Abstract
Background
The rice small GTPase OsRac1 is a molecular switch in rice innate immunity. The Receptor for Activated Kinase C-1 (RACK1) interacts with OsRac1 to suppress the growth of the rice blast fungus, Magnaporthe oryzae. RACK1 has two homologs in rice, RACK1A and RACK1B. Overexpressing RACK1A enhances resistance to the rice blast fungus. However, RACK1A downstream signals are largely unknown.
Results
Here, we report the identification of OsRap2.6, a transcription factor that interacts with RACK1A. We found a 94% similarity between the OsRap2.6 AP2 domain and Arabidopsis Rap2.6 (AtRap2.6). Bimolecular fluorescence complementation (BiFC) assays in rice protoplasts using tagged OsRap2.6 and RACK1A with the C-terminal and N-terminal fragments of Venus (Vc/Vn) indicated that OsRap2.6 and RACK1A interacted and localized in the nucleus and the cytoplasm. Moreover, OsRap2.6 and OsMAPK3/6 interacted in the nucleus and the cytoplasm. Expression of defense genes PAL1 and PBZ1 as well as OsRap2.6 was induced after chitin treatment. Disease resistance analysis using OsRap2.6 RNAi and overexpressing (Ox) plants infected with the rice blast fungus indicated that OsRap2.6 RNAi plants were highly susceptible, whereas OsRap2.6 Ox plants had an increased resistance to the compatible blast fungus.
Conclusions
OsRap2.6 contributes to rice innate immunity through its interaction with RACK1A in compatible interactions.
Electronic supplementary material
The online version of this article (doi:10.1186/1939-8433-5-35) contains supplementary material, which is available to authorized users.
Keywords: OsRac1, OsRap2.6, RACK1, OsMAPK3/6, BiFC
Background
Rice production is constrained by various diseases, the rice blast fungus, Magnaporthe oryzae being among the most prominent (Ribot et al 2008; Couch et al 2005; Valent and Chumley 1991). This fungus accounts for major losses in crops and grain yields (Wilson and Talbot 2009). M. oryzae produces asexual spores that are dispersed rapidly by wind or by other means. Breeding for resistance is one of the safest ways to counteract M. oryzae; however, understanding the resistance mechanisms for blast fungus is still a challenge (Ribot et al 2008; Valent and Chumley 1991).
Pathogen associated molecular pattern (PAMP) triggered immunity (PTI) and effector triggered immunity (ETI) responses occur to evade pathogens such as the blast fungus (He et al 2007; Bent and Mackey 2007; Chisholm et al 2006; Jones et al 2006). PTI is the first line of defense, which requires membrane receptor proteins known as pattern recognition receptors stimulated by chitin, flagellin or elicitors. ETI is the second line of defense that requires intracellular receptors of pathogen virulence molecules called effectors, whose recognition induces ETI and is triggered by resistance (R) proteins (Kawano et al 2010; Zipfel 2008; Dangl and Jones 2001).
The PTI response occurs in seconds to minutes, leading to calcium ion fluxes and oxidative bursts, whereas structural responses such as callose deposition may take hours to days (Boudsocq et al 2010, Boller and Felix 2009). These secondary responses could result in hypersensitive responses (HR) (Nimchuk et al 2003), nitric oxide (NO) (Heath 2000) and reactive oxygen species (ROS) production (Mittler et al 1999; Jabs et al 1996). The mechanisms that link PTI to downstream signals are, however, unclear.
The OsRac1 small GTPase is a molecular switch in rice innate immunity (Ono et al 2001; Kawasaki et al 1999). Endogenous GTPase activity of OsRac1 hydrolyzes active guanosine triphosphate (GTP) to inactive guanosine diphosphate (GDP). On the other hand, guanine nucleotide exchange factors (GEFs) catalyze the exchange of inactive GDP to active GTP. The GEFs act as positive regulators leading to activation of downstream signals and, ultimately, resistance to pathogens (Paduch et al. 2001).
OsRac1 interacts with Pit through the nucleotide-binding (NB-ARC) (ARC: APAF-1, certain R gene products and CED-4) domain and is involved in the ETI response (Kawano et al 2010). The components in rice involved in PTI and ETI form part of the “defensome network” and include mitogen-activated protein kinase 3 and 6 (OsMAPK 3/6), nicotinamide adenine dinucleotide phosphate (NADPH) oxidase and co-chaperones RAR1, SGT1 and heat shock proteins 90 and 70 (Hsp90 and Hsp70) among others (Kim et al 2012; Chen et al 2010a; Kawano et al 2010; Nakashima et al 2008; Shirasu 2009Thao et al 2007; Wong et al 2007; Kawasaki et al 2006; Lieberherr et al 2005; Ono et al 2001).
To understand the role of OsRac1, transgenic rice plants expressing constitutively active (CA) (GTP-bound) or dominant negative (DN) (GDP-bound) OsRac1s were infected with the compatible (race 007) and incompatible rice blast fungus (race 031). The CA mutant had increased resistance and ROS production in when infected with the compatible race, whereas the DN mutant suppressed resistance and reduced ROS production when infected with the incompatible race (Chen et al 2010b; Ono et al 2001). These observations show the importance of OsRac1 as a signal transducer in rice and a positive regulator of disease resistance (Chen et al 2010b; Kawano et al 2010; Berken 2006; Suharsono et al 2002; Ono et al 2001; Kawasaki et al 1999).
Receptor for Activated Kinase C-1 (RACK1) was identified as a downstream target of OsRac1 (Nakashima et al 2008). RACK1, a 36-kDa protein is similar to the G-protein ß-subunit highly conserved in diverse species including plants (Adams et al 2011; McCahill et al 2002; Sondek and Siderovski 2001; Kwak et al 1997). RACK1 serves as a scaffold protein and binds phosphatases and transcription factors as well as membrane receptors (Adams et al 2011; Chen et al 2010a; Chen et al 2006). Rice has two RACK1 homologs annotated as RACK1A and RACK1B (Nakashima et al 2008). Constitutively active-OsRac1 interacts more strongly with RACK1A than the dominant negative-OsRac1. RACK1A over-expressing rice enhances the resistance to the compatible race of rice blast fungus (007) (Nakashima et al 2008). In rice, RACK1A interacts directly with OsRac1 and co-chaperones RAR1 and SGT1 and indirectly with Hsp90 and Hsp70 (Thao et al 2007). Hsp90 also specifically interacts with SGT1 (Takahashi et al 2003).
In this work, we identified a protein, OsRap2.6 that interacted with RACK1A in yeast two-hybrid assays. We also demonstrated that OsRap2.6 interacts with RACK1A and OsMAPK3/6 in the nucleus and the cytoplasm, the same place they localized. Expression of the defense genes Phenylalanine ammonia lyase 1 (PAL1) and Probenazole-inducible gene 1 (PBZ1) as well as OsRap2.6 was induced in suspension cells treated with chitin. OsRap2.6 RNAi plants had high susceptibility, whereas OsRap2.6 overexpressing (Ox) plants had increased resistance to the compatible race (007) of rice blast fungus. However, no significant differences were found in OsRap2.6 RNAi or Ox plants when challenged by the incompatible race (031). These results demonstrate that OsRap2.6 contributes to resistance towards the compatible race (007) of rice blast fungus.
Results and discussion
RACK1A interacts specifically with OsRap2.6 in yeast two-hybrid assays
Proteins that interacted with RACK1A in the rice cDNA library were screened in yeast two-hybrid (Y2H) assays. The primary candidate gene (Os04g0398000 or AK101501) had an AP2/ERF domain whose sequence shared 94% amino acid identity with Arabidopsis Rap2.6 (AtRap2.6) (shadowed regions in Figure 1A). We, therefore, named it as Oryza sativa Rap2.6 (OsRap2.6) and selected it for further analysis. The other candidate genes included hypothetical proteins with a MATH domain (Os01g0775300), a CaMKII association domain (Os01g0753200), or a ToIA/TF11B domain (Os12g0112600); Universal stress protein (USP) (Os5g0453700) containing a USP domain; and a V1P1-like protein whose domain was unknown (Os01g0698000) (Table 1).
Figure 1.

OsRap2.6 AP2/ERF domain resembles Arabidopsis Rap2.6 and interacts with RACK1A. (A) Comparison of amino acids sequences of rice and Arabidopsis Rap2.6. (B) Interaction of OsRap2.6 with RACK1A in yeast two-hybrid assays. OsRac1 (WT), constitutively active (CA) and dominant negative (DN)-OsRac1 mutants were examined. (C) Interaction of RACK1 with OsRap2.6 at WD repeat 1–2 in yeast two-hybrid assays.
Table 1.
RACK1A interacting proteins
| Protein name | Clone identity | Number of clones | Domain |
|---|---|---|---|
| OsRap2.6 | Os04g0398000 | 1 | AP2 domain |
| Hypothetical protein | Os01g0775300 | 1 | MATH domain |
| Hypothetical protein | Os01g0753200 | 4 | CaMKII association domain |
| Hypothetical protein | Os12g0112600 | 1 | ToIA/TF11B domain |
| Universal stress protein | Os5g0453700 | 17 | USP domain |
| V1P1 like protein | Os01g0698000 | 1 | - |
Proteins that interact with RACK1A identified in yeast two-hybrid assays.
The bait constructs RACK1A, OsRac1 (WT) and (CA and DN) were fused with the pBTM116ss vector. The OsRap2.6 coding region was ligated into the pVP16 prey vector. The negative controls were pBTM116ss and pVP16-Empty. The paired plasmids were transformed into the yeast Saccharomyces cerevisiae (L40). Positive transformants were selected based on the ability to activate transcription of the histidine 3 (HIS3) reporter gene. We found a strong interaction between RACK1A and OsRap2.6; however, there was no observed interaction in the (WT) or the (CA and DN) OsRac1 mutants. There was no growth of colonies in the negative controls, pBTM116ss and pVP16 as expected (Figure 1B). These results demonstrated that OsRap2.6 interacts specifically with RACK1A in Y2H assays. We, therefore, hypothesised that OsRap2.6 may be functionally similar to AtRap2.6 or most members in the AP2/ERF family.
Rap2.6 is a single copy gene in the Arabidopsis genome with one AP2 domain located at the N-terminus (Nakano et al 2006). This domain has about 60 amino acids and is useful for binding DNA sequences (Magnani et al 2004). AP2/ERFs bind DNA sequences with cis elements such as the GCC box (AGCCGCC) and CE1 that regulates plant-pathogen interactions (Ohme-Takagi and Shinshi et al 1995). In general, AP2/ERFs are the most diverse transcription factors in plants (Riechmann and Ratcliffe 2000Ohme-Takagi and Shinshi et al. Ohme-Takagi and Shinshi 1995). AP2/ERF transcription factors are important in plant responses to abiotic and biotic stresses (Agrawal et al 2006). Arabidopsis has 145 members including Rap2.6 (Sharoni et al 2011; Sakuma et al 2002, Riechmann and Ratcliffe 2000) that confers resistance to Pseudomonas syringae DC3000 (He et al 2007).
OsRap2.6 specifically interacts with RACK1A at WD repeats 1 and 2
We further analyzed the interaction between OsRap2.6 and tryptophan-aspartate (WD) repeats of RACK1A in Y2H assays. RACK1 interacts with co-chaperones, phosphatases and transcription factors through its seven WD (1–7) repeats (Adams et al 2011). We found strong interactions between OsRap2.6 and WD repeats 1 and 2 (Figure 1C). Thus, WD 1 and 2 repeats may be a common binding site for OsRac1 and OsRap2.6 and may possibly act as a potential interaction site or bridge for the three proteins. In another study, when constitutively active OsRac1 (CA-OsRac1) was expressed, it bound RACK1A at WD repeat 1 and 2, enabling OsRac1 to regulate RAR1 and RACK1A at the post-transcriptional levels (Nakashima et al 2008). RACK1 forms homodimers (Liu et al 2007; Thornton et al 2004; Yaka et al 2003) and heterodimers with the remaining WD repeat motifs (3–7) (Chen et al 2004). RACK1 anchors at amino acids 39 and 40 on the 18S ribosomal RNA subunit, constantly mediated by WD repeats 1 and 2 and their associated loops (Adams et al 2011).
OsRap2.6 localizes in the nucleus and the cytoplasm in rice protoplasts
To determine the intracellular localization of OsRap2.6 protein, we tagged OsRap2.6 to a variant of yellow fluorescent protein (Venus) at the N-terminus (Venus-OsRap2.6) and expressed the fusion protein in rice protoplasts with the internal positive controls mCherry and mCerulean with a nuclear localization signal (NLS-mCerulean). OsRap2.6 localized with mCherry in the nucleus and the cytoplasm (94%) and the nucleus alone (6%) (Figure 2A). A further comparison using the NLS-mCerulean marker showed 90% of the marker localized within the nucleus and the cytoplasm and 10% in the nucleus alone (Figure 2B). Altogether, OsRap2.6 localized in the nucleus and the cytoplasm in more than 90% of the transformed rice protoplasts.
Figure 2.
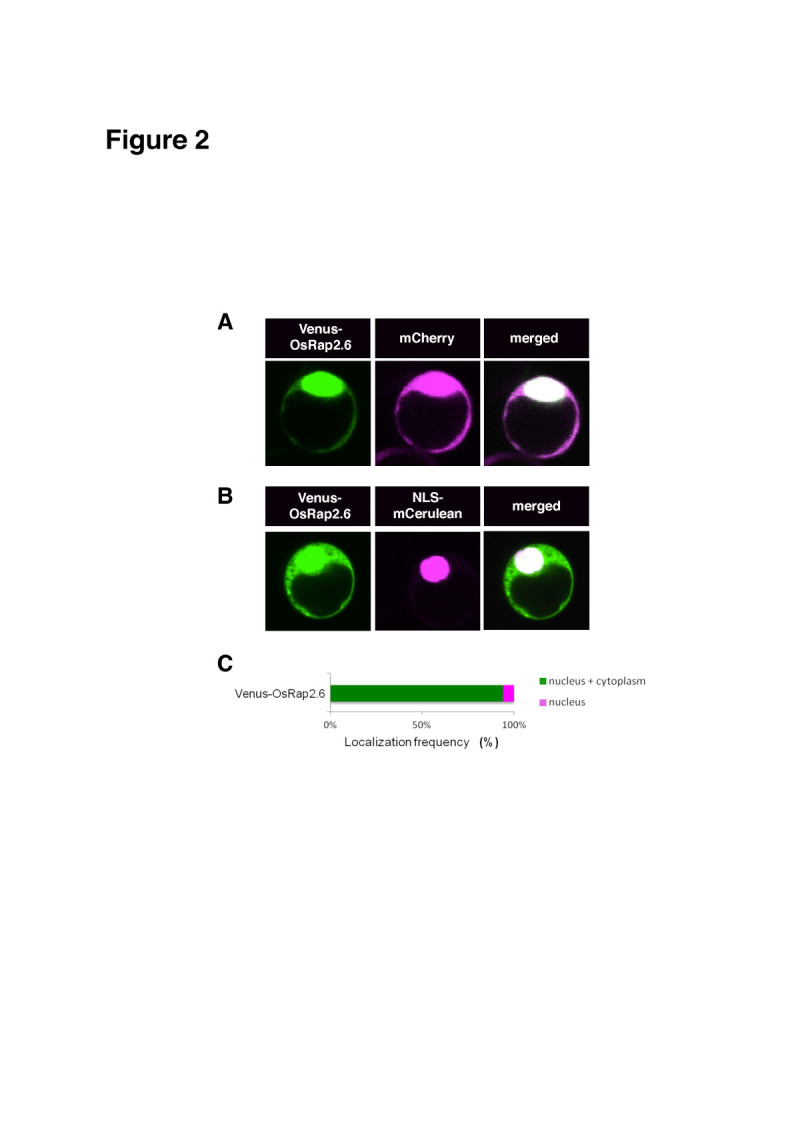
Subcellular localization of OsRap2.6. Rice protoplasts were transformed with known fluorescent proteins mCherry and NLS-mCerulean. Fluorescence was detected using a CCD camera connected to a confocal microscope. The localization frequency of the cells was analyzed in 50–100 cells expressing YFP/CFP as compared to the positive controls using Excel. Means and standard deviations were separated using Student’s t -test (p<0.05). (A) Subcellular localization of Venus-OsRap2.6 with the mCherry. (B) Subcellular localization of Venus-OsRap2.6 with the NLS-mCerulean. (C) Localization frequency of Venus-OsRap2.6.
In Arabidopsis, Rap2.6-YFP and Rap2.6L-YFP localize to the nucleus. In our study, we found a discrepancy between the protein localizations in rice and in Arabidopsis, although both proteins shared a similar NLS. The likely reason for the difference was that Rap2.6 was tagged to the C-terminus, whereas OsRap2.6 was tagged to the N-terminus. Rap2.6 acts as a trans-activator in yeast and localizes in the nucleus of onion epidermal cells. A putative nuclear localization signal sequence (RPPKKYRGY), which indicates a possible nuclear localization, is found near the AP2 domain (Zhu et al 2010). A recent report indicates that Rac Immunity 1 (RAI1), a bHLH transcription factor which plays a role in OsRac1-mediated immunity, localizes mainly in the nucleus and partly in the cytoplasm (Kim et al 2012).
RACK1A localizes in the nucleus and the cytoplasm in rice protoplasts
We expressed RACK1A with the Venus tag at the C-terminus and examined the intracellular localization of this construct with the internal positive controls mCherry, a rice PAMP receptor OsCERK1-GFP (ER and plasma membrane marker) and OsGenL-CFP (nuclear marker). RACK1A-Venus localized mainly in the nucleus and the cytoplasm (CN) (90%) with mCherry (Figure 3A). Moreover, in an independent experiment, RACK1A-Venus co-localized with OsGenL-CFP in the cytoplasm and the nucleus (88%) with the remainder of the CFP localized in the nucleus (12%) (Figure 3B). However, a small proportion (6%) was associated with OsCERK1-GFP at the plasma membrane (PM) and ER (3%) (Figure 3C). Our findings further confirmed the ability of RACK1A to localize as a scaffold protein to different parts of the cell. According to an earlier report, RACK1A localizes in the cytoplasm in rice protoplasts (Nakashima et al 2008). In another report, RACK1A and OsRac1 shifted to detergent-resistant membranes (DRM), regions near PM after elicitation with chitin (Fujiwara et al 2009). Moreover, RACK1A associated with heterotrimeric G-protein’s γ-subunit 2 (RACK1A-AGG2) and localized at PM, the same cellular component where AGG2 is apparently localized; however, RACK1A associated with the γ-subunit 1 (RACK1A-AGG1) throughout the cell (Kamil et al. 2011; Adjobo-Hermans et al 2006). RACK1A modulates its defense responses at posttranscriptional levels through its interaction with OsRac1 in the cytoplasm (Nakashima et al 2008).
Figure 3.
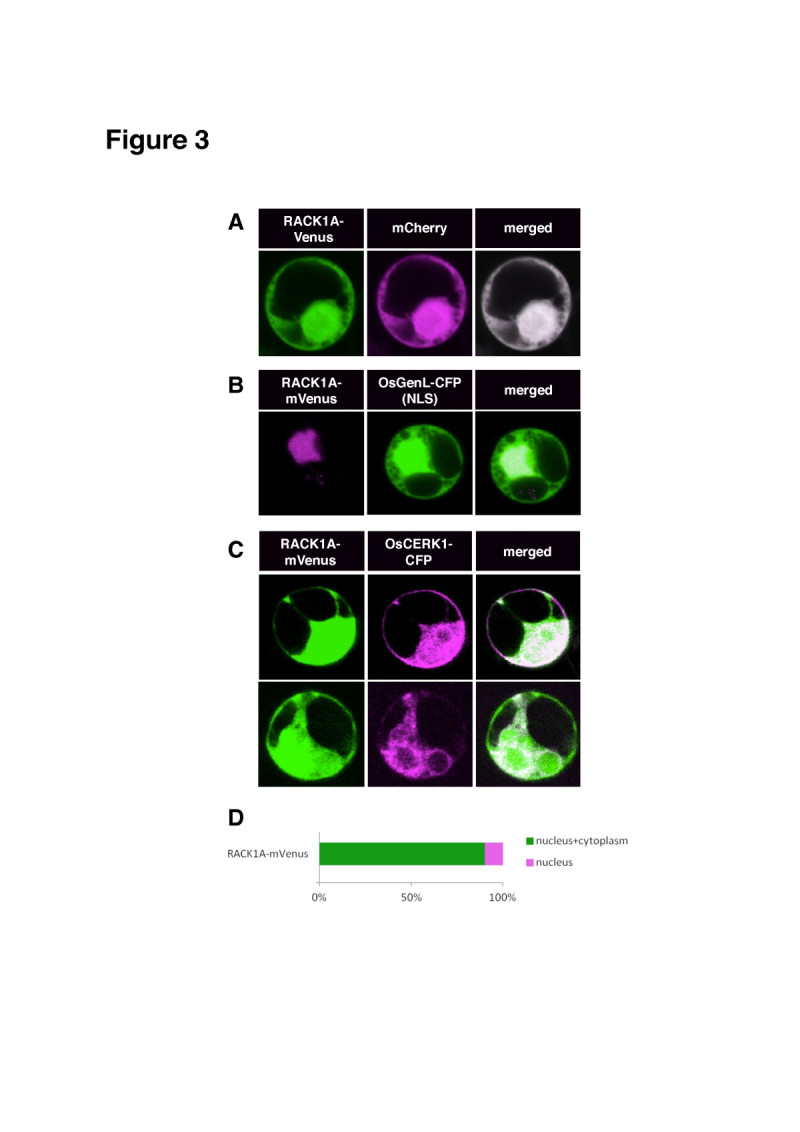
Subcellular localization of RACK1A in rice protoplasts. Rice protoplasts were transformed with a plasmid harboring the RACK1A-Venus construct. Protoplasts transformed with known fluorescent proteins mCherry, OsCERK1-GFP and OsGenL-CFP were used as positive controls. Conditions for microscopy and data analysis were identical to those outlined in the legend to Figure 1. (A) Subcellular localization of RACK1A-mVenus with mCherry. (B) Subcellular localization of RACK1A-Venus with OsGenL-CFP. (C) Subcellular localization of RACK1A-Venus with OsCERK1-GFP. (D) Localization frequency (%) of RACK1A-Venus.
OsRap2.6 and RACK1A interact in the nucleus and the cytoplasm in rice protoplasts
OsRap2.6 and RACK1A localized to the nucleus and the cytoplasm, but the next question was whether the two proteins interact within the same subcellular region or not. We confirmed the in vivo interaction using Bimolecular Fluorescence Complementation (BiFC) methods that detect interactions between two proteins in living cells. The absence of an interaction prevents reassembly of the fluorescent protein and results in background fluorescence (Kerppola 2009). We split the Venus fluorescent protein into two halves (Vn/Vc) and tagged OsRap2.6 with Vn and RACK1A with Vc at N and C-termini, respectively. GUS was used as a negative internal control. The paired constructs (Vn-OsRap2.6 + RACK1A-Vc) and (Vn-OsRap2.6 + GUS-Vc) and their controls mCherry and OsGenL-CFP were transfected into rice protoplasts. We found a strong interaction between OsRap2.6 and RACK1A in the cytoplasm and the nucleus and the rest remaining signal in the nucleus with mCherry (Figure 4A). The negative control, GUS, had less than 10% fluorescence in all cells (Figure 4A and C). A further comparison with OsGenL-CFP gave a similar finding (Figure 4B). Our results further confirmed the potential of RACK1A to interact with OsRap2.6 in vivo.
Figure 4.
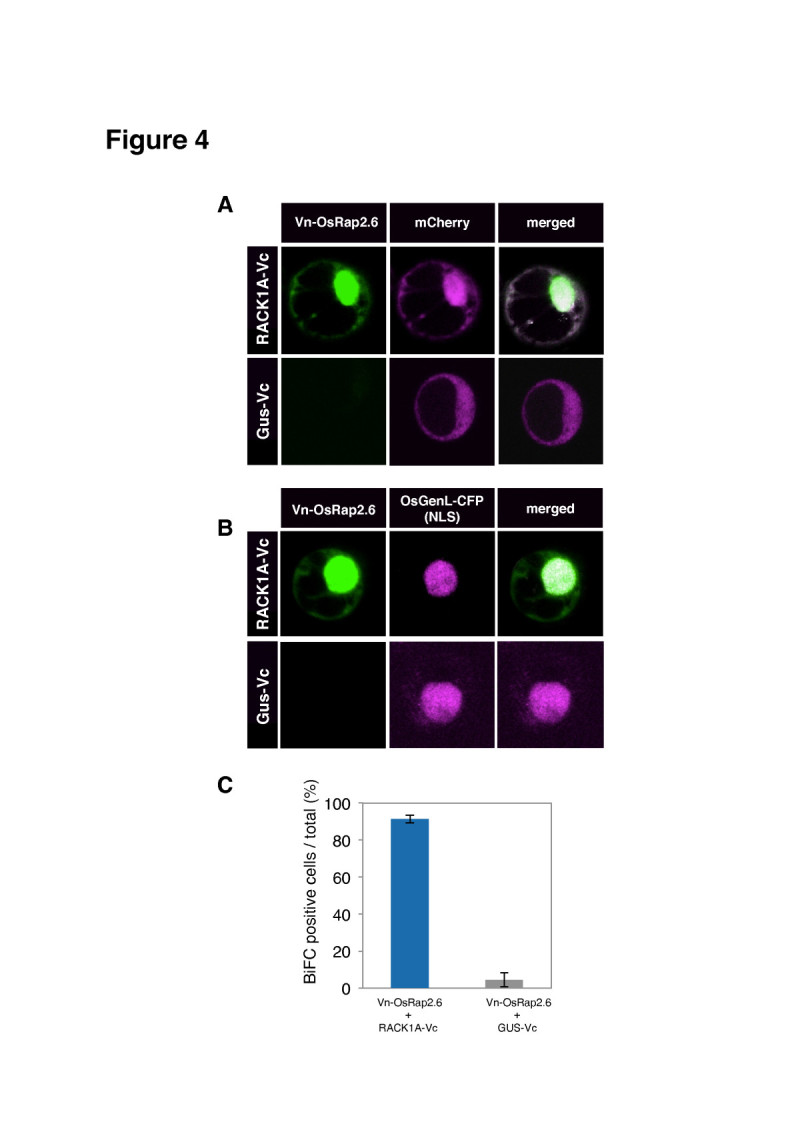
Interaction of OsRap2.6 with RACK1A in rice protoplasts. Rice protoplasts were co-transfected with the fluorescent construct (Vn-OsRap2.6 + RACK1A-Vc/GUS-Vc). Conditions for microscopy and data analysis were identical to those outlined in the legend to Figure 1. (A) Interaction of OsRap2.6 with RACK1A compared with the mCherry. (B) Interaction of OsRap2.6 with RACK1A compared with the OsGenL-CFP. (C) Quantitative analysis of BiFC positive cells.
As a scaffold protein, RACK1A translocates to different parts of the cell and interacts with different phosphatases and transcription factors (Adams et al 2011). According to a recent report, RACK1A interacts with Arabidopsis Nudix hydrolase (AtNUD7) in the nucleus and the cytoplasm. AtNUD7 expression is induced rapidly in response to an avirulent bacteria and abiotic stresses (Olejnik et al 2011; Kamil et al. 2011). RACK1A forms an interactive complex including OsRac1, RAR1 and SGT1 and maintains an effective conformation, which is able to activate the downstream effectors leading to an immune response (Nakashima et al 2008; Thao et al 2007).
OsRap2.6 interacts with OsMAPK3 and OsMAPK6 in the nucleus and the cytoplasm
Mitogen-activated kinase cascades respond to pathogens or pathogen-derived elicitors, for example OsMAPK6 is activated in response to sphingolipid elicitors in rice cell cultures (Kim et al 2012; Lieberherr et al 2005). Furthermore, OsMAPK3 and OsMAPK6 are involved in defense responses in rice (Kim et al 2012; Kishi-Kaboshi et al 2010, Lieberherr et al 2005). We, therefore, investigated whether OsRap2.6 interacts with OsMAPK3 and OsMAPK6. The paired constructs (Vn-OsRap2.6 + Vc-OsMAPK6), (Vn-OsRap2.6 + Vc-OsMAPK3) and their negative control (Vn-OsRap2.6 + GUS-Vc) were transfected into rice protoplasts with mCherry. We found an interaction between OsRap2.6 and OsMAPK6 in the cytoplasm and the nucleus (76%) and (24%) in the cytoplasm. The negative controls had less than 10% fluorescence signal (Figure 5A). In addition, OsRap2.6 and OsMAPK3 interacted in the cytoplasm and the nucleus (72%) and the remainder (28%) of the signal was in the cytoplasm (Figure 5B). Together, these results indicate that OsRap2.6 interacts with OsMAPK6 and OsMAPK3 mainly in the nucleus and the cytoplasm with more than 70% of the cells fluorescencing at the same location.
Figure 5.

Interaction of OsRap2.6 with OsMAPK6/3 in rice protoplasts. Rice protoplasts were co-transfected with the fluorescent constructs (Vn-osRap2.6 + Vc-OsMAPK6) and (Vn-OsRap2.6 + Vc-OsMAPK3) and examined under fluorescence, bright field and overlay. Known fluorescent proteins mCherry, and OsGenL-CFP (nuclear marker) were used as markers of localizations. Conditions for microscopy and data analysis were identical to those outlined in the legend to Figure 1. (A) Interaction between OsRap2.6 and OsMAPK6 compared with the mCherry construct. (B) Quantitative analysis of BiFC positive cells from (A) (OsRap2.6 + OsMAPK6). (C) Interaction between OsRap2.6 and OsMAPK3 compared with the mCherry. (D) Quantitative analysis of BiFC positive cells from (C) (OsRap2.6 + OsMAPK3). (E) Frequency (%) of interactions between OsRap2.6 and OsMAPKs in cells.
OsMAPK6 indirectly interacts with CA-OsRac1 in a complex but not with DN-OsRac1 (Lieberherr et al 2005). A complete MAPK cascade (comprised of MEKK1, MKK4/MKK5 and MPK3/MPK6) was proposed to be downstream of the flagellin receptor kinase, FLS2, in Arabidopsis. This signaling cascade activates WRKY22 and WRKY29 transcription factors (Asai et al 2002). Suppression of OsMAPK6 expression by RNAi decreased PAL1 mRNA levels (Lieberherr et al 2005). RAI1 transcription factor interacts with OsMAPK3 and OsMAPK6 proteins in vivo and in vitro. Moreover, OsMAPK3/6 and OsMKK4-dd phosphorylate RAI1 in vitro. OsBWMK1 is activated in rice leaves after infection with rice blast fungus, elicitor treatment, and wounding (Cheong et al 2003; He et al 1999). OsBWMK1 localizes in the nucleus and phosphorylates OsEREBP1, an ERF transcription factor (Cheong et al 2003). From our findings, we hypothesised that OsRap2.6 may be phosphorylated by OsMAPK3/6 to carry out its transcriptional regulation.
Chitin elicitor in rice suspension cells induces OsRap2.6 expression
Suspension cells derived from wild-type japonica cv. Kinmaze rice were treated with chitin (2 μg/ml), and the expression of potential downstream genes, PAL1, PBZ1 as well as OsRap2.6 was examined by reverse transcription qPCR. Ubiquitin was used as an internal control. OsRap2.6 transcripts were rapidly increased after chitin treatment and after 3 hr they were not further increased. In contrast to OsRap2.6, PAL transcripts peaked at 1 hr after chitin treatment and PBZ1 transcripts started to increase at 3 hr after chitin treatment. (Figure 6A, B and C).
Figure 6.
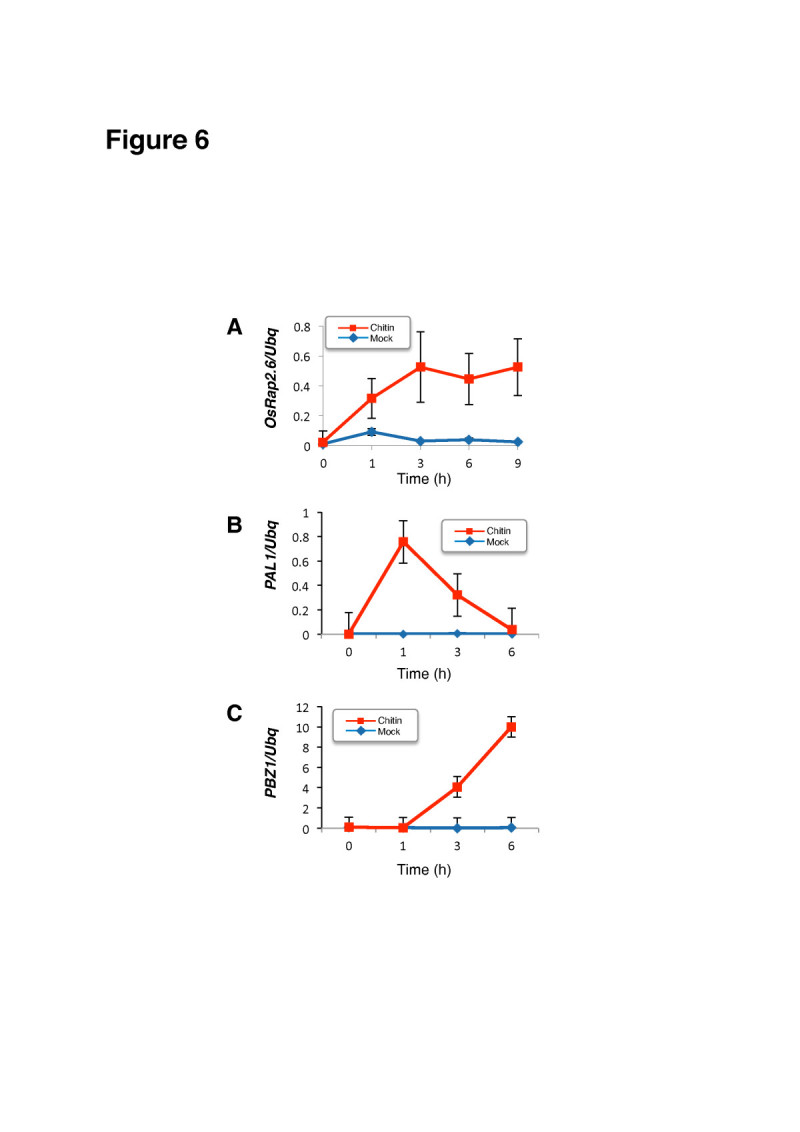
Induction of OsRap2.6 , PAL1 , and PBZ1 expression induced by chitin in rice suspension cells. (A) OsRap2.6 expression in WT suspension cells after chitin treatment measured by reverse transcription qPCR. Ubiquitin was used as an internal control. (B) PAL1 expression in WT suspension cells after chitin treatment measured by reverse transcription qPCR. (C) PBZ1 expression in WT suspension cells after chitin treatment measured by reverse transcription qPCR.
These data agree with recent findings about Rac Immunity1 (RAI1), a bHLH protein, where a gradual increase in PAL1 and OsWRKY19 was noted after OsMAPK6 and OsMAPK3 were overexpressed in rice protoplasts (Kim et al 2012). Defense genes PAL1 and PBZ1 are rapidly induced by infection with rice blast fungus as previously reported (Chen et al 2010a; Kawano et al 2010; Nakashima et al 2008; Kawasaki et al 1999).
OsRap2.6 RNAi plants are susceptible to M. oryzae compatible race 007
We tested if OsRap2.6 contributes to defense responses in rice by constructing OsRap2.6 RNAi and over-expression (Ox) plants. OsRap2.6 transcripts from three independent RNAi lines were confirmed by reverse transcription qPCR (R1, R5 and R10) (Figure 7A). The RNAi and Ox plants were grown in the greenhouse for two months and inoculated with M. oryzae compatible (virulent) Ina 86–137 (race 007) and incompatible (avirulent) TH67-22 (race 031) fungal spore suspension. OsRap2.6 RNAi plants showed high susceptibility characterised by larger disease lesions when infected with the compatible race (007) as compared to non-transformed plants (WT) (Figure 7B), qPCR anlysis of fungal growth (p ≤ 0.01, n=48) (Figure 7C) and lesion lengths (p ≤ 0.01, n=48) (Figure 7D). The PAL1 transcripts were down regulated (p ≤ 0.01) in selected OsRap2.6 RNAi plants (Figure 7E). Together, these results could indicate that fungal growth was enhanced in OsRap2.6 RNAi plants as compared to the non-transformed plants. These results demonstrated that OsRap2.6 contributes to defense responses towards compatible rice blast fungus.
Figure 7.
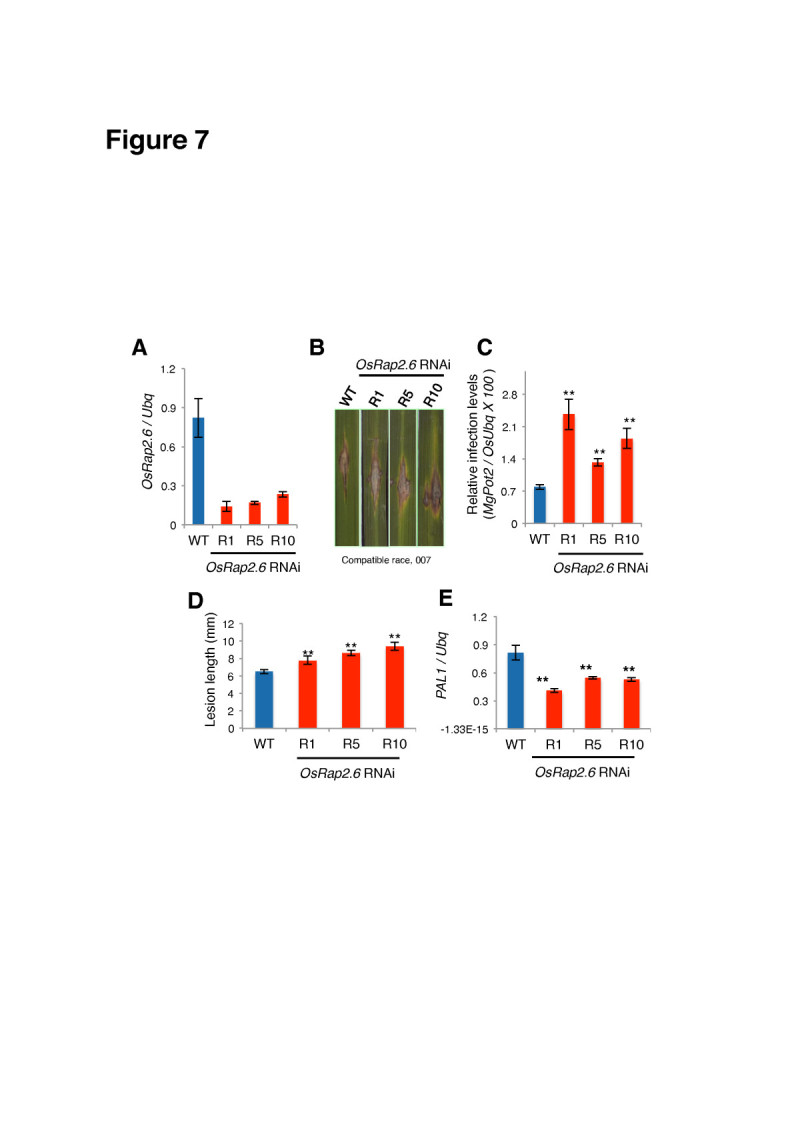
OsRap2.6 RNAi plants are susceptible to a compatible race of M. oryzae. OsRap2.6 transcript levels in RNAi plants were measured by reverse transcription qPCR for three independently transformed lines, R1, R5 and R10. The RNAi plants were grown in the greenhouse for two months and inoculated with M. oryzae compatible (virulent) Ina 86–137 (race 007) fungal spore suspensions. (A) Expression levels of OsRap2.6 transcripts in T1 OsRap2.6 RNAi plants before infection with rice blast fungus. (B) Photographs showing lesions in leaf blades in WT and OsRap2.6 RNAi plants. (C) Quantitative analysis of fungal growth showing increased susceptibility in OsRap2.6 RNAi plants 7 days after infection with a compatible race (007) of rice blast fungus. Rice Ubiquitin was used as an internal control. Bars represent the means ± SE calculated using four biological replicates where each consists of three independent technical replicates (p ≤ 0.01, n=48). (D) Lesion length of OsRap2.6 RNAi plants compared to WT (p ≤ 0.01, n=48). (E) Expression of PAL1 mRNA in OsRap2.6 RNAi plants after rice blast infection. Levels of PAL1 mRNA were down regulated as measured by reverse transcription qPCR (p ≤ 0.01).
We also investigated if OsRap2.6 RNAi contributes to increased susceptibility to an incompatible blast fungus race (031) in a similar approach as described for the compatible race. From our findings, the susceptibility to the incompatible blast fungus in RNAi plants was not significant as shown in the photograph (Additional file 1: Figure S1A), qPCR analysis of fungal growth (p ≥ 0.05, n= 48) (Additional file 1: Figure S1B) and relative lesion length (p ≥ 0.05, n= 48) (Additional file 1: Figure S1C). Expression of the PAL1 gene was not significantly reduced in OsRap2.6 RNAi plants (p ≥ 0.05) (Additional file 1: Figure S1D). Therefore, our results indicate that OsRap2.6 RNAi does not contribute to increased susceptibility to M. oryzae incompatible interactions.
In a study on rice disease resistance, transcription factors including Mybs, WYKYs, NACs and AP2s were induced in leaves infected with blast fungus, indicating the occurrence of transcriptional reprogramming in rice plants after infection (Ribot et al 2008). AP2/EREBPs are also involved in rice viral infections, for example, rice stripe virus (RSV), rice tungro spherical virus (RTSV) and rice dwarf virus (RDV) (Sharoni et al 2011).
PAL1 is among the 10 most repressed or induced genes in response to M. oryzae susceptible interactions (Jantasuriyarat et al 2005). The most highly induced genes in a compatible interaction are PR-1 and PR-5 (thaumatin-like proteins), PBZ1 (PR-10), class 11 chitinase (PR-1a) and PAL1 (Kim et al 2012; Chen et al 2010b; Kawano et al 2010; Kim et al 2001). A recent report on RAl1 indicates that PAL1 and OsWRKY19 expression increased at 12 and 24 hours in the wild type (control) leaves infected with the compatible rice blast fungus (Kim et al 2012).
OsRap2.6 Ox plants have increased resistance to a compatible race of M. oryzae
OsRap2.6 mRNA transcript levels from three independent Ox plants were measured by reverse transcription qPCR (P4, P6 and P14) (Figure 8A). The plants were infected with the rice blast fungus compatible race, 007. From our findings, smaller disease lesions were observed in OsRap2.6 Ox plants as compared to the WT as shown in the photograph (Figure 8B), qPCR anlysis of fungal growth (p ≤ 0.01, n= 48) (Figure 8C) and relative lesion length (p ≤ 0.01, n= 48) (Figure 8D). The PAL1 gene was up regulated (p ≤ 0.01) (Figure 8E). Therefore, OsRap2.6 Ox showed increased resistance to rice blast fungus compatible interactions.
Figure 8.

OsRap2.6 Ox plants are resistant to a compatible race (007) of M. oryzae. OsRap2.6 mRNA transcript levels from three independent Ox plants (P1, P6 and P14) were measured by qPCR. The Ox plants were grown in the greenhouse for two months and inoculated with M. oryzae compatible (virulent) Ina 86–137 (Race 007) fungal spore suspension. (A) Expression levels of OsRap2.6 transcripts in T1 OsRap2.6 Ox plants before infection with rice blast fungus. (B) Photographs showing lesions in leaf blades in WT and OsRap2.6 Ox plants after infection. (C) Quantitative analysis of fungal growth showing increased resistance in OsRap2.6 Ox plants 7 days after infection with rice blast fungus compatible race (007). Ubiquitin was used as an internal control. Bars represent the means ± SE calculated using four biological replicates where each replicate consists of three independent technical replicates (p ≤ 0.01, n=48). (D) Lesion length of OsRap2.6 Ox plants showing increased resistance to blast fungus as compared to WT as shown by increased lesion length after infection (p ≤ 0.01, n=48). (E) Expression of PAL1 mRNA in OsRap2.6 Ox plants after infection with the compatible race 007. Levels of PAL1 mRNA were up regulated as measured by reverse transcription qPCR (p ≤ 0.01, n=48).
We also investigated if OsRap2.6 Ox plants are resistant to the incompatible rice blast fungus race, 031. No significant resistance was noted as shown in the photograph (Additional file 2: Figure S2A), qPCR anlysis of fungal growth (p ≥ 0.05, n= 48) (Additional file 2: Figure S2B), and relative lesion length measurements (p ≥ 0.05, n= 48) (Additional file 2: Figure S2C). Expression of the PAL1 gene did not significantly increase after infection (p ≥ 0.05) (Additional file 2: Figure S2D). Therefore, OsRap2.6 Ox does not contribute to disease resistance in incompatible interactions.
These data agreed with other findings; for instance, in response to M. oryzae, RACK1A interacted with OsRac1 (Nakashima et al 2008) at the N-terminus of Rboh, leading to ROS production (Wong et al 2007). RACK1A-Ox showed increased resistance to a compatible race 007 as compared to the wild type (Nakashima et al 2008). Our findings demonstrated that OsRap2.6 Ox contributes to increased resistance in compatible interactions.
Conclusions
Our study confirmed the role of OsRap2.6 in disease resistance to rice blast fungus, and its localization and interaction with RACK1A and OsMAPK3/6 in rice protoplasts. OsRap2.6 possibly localizes in the nucleus when cells are active, during transcriptional regulation, and in the cytoplasm after a stimulus like chitin or a fungus is sensed. OsRap2.6 localization in the nucleus is essential as a transcription factor; furthermore, its interaction with RACK1A is likely to enable it be involved in disease resistance in rice. The interaction with OsMAPK3/6 could potentially lead to phosphorylated OsRap2.6 for transcriptional regulation, a step that is yet to be confirmed. We found OsRap2.6 to be a positive regulator in M. oryzae compatible interactions possibly as a downstream signal of RACK1A. This study has opened up other areas for further research such as the analysis of OsRap2.6 target genes in the defense response pathway.
Methods
Comparison of predicted amino acid sequences of OsRap2.6
To identify OsRap2.6-related genes, the sequence of OsRap2.6 was used as a query for BLAST searches in the rice and Arabidopsis genome databases (http://www.ncbi.nlm.nih.gov/nuccore). Highly similar amino acid sequences were aligned with the OsRap2.6 sequence using Genetyx software for Mac-Pro, Version 10 (Genetyx, USA).
Yeast two-hybrid assays
The bait constructs, RACK1A, OsRac1 (WT) and (CA and DN) coding regions were ligated to the pBTM116 vector, and OsRap2.6 was ligated to the prey vector, pVP16 as described previously (Nakashima et al 2008; Kawasaki et al 1999). The negative controls were pBTM116ss and pVP16. The vectors concentrations ranged between 150–200 ng/μl hosted by the yeast Saccharomyces cerevisiae L40 (25 μl). The cells were cultured on synthetic complete medium lacking uracil and tryptophan, either with histidine (SC-UW) or without histidine (SC-UWLH). The inhibitor 3-amino-1, 2, 4-triazole (3-AT) (3 mM), was included in the SC-UWLH media.
OsRap2.6, RACK1A and OsMAPK3/6 constructs
An entry clone, pENTR-OsRap2.6 was amplified from pVP16-OsRap2.6 (0.5 μl) with forward (5’-CACCATGGTCACCGCGCTAGCCACGT-3’) and reverse (5’-TCACGACGACGAATCCTTCTTCTTG-3’) primers. The blunt-end PCR product was cloned into pENTR-D/TOPO as per the manufacturer’s instructions (Invitrogen, USA). Colonies were selected with M13 forward (5’-TGTAAAACGACGGCCAGT-3’) and reverse (5’-CAGGAAACAGCTATGAC-3’) primers. The pENTR-OsRap2.6 was ligated into Gateway destination vectors (GW) with the LR clonase II enzyme (0.5 μl) (Invitrogen, USA) whose expression was driven by the 35S-Cauliflower mosaic virus promoter (35S-Vn-OsRap2.6).
For the subcellular localization studies, we used BiFC in which Venus, a variant of YFP, was split into the N-terminal and C-terminal halves (Vn/Vc). The N-terminal half of Venus (Vn) was tagged to OsRap2.6 (Vn-OsRap2.6) and sequenced with pB12221-35S forward primer and the nos terminator as the reverse primer as listed in Table 2. RACK1A-Venus constructs were provided from our laboratory stocks. The half Venus constructs (Vc-OsMAPK3) and (Vc-OsMAPK6) were described previously (Kim et al 2012). The DNA sequence of all plasmids was confirmed in using an ABI-PRISM Big Dye Terminator Cycle Sequencing Kit with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, USA). Data were analysed using Genetyx software for the Mac-Pro, Version 10 (Genetyx, USA).
Table 2.
Primers used to sequence RNAi and Ox constructs
| Primer | Sequence |
|---|---|
| Ubq 1st intron forward | 5’-GCTCTAACCTTGAGTACCTATCTA-3’ |
| Ubq 1st intron forward | 5’-TTATCGCATACTTCCGTCCCGAT-3’ |
| Nos terminator reverse | 5’-CCATCTCATAAATAACGTCATGCAT-3’ |
| Nos terminator reverse | 5’- AGACAACTTAATGCAATTCGTACAT-3’ |
| GUS linker forward | 5’-CGTCGGTGAACAGGTATGGAATT-3’ |
| GUS linker forward | 5’-TTATACGGAACGCTCCAGCGTT-3’ |
| GUS linker reverse | 5’-CACGTAAGTCCGCATCTTCATGA-3’ |
| GUS linker reverse | 5’-CGCTTGTCAAGGACTAATTGGTG-3’ |
| attB1 forward | 5’-AGTTTGTACAAAAAAGCAGGCTCC-3’ |
| attB2 reverse | 5’-GCTGGGTCGAAAGAACATGTTTCA-3’ |
| Real time PAL forward | 5’-TGAATAACAGTGGAGTGTGGAG-3’ |
| Real time PAL reverse | 5’-AACCTGCCACTCGTACCAAG-3’ |
| Real time PBZ1 forward | 5’-ATGAAGCTTAACCCTGCCGC-3’ |
| Real time PBZ1 reverse | 5’-GTCTCCGTCGAGTGTGACTTG-3’ |
| Real time ubiquitin forward | 5’-AACCAGCTGAGGCCCAAGA-3’ |
| Real time ubiquitin reverse | 5’-ACGATTGATTTAACCAGTCCATGA-3’ |
| M. grisea Pot2 forward | 5’-ACGACCCGTCTTTACTTATTTGG-3’ |
| M. grisea Pot2 reverse | 5’-AAGTAGCGTTGGTTTTGTTGGAT-3’ |
| OsRap2.6 forward | 5’-CACCCGGCACCTGGACAGAACAGATCA-3’ |
| OsRap2.6 reverse | 5’ -AGAATCCTCTCTCTTGCTTTACTTGGAC-3’ |
| Rap2.6 forward | 5’-GAGCCTGACCTATTGCATCTCC-3’ |
| Rap2.6 reverse | 5’-GGCCTCCAGAAGAAGATGTTGG-3’ |
| pB12221-35S forward | 5’-ACTGACGTAAGGATGACGC-3’ |
| nos terminator | 5’-GATAATCATCGCAAGACCG-3’ |
| OsRap2.6 Ox forward | 5’-CACCATGGTCACCGCGCTAGCCCACGTCA-3’ |
| OsRap2.6 Ox reverse | 5’-GAACGATCGGGGAAATTCGAGCTC-3’ |
Primer names and oligonucleotide sequences used for sequencing.
Isolation of rice protoplasts, transfection and BiFC
For effective protoplast isolation, suspension cells were crushed from primary calli into small pieces prior to enzyme treatment. The protoplasts were adjusted to a density of 1.5-2 x 107 cells/ml (Kyozuka et al 1987). For intracellular localization studies, 100 μl of protoplasts were transfected with 9–10 μg plasmids (Venus-OsRap2.6) or (RACK1A-Venus) and/or control plasmids Cherry, NLS-Cerulean and OsGenL-CFP (nuclear marker). The BiFC system used in this study was as described previously with slight modifications (Chen et al 2010a; Kawano et al 2010; Kakita et al. 2007). For the interaction studies, protoplasts (100 μl) (1.5-2 × 106 cells) were transformed with 2.5-5 μg of each paired construct (Vn-OsRap2.6 + RACK1A-Vc), (Vn-OsRap2.6 + Vc-OsMAPK3/6) and a negative control (Vn-OsRap2.6 + GUS-Vc) by the polyethylene glycol (PEG) method with minor modifications (Yoo et al 2007). The protoplasts were incubated at 30°C for 15 hours. The localization or co-localization of YFP/CFP proteins and their markers was assessed with a confocal microscope (Leica TCS SP5) in sequential scanning mode as described in the next section. Quantitative assays were accomplished using a method described previously where 50–100 cells of each construct were randomly scanned and categorized according to their plasma membrane (PM), cytoplasm (C), nuclear (N), or cytoplasm and nuclear (CN) localization patterns.
Confocal scanning microscopy
Confocal scanning microscope was used to image the rice protoplasts expressing fluorescent proteins. The microscope was equipped with the Leica confocal software (LCS), a 100mW multi-line Argon laser (458nm, 476nm, 488nm, 496nm and 514nm), diode pumped solid state laser (DPSS) (442nm), a 10mW DPSS (561nm), a 10mW He-Ne Laser (633nm) and a 50mW UV laser (351nm-364nm) as excitation sources. The SP scanner collected the FP signal at various wavelengths, and the auto fluorescence of the protoplasts was measured between 440 nm and 650 nm. LCS carried out the image maximal projection. Images were acquired using the 10x/0.4 HC PLAPO CS object lens and the 40x/0.85 HCX PLAPO CS object lens. The 63x/1.2 HCX PLAPO CS and 40x 1.25-0.70 HCX PL APO CS object lenses were used to obtain images when fluorescent proteins were targeted to any location.
RNAi, Ox constructs and rice transformation
To generate an RNAi construct for gene suppression, a 300 base pair fragment was amplified by PCR from OsRap2.6 with the OsRap2.6 RNAi primers listed in Table 2. The open reading frame (ORF) of the OsRap2.6 construct was amplified using the OsRap2.6 Ox primers listed in Table 2. The PCR fragments were cloned into the Gateway pENTR/D-TOPO cloning vector. Subsequently, the derived fragments were transferred to the pANDA destination vector by recombinase (LR) reactions. The pANDA vector has kanamycin and hygromycin resistance markers for transformation (Miki and Shimamoto 2004). The insert and vector sequences were confirmed by PCR using the first intron of Ubiquitin, the nos terminator, GUS linker and attribute B1 and B2 primers listed in Table 2.
Suspension cells, RNAi and over-expressing plants
OsRap2.6 RNAi and Ox calli were derived from japonica rice cv. Kimnaze. The seeds were surface sterilised with 1.2% sodium hypochlorite for 45 min, washed in distilled water and placed on Murashige and Skoog (MS) medium supplemented with 2 mg/L, 2, 4-dichlorophenoxyacetic acid (2,4-D) (Murashige and Skoog 1962). Plants were generated by Agrobacterium tumifaciens-mediated transformation of rice callus as described previously (Miki and Shimamoto 2004; Hiei et al 1994). The transformed callus was selected with forward (5’-TGGCGGCTACTACCCCTCGTCGT-3’) and reverse (5’-GAACGATCGGGGAAATTCGAGCTC-3’) primers. The suspension culture derived from transformed callus was maintained in R2S medium (Ohira et al 1973). OsRap2.6 RNAi plants were screened using PCR with Rap2.6 primers listed in Table 2.
RNA extraction and reverse transcription PCR
For the analysis of gene expression, rice calli from the WT suspension cells were treated with 2 μg/ml chitin (Hepta-N-acetylchitoheptaose, Sigma) and harvested at different time intervals (Lieberherr et al 2005). The samples were frozen in liquid nitrogen and stored at –80°C. Briefly, RNA was extracted by the TRIzol method (Nacalai tesque, Japan). The samples were digested with DNaseI (Takara, Shiga, Japan). Electrophoresis was done in 1.5% agarose gels in 1 X TBE buffer, at 100V for 30 min. The gels were stained with ethidium bromide for 15 min. Bands were visualized under UV light.
Infection of rice plants with M. oryzae
OsRap2.6 RNAi and Ox plants were infected with the compatible race (007) or the incompatible race (031) of rice blast fungus (M. oryzae). The fungal growth conditions and the punch infection method were done as described previously with minor modifications (Kim et al 2012; Chen et al 2010a; Kawano et al 2010). The spores were estimated to contain ~ 1 × 105 spores per ml. The spore suspension was inoculated on leaf blades and kept at 23~30°C in the greenhouse. Disease lesions sizes were measured 7 days after inoculation. Briefly, the two youngest leaf blades were selected for infection. Six holes were punched per blade in 4 plants giving a total of 48 infected sampling points. Lesion length was measured quantitatively with a digital calliper. The resistance and susceptibility of each plant was compared with the wild type using four cv. Kinmaze plants. The experiment was repeated three times. The data were analysed for statistical significance using the Excel program (Microsoft). The means and standard errors were analysed and the p-values were determined by a standard t -test (p<0.05).
Quantitative PCR (qPCR)
We used the standard curve quantification method that absolute values were derived from known quantities. The qPCR mixture (20 μl) was loaded into Ultra AMP PCR plates and analysed in an ABI StepOne Real-Time PCR System for 2.5 hours. To detect M. grisea and rice DNAs, two sets of primers against M. grisea Pot2 and rice Ubiquitin were used in qPCR (Beruyer et al 2006). The DNA representing the relative number of fungus cells was quantified per plant cell from the infected rice tissues by calculating an infection ratio with the formula (N: Mgpot2/ N: Osubiquitin x 100) as described previously (Kawano et al 2010). DNA was extracted from the infected lesions and analysed further qualitatively in qPCR (ABI StepOne Real-Time PCR System) using M. grisea Pot2, PAL1, PBZ1 and Ubiquitin primers listed in Table 2.
Electronic supplementary material
Additional file 1:Figure S1. OsRap2.6 RNAi plants are not susceptible to M. oryzae, incompatible race, 031. (A) Fungal infections on leaf blades of WT and OsRap2.6 RNAi after infection with 031, an incompatible race of rice blast fungus. R1, R5 and R10 are independently transformed lines. (B) Quantitative analysis of fungal growth in OsRap2.6 RNAi, 7 days after infection. Ubiquitin was used as an internal control. Bars represent the means ± SD calculated using four biological replicates where each consists of three independent technical replicates (p ≥ 0.05, n=48). (C) Lesion length of OsRap2.6 RNAi plants showing susceptibility to blast fungus incompatible race, 031 as compared to WT (p ≥ 0.05, n=48). (D) Expression of PAL1 mRNA in OsRap2.6 RNAi after rice blast infection with the incompatible race, 031. Levels of PAL1 mRNA were measured by reverse transcription qPCR (p ≥ 0.05, n=48). (PDF 240 KB)
Additional file 2:Figure S2. OsRap2.6 Ox plants were not resistant to M. oryzae, incompatible race, 031. (A) Fungal infections on leaf blades of WT and OsRap2.6 Ox plants in incompatible race, 031. P4, P6, and P14 are independently transformed lines. (B) Quantitative analysis of fungal growth in OsRap2.6 Ox plants 7 days after infection with the incompatible race (031) of rice blast fungus. Ubiquitin was used as an internal control. Bars represent the means ± SD calculated using four biological replicates where each consists of three independent technical replicates (p ≥ 0.05, n=48). (C) Lesion length of OsRap2.6 Ox showing susceptibility to blast fungus incompatible race, 031 as compared to WT. (D) Expression of PAL1 mRNA in OsRap2.6 Ox plants after infection (p ≥ 0.05). (PDF 2 MB)
Below are the links to the authors’ original submitted files for images.
Acknowledgements
We thank Prof. Ian Smith for constructive guidance throughout the study. We thank Letian Chen for OsGenL-CFP and OsCERK1-GFP and Kim Sung Hyun for Vc-MAPK3 and Vc-MAPK6 constructs. We also thank the laboratory and technical staff of the Shimamoto laboratory. This research was supported by Grants-in-Aid from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, PMI-0007) and the Japan Society for Promotion of Science (13G0023) to K.S.
Abbreviations
- AP2
Apetala2
- ERF
Ethylene-responsive element-binding proteins
- bHLH
Basic Helix Loop Helix
- BiFC
Bimolecular fluorescence complementation
- CA-OsRac1
Constitutively active-OsRac1
- CCR1
Cinnamoyl CoA reductase 1
- CC-NB-LRR
Coiled-coil NB-LRR
- CERK1
a LysM receptor kinase essential for chitin elicitor signaling
- CFP
Cyan fluorescent protein
- DN-OsRac1
Dominant negative-OsRac1
- DREB
Dehydration-responsive element-binding protein
- DRMs
Detergent resistant membranes
- ER
Endoplasmic reticulum
- ERF
Ethylene-responsive factor
- ETI
Effector triggered immunity
- FLS2
Flagellin sensing 2
- GAP
GTPase-activating protein
- GTP
Guanosine triphosphate
- GDP
Guanosine diphosphate
- GEF
Guanine exchange factors
- GFP
Green fluorescent protein
- HR
Hypersensitive responses
- Hsp70
Heat shock protein 70
- Hsp90
Heat shock protein 90
- LCM
Leica confocal software
- MAPK 3/6
Mitogen-activated protein kinases 3 and 6
- NADPH
Nicotinamide adenine dinucleotide phosphate
- NB-LRR
(NB)-leucine rich repeat (LRR)
- NB-ARC
ARC: APAF-1, certain R gene products and CED-4
- NLS
Nuclear localisation signal
- NO
Nitric oxide
- OsRac1
Rice small GTPase, Rac1
- OsRACK1
A rice Receptor for Activated Kinase C1 (RACK1)
- OsRap2.6
A rice transcription factor, Rap2.6
- Ox
Overexpressing
- PAL1
Phenylalanine ammonia-lyase 1
- PBZ1
Probenazole-induced protein 1
- PAMP
Pathogen associated molecular pattern
- PKC
Protein kinase C
- PM
Plasma membrane
- PR genes
Pathogen related genes
- qPCR
Real time polymerase chain reaction
- R protein
Resistance protein
- RAl1
Rac immunity 1
- RACK1
Receptor for activated Kinase C-1
- RDV
Rice dwarf virus
- RLK
Receptor-like kinases
- ROS
Reactive oxygen species
- RSV
Rice stripe virus
- RTSV
Rice tungro spherical virus
- SA
Salicyclic acid
- TIR
Toll/ interleukin-1 receptor (TIR) or
- Vn/Vc
Venus N terminus and C terminus
- YFP
Yellow fluorescent protein
- Y2H
Yeast two-hybrid.
Footnotes
Competing interests
The authors declare no competing interests.
Authors’ contributions
MJW carried out the laboratory experiments and co-wrote the manuscript. MJW, RY and YK performed the yeast two-hybrid screening. HLW and TK provided critical guidance during the research. KS designed the experiments and co-wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Mwathi Jane Wamaitha, Email: j-mwathi@bs.naist.jp.
Risa Yamamoto, Email: r-yamamoto@bs.naist.jp.
Hann Ling Wong, Email: hlwong@utar.edu.my.
Tsutomu Kawasaki, Email: t-kawasaki@nara.kindai.ac.jp.
Yoji Kawano, Email: y-kawano@bs.naist.jp.
Ko Shimamoto, Email: simamoto@bs.naist.jp.
References
- Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Comm Signal. 2011;9(22):823–830. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjobo-Hermans MJ, Goedhart J, Gadella TW., Jr Plant G protein heterotrimers require dual lipidation motifs of G alpha and G gamma and do not dissociate upon activation. J Cell Sci. 2006;119:5087–5097. doi: 10.1242/jcs.03284. [DOI] [PubMed] [Google Scholar]
- Agrawal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase-signaling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopath. 2007;45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- Berken A. Rops in the spotlight of plant signal transduction. Cell Mol Life Sci. 2006;63:2446–2459. doi: 10.1007/s00018-006-6197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beruyer R, Poussier S, Kankanala P, Mosquera G, Valent B. Quantitative and qualitastive influence of inoculation methods on in planta growth of rice blast fungus. Phytopathology. 2006;96:346–355. doi: 10.1094/PHYTO-96-0346. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. Differential innate immune signalling via Ca (2+) sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Dell EJ, Lin F, Sai J, Hamm HE. RACK1 regulates specific functions of G beta gamma. J Biol Chem. 2004;279:17861–17868. doi: 10.1074/jbc.M313727200. [DOI] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Temple B, Liang J, Guo J, Alonso JM, Ecker JR, Jones AM. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot. 2006;57:2697–2708. doi: 10.1093/jxb/erl035. [DOI] [PubMed] [Google Scholar]
- Chen L, Hamada S, Fujiwara M, Zhu T, Thao NP, Wong HL, Krishna P, Ueda T, Kaku H, Shibuya N, Kawasaki T, Shimamoto K. The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe. 2010;7:185–196. doi: 10.1016/j.chom.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Chen L, Shiotani K, Togashi T, Miki D, Aoyama M, Wong HL, Kawasaki T, Shimamoto K. Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant Cell Physiol. 2010;51:585–595. doi: 10.1093/pcp/pcq024. [DOI] [PubMed] [Google Scholar]
- Cheong YH, Moon BC, Kim JK, Kim CY, Kim MC, Kim IH, Park CY, Kim JC, Park BO, Koo SC. BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 2003;132:1961–1972. doi: 10.1104/pp.103.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Couch BC, Fudal I, Lebrun MH, Tharreau D, Valent B, van Kim P, Notteghem J-L, Kohn LM. Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics. 2005;170:613–630. doi: 10.1534/genetics.105.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Hamada S, Hiratsuka M, Fukao Y, Kawasaki T, Shimamoto K. Proteome analysis of detergent-resistant membranes (DRMs) associated with OsRac1-mediated innate immunity in rice. Plant Cell Physiol. 2009;50:1191–1200. doi: 10.1093/pcp/pcp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Fong SH, Yang D, Wang GL. BWMK1, a novel MAP kinase induced by fungal infection and mechanical wounding in rice. Mol Plant Microbe Interact. 1999;12:1064–1073. doi: 10.1094/MPMI.1999.12.12.1064. [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Sheen J. Elicitation and suppression of microbe-associated molecular pattern-triggered immunity in plant-microbe interactions. Cell Microbiol. 2007;9:1385–1396. doi: 10.1111/j.1462-5822.2007.00944.x. [DOI] [PubMed] [Google Scholar]
- Heath MC. Hypersensitive response related death. Plant Mol Biol. 2000;44:321–334. doi: 10.1023/A:1026592509060. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL. Initiation of run away cell death in an Arabidopsis mutant by extracellular superoxide. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- Jantasuriyarat C, Gowda M, Haller K, Hatfield J, Lu G, Stahlberg E, Zhou B, Li H, Kim H, Yu Y, Dean RA, Wing RA, Soderlund C, Wang GL. Large-scale identification of expressed sequence tags involved in rice and rice blast fungus interaction. Plant Physiol. 2005;138:105–115. doi: 10.1104/pp.104.055624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL, Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem. 2006;285:13471–13479. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakita M, Murase K, Iwano M, Matsumoto T, Watanabe M, Shiba H, Isogai A, Takayama S. Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell. 2007;19:3961–73. doi: 10.1105/tpc.106.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong HL, Kawasaki T, Shimamoto K. Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe. 2010;7:362–375. doi: 10.1016/j.chom.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K. The small GTP-binding protein Rac is a regulator of cell death in plants. Proc Natl Acad Sci USA. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Koita H, Nakatsubo T, Hasegawa K, Wakabayashi K, Takahashi H, Umemura K, Umezawa T, Shimamoto K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc Natl Acad Sci USA. 2006;103:230–235. doi: 10.1073/pnas.0509875103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev. 2009;38:2876–2886. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Il-Pyung A, Lee YH. Analysis of genes expressed during rice Magnaporthe grisea interactions. Mol Plant Microbe Interact. 2001;14:1340–1346. doi: 10.1094/MPMI.2001.14.11.1340. [DOI] [PubMed] [Google Scholar]
- Kim S-H, Oikawa T, Kyozuka K, Wong H-L, Umemura K, Kishi-Kaboshi M, Takahashi A, Kawano Y, Kawasaki T, Shimamoto K. The bHLH Rac Immunity1 (RAI1) is activated by OsRac1 via OsMAPK3 and OsMAPK6 in rice immunity. Plant Cell Physiol. 2012;53(4):740–754. doi: 10.1093/pcp/pcs033. [DOI] [PubMed] [Google Scholar]
- Kishi-Kaboshi M, Okada K, Kurimoto L, Murakami S, Umezawa T, Shibuya N, Yamane H, Miyao A, Takatsuji H, Takahashi A, Hirochika H. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 2010;63:599–612. doi: 10.1111/j.1365-313X.2010.04264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Kim SA, Lee SK, Oh SA, Byoun CH, Han JK, Nam HG. Insulin-induced maturation of Xenopus oocytes is inhibited by microinjection of a Brassica napus cDNA clone with high similarity to a mammalian receptor for activated protein kinase C. Planta. 1997;201:245–251. doi: 10.1007/s004250050063. [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Hayashi Υ, Shimamoto Κ. High frequency plant regeneration from rice protoplasts by novel nurse culture methods. Mol Gen Genet. 1987;206:408–413. doi: 10.1007/BF00428879. [DOI] [Google Scholar]
- Lieberherr D, Thao NP, Nakashima A, Umemura K, Kawasaki T, Shimamoto K. A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice. Plant Physiol. 2005;138:1644–1652. doi: 10.1104/pp.104.057414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YV, Hubbi ME, Pan F, McDonald KR, Mansharamani M, Cole RN, Liu JO, Semenza GL. Calcineurin promotes hypoxia-inducible factor 1 alpha expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J Biol Chem. 2007;282:37064–37073. doi: 10.1074/jbc.M705015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E, Sjolander K, Hake S. From endonucleases to transcription factors: Evolution of the AP2 DNA binding domain in plants. Plant Cell. 2004;16:2265–2277. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharm. 2002;62:1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Mittler R, Lam E, Shulaev V, Cohen M. Signals controlling the expression of cytosolic ascorbate peroxidase during pathogen-induced programmed cell death in tobacco. Plant Mol Biol. 1999;39:1025–1035. doi: 10.1023/A:1006110223774. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Plant Physiol. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Chen L, Thao NP, Fujiwara M, Wong HL, Kuwano M, Umemura K, Shirasu K, Kawasaki T, Shimamoto K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell. 2008;20:2265–2279. doi: 10.1105/tpc.107.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF, 3rd, Dangl JL. Recognition and response in the plant immune system. Annu Rev Genet. 2003;37:579–609. doi: 10.1146/annurev.genet.37.110801.142628. [DOI] [PubMed] [Google Scholar]
- Ohira K, Ojima K, Fujiwara A. Studies on the nutrition of rice cell culture 1. A simple, defined medium for rapid growth in suspension culture. Plant Cell Physiol. 1973;14:1113–1121. [Google Scholar]
- Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejnik K, Bucholc M, Anielska-Mazur A, Lipko A, Kujawa M, Modzelan M, Augustyn A, Kraszewska E. Arabidopsis thaliana Nudix hydrolase AtNUDT7 forms complexes with the regulatory RACK1A protein and Ggamma subunits of the signal transducing heterotrimeric G protein. Acta Biochem Pol. 2011;58:609–616. [PubMed] [Google Scholar]
- Olejnik K, Bucholc M, Anielska-Mazur A, Lipko A, Kujawa M, Modzelan M, Augustyn A, Kraszewska E. Arabidopsis thaliana Nudix hydrolase AtNUDT7 forms complexes with the regulatory RACK1A protein and Ggamma subunits of the signal transducing heterotrimeric G protein. Acta Biochimica Polonica. 2011;58(4):609–616. [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA. 2001;98:759–764. doi: 10.1073/pnas.98.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paduch M, Jeleń F, Otlewski J. Structure of small G proteins and their regulators. Acta Biochim Pol. 2001;48:829–50. [PubMed] [Google Scholar]
- Ribot C, Hirsch J, Balzergue S, Tharreau D, Notteghem JL, Lebrun MH, Morel JB. Susceptibility of rice to the blast fungus, Magnaporthe grisea. J Plant Physiol. 2008;165:114–124. doi: 10.1016/j.jplph.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Curr Opin Plant Biol. 2000;3:423–434. doi: 10.1016/S1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Comm. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Sharoni AM, Nuruzzaman M, Satoh K, Shimizu T, Kondoh H, Sasaya T, Choi IR, Omura T, Kikuchi S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011;52:344–360. doi: 10.1093/pcp/pcq196. [DOI] [PubMed] [Google Scholar]
- Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- Sondek J, Siderovski DP. G gamma-like (GGL) domains: new frontiers in G-protein signaling and beta-propeller scaffolding. Biochem Pharm. 2001;61:1329–1337. doi: 10.1016/S0006-2952(01)00633-5. [DOI] [PubMed] [Google Scholar]
- Suharsono U, Fujisawa Y, Kawasaki T, Iwasaki Y, Satoh H, Shimamoto K. The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA. 2002;99:13307–13312. doi: 10.1073/pnas.192244099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:11777–11782. doi: 10.1073/pnas.2033934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao NP, Chen L, Nakashima A, Hara S, Umemura K, Takahashi A, Shirasu K, Kawasaki T, Shimamoto K. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell. 2007;19:4035–4045. doi: 10.1105/tpc.107.055517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton C, Tang KC, Phamluong K, Luong K, Vagts A, Nikanjam D, Yaka R, Ron D. Spatial and temporal regulation of RACK1 function and N-methyl-D-aspartate receptor activity through WD40 motif-mediated dimerization. J Biol Chem. 2004;279:31357–31364. doi: 10.1074/jbc.M402316200. [DOI] [PubMed] [Google Scholar]
- Valent B, Chumley FG. Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu Rev Phytopath. 1991;29:443–467. doi: 10.1146/annurev.py.29.090191.002303. [DOI] [PubMed] [Google Scholar]
- Wilson RA, Talbot NJ. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 2009;7:185–195. doi: 10.1038/nrmicro2032. [DOI] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima C, Yoshioka H, Iba K, Kawasaki T, Shimamoto K. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell. 2007;19:4022–4034. doi: 10.1105/tpc.107.055624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaka R, He DY, Phamluong K, Ron D. Pituitary adenylate cyclase-activating polypeptide (PACAP (1–38)) enhances N-methyl-D-aspartate receptor function and brain-derived neurotrophic factor expression via RACK1. J Biol Chem. 2003;278:9630–9638. doi: 10.1074/jbc.M209141200. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protocol. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, Zhang H. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene. 2010;457:1–12. doi: 10.1016/j.gene.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Zipfel C. Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol. 2008;20:10–16. doi: 10.1016/j.coi.2007.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1:Figure S1. OsRap2.6 RNAi plants are not susceptible to M. oryzae, incompatible race, 031. (A) Fungal infections on leaf blades of WT and OsRap2.6 RNAi after infection with 031, an incompatible race of rice blast fungus. R1, R5 and R10 are independently transformed lines. (B) Quantitative analysis of fungal growth in OsRap2.6 RNAi, 7 days after infection. Ubiquitin was used as an internal control. Bars represent the means ± SD calculated using four biological replicates where each consists of three independent technical replicates (p ≥ 0.05, n=48). (C) Lesion length of OsRap2.6 RNAi plants showing susceptibility to blast fungus incompatible race, 031 as compared to WT (p ≥ 0.05, n=48). (D) Expression of PAL1 mRNA in OsRap2.6 RNAi after rice blast infection with the incompatible race, 031. Levels of PAL1 mRNA were measured by reverse transcription qPCR (p ≥ 0.05, n=48). (PDF 240 KB)
Additional file 2:Figure S2. OsRap2.6 Ox plants were not resistant to M. oryzae, incompatible race, 031. (A) Fungal infections on leaf blades of WT and OsRap2.6 Ox plants in incompatible race, 031. P4, P6, and P14 are independently transformed lines. (B) Quantitative analysis of fungal growth in OsRap2.6 Ox plants 7 days after infection with the incompatible race (031) of rice blast fungus. Ubiquitin was used as an internal control. Bars represent the means ± SD calculated using four biological replicates where each consists of three independent technical replicates (p ≥ 0.05, n=48). (C) Lesion length of OsRap2.6 Ox showing susceptibility to blast fungus incompatible race, 031 as compared to WT. (D) Expression of PAL1 mRNA in OsRap2.6 Ox plants after infection (p ≥ 0.05). (PDF 2 MB)


