13), and cardiac endothelial cells (42). No toxicities associated with targeting WT1 on normal cells have been reported in vaccine trials. However, this may be due to the recruitment of T cells with low avidity TCRs that survived negative selection in the thymus (22). Some reports have suggested that WT1 reactive T cells can distinguish between tumor cells that overexpress WT1 and normal tissues with lower levels of expression (24, 25). However, caution seems warranted when using a high avidity WT1 reactive TCR since adoptively transferred T cells reactive with epitopes presented on normal tissues, even at very low levels, may induce severe toxicities (5, 9, 10).
Supplementary Material
Acknowledgments
The authors thank Dr. Terry Fry, head of the Hematologic Malignancies Section in the Pediatric Oncology Branch of the NCI, for kindly providing bone marrow aspirates from patients with leukemia. The authors also thank Arnold Mixon and Shawn Farid for conducting FACS experiments.
Reference List
- 1.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–7. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–46. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kochenderfer JN, Dudley ME, Kassim SH, Somerville RP, Carpenter RO, Stetler-Stevenson M, et al. Chemotherapy-Refractory Diffuse Large B-Cell Lymphoma and Indolent B-Cell Malignancies Can Be Effectively Treated With Autologous T Cells Expressing an Anti-CD19 Chimeric Antigen Receptor. J Clin Oncol. 2015;33:540–9. doi: 10.1200/JCO.2014.56.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–9. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkhurst MR, Yang JC, Langan RC, Dudley ME, Nathan DA, Feldman SA, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther. 2011;19:620–6. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brentjens RJ, Riviere I, Park JH, Davila ML, Wang X, Stefanski J, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011;118:4817–28. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen MC, Popplewell L, Cooper LJ, DiGiusto D, Kalos M, Ostberg JR, et al. Antitransgene rejection responses contribute to attenuated persistence of adoptively transferred CD20/CD19-specific chimeric antigen receptor redirected T cells in humans. Biol Blood Marrow Transplant. 2010;16:1245–56. doi: 10.1016/j.bbmt.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riddell SR, Sommermeyer D, Berger C, Liu LS, Balakrishnan A, Salter A, et al. Adoptive therapy with chimeric antigen receptor-modified T cells of defined subset composition. Cancer J. 2014;20:141–4. doi: 10.1097/PPO.0000000000000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther. 2010;18:843–51. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother. 2013;36:133–51. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugiyama H. Wilms’ tumor gene WT1: its oncogenic function and clinical application. Int J Hematol. 2001;73:177–87. doi: 10.1007/BF02981935. [DOI] [PubMed] [Google Scholar]
- 12.Sugiyama H. WT1 (Wilms’ tumor gene 1): biology and cancer immunotherapy. Jpn J Clin Oncol. 2010;40:377–87. doi: 10.1093/jjco/hyp194. [DOI] [PubMed] [Google Scholar]
- 13.Baird PN, Simmons PJ. Expression of the Wilms’ tumor gene (WT1) in normal hemopoiesis. Exp Hematol. 1997;25:312–20. [PubMed] [Google Scholar]
- 14.Buckler AJ, Pelletier J, Haber DA, Glaser T, Housman DE. Isolation, characterization, and expression of the murine Wilms’ tumor gene (WT1) during kidney development. Mol Cell Biol. 1991;11:1707–12. doi: 10.1128/mcb.11.3.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oka Y, Elisseeva OA, Tsuboi A, Ogawa H, Tamaki H, Li H, et al. Human cytotoxic T-lymphocyte responses specific for peptides of the wild-type Wilms’ tumor gene (WT1) product. Immunogenetics. 2000;51:99–107. doi: 10.1007/s002510050018. [DOI] [PubMed] [Google Scholar]
- 16.Ohminami H, Yasukawa M, Fujita S. HLA class I-restricted lysis of leukemia cells by a CD8(+) cytotoxic T-lymphocyte clone specific for WT1 peptide. Blood. 2000;95:286–93. [PubMed] [Google Scholar]
- 17.Bellantuono I, Gao L, Parry S, Marley S, Dazzi F, Apperley J, et al. Two distinct HLA-A0201-presented epitopes of the Wilms tumor antigen 1 can function as targets for leukemia-reactive CTL. Blood. 2002;100:3835–7. doi: 10.1182/blood.V100.10.3835. [DOI] [PubMed] [Google Scholar]
- 18.Asemissen AM, Keilholz U, Tenzer S, Muller M, Walter S, Stevanovic S, et al. Identification of a highly immunogenic HLA-A*01-binding T cell epitope of WT1. Clin Cancer Res. 2006;12:7476–82. doi: 10.1158/1078-0432.CCR-06-1337. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, Niiya H, Azuma T, Uchida N, Yakushijin Y, Sakai I, et al. Direct recognition and lysis of leukemia cells by WT1-specific CD4+ T lymphocytes in an HLA class II-restricted manner. Blood. 2005;106:1415–8. doi: 10.1182/blood-2005-01-0413. [DOI] [PubMed] [Google Scholar]
- 20.Fujiki F, Oka Y, Tsuboi A, Kawakami M, Kawakatsu M, Nakajima H, et al. Identification and characterization of a WT1 (Wilms Tumor Gene) protein-derived HLA-DRB1*0405-restricted 16-mer helper peptide that promotes the induction and activation of WT1-specific cytotoxic T lymphocytes. J Immunother. 2007;30:282–93. doi: 10.1097/01.cji.0000211337.91513.94. [DOI] [PubMed] [Google Scholar]
- 21.Doubrovina E, Carpenter T, Pankov D, Selvakumar A, Hasan A, O'Reilly RJ. Mapping of novel peptides of WT-1 and presenting HLA alleles that induce epitope-specific HLA-restricted T cells with cytotoxic activity against WT-1(+) leukemias. Blood. 2012;120:1633–46. doi: 10.1182/blood-2011-11-394619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van DA, Berneman ZN, Van Tendeloo VF. Active specific immunotherapy targeting the Wilms’ tumor protein 1 (WT1) for patients with hematological malignancies and solid tumors: lessons from early clinical trials. Oncologist. 2012;17:250–9. doi: 10.1634/theoncologist.2011-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapuis AG, Ragnarsson GB, Nguyen HN, Chaney CN, Pufnock JS, Schmitt TM, et al. Transferred WT1-reactive CD8+ T cells can mediate antileukemic activity and persist in post-transplant patients. Sci Transl Med. 2013;5:174ra27. doi: 10.1126/scitranslmed.3004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao L, Bellantuono I, Elsasser A, Marley SB, Gordon MY, Goldman JM, et al. Selective elimination of leukemic CD34(+) progenitor cells by cytotoxic T lymphocytes specific for WT1. Blood. 2000;95:2198–203. [PubMed] [Google Scholar]
- 25.Asai H, Fujiwara H, Kitazawa S, Kobayashi N, Ochi T, Miyazaki Y, et al. Adoptive transfer of genetically engineered WT1-specific cytotoxic T lymphocytes does not induce renal injury. J Hematol Oncol. 2014;7:3. doi: 10.1186/1756-8722-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapiro J, Claverol S, Piette F, Ma W, Stroobant V, Guillaume B, et al. Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J Immunol. 2006;176:1053–61. doi: 10.4049/jimmunol.176.2.1053. [DOI] [PubMed] [Google Scholar]
- 27.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 2006;66:8878–86. doi: 10.1158/0008-5472.CAN-06-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guillaume B, Stroobant V, Bousquet-Dubouch MP, Colau D, Chapiro J, Parmentier N, et al. Analysis of the processing of seven human tumor antigens by intermediate proteasomes. J Immunol. 2012;189:3538–47. doi: 10.4049/jimmunol.1103213. [DOI] [PubMed] [Google Scholar]
- 29.Basler M, Kirk CJ, Groettrup M. The immunoproteasome in antigen processing and other immunological functions. Curr Opin Immunol. 2013;25:74–80. doi: 10.1016/j.coi.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Turcotte S, Gros A, Tran E, Lee CC, Wunderlich JR, Robbins PF, et al. Tumor-reactive CD8+ T cells in metastatic gastrointestinal cancer refractory to chemotherapy. Clin Cancer Res. 2014;20:331–43. doi: 10.1158/1078-0432.CCR-13-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–37. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arber C, Feng X, Abhyankar H, Romero E, Wu MF, Heslop HE, et al. Survivin-specific T cell receptor targets tumor but not T cells. J Clin Invest. 2015;125:157–68. doi: 10.1172/JCI75876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosati SF, Parkhurst MR, Hong Y, Zheng Z, Feldman SA, Rao M, et al. A novel murine T-cell receptor targeting NY-ESO-1. J Immunother. 2014;37:135–46. doi: 10.1097/CJI.0000000000000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkhurst MR, Fitzgerald EB, Southwood S, Sette A, Rosenberg SA, Kawakami Y. Identification of a shared HLA-A*0201-restricted T-cell epitope from the melanoma antigen tyrosinase-related protein 2 (TRP2). Cancer Res. 1998;58:4895–901. [PubMed] [Google Scholar]
- 35.Thomas S, Xue SA, Cesco-Gaspere M, San JE, Hart DP, Wong V, et al. Targeting the Wilms tumor antigen 1 by TCR gene transfer: TCR variants improve tetramer binding but not the function of gene modified human T cells. J Immunol. 2007;179:5803–10. doi: 10.4049/jimmunol.179.9.5803. [DOI] [PubMed] [Google Scholar]
- 36.Kuhn DJ, Orlowski RZ. The immunoproteasome as a target in hematologic malignancies. Semin Hematol. 2012;49:258–62. doi: 10.1053/j.seminhematol.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka K. Role of proteasomes modified by interferon-gamma in antigen processing. J Leukoc Biol. 1994;56:571–5. doi: 10.1002/jlb.56.5.571. [DOI] [PubMed] [Google Scholar]
- 38.Xue SA, Gao L, Hart D, Gillmore R, Qasim W, Thrasher A, et al. Elimination of human leukemia cells in NOD/SCID mice by WT1-TCR gene-transduced human T cells. Blood. 2005;106:3062–7. doi: 10.1182/blood-2005-01-0146. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Q, Ahmed M, Tassev DV, Hasan A, Kuo TY, Guo HF, et al. Affinity maturation of T-cell receptor-like antibodies for Wilms tumor 1 peptide greatly enhances therapeutic potential. Leukemia. 2015;29:2238–47. doi: 10.1038/leu.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stromnes IM, Schmitt TM, Chapuis AG, Hingorani SR, Greenberg PD. Re-adapting T cells for cancer therapy: from mouse models to clinical trials. Immunol Rev. 2014;257:145–64. doi: 10.1111/imr.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doubrovina ES, Doubrovin MM, Lee S, Shieh JH, Heller G, Pamer E, et al. In vitro stimulation with WT1 peptide-loaded Epstein-Barr virus-positive B cells elicits high frequencies of WT1 peptide-specific T cells with in vitro and in vivo tumoricidal activity. Clin Cancer Res. 2004;10:7207–19. doi: 10.1158/1078-0432.CCR-04-1040. [DOI] [PubMed] [Google Scholar]
- 42.Duim SN, Kurakula K, Goumans MJ, Kruithof BP. Cardiac endothelial cells express Wilms’ tumor-1: Wt1 expression in the developing, adult and infarcted heart. J Mol Cell Cardiol. 2015;81:127–35. doi: 10.1016/j.yjmcc.2015.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.