Abstract
Background
A number of QTL studies reported that one genomic region was associated with several traits, indicating linkage and/or pleiotropic effects. The question of pleiotropy versus tight linkage in these studies should be solved using a large-size population combined with high-density mapping. For example, if each of the 2 parents has a TGW-increasing or SPP-increasing QTL that is tightly linked, complementary combination of the 2 beneficial QTLs by using molecular markers could produce higher yields compared to the 2 parents. However, a pleiotropic QTL with opposite effects on the SPP and 1,000-grain weight (TGW) is complicated and challenging in terms of its application to rice improvement.
Results
In this study, using a series of BC5F4 nearly isogenic lines (NILs) that were derived from a cross between the Korean japonica cultivar Hwayeongbyeo and Oryza rufipogon, we demonstrated that 2 QTLs, qSPP5 for spikelets per panicle (SPP) and qTGW5 for grain weight (TGW), are tightly linked on chromosome 5. Alleles from the O. rufipogon parent increased the SPP and decreased TGW in the Hwayeongbyeo background. qSPP5 was located within a 803-kb interval between the simple sequence repeat (SSR) markers INDEL3 and RM18076. Based on the map position, qTGW 5 seemed to be the same gene as qSW5, which controls grain morphology. The additive effect of the O. rufipogon allele at qSPP5 was 10–15 SPP, and 33.0% of the phenotypic variance could be explained by the segregation of the SSR marker RM18058. Yield trials with BC5F4 NILs showed that lines that contained a homozygous O. rufipogon introgression at the qSPP5 region out-yielded sibling NILs that contained Hwayeongbyeo DNA by 15.3% and out-yielded the Hwayeongbyeo parent by 7.3%.
Conclusion
Based on the finding that the O. rufipogon allele for the SPP was beneficial in the japonica and indica cultivar backgrounds, the qSPP5 allele could be valuable for improving rice yields. In addition, the NIL populations and molecular markers are useful for cloning qSPP5.
Electronic supplementary material
The online version of this article (doi:10.1186/1939-8433-6-33) contains supplementary material, which is available to authorized users.
Keywords: Rice; Spikelets per panicle; 1,000-grain weight; QTL; Linkage; Near isogenic lines
Background
Asian cultivated rice (Oryza sativa L.) originated from common wild rice (Oryza rufipogon Griff.), and their morphological, biochemical and genetic relationships have been analyzed in many studies (Sun et al., 2001; Cai & Morishima 2002). Much of its genetic architecture and phenotypic construction changed during domestication from wild rice. In general, Oryza sativa is different from O. rufipogon in terms of a number of traits such as plant height, number of spikelets per panicle (SPP), 1000-grain weight, grain shape, and awn. Among these agronomic traits, the SPP and 1000-grain weight are determinants of grain yield (YD).
The number of primary and secondary branches (SBs) strongly influences the average number of SPP (Yamagishi et al., 2002). QTLs for the SPP have been detected using various segregating populations (Kobayashi et al., 2004). Several QTLs for the SPP have also been identified in wild relatives (Thomson et al., 2003; Suh et al., 2005; Onishi et al., 2007). These QTLs are located across the chromosomes and provide valuable information on the genes that control the SPP in different populations. In addition, SPP QTLs have been mapped as a single Mendelian factor (Zhang et al., 2006,2009) and were rarely found on chromosomes 5 and 10 (Thomson et al., 2003; Tan et al., 2008). And these studies showed that the wild rice allele leads to increased or decreased number of SPP.
Increase of the grain weight is a method for increasing rice yield. Genes that affect the grain size have been identified in inter-specific crosses (Xiao et al., 1998; Thomson et al., 2003; Li et al., 2004; Aluko et al., 2004; Brondani et al., 2002). In most cases, wild-type alleles were associated with small grain, whereas cultivar alleles were associated with large grains. Usually, grain size is determined by grain length (GL), width, and thickness. These 3 traits are quantitatively inherited under the control of several or many genes. To date, 5 key genes controlling seed size have been isolated in rice: GS 3, GW 2, qSW 5 or GW 5, GIF1 and GS5. (Fan et al., 2006; Song et al., 2007; Shomura et al., 2008; Weng et al., 2008; Li et al., 2011). GS 3 has a major effect on seed length, whereas qSW5/GW5 and GW2 confer both the seed or grain width (GW) and weight in rice. GIF1 encodes a cell-wall invertase that is required for carbon partitioning during early grain filling, and the over-expression of GIF1 by using its native promoter leads to large grains (Wang et al., 2008). Shomura et al. (2008) found that a deletion in qSW 5 was associated with grain size owing to an increase in the cell number in the outer glume of the rice spikelet.
A number of QTL studies showed that one genomic region was associated with several traits, especially yield component traits, indicating linkage and/or pleiotropic effects (Xiao et al., 1996; Tian et al., 2006; Tan et al., 2008; Liu et al., 2010). The question of pleiotropy versus tight linkage in these studies should be solved using a large-size population combined with high-density mapping, because its implication is important for improving rice quality and yield. For example, if each of the 2 parents has a TGW-increasing or SPP-increasing QTL that is tightly linked, complementary combination of the 2 beneficial QTLs by using molecular markers could produce higher yields compared to the 2 parents. However, a pleiotropic QTL with opposite effects on the SPP and 1,000-grain weight (TGW) is complicated and challenging in terms of its application to rice improvement.
We conducted this study to characterize the QTL, qSPP5 in terms of the SPP and to determine its linkage relationship with the grain weight gene, qTGW5 by using near-isogenic lines that were derived from a cross between Hwayeongbyeo (O. sativa) and W1944 (O. rufipogon).
Methods
Population development
In previous studies, the QTLs for the SPP and GW were detected near the SSR markers RM413 and RM194 on chromosome 5 (Lee et al., 2005; Yuan et al., 2009). The scheme that we used to develop the genetic material is shown in Figure 1. To analyze these QTLs, we selected the BC3F4 introgression line CR6 as the basis for fine-mapping for the following reasons: (a) it had an O. rufipogon introgression across the target region as identified by markers RM413 and RM194 on chromosome 5; (b) it was associated with increased SPP and decreased grain weight; and (c) it had only 4 non-target O. rufipogon segments (Figure 2). CR6 was backcrossed to Hwayeongbyeo and then allowed to self to generate a near isogenic line (NIL)-derived BC4F2 population (457 plants), which showed segregation in the target region on chromosome 5. A single BC4F2 plant was selected from this population using the same criteria as mentioned above, and the plant was heterozygous across the target region with respect to markers RM413 and RM194 on chromosome 5. The plant was selfed to produce 434 BC4F3 plants. The QTLs for the SPP and TGW were validated in both of the populations. To further fine map qTGW 5, one BC4F3 plant, CR7111-30, which carried the W1944 homozygous segment for the target region at qTGW5 locus, was crossed with Hwayeongbyeo to produce a BC5F2 population with 326 plants. CR7111-30 had no O. rufipogon introgression at the non-target regions. Among 326 plants, 127 BC5F2 plants were evaluated and used for QTL analysis. 26 BC5F2 plants with informative recombination breakpoints between RM18003 and RM249 were selfed to produce 26 BC5F3 lines for substitution mapping. Finally, 18 BC5F3 lines were selected and selfed to produce BC5F4 lines.
Figure 1.
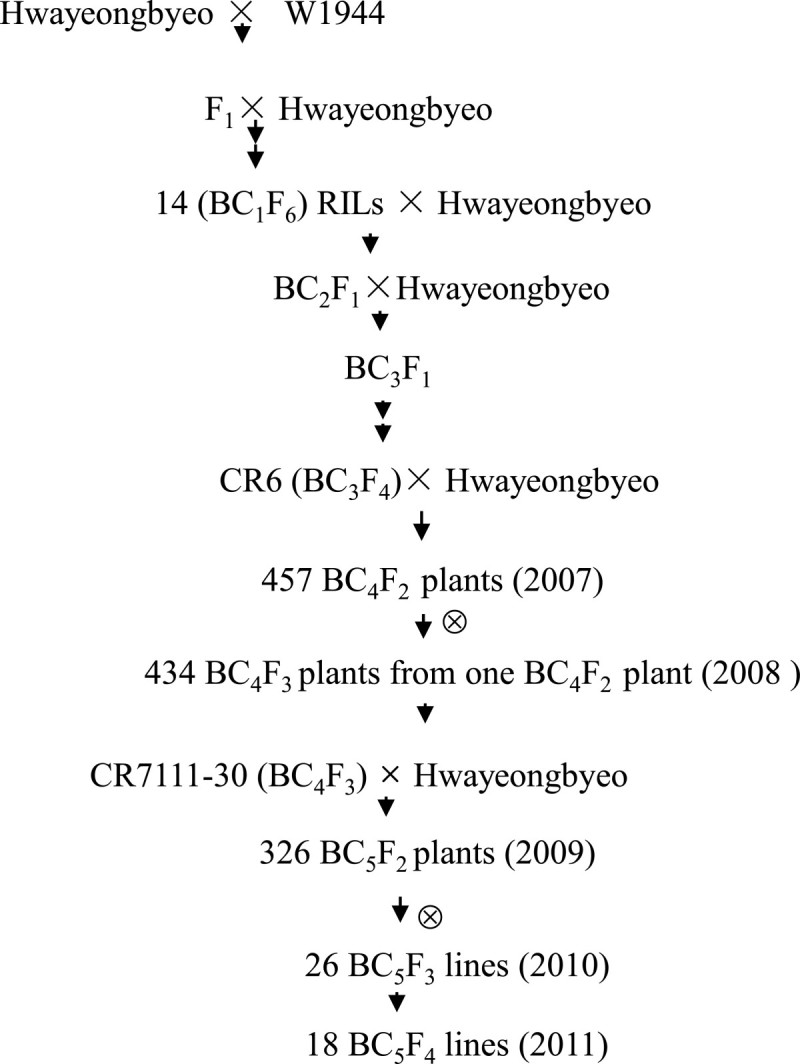
Development of genetic materials that were used in this study.
Figure 2.
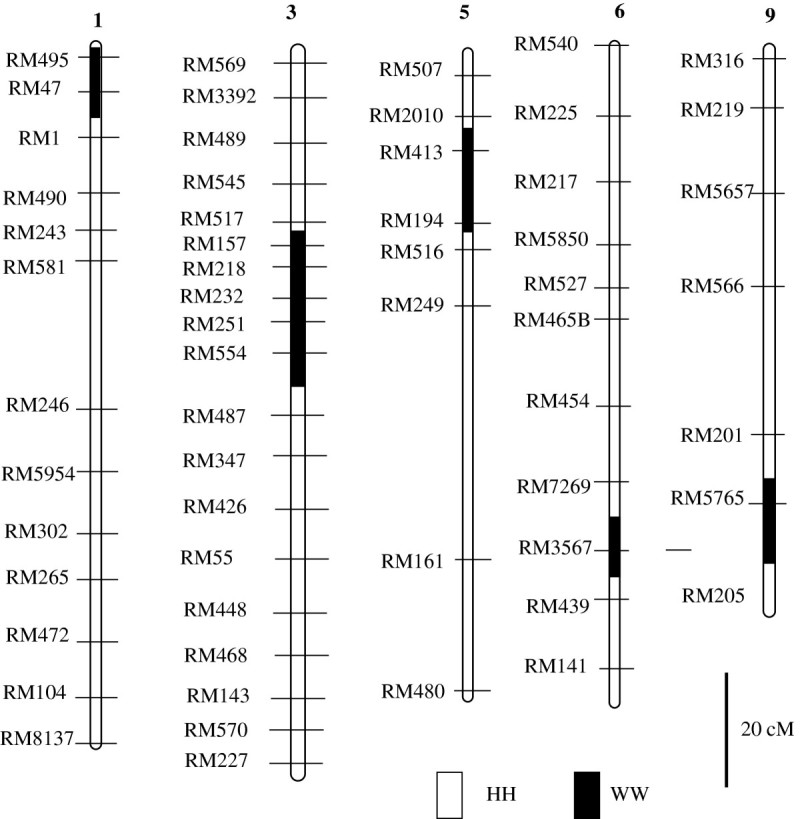
Graphical genotype of the BC3F4line, CR6. CR6 had, in total, 5 introgressed segments including the target segment on chromosome 5. HH: Hwayeongbyeo homozygote; WW: W1944 homozygote.
Phenotypic evaluation
Two populations (BC4F3 and BC5F2), 26 BC5F3 lines, 18 BC5F4 lines, and the parent Hwayeongbyeo, were grown in the field during the summers of 2008–2011 at the Chungnam National University (36°22′ N, 127°22′ E), Daejeon, Korea. Each plant in BC4F3 and BC5F2 was planted 15 cm from the next plant and was spaced at 30 cm between rows. Each line with 25 plants in BC5F3 and BC5F4 was represented by a single row of 30-day-old seedlings that were planted 15 cm from the next plant and spaced at 30 cm between rows. The BC5F4 lines were planted in a completely randomized block design with 3 replications.
Agronomic traits
The culm length (CL), panicle length, primary branch (PB), secondary branch (SB), SPP, TGW, grain length (GL), grain width (GW), grain thickness (GT), and yield per plant (YD) were evaluated for each plant and line as follows. Five plants from the middle of each line were selected to evaluate the CL and panicle length, and the 2 biggest panicles of 5 plants were selected to evaluate the PB, SB, and SPP. Grains that had hulls were allowed to dry naturally after harvesting, and partial or un-filled seeds were removed by soaking the grains in water. Fully filled seeds were re-dried in an oven at 30°C for 24 h. The TGW was evaluated by measuring the weight of 100 randomly selected, fully filled grains: this method was performed in triplicate and the values were averaged to yield a single mean. The GL, GW, and GT of 100 grains that were fully filled were measured in triplicate using a 150-mm vernier caliper (Mitutoyo Corp., Japan). The YD, which was measured in grams of seed per plant, was determined for 15 plants that were harvested from the middle of 1 plot per block. The TGW and yield per plant were corrected for 12% grain moisture content.
DNA extraction and simple sequence repeat analysis
DNA was extracted from the fresh leaves of BC4F3 plants, BC5F2 plants, and BC5F4 lines by using the CTAB method described by Causse et al. (1994). SSR primers were synthesized according to an available public rice genomic sequence (http://www.gramene.org/markers/). One primer, INDEL3, in the target region, was designed using primer 3.0 (forward: 5′CATCACTTTCTCTCCTTCCGTTA3′, reverse: 5′TACAGTGTACAGAAAGCTGGTTG3′). A total volume of 20 μL of reaction mixture was composed of 5.0 μL (5 ng/μL) of template DNA, 0.1 μL of Taq polymerase (5 Unit/μL), 0.8 μL of dNTP (2.5 mM each), 1 μL of forward + reverse primer (10 pmol each), 2.0 μL of 10× PCR buffer (10 mM Tris–HCl PH 8.3, 50 mM KCl, 1.5 mM MgCl2, and 0.1% Gelatin), and 11.1 μL of triply distilled water. Amplification was achieved using a Thermo Cycler (Bio-Rad) according to the step-cycle program of denaturation at 94°C for 5 min and then subsequent denaturation performed at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. Steps 2 through 4 were repeated for 35 cycles, in all, followed by a final extension step at 72°C for 5 min. The PCR products were run on a 4% polyacrylamide denaturing gel for 1–2 h at 1800–2000 V, and marker bands were revealed by silver staining (Panaud et al., 1996). 11 SSR markers failed to detect polymorphism in the region between INDEL3 and RM18058 due to genetic similarity between the parents (Lee et al., 2005), and additional genotyping of BC5F4 lines was conducted with targeted SNP markers. The polymorphism was assayed by direct sequencing of 441-bp (5,697,197 - 5,697,637th position) and 1,162-bp (5,892,883 -5,894,044th position) PCR product generated by two primer pairs in Solgent Co., Korea (http://www.solgent.com). The first (F 5′-gattgacttatatttggacctcc-3′ and R 5′- gtaaacggtagtgttgactgca-3′) and second (F 5′- caaaatgaatcggccgaagcac -3′ and R 5′- cagaccagtgtgaagaggagg -3′) primers were designed according to the sequences of the O. rufipogon and Nipponbare (http://rgp.dna.affrc.go.jp/E/IRGSP/Build5/build5.html). The sequence information of O. rufipogon in the target region was provided by Dr. S.R. McCouch at Cornell University. The first SNP, hereafter referred to as SNP-1 which occurs at the 5,697,388th position based on the Nipponbare sequence (http://rgp.dna.affrc.go.jp/E/IRGSP/Build5/build5.html) is characterized by nucleotide T in Hwayeongbyeo but nucleotide C in O. rufipogon. The second SNP, SNP-2 which occurs at the 5,893,072th position based on the Nipponbare sequence (http://www.gramene.org) is characterized by nucleotide thymine (T) in Hwayeongbyeo but nucleotide cytosine (C) in O. rufipogon.
Statistical analysis
One-way ANOVA was performed to determine the effect of each marker on each of the traits. Phenotypic means of 3 genotypes- Hwayeongbyeo and W1944 homozygotes and heterozygotes- were compared using Student’s t-test, and a probability level of 0.5% was used as the threshold for detecting a QTL. The proportion of total phenotypic variance that was explained by each QTL was calculated as an R2 value by carrying out regression analysis using each marker/phenotype combination. QTLs were fine-mapped by comparing the phenotypic means of 3 genotypes of recombinants within the target region by using the SAS statistical software package (SAS Institute, Cary, NC, USA).
Results
Characteristics of CR6
Two parents, CR6 and Hwayeongbyeo, showed significant differences in 6 traits (Table 1). Hwayeongbyeo exhibited less number of SPP but higher TGW than CR6 did. The GW of Hwayeongbyeo was larger than that of CR6, whereas no significant differences in the GT and GL were detected between the 2 parents (data not shown). Moreover, no significant difference was observed for days to heading and spikelet fertility (data not shown).
Table 1.
Comparison of 6 agronomic traits between Hwayeongbyeo and CR6
| Trait# | Hwayeongbyeo | CR6 | Difference@ |
|---|---|---|---|
| SPP | 118.2 ± 12.3 | 142.4 ± 15.1 | ** |
| TGW | 25.5 ± 1.8 | 23.1 ± 1.3 | ** |
| GW | 1.58 ± 0.19 | 1.45 ± 0.18 | ** |
| SB | 20.4 ± 5.2 | 27.2 ± 5.9 | ** |
| PL | 20.3 ± 2.6 | 22.1 ± 2.9 | * |
| CL | 83 ± 5.2 | 87 ± 4.2 | ** |
#SPP, TGW, GW, SB, PL, and CL: spikelets per panicle, 1,000-grain weight, grain width, secondary branches per panicle, panicle length, and culm length, respectively.
@ *, **: Significant at P = 0.05 and 0.01, respectively.
Frequency distribution of the BC5F2 population
Frequency distributions of phenotypes for the TGW, SPP, SB, and CL of the BC5F2 population are shown in Figure 3. The TGW showed a bimodal distribution with 23.5 as the trait value boundary. The other 3 traits exhibited continuous and normal distributions. The distribution indicated that the O. rufipogon segment was associated with increases in the SB and SPP, and decreases in the TGW in the Hwayeongbyeo background. The genotypes of the BC5F2 plants were determined at RM194 and the phenotypic variances that were explained by the marker were 37.0%, 13.9%, 9%, and 20.0%, respectively.
Figure 3.
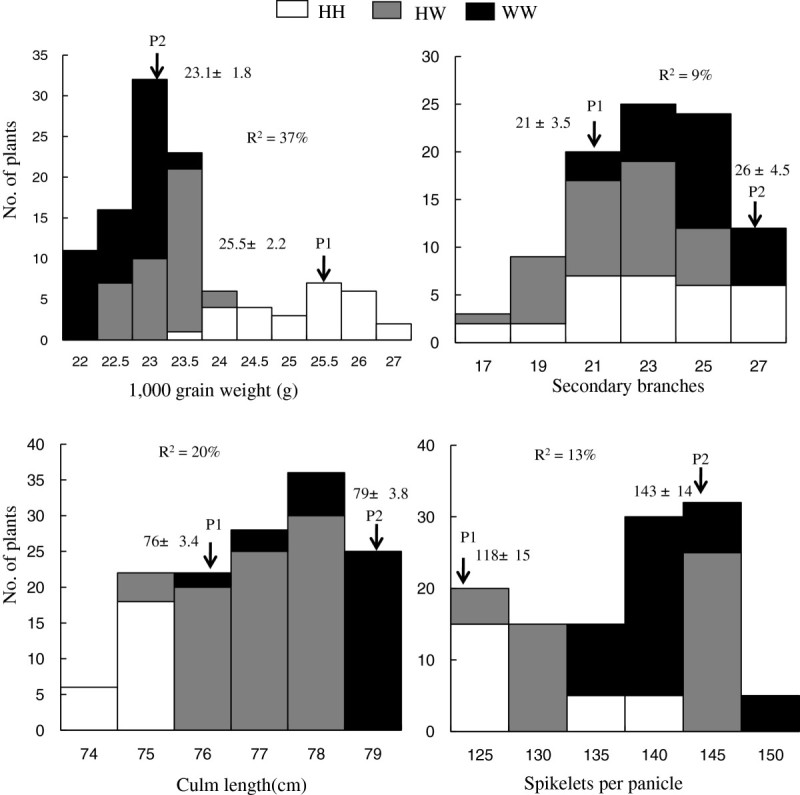
Frequency distribution of 4 traits in the BC5F2population. Three genotypes of the Hwayeongbyeo homozygous and heterozygous and O. rufipogon homozygous classes were identified using the simple sequence repeat marker RM194. HH: Hwayeongbyeo homozygote; WW: W1944 homozygote; and HW: heterozygote. P1 and P2 denote Hwayeongbyeo and CR7111-30, respectively.
QTLs in BC4F3, BC5F2, BC5F3, and BC5F4
The possibility of the effect of non-target regions on SPP and other traits can be excluded because CR7111-30 had no O. rufipogon introgression at the non-target regions. Two SSR markers, RM413 and RM194, were used to genotype the BC4F3 and BC5F2 generations. The QTLs for the TGW, SPP, CL, PL, SB, and GW were all linked to RM194 (Table 2). Seven markers were used to genotype the BC5F3 and BC5F4 populations. The orientations and distances between the markers were based on Nipponbare sequence information (http://www.gramene.org/markers/microsat/). QTL analysis for 5 traits revealed that there was a significant peak near the marker RM194 for the TGW, CL, SPP, SB, and GW, and a peak near RM18076 (Table 2). The phenotypic variance that was explained by each QTL was 9.4-79.0%. This result indicates that this region was a QTL cluster. The SPP for the Hwayeongbyeo homozygous class (HH), the heterozygous class (HW), and the O. rufipogon class (WW) were 119, 147, and 141 at RM18076 in BC5F4, respectively. TGW for the Hwayeongbyeo HH, HW, and the O. rufipogon class WW were 27.1, 25.4, and 24.2 respectively.
Table 2.
QTLs detected in the BC 4 F 3, BC 5 F 2, BC 5 F 3, and BC 5 F 4 generations
| Trait$ | QTL | Marker | Pop. | P | R2 | Phenotypic mean ± s.d.% | ||
|---|---|---|---|---|---|---|---|---|
| HH | HW | WW | ||||||
| TGW | qTGW5 | RM194 | BC4F3 | 0.0001 | 45.9 | 26 ± 1.3(108)# | 24 ± 1.7(210) | 23 ± 1.2(113) |
| RM194 | BC5F2 | 0.0001 | 37.1 | 26 ± 0.8(28) | 24 ± 1.1(55) | 23 ± 0.8(44) | ||
| RM194 | BC5F3 | 0.0001 | 64.8 | 25 ± 0.4(5) | 24 ± 0.6(11) | 23 ± 0.4(10) | ||
| RM194 | BC5F4 | 0.0001 | 79.0 | 25 ± 0.7(6) | 24 ± 0.9(6) | 23 ± 0.6(6) | ||
| SPP | qSPP5 | RM194 | BC4F3 | 0.01 | 9.7 | 123 ± 22 | 145 ± 25 | 144 ± 21 |
| RM194 | BC5F2 | 0.01 | 13.0 | 126 ± 17 | 140 ± 17 | 140 ± 16 | ||
| RM194 | BC5F3 | 0.01 | 19.5 | 133 ± 11 | 148 ± 12 | 146 ± 12 | ||
| RM18058 | BC5F4 | 0.005 | 33.0 | 125 ± 10 | 148 ± 12 | 150 ± 12 | ||
| SB | qSB5 | RM194 | BC4F3 | 0.01 | 9.0 | 24 ± 0.8 | 25 ± 1.2 | 26 ± 1.1 |
| RM194 | BC5F2 | 0.005 | 9.0 | 23 ± 0.8 | 26 ± 1.0 | 26 ± 0.9 | ||
| RM194 | BC5F3 | 0.005 | 20.9 | 23 ± 0.7 | 24 ± 1.0 | 25 ± 0.8 | ||
| RM194 | BC5F4 | 0.0001 | 35.7 | 23 ± 0.8 | 26 ± 1.0 | 27 ± 0.9 | ||
| CL | qCL5 | RM194 | BC4F3 | 0.01 | 9.4 | 77 ± 3.1 | 78 ± 3.2 | 79 ± 2.8 |
| RM194 | BC5F2 | 0.005 | 20.0 | 75 ± 2.6 | 78 ± 3.0 | 78 ± 2.9 | ||
| RM194 | BC5F4 | 0.005 | 21.5 | 79 ± 1.9 | 83 ± 2.0 | 83 ± 1.9 | ||
| GW | qGW5 | RM194 | BC5F4 | 0.0001 | 62.0 | 1.58 ± 0.10 | 1.49 ± 0.10 | 1.45 ± 0.11 |
$TGW: 1,000-grain weight; SPP: number of spikelets; SB: number of secondary branches; CL: culm length; and GW: grain width. %HH: Hwayeongbyeo homozygotes; HW: heterozygotes; and WW: O. rufipogon homozygotes. #Numbers in parenthesis indicate the number of plants or lines.
A strong positive correlation (r = 0.845, P < 0.001) was observed between the GW and TGW in BC5F4, indicating that the variation in the GW was associated with that in the TGW at this locus (data not shown).
Substitution mapping
Substitution mapping was carried out for qTGW5 and qSPP5 by using the BC5F3 and BC5F4 populations (Figure 4). Seven markers were used to screen 26 BC4F3 lines, and these lines were evaluated for the TGW and SPP. The 26 lines were classified into 8 groups based on the genotypes of the SSR markers. The mean phenotypic values of the TGW and SPP for each group were compared to those of the controls, Hwayeongbyeo and CR7111-30. A comparison of the genotypes of recombinants delimited the qTGW5 locus between markers INDEL3 and RM18003 based on the finding that the TGW of the B5 lines with a recombination breakpoint between RM18003 and RM3419 did not significantly differ from that of Hwayeongbyeo but was higher than that of CR7111-30. Moreover, the TGW of B8 lines with a recombination breakpoint between INDEL3 and RM194 did not significantly differ from that of CR7111-30 but was lower than that of Hwayeongbyeo.
Figure 4.
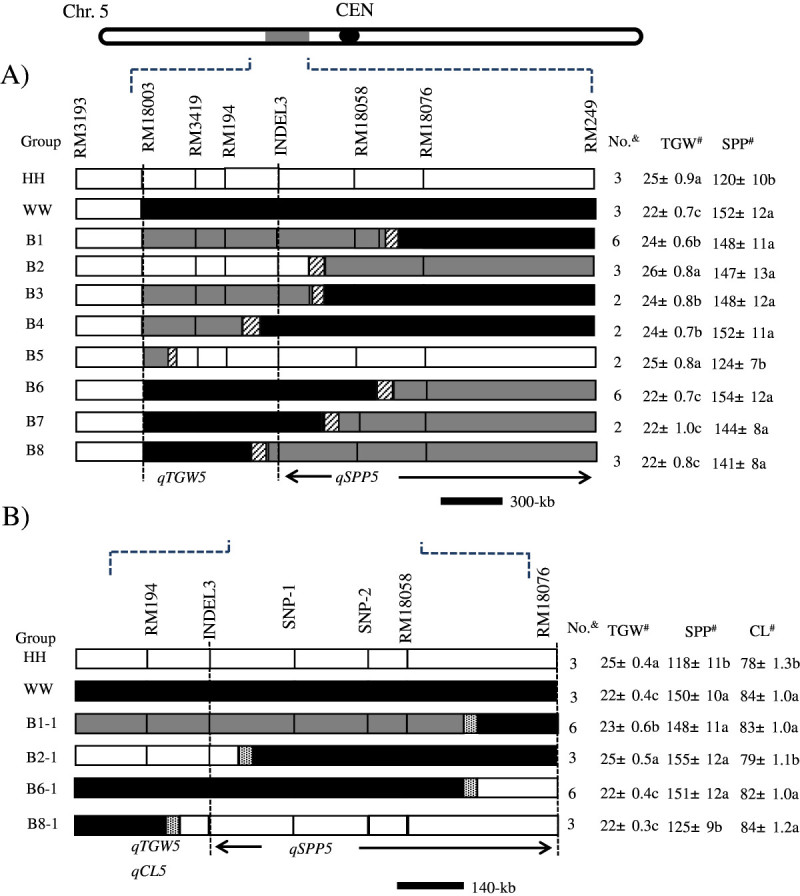
Substitution mapping of qSPP5 and qTGW5 using two populations. A) Graphical genotypes of the BC5F3 lines that were used for the substitution mapping of qSPP5 and qTGW5. The white portions of the graph indicate homozygous Hwayeongbyeo chromosome segments, the black regions indicate homozygous O. rufipogon chromosomes, the gray areas indicate heterozygous regions, and the slashed areas are regions where crossing-over occurred. The table on the right of the graphical genotypes shows the mean values of 2 traits for each genotype. The broken vertical lines define the interval that contained 2 QTLs. &Number of lines in each group. #The numbers that are followed by different letters in each column were significantly different according to Tukey’s HSD test at 5%. B) Graphical genotypes of the BC5F4 lines that were used for the substitution mapping of qSPP5, qTGW5 and qCL5. HH and WW in the Group indicate Hwayeongbyeo and CR7111-30, respectively.
For the qSPP5 locus, group B5 had a significantly lower SPP than CR7111-30 did. The SPP of group B2 with a breakpoint between markers RM18058 and INDEL3 did not significantly differ from that of CR7111-30 but was significantly higher than that of Hwayeongbyeo. These results imply that qSPP5 was located downstream of INDEL3. The parents and the heterozygote class (B1 and B3) showed significant differences in the TGW, indicating that the TGW gene was under additive genetic control. The heterozygote classes (B6 and B7) and CR7111-30 showed significantly higher SPP than Hwayeongbyeo did, indicating that the SPP gene was under dominant genetic control.
To further define the linkage relationship between qSPP5 and qTGW5, we self-crossed 18 BC5F3 plants that were selected from B1, B2, B6, and B8 to produce 18 BC5F4 lines and evaluated them in terms of the TGW, SPP and CL (Figure 4B). Also, BC5F4 lines were genotyped with two SNP markers, SNP-1 and SNP-2. The mean phenotypic values of the SPP and TGW for each group in BC5F4 were compared to those of the controls, Hwayeongbyeo and CR7111-30. The TGW of B2-1 was significantly higher than that of B1-1 and B6-1, which suggests that the qTGW5 allele was located in the upstream region of SNP-1. The TGW of B8-1 was significantly lower than that of Hwayeongbyeo, which suggests that the qTGW5 allele was located in the upstream region of INDEL3. For qSPP5, B2-1 significantly differed from Hwayeongbyeo in SPP, which indicated that the qSPP5 allele was located in the downstream region of INDEL3. The number of SPP of B6-1 was significantly higher than that of Hwayeongbyeo, which suggests that qSPP5 was located in the upstream region of RM18076. The group B8-1 did not show difference in the number of SPP compared to Hwayeongbyeo, and this indicated that the QTLs for the SPP and TGW were different. We found that qTGW5 was located in the upstream region of INDEL3, whereas qSPP5 was located in about 860-kb interval between INDEL3 and RM18076 based on the Nipponbare sequence (http://www.gramene.org). To map the qCL5, the same procedure was applied and qCL5 was located in the upstream of INDEL3.
O. rufipogon contains Kasalath-type qSW5
The qTGW 5 seemed to be the same gene as qSW5 based on its position (Shomura et al., 2008). Three allelic types at the qSW5 locus exist: Kasalath-type, Indica II-type, and Nipponbare-type. Of these, the Kasalath-type allele is functional and the Nipponbare-type is a loss-of-function allele. A 1212-bp deletion at the qSW5 locus in Nipponbare was associated with an increase in the GW, as compared to Kasalath. One hundred eighty rice cultivars were genotyped at the qSW5 locus by using the primers, and they were divided into 3 types: Kasalath-type, Indica II-type, and Nipponbare-type (Song et al., 2011). To determine the allele type of O. rufipogon at qGW5, we genotyped O. rufipogon by using the N1212del. The results showed that Hwayeongbyeo and W1944 had the Nipponbare-type and Kasalath-type alleles, respectively (data not shown). This result seemed to confirm that qTGW5 in this study was the same gene as qSW5.
Impact of the QTL cluster on the YD per plant
Two BC5F4 NILs, B8-1 (O. rufipogon homozygous at qTGW5 and Hwayeongbyeo homozygous at qSPP5) and B2-1 (Hwayeongbyeo homozygous at qTGW5 and O. rufipogon homozygous at qSPP5), were used for yield trials together with the parental controls in 2011. The trials were conducted using a completely randomized block design with 3 repetitions. The results show that the average YD per plant of B2-1 was 15.3% higher than that of B8-1 (P ≤ 0.02). The average YD per plant of B2-1 was 7.3% higher than that of Hwayeongbyeo (P = 0.06), although the difference was not significant at P = 0.05 (Table 3).
Table 3.
Comparison of grain yield per plant between 2 QTL-NILs and their parents
| Line | Trait mean ± s.d.@ | ||
|---|---|---|---|
| DTH | CL | YD | |
| Hwayeongbyeo | 98a+, a# | 83b, b | 26.0 ± 1.3 ab, b |
| CR7111-30 | 98a, a | 86a, a | 25.7 ± 1.4 bc, b |
| B2-1 | 98a, a | 82b, b | 27.9 ± 1.6 a, a |
| B8-1 | 97a, a | 86a, a | 24.0 ± 1.5 d, c |
@DTH: days to heading; CL: culm length; and YD: yield per plant.
+, #The numbers that are followed by the same letters were not significantly different according to Tukey’s HSD test at 5% (+) and 10% (#), respectively.
Discussion
The original target of this study was the QTL for the TGW, which was qTGW5 mapped on chromosome 5 (,2005). During the process of fine-mapping this trait, the QTLs for the SPP, SB, and CL were consistently detected in the same region. The QTL for the SPP was detected near the SSR markers RM413 and RM194 on chromosome 5, and the coefficient of determination was low being 3.7% (Lee et al., 2005). However, the effect of qSPP5 was not strong to be detected by both interval mapping and single-point analysis near the same SSR markers (Yuan et al., 2009). It is likely that qSPP5 is a minor QTL and not stable. Substitution lines confirmed that the QTL for TGW resided in the 165-kb region and that the additional 4 QTLs were co-localized near qTGW5.
A number of QTLs for the SPP have been identified using inter- (Thomson et al., 2003; Tian et al., 2006) and intra-specific populations (Cui et al., 2003; Lu et al., 1997), and these QTLs were located on all of the rice chromosomes. However, a few studies reported on a QTL that is associated with the SPP and is located on chromosome 5 by using inter-specific populations (Lee et al., 2005; Tian et al., 2006; Tan et al., 2008). Based on the finding that the wild alleles increased the number of SPP and decreased the TGW, and their map position, it appears that qGPA 5 reported by Tian et al. (2006) and spp5.1 detected by Tan et al. (2008) are allelic to qSPP5 in this study. It is interesting that the QTL for the SPP was detected exclusively using introgression lines from crosses between cultivars and Asian common wild rice (Lee et al., 2005; Tian et al., 2006; Tan et al., 2008). One possible reason is that the effect of these QTLs was not so strong that they could not be detected in primary mapping populations such as F2 and RILs (Xiao et al., 1996) because qGPA 5 (Tian et al., 2006), spp5.1 (Tan et al., 2008), and qSPP5 in this study were detected in the introgression lines population. Because the SPP is inherited quantitatively, this trait is tractable to genetic analysis via the development of high-resolution NILs. NILs that block genetic background noise would be useful for validating minor QTLs and mapping them as a single Mendelian factor (Xie et al., 2008). As documented in this study, the R2 values steadily increased with progressive generations of backcrossing from 9.7% for the BC4F3 generation to 33.0% for the BC5F4 generation of NILs. As the number of spurious donor (i.e., O. rufipogon) introgressions in the genetic background decreased and the linkages between the markers and the target gene(s) increased, the proportion of phenotypic variation that could be explained by the markers greatly enhanced.
Whether similar genomic locations of QTLs that affect different traits are attributable to the pleiotropy of a single gene or the tight linkage of several genes that individually influence specific traits has been a topic of debate. In a previous study by Xiao et al. (1996), pleiotropy was suggested for 3 chromosomal regions that were simultaneously associated with the TGW and grains per plant or the TGW and grains per panicle. These yield components showed highly negative correlations, and 3 significant QTLs that were associated with the TGW were mapped to the same positions as 3 QTLs that affect grains per plant and grains per panicle. In this study, one genomic region was associated with more than one trait, which indicated the existence linkage and/or pleiotropic effects. Liu et al. (2010) mapped the QTLs for grain weight TGW3b and the SPP SPP3b to a 2.6-cM interval between RM15885 and W3D16. At this QTL region, the Teqing allele was associated with an increase in the SPP and a decrease in the TGW, and no conclusion could be drawn about whether one pleiotropic QTL or two linked QTLs were located within the interval. Bai et al. (2011) also reported that 2 QTLs, qssp8 and tgw8, which are located between RM502 and RM264, might be the same gene. In our study, we demonstrated that 2 tightly linked QTLs, qSPP5 and qTGW5, control the SPP and grain weight, respectively. In this regard, the question of pleiotropy versus tight linkage in these studies remains to be resolved using larger populations and high-density mapping.
A high YD is one of the most important goals of rice breeding programs. Much attention has been focused on the genetic bases of the SPP and TGW because of their importance in determining rice yield. In this study, the effect of the detected QTL qSPP5 was confirmed by the increase in the SPP of the NILs. qSPP5 is a minor QTL that exhibits a small additive effect of approximately 10–15 spikelets. The high number of SPP in the NIL was mainly attributed to the increased number of SBs. The finding that yield per Hwayeongbyeo plant could be improved by introgressing qSPP5, which is a QTL for the SPP from O. rufipogon, demonstrates the existence of a complementary combination between 2 linked QTLs, qTGW5 and qSPP5, with the aid of molecular markers. Specifically, the pyramiding of the Hwayeongbyeo allele at qTGW5 and the O. rufipogon allele at qSPP5 should produce a higher yield compared to the parental genotypes. As expected, the NIL with the wild allele at qTGW5 and the Hwayeongbyeo allele at qSPP5 had lower yields compared to Hwayeongbyeo. The data presented in this study clearly indicate the linkage of qSPP5 and qTGW5 although additional experiments using lines from a cross between two separate lines each segregating at one QTL region but fixed at another QTL might be necessary to further confirm their linkage. Based on the finding that the O. rufipogon alleles for the SPP are beneficial in the japonica and indica cultivar backgrounds (Lee et al., 2005; Tian et al., 2006; Tan et al., 2008), the qSPP5 allele could be valuable gene (s) for improving rice yields.
QTL mapping indicated the existence of five QTLs in this region across different generations and substitution mapping confirmed the linkage of QTLs for SPP and TGW. The finding that the gene (s) affecting two traits, SPP and SP were mapped to the same region and the same direction of the genetic effect with O. rufipogon alleles increasing trait values across different generations implies that this locus was associated with panicle structure with pleiotropic effects. Similar results were reported in the study by Ohsumi et al. (2011) that Habataki alleles of qSBN1 and qPBN6 increased spikelet number on secondary rachis branches and primary rachis branches in the Sasanishiki genetic background. A strong positive correlation (r = 0.845, P < 0.001) between the GW and TGW in BC5F4 seems to suggest that the variation in the GW was associated with that in the TGW at this locus which controls grain morphology traits (data not shown). This result is also consistent with the report by Weng et al. (2008) that GW5 is associated with rice grain width and weight.
Several QTLs that control the SPP have been cloned using NILs (Xue et al. 2008; Miura et al. 2010). In the present study, qSPP5 was responsible for 33.0% of the phenotypic variance. No QTL around the qSPP5 region has been cloned to date. It would be interesting to clone qSPP5 to examine the functional relationships of the genes that control the SPP and to determine how they interact with other genes/alleles in various genetic backgrounds. The BC5F4 NILs that were developed in this study could be good materials for further fine mapping and cloning of qSPP5.
Conclusion
In this study, we demonstrated that 2 QTLs, qSPP5 for spikelets per panicle (SPP) and qTGW5 for grain weight (TGW), are tightly linked on chromosome 5. Based on the finding that the O. rufipogon allele for the SPP was beneficial in the japonica and indica cultivar backgrounds, the qSPP5 allele could be valuable for improving rice yields. In addition, the NIL populations and molecular markers are useful for cloning qSPP5.
Acknowledgements
This work was supported by the Rural Development Administration under the “Cooperative Research Program for Agriculture Science & Technology Development” (Project No. PJ906910) and the Next-Generation Biogreen 21 Program (Plant Molecular Breeding Center No. PJ008136), Rural Development Administration, Republic of Korea.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Footnotes
Xiao Luo, Shi-Dong Ji contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XL and SJ participated in phenotyping, genotyping, drafting the manuscript. PY, and SB participated in phenotyping. HL participated in primer design and genotyping. DK and JK participated in experimental design and coordination. SA conceived of the study, drafted proposal and corrected manuscript. All authors have read and approved the manuscript.
Contributor Information
Xiao Luo, Email: jlluoxiao@hotmail.com.
Shi-Dong Ji, Email: jishidong2000@126.com.
Ping-Rong Yuan, Email: yuanpr2003@yahoo.cn.
Hyun-Sook Lee, Email: leehs0107@gmail.com.
Dong-Min Kim, Email: acekdm@naver.com.
Sangshetty Balkunde, Email: sangu_agr@yahoo.com.
Ju-Won Kang, Email: hgorilla@gmail.co.
Sang-Nag Ahn, Email: ahnsn@cnu.ac.kr.
References
- Aluko G, Martinez C, Tohme J, Castano C, Bergman C, Oard HJ. QTL mapping of grain quality traits from the interspecific cross Oryza sativaxO. glaberrima. Theor Appl Genet. 2004;109:630–639. doi: 10.1007/s00122-004-1668-y. [DOI] [PubMed] [Google Scholar]
- Bai XF, Luo LJ, Yan WH, Kovi MR, Xing YZ. Quantitative trait loci for rice yield-related traits using recombinant inbred lines derived from two diverse cultivars. J Genet. 2011;90:209–215. doi: 10.1007/s12041-011-0057-y. [DOI] [PubMed] [Google Scholar]
- Brondani C, Rangel PHN, Brondani RPV, Ferreira ME. QTL mapping and introgression of yield-related traits from Oryza glumaepatula to cultivated rice (Oryza sativa) using microsatellite markers. Theor Appl Genet. 2002;104:1192–1203. doi: 10.1007/s00122-002-0869-5. [DOI] [PubMed] [Google Scholar]
- Cai HW, Morishima H. QTL clusters reflect character associations in wild and cultivated rie. Theor Appl Genet. 2002;104:1217–1228. doi: 10.1007/s00122-001-0819-7. [DOI] [PubMed] [Google Scholar]
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwogse J, Wu K, Xiao J, Yu Z, Ronald PC, Harrington SE, Second G, McCouch SR, Tanksley SD. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics. 1994;138:1251–1374. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui KH, Peng SB, Xing YZ, Yu SB, Xu CG, Zhang Q. Molecular dissection of the genetic relationships of source-sink and transport tissue with yield traits in rice. Theor Appl Genet. 2003;106:649–658. doi: 10.1007/s00122-002-1113-z. [DOI] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Fukuta Y, Yagi T, Sato T, Osaki M, Khush GS. Identification and characterization of quantitative trait loci effecting spikelet number per panicle in rice (Oryza sativa L.) Field Crops Res. 2004;89:253–262. doi: 10.1016/j.fcr.2004.02.004. [DOI] [Google Scholar]
- Lee SJ, Oh CS, Suh JP, McCouch SR, Ahn SN. Identification of QTLs for domestication-related and agronomic traits in an Oryza sativax O. rufipogon BC1F7 population. Plant Breed. 2005;124:209–219. doi: 10.1111/j.1439-0523.2005.01092.x. [DOI] [Google Scholar]
- Li JM, Thomson M, McCouch SR. Fine mapping of a grain weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics. 2004;168:2187–2195. doi: 10.1534/genetics.104.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, He Y, Zhang Q. Nat Genet. 2011. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. [DOI] [PubMed] [Google Scholar]
- Liu T, Shao D, Kovi MR, Xing Y. Mapping and validation of quantitative trait loci for spikelets per panicle and 1,000-grain weight in rice (Oryza sativa L.) Theor Appl Genet. 2010;120(5):933–942. doi: 10.1007/s00122-009-1222-z. [DOI] [PubMed] [Google Scholar]
- Lu CF, Shen LS, Tan ZB, Xu YB, He P, Chen Y, Zhu LH. Comparative mapping of QTLs for agronomic traits of rice across environments by using a doubled-haploid population. Theor Appl Genet. 1997;94:145–150. doi: 10.1007/s001220050393. [DOI] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- Ohsumi A, Takai T, Ida M, Yamamoto T, Arai-Sanoh Y, Yano M, Ando T, Kondo M. Evaluation of yield performance in rice near-isogenic lines with increased spikelet number. Field Crops Res. 2011;120:68–75. doi: 10.1016/j.fcr.2010.08.013. [DOI] [Google Scholar]
- Onishi K, Horiuchi Y, Ishigoh-Oka N, Takagi K, Ichikawa N, Maruoka M, Sano Y. A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed Sci. 2007;57:7–16. doi: 10.1270/jsbbs.57.7. [DOI] [Google Scholar]
- Panaud O, Chen X, McCouch SR. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.) Mol Gen Genet. 1996;252:597–607. doi: 10.1007/BF02172406. [DOI] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40(8):1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Song Y, Guihua Z, Sujuan L, Hua W, Heqin L, Guowei Z, Peng G, Hongmiao S, Changjie Y, Yuezhi T. Seed size is determined by the combinations of the genes controlling different seed characteristics in rice. Theor Appl Genet. 2011;123:1173–1181. doi: 10.1007/s00122-011-1657-x. [DOI] [PubMed] [Google Scholar]
- Suh J, Ahn SN, Cho YC, Kang KH, Choi IS, Kim YG, Suh HS, Hwang HG. Mapping for QTLs for yield traits using an advanced backcross population from a cross between Oryza sativa and O. glaberrima. Korean J Breed. 2005;37:214–220. [Google Scholar]
- Sun CQ, Wang XK, Yoshimura A, Iwata N. Comparison of the genetic diversity of common wild rice (Oryza rufipogon Griff.) and cultivated rice (O. sativa L.) using RFLP markers. Theor Appl Genet. 2001;102:157–162. doi: 10.1007/s001220051631. [DOI] [Google Scholar]
- Tan L, Zhang P, Liu F, Wang G, Ye S, Zhu Z, Fu Y, Cai H, Sun C. Quantitative trait loci underlying domestication and yield-related traits in an Oryza sativax Oryza rufipogon advanced backcross population. Genome. 2008;51:692–704. doi: 10.1139/G08-054. [DOI] [PubMed] [Google Scholar]
- Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, Lobos KB. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet. 2003;107:479–493. doi: 10.1007/s00122-003-1270-8. [DOI] [PubMed] [Google Scholar]
- Tian F, Li DJ, Fu Q, Zhu ZF, Fu YC, Wang XK, Sun CQ. Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor Appl Genet. 2006;112:570–580. doi: 10.1007/s00122-005-0165-2. [DOI] [PubMed] [Google Scholar]
- Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, Ma H, Zhang G, He Z. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008;40:1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- Weng JF, Gu SH, Wan XY, Gao H, Guo T, Su N, Lei CL, Zhang X, Cheng ZJ, Guo XP, Wang JL, Jiang L, Zhai HQ, Wan JM. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li J, Yuan L, Tanksley SD. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet. 1996;92:230–244. doi: 10.1007/BF00223380. [DOI] [PubMed] [Google Scholar]
- Xiao JH, Li JM, Crandillo S, Ahn SN, Yuan LP, Tanksley SD, McCouch SR. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics. 1998;150:899–909. doi: 10.1093/genetics/150.2.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XB, Jin FX, Song MH, Suh JP, Hwang HG, Kim YG, McCouch SR, Ahn SN. Fine mapping of a yield-enhancing QTL cluster associated with transgressive variation in an Oryza sativa × O. rufipogon cross. Theor Appl Genet. 2008;116:613–622. doi: 10.1007/s00122-007-0695-x. [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Takeuchi Y, Kono I, Yano M. QTL analysis for panicle characteristics in temperate japonica rice. Euphytica. 2002;128:219–224. doi: 10.1023/A:1020893731249. [DOI] [Google Scholar]
- Yuan PR, Kim HJ, Chen QH, Ju HG, Lee SJ, Ji SD, Ahn SN. QTL dissection of agronomic and domestication trait using introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (O. sativa L.) background. J Crop Sci Biotechnol. 2009;12:241–248. doi: 10.1007/s12892-009-0144-2. [DOI] [Google Scholar]
- Zhang YS, Luo LJ, Xu CG, Zhang QF, Xing YZ. Quantitative trait loci for panicle size, heading date and plant height co-segregating in trait-performance derived near-isogenic lines of rice (Oryza sativa) Theor Appl Genet. 2006;113:361–368. doi: 10.1007/s00122-006-0305-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Luo L, Liu T, Xu C, Xing Y. Four rice QTLs controlling number of spikelets per panicle expressed the characteristics of single Mendelian gene in near isogenic backgrounds. Theor Appl Genet. 2009;118:1035–1044. doi: 10.1007/s00122-008-0960-7. [DOI] [PubMed] [Google Scholar]


