Abstract
Altered expression of the E3 ubiquitin ligase UBE3A, which is involved in protein degradation through the proteasome-mediated pathway, is associated with neurodevelopmental and behavioral defects observed in Angelman syndrome (AS) and autism. However, little is known about the neuronal function of UBE3A and the pathogenesis of UBE3A-associated disorders. To understand the in vivo function of UBE3A in the nervous system, we generated multiple mutations of ube3a, the Drosophila ortholog of UBE3A. We found a significantly increased number of total boutons and satellite boutons in conjunction with compromised endocytosis in the neuromuscular junctions (NMJs) of ube3a mutants compared to the wild type. Genetic and biochemical analysis showed upregulation of bone morphogenetic protein (BMP) signaling in the nervous system of ube3a mutants. An immunochemical study revealed a specific increase in the protein level of Thickveins (Tkv), a type I BMP receptor, but not other BMP receptors Wishful thinking (Wit) and Saxophone (Sax), in ube3a mutants. Ube3a was associated with and specifically ubiquitinated lysine 227 within the cytoplasmic tail of Tkv, and promoted its proteasomal degradation in Schneider 2 cells. Negative regulation of Tkv by Ube3a was conserved in mammalian cells. These results reveal a critical role for Ube3a in regulating NMJ synapse development by repressing BMP signaling. This study sheds new light onto the neuronal functions of UBE3A and provides novel perspectives for understanding the pathogenesis of UBE3A-associated disorders.
Author Summary
Angelman syndrome (AS), characterized by severe mental retardation, developmental delay, ataxia, seizures, speech impairment, and happy disposition, is caused by mutation of E3 ubiquitin ligase UBE3A; a critical enzyme involved in proteasome-mediated protein degradation. Increasing evidence demonstrates that overexpression or hyperactivation of UBE3A is associated with autism. Thus, both loss and gain of UBE3A functions result in neurodevelopmental and cognitive defects. However, the neuronal functions of UBE3A and the mechanism by which altered expression of UBE3A leads to developmental and cognitive defects are poorly understood. Using Drosophila melanogaster as a model system in conjunction with an array of biochemical and physiological assays, we showed that mutants of ube3a had excess synaptic boutons and endocytic defects at the neuromuscular junction terminals due to an elevated level of bone morphogenetic protein (BMP) signaling. Specifically, Ube3a directly binds and ubiquitinates the BMP receptor Thickveins for proteasomal degradation; a function that is conserved in mammalian cells. Negative regulation of BMP signaling by UBE3A suggests a previously unknown molecular mechanism that underlies the pathogenesis of UBE3A-associated AS and autism.
Introduction
Angelman syndrome (AS) is a neurodevelopmental disorder characterized by severe mental retardation, developmental delay, ataxia, seizures, speech impairment, and happy disposition [1, 2]. It is caused by disruption of the ubiquitin protein ligase E3A (UBE3A), which is expressed in the brain primarily from the maternal allele as a result of tissue-specific genomic imprinting [3, 4]. While loss of UBE3A causes AS, a maternal 15q11–13 duplication encompassing UBE3A results in autism [5–7]. A point mutation of UBE3A identified in an autism proband disrupts its phosphorylation by protein kinase A and increases its ligase activity [8]. Thus, both loss and gain of UBE3A function are associated with neurodevelopmental and cognitive defects.
UBE3A is involved in ubiquitin–proteasome-mediated protein degradation. Although several substrates of UBE3A have been identified, few have been implicated in neural development and function [9]. Ube3a mutant mice, a valid model of AS, show impairment in hippocampal long-term potentiation (LTP) [10, 11] and experience-dependent synaptic plasticity in the visual cortex [12, 13], together with imbalanced excitatory/inhibitory neurotransmission [14] and altered intrinsic membrane properties [15]. Moreover, Ube3a has been firmly implicated in the regulation of dendrites and spine development in neurons [16–18]. While the underpinnings of UBE3A-associated disorders remain to be elucidated, a recent study has shown that Ube3a targets the small-conductance potassium channel SK2 for degradation, and decreased NMDA receptor activation due to an elevated SK2 protein level underlies the impaired LTP in Ube3a mice [11]. Abnormal development and function of excitatory synapses in conjunction with a reduced number of AMPA receptors in Ube3a mutant mice are attributed to decreased Ube3a-mediated proteasomal degradation of RhoA guanine exchange factor (GEF) Ephexin5 and synaptic protein Arc [19, 20]. However, later studies showed that Arc is not a direct substrate of UBE3A [21, 22]. Thus, the molecular mechanisms by which UBE3A regulates synapse development and function remain unclear.
Null mutants of ube3a, the Drosophila homolog of human UBE3A [23], show defects in locomotive behavior, circadian rhythms, and long-term memory [24, 25], as well as reduced dendritic branching and growth of terminal dendritic processes of sensory neurons [26]. Similar to ube3a loss-of-function mutants, overexpression of wild-type ube3a also leads to motor abnormalities, circadian rhythm defects, and decreased dendritic branching [24–26]. So far, it has not been shown if Drosophila ube3a regulates synapse development. No direct targets of Ube3a-involved proteasomal degradation have been definitely identified in Drosophila, despite a series of proteomic studies [23, 27, 28].
Bone morphogenetic protein (BMP) pathway plays a crucial role in neural development throughout evolution [29–31]. In this study, we show that Drosophila Ube3a regulates synapse development and function at neuromuscular junctions (NMJs) by ubiquitinating and promoting proteasome-mediated degradation of the type I BMP receptor Tkv, and that the negative regulation of Tkv by Ube3a is conserved in mammalian cells. These results offer novel insight into the intricate regulation of BMP signaling in the nervous system, and suggest a potential intervention strategy for UBE3A-associated disorders by manipulating BMP signaling via genetic and pharmacological means.
Results
ube3a mutants are viable and fertile
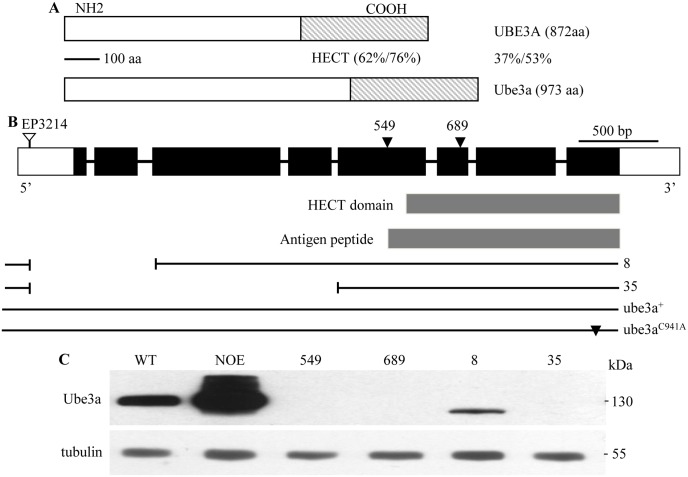
To examine the functions of Ube3a, we generated ube3a mutants through P-element-mediated excisions of EP3214, an insertion in the first exon. Multiple imprecise excisions (e.g., ube3a8 and ube3a35) were obtained (Fig 1B). In addition, two nonsense mutations (ube3a549 and ube3a689) induced by the chemical mutagen ethyl methanesulfonate (EMS) were identified via a TILLING service (see Materials and Methods) (Fig 1B). Western blotting using a mouse monoclonal antibody against the C-terminal peptide containing the HECT (Fig 1B) confirmed the absence of Ube3a protein in hemizygous ube3a549/Df(3L)ED4470 (which removes the ube3a locus completely) and ube3a689/Df(3L)ED4470 mutants and in homozygous ube3a35 mutants, whereas a truncated protein of 110 kDa was detected in ube3a8 mutants (Fig 1C). Immunostaining of larval and adult tissues using the monoclonal Ube3a antibody 8F7 showed that the endogenous Ube3a was cytoplasmic and widely expressed in various tissues including ventral ganglia, muscles, wing discs, ovaries and testes with an obvious enrichment at presynaptic NMJ terminals (S1 and S2 Figs). Both homozygous and hemizygous ube3a35 mutants, as well as independent null mutants generated in other laboratories [24, 26], were fully viable with no apparent developmental abnormalities, and both male and female adults were fertile.
Fig 1. Characterization of ube3a mutations.
(A) The percent amino acid identity/similarity between Drosophila Ube3a and human UBE3A. (B) Intron-exon organization and various mutations of ube3a. Coding and non-coding exons are represented by black and empty rectangles, respectively. Triangles indicate nonsense mutations; the HECT domain and the peptide used for antibody production are indicated. Two deletions, ube3a8 and ube3a35, produced by imprecise excision of the P-element insertion EP3214 are depicted by interrupted lines. Genomic transgenes carrying wild-type (ube3a+) and mutant (ube3aC941A) containing a missense mutation in the codon encoding the catalytic cysteine residue ube3a are shown. (C) Western results of adult head extracts from various genotypes using the monoclonal antibody 8F7 against Ube3a. NOE denotes elav-Gal4/UAS-ube3a. No Ube3a expression was observed in hemizygous ube3a549 (ube3a549/Df(3L)ED4470) and ube3a689 (ube3a689/Df(3L) ED4470) mutants as well as homozygous ube3a35 mutants. A truncated Ube3a of ~110 kDa was observed in ube3a8 homozygous mutants. Tubulin was used as a loading control.
ube3a regulates NMJ development in presynaptic neurons
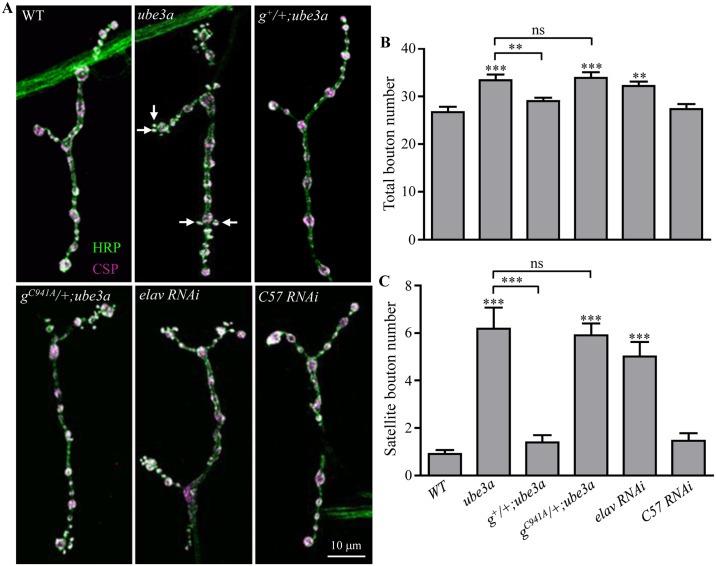
Given the synaptic localization of Ube3a in cultured hippocampal neurons [16], the impaired LTP and defective synaptic transmission and plasticity observed in Ube3a mutant mice [10, 11, 14], and the presynaptic enrichment of Ube3a (S1 Fig), we hypothesized that Drosophila ube3a may play an important role at synapses. To test this possibility, we examined the NMJs of wandering third instar larvae by staining them with anti-cysteine string protein (CSP), a synaptic vesicle marker, and anti-horseradish peroxidase (HRP), which labels the presynaptic neuronal membrane. In ube3a35 null mutants, the total bouton number for muscle 4 NMJs in abdominal segments A2 and A3 was 33.4 ± 1.2, which was significantly higher than 26.8 ± 1.1 in wild-type (WT) larvae (P < 0.001) (Fig 2A and 2B). Similar to homozygous ube3a35 mutants, allelic combination of ube3a35/ube3a549 and hemizygous ube3a35/Df(3L)ED4470 and ube3a549/Df(3L)ED4470 also showed more boutons than the wild type. The number of satellite boutons budding from a parental bouton in ube3a35 mutants was also significantly higher than in wild-type larvae; these satellites were more frequent at the distal ends of NMJ terminals (Fig 2A and 2C). To confirm that the observed phenotype was attributable to loss of ube3a, we conducted rescue experiments using a genomic transgene. One copy of wild-type ube3a restored the number of total and satellite boutons in ube3a35 mutants (Fig 2A, 2B and 2C). However, a ube3a genomic transgene carrying the missense mutation C941A disrupting the thioester bond with ubiquitin did not (Fig 2A, 2B and 2C), although it was expressed at a level comparable to the wild-type genomic transgene.
Fig 2. ube3a regulates NMJ synapse growth.
(A) Muscle 4 NMJ synapses were double-stained with anti-HRP (green) and anti-CSP (magenta). The precise excision line P95 was used as the wild-type control. ube3a35 homozygotes showed more total boutons and satellite boutons (indicated by arrows) than wild-type larvae. A copy of wild-type genomic ube3a transgene (g+) rescued the NMJ phenotype of ube3a35 mutants, however, a copy of ube3a transgene harboring the missense mutation C941A (gC941A) did not. elav-Gal4-driven Thu3266 RNAi against ube3a in presynaptic neurons led to increased numbers of total and satellite boutons, while C57-Gal4-driven postsynaptic knockdown of Ube3a had no effect on NMJ growth. Scale bar = 10 μm. (B, C) Quantification of total bouton (B) and satellite bouton (C) number in different genotypes including wild type, ube3a35, genomic ube3a+/+; ube3a35, genomic ube3aC941A/+; ube3a35, elav-Gal4/+; RNAi/+, and RNAi/+; C57-Gal4/+. n ≥ 18 NMJs; **P < 0.01, ***P < 0.001; ns, not significant. Error bars represent SEM.
To determine whether ube3a exerts its effect on the pre- or postsynaptic side, we knocked down Ube3a expression by RNA interference (RNAi) in a tissue-specific manner. Targeted knockdown of ube3a in presynaptic neurons by elav-Gal4-driven Thu3266 RNAi resulted in a modest but significant increase in the number of total boutons (20.15%) and a dramatic increase in satellite boutons (371.7%) compared to wild-type larvae (Fig 2). Motoneuron specific knockdown of ube3a by OK6-Gal4 also showed overgrown NMJs (S3 Fig), whereas knockdown of ube3a in postsynaptic muscles using C57-Gal4 had no effect on bouton number (P > 0.05 compared to wild type; Fig 2). Similar results were observed using an independent RNAi line VDRC45876. These findings together demonstrate that Ube3a regulates NMJ synapse growth presynaptically.
Synaptic endocytosis is impaired in ube3a mutants
An increased number of satellite boutons at NMJs is closely associated with a defect in endocytosis [32–34]. To find out if ube3a mutant NMJs exhibited an endocytic defect, we first examined NMJ synapses by electron microscopy. The critical synaptic structures such as active zones and subsynaptic reticulum appeared normal in ube3a mutant boutons. Specifically, the mean density of synaptic vesicles (SVs) within a 200 nm radius of the active zone of transmitter release was normal in ube3a mutants compared to wild type (S4 Fig), as reported previously [35]. However, significantly more enlarged vesicles > 60 nm in diameter (also known as cisternae, presumably endosomes) per bouton cross-section were observed in ube3a mutants (5.06 ± 0.90 in ube3a vs 1.19 ± 0.21 in wild type; n ≥ 38 boutons from ≥ 4 larvae, ***p < 0.001; S4 Fig), suggesting a vesicle formation defect.
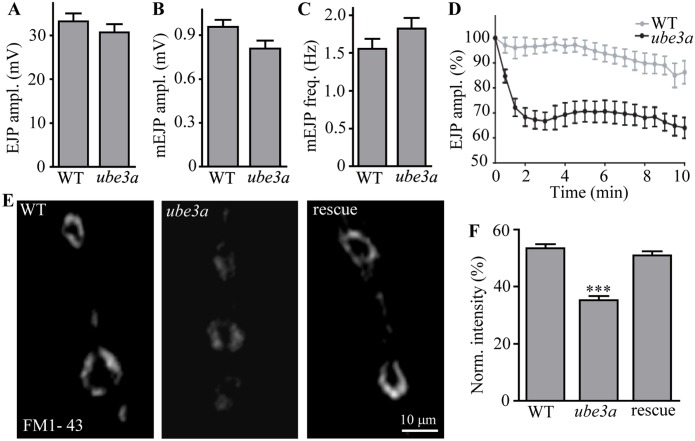
We then examined both evoked and spontaneous synaptic glutamate release at ube3a35 mutant NMJs using intracellular recordings. We first stimulated motor neurons at a low frequency of 0.3 Hz in the presence of 0.5 mM Ca2+. The amplitudes of excitatory junctional potentials (EJPs) and miniature EJPs (mEJPs) in ube3a35 mutants did not differ from those in the controls (Fig 3A and 3B). The mEJP frequency in ube3a mutants was similar to that in wild-type larvae (Fig 3C). Representative traces of EJP and mEJP of wild type (A) and ube3a35 mutant (B) are shown in S5 Fig. Thus, the basal transmission appeared normal in ube3a mutants. We then examined the ability of ube3a mutants to maintain neurotransmitter release during intense stimulation. The EJP amplitudes in ube3a mutants under high-frequency stimulation of 10 Hz in the presence of 0.5 mM extracellular Ca2+ for 10 min declined much faster and remained at 64% of the initial response (Fig 3). In contrast, control animals sustained release at about 85% of the initial amplitude of EJP (Fig 3D). The inability of ube3a mutants to maintain normal levels of transmission during intense activity is consistent with a specific defect in vesicle trafficking or recycling.
Fig 3. Synaptic endocytosis is impaired in ube3a mutants.
(A–C) Statistical results of EJP amplitudes (A), mEJP amplitudes (B), and mEJP frequencies (C) of wild type and ube3a35 mutants. (D) Relative EJP amplitudes (%) during high-frequency stimulation (10 Hz) for 10 min in wild type and ube3a35 mutants. n ≥ 11 larvae; error bars indicate SEM. (E) Loss of ube3a causes synaptic endocytic defects. FM1-43 dye uptake in wild type, ube3a35 mutants, and rescued animals (gube3a+/+; ube3a35). Scale bar: 10 μm. (F) Normalized fluorescence intensity (%) of endocytosed FM1-43 for wild type, ube3a35 and rescued mutants (gube3a+/+; ube3a35). n ≥ 20 NMJs, error bars indicate SEM, *** indicates p < 0.001.
Finally, to examine whether endocytosis is affected in ube3a mutant NMJ synapses, we performed fluorescent dye FM1-43 uptake experiments. There was intense labeling of FM1-43 at wild-type synaptic boutons, whereas ube3a35 mutants showed significantly reduced uptake of FM1-43 by 34.1% compared to wild type (p < 0.001; Fig 3E and 3F). Importantly, the endocytosis defect could be fully rescued by one copy of the genomic ube3a transgene (Fig 3E and 3F). The failure to maintain neurotransmission under tetanic stimulation and reduced dye uptake support that ube3a is required for efficient synaptic vesicle endocytosis.
ube3a regulates NMJ growth through the BMP signaling pathway
ube3a mutations resulted in more satellite boutons and endocytic defects (Figs 2 and 3). We investigated the underlying mechanism of the synaptic defects in ube3a mutants. Synapse formation requires the coordinated activity of several signaling cascades. At the Drosophila NMJ, the BMP pathway acts in a retrograde manner from muscles to motor neurons and promotes NMJ growth [30, 34, 36–39]. We hypothesized that the overgrowth of synaptic boutons in ube3a mutants may be caused by elevated BMP signaling. To test this possibility, we examined genetic interactions between ube3a and the major genes of the BMP signaling pathway, tkv and mad.
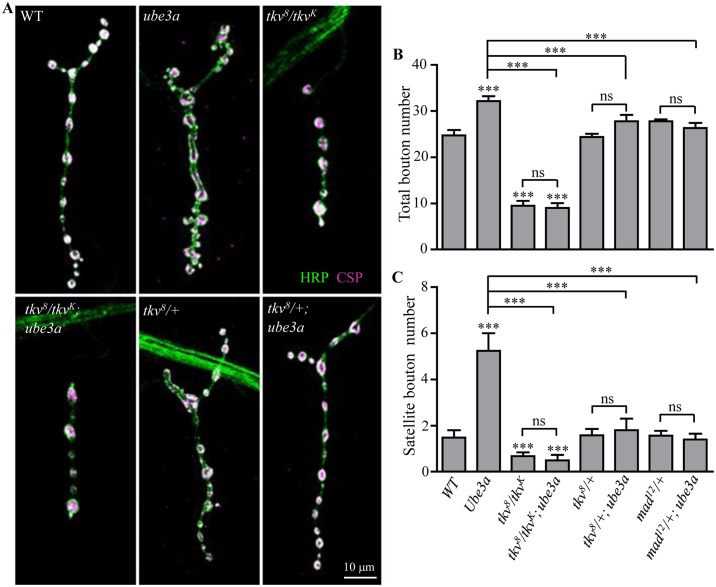
Compared to wild type, muscle 4 NMJ terminals were overgrown in ube3a35 mutants as described above, but severely underdeveloped in tkv8/tkvK16713 mutants (Fig 4A and 4B). In ube3a tkv double mutants, bouton numbers were similar to that of tkv single mutants (Fig 4A and 4B), suggesting that the overgrowth of NMJ synapses produced by loss of ube3a may be mediated by alterations in BMP signaling. Furthermore, NMJ overgrowth in ube3a mutants was dependent on the level of Tkv and the BMP effector Mad. Loss of one copy of tkv, which alone had no effect on bouton number, reversed the increase in satellite bouton number caused by loss of Ube3a (Fig 4A, 4B and 4C). Mutating one copy of mad also suppressed the overgrowth of NMJ synapses in ube3a mutants (Fig 4B and 4C). These results indicate that the excessive synaptic growth induced by loss of ube3a is mediated by elevated levels of the BMP signaling components Tkv, Mad, or both.
Fig 4. ube3a interacts genetically with BMP signaling pathway components tkv and mad in regulating NMJ growth.
(A) Confocal images of muscle 4 NMJ synapses co-stained with anti-CSP (magenta) and anti-HRP (green) to reveal synaptic vesicles and presynaptic membrane, respectively. In ube3a tkv double mutants, bouton numbers were similar to those of tkv8/tkvK16713 single mutants. One copy of tkv8 rescued the synapse overgrowth in ube3a35 mutants. Scale bar = 10 μm. (B, C) Quantification of total bouton number (B) and satellite bouton number (C) of various genotypes including wild type, ube3a35, tkv8/tkvK16713, tkv8/ tkvK16713; ube3a35, tkv8/+, tkv8/+; ube3a35, mad12/+, mad12/+; ube3a35. n ≥ 18 NMJ terminals, one-way ANOVA test, ***P < 0.001, ns, not significant,. Error bars represent SEM.
Increased BMP signaling in ube3a mutants
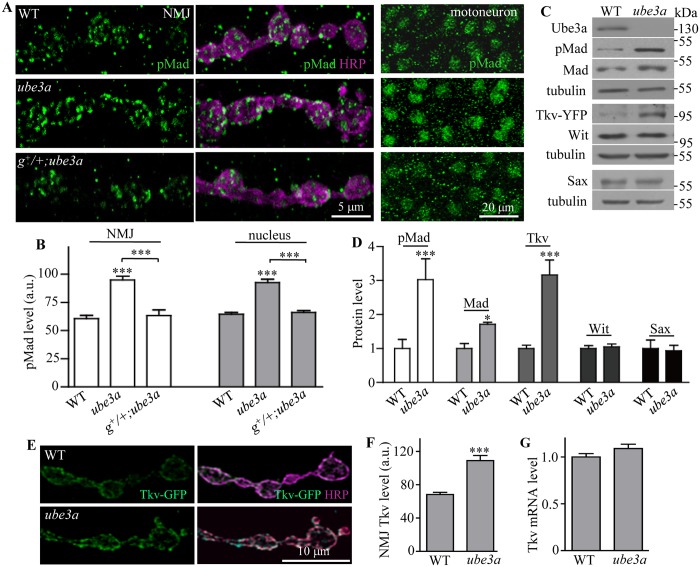
BMP receptor activation leads to phosphorylation of Mad (pMad), which then translocates to the nucleus to activate target gene transcription. Elevated pMad levels are thus indicative of active BMP signaling [34, 40, 41]. To determine if ube3a loss-of-function does indeed lead to NMJ overgrowth through upregulated BMP signaling, we quantified the level of pMad in NMJ synapses and motoneuron nuclei. pMad staining intensity at NMJ synapses was significantly increased by 56.5% in ube3a35 mutants compared to wild type (P < 0.001; Fig 5A and 5B). The intensity of pMad immunofluorescence was also significantly higher in motoneuron nuclei of ube3a mutants than in wild types (P < 0.001; Fig 5A and 5B). Furthermore, pMad expression was elevated in larval brain extracts as measured by western analysis (Fig 5C and 5D). One copy of the genomic ube3a transgene fully reversed the increased pMad levels at both NMJs and motoneuron nuclei of ube3a mutants (Fig 5A and 5B). These results show that ube3a negatively regulates pMad expression at both NMJs and motoneuron nuclei. Remarkably, the increased pMad staining, together with reduced dye loading and faster rundown of EJP amplitudes, in ube3a mutants is fully rescued by reducing the dose of Tkv by half (S6 Fig).
Fig 5. Levels of pMad and Tkv protein are increased in ube3a mutants.
(A) Increased level of pMad at the NMJ synapses and in the motoneuron nuclei of ube3a35 mutants was rescued by one copy of the wild-type genomic ube3a transgene (g+). (B) Statistical analysis of mean fluorescence intensities in arbitrary units (a.u.) for pMad in NMJs and motoneuron nuclei of different genotypes (n = 4, one-way ANOVA test, mean ± SEM, ***P < 0.001). (C) Increased levels of pMad, Mad, and Tkv-YFP, but not the endogenous Wit and Sax protein, in larval brains of ube3a35 mutants. Tkv protein level was examined from a gene trap line expressing the endogenous Tkv tagged with YFP. α-Tubulin was used as a loading control. (D) Statistical analysis of different protein levels in ube3a35 mutant brains (n = 4, t test, mean ± SEM, *P < 0.05, ***P < 0.001). (E) An increased level of elav-Gal4-driven Tkv-GFP in ube3a35 mutant NMJs compared with wild type. Scale bar = 10 μm. (F) Quantitative analysis of Tkv-GFP fluorescence intensities in a.u. normalized to HRP-positive area at NMJs of wild-type and ube3a35 mutant larvae (n = 4, t test, mean ± SEM, ***P < 0.001). (G) Normal level of tkv mRNA in ube3a35 mutant larval brains examined by quantitative PCR. tkv mRNA level was normalized to actin mRNA (n = 3, t test, mean ± SEM).
Given that Ube3a is an E3 ubiquitin ligase, we examined the protein expression levels of BMP signaling components in ube3a mutants to determine whether any are Ube3a substrates. Western blotting of larval brain extracts showed that pMad level was significantly increased by 203%, while Mad level was increased by 71% in ube3a mutants, consistent with the staining results (Fig 5A–5D). As a high quality of antibody against Tkv was not readily available, we took advantage of a yellow fluorescent protein (YFP) gene trap line that expresses functional YFP-tagged Tkv (Tkv-YFP) in the ventral nerve cord, including motoneurons under the control of the endogenous promoter (S7 Fig). Western blotting with an anti-GFP antibody that also recognizes YFP, showed that the protein level of Tkv-YFP was increased by 216% in ube3a mutants compared to wild type (P < 0.001), while the level of the endogenous Sax and Wit, the other type I and type II BMP receptors, respectively, was unaltered (Fig 5C and 5D). This suggests that Tkv is a primary target of Ube3a-mediated degradation. Indeed, the fluorescence intensity of elav-Gal4-driven Tkv-GFP was elevated by 59.2% at NMJ synapses of ube3a mutants compared to wild type (Fig 5E and 5F). Quantitative real-time PCR, however, detected similar levels of tkv mRNA in the brains of ube3a35 mutant and wild-type larvae (Fig 5G), indicating that the increase in Tkv protein expression in ube3a mutants is likely due to post-transcriptional regulation.
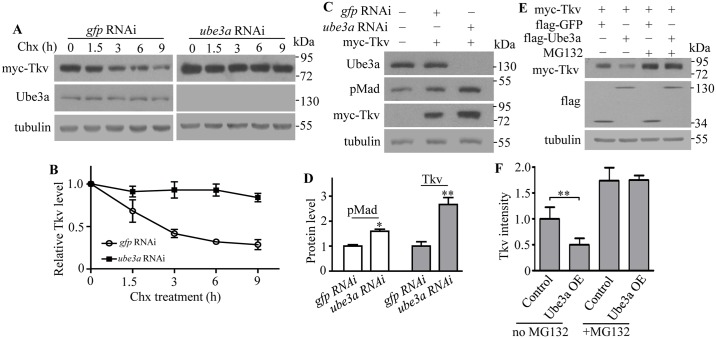
As Tkv acts upstream of Mad, and Tkv protein level is obviously upregulated in ube3a mutants, we focused on the mechanisms mediating negative regulation of Tkv by Ube3a by examining the stability of Tkv protein in S2 cells at various time points after treatment with the protein synthesis inhibitor cycloheximide. In control cells transfected with double-stranded RNAs (dsRNAs) against GFP, Tkv level decreased markedly from 1.5 h to 9 h upon drug treatment (Fig 6A). In Ube3a knockdown cells, however, a relatively stable level of Tkv was observed (Fig 6A and 6B). Ube3a knockdown resulted in a significantly elevated level of pMad (1.6 times the control) and Tkv (2.7 times the control) at 48 h after dsRNA transfection (Fig 6C and 6D). This effect was confirmed by using a distinct dsRNA targeting a different sequence of ube3a mRNA, demonstrating the specificity of Tkv downregulation by Ube3a.
Fig 6. Ube3a negatively regulates Tkv protein levels in S2 cells.
(A) Tkv protein levels in S2 cells treated with dsRNAs targeting ube3a or gfp (control) at various times after blocking protein synthesis by cycloheximide treatment. (B) A slowed decline in Tkv protein levels in S2 cells treated with cycloheximide after knockdown of ube3a by dsRNA. Values are shown as the ratio of Tkv intensity to tubulin control, normalized to the untreated cells at time zero (n = 3, mean ± SEM). (C) An increase in Tkv protein level after Ube3a knockdown in S2 cells. dsRNA against GFP was used as a control. (D) Statistical analysis of pMad and Tkv protein levels in S2 cells expressing a reduced level of Ube3a by RNAi (n = 4, one-way ANOVA test, mean ± SEM, *P < 0.05, **P < 0.01). (E) A decrease in Tkv protein level was observed in Ube3a-overexpressing S2 cells; the effect was abolished by treatment with the 26S proteasome inhibitor MG132. The experiment was repeated three times with similar results. (F) Statistical analysis of relative Tkv protein level normalized to the loading control tubulin in S2 cells co-expressing Ube3a with or without MG132 treatment (n = 3, one-way ANOVA test, **P < 0.01, mean ± SEM).
To determine whether Tkv stability is regulated by the proteasomal degradation pathway, Tkv protein level was measured in Ube3a-overexpressing S2 cells treated with the 26S proteasome inhibitor MG132. In contrast to the upregulation of Tkv observed upon Ube3a knockdown, Tkv levels were significantly reduced by 50.17% in Ube3a-overexpressing cells compared with GFP-overexpressing control cells, and this effect was abolished by MG132 treatment (Fig 6E). These results indicate that Ube3a negatively regulates Tkv protein levels by targeting the protein for proteasome-mediated degradation.
Tkv is a direct substrate of Ube3a
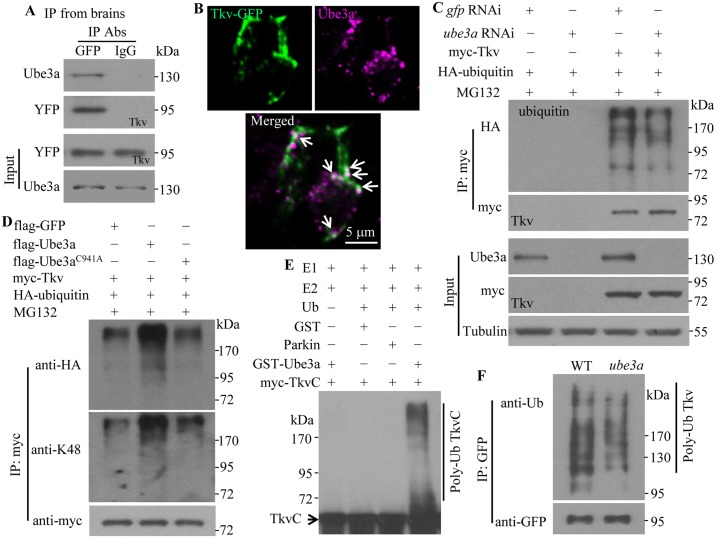
Given the increased stability of Tkv in Ube3a knockdown cells and the involvement of the proteasome in Tkv degradation (Fig 6), Tkv may be a direct target for Ube3a-mediated ubiquitination and ensuing proteasome-mediated degradation. This model predicts a physical interaction between the two, and direct ubiquitination of Tkv by Ube3a. We investigated the physical interaction between Ube3a and Tkv by co-immunoprecipitation (co-IP). In third instar larval brain extracts, endogenous Ube3a co-precipitated with Tkv-YFP using an antibody against GFP, but was absent in control immunoprecipitates using anti-IgG (Fig 7A). We defined the regions mediating the interaction in S2 cells and found that the N-terminal 201–400 and 400–640 amino acid fragments of Ube3a each interacted with Tkv, while the STYKc domain of Tkv mediated its interaction with Ube3a (S8 Fig). Co-localization of Tkv-GFP and Ube3a driven by elav-Gal4 was observed in the soma of motoneurons in the ventral ganglion (6.3 puncta positive for both Tkv-GFP and Ube3a per neuron, n = 16; Fig 7B), further supporting a physical interaction between Ube3a and Tkv.
Fig 7. Ube3a interacts with and ubiquitinates Tkv in vivo and in vitro.
(A) Ube3a interacts with Tkv in larval brains as detected by co-IP. Ube3a was co-immunoprecipitated by anti-GFP from larval brain lysates of a gene trap line expressing endogenous Tkv tagged with YFP. (B) Co-localization of Tkv-GFP and Ube3a driven by elav-Gal4 in motoneuron soma. Images from two consecutive single slices are presented. Arrows indicate co-localized puncta. (C) Decreased Tkv ubiquitination by RNAi knockdown of Ube3a in S2 cells. Cells were co-transfected with plasmids encoding myc-tagged Tkv and HA-tagged ubiquitin, and treated with dsRNA against Ube3a or GFP dsRNA as a control. Anti-HA antibody was used to detect ubiquitinated Tkv. Tubulin was used as a loading control. (D) Increased Tkv poly-ubiquitination by overexpression of wild-type but not C941A mutant Ube3a in S2 cells. Poly-ubiquitination was detected using anti-HA and anti-K48 antibodies. (E) GST-Ube3a ubiquitinates myc-TkvC in vitro. The reaction was reconstituted with the components E1, E2, HA-Ub, the potential substrate myc-TkvC, together with the GST control, the E3 ligase Parkin control, or GST-Ube3a. Both poly-ubiquitinated and non-ubiquitinated TkvC are indicated. (F) In vivo ubiquitination of Tkv. Tkv-GFP from larval brain lysates was immunoprecipitated with anti-GFP and the level of ubiquitinated Tkv was detected by anti-ubiquitin. A decreased level of Tkv ubiquitination was observed in ube3a35 mutants compared with wild type.
To establish whether Tkv is a direct substrate of Ube3a-mediated ubiquitination, we measured the level of unmodified and ubiquitinated Tkv in S2 cells. In Ube3a knockdown cells, Tkv poly-ubiquitination was reduced (Fig 7C). Conversely, overexpression of Ube3a caused an increase in Tkv poly-ubiquitination (Fig 7D). Poly-ubiquitination occurred at lysine 48 of ubiquitin (Fig 7D); a form of poly-ubiquitination that targets proteins for proteasomal degradation [42]. An in vitro ubiquitination assay further revealed that GST-Ube3a induced poly-ubiquitination of myc-TkvC, while neither GST alone, the control E3 ligase Parkin, nor a cocktail missing the essential component ubiquitin, led to TkvC poly-ubiquitination (Fig 7E). To verify if the negative of BMP signaling by Ube3a occurred in vivo, we treated dissected larvae with MG132 for 4 h and observed a significantly increased level of pMad at both NMJs and motoneuron nuclei (S9 Fig). Blocking proteasome function by expressing a dominant negative proteasomal subunit DTS5 in presynaptic neurons by elav-Gal4 also resulted in a similar increase of pMad staining (S9 Fig). Western blotting of larval brain lysates showed a decreased level of ubiquitinated Tkv in ube3a mutants (Fig 7F). These results demonstrate that Ube3a promotes ubiquitin-mediated degradation of Tkv.
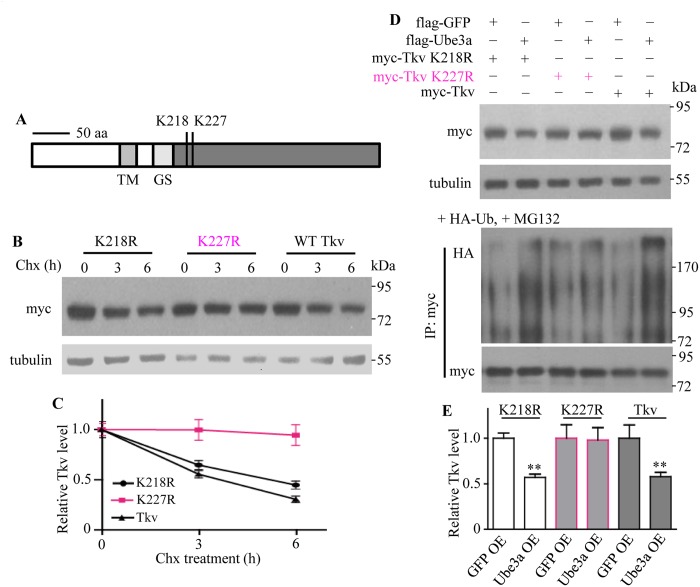
Given that Ube3a ubiquitinated Tkv and promoted its degradation by proteasome pathway (Figs 6 and 7), we aimed to identify specific amino acids of Tkv targeted by UBE3A. Mass spectrometry revealed two potential ubiquitinated residues, lysine 218 and 227, in the C-terminal STYKc region of Tkv (Fig 8A). To verify which specific sites of Tkv were ubiquitinated by Ube3a, we mutated lysine to arginine at two individual sites, namely, K218R and K227R. We compared the stability of wild-type and mutant Tkv protein in S2 cells by western blotting following treatment with cycloheximide to inhibit protein synthesis for 0, 3 and 6 h. We found that the stability of K218R was comparable to that of wild-type Tkv, the levels of both decreased with time, while the amount of K227R remained stable within 6 h treatment of cycloheximide (Fig 8B and 8C). Ubiquitination analysis showed that K218R was as sensitive as wild-type Tkv for ubiquitination in S2 cells by Ube3a in the presence of HA-Ub and MG132 (Fig 8D and 8E). There was a concomitant significant reduction in wild-type and mutant Tkv protein level, while K227R was stable and ubiquitination resistant (Fig 8D and 8E). These results demonstrate that K227 is a key target residue for ubiquitination by Ube3a.
Fig 8. Identification of Tkv ubiquitination sites targeted by Ube3a.
(A) Lysine K218 and K227 in the cytoplasmic region of Tkv were identified as ubiquitination sites by mass spectrometry. Scale bar indicates 50 amino acids. TM indicated transmembrane domain; GS denotes Gly-Ser rich motif. (B) K227R Tkv was more stable than K218R or wild-type Tkv. S2 cells transfected with Ube3a and wild-type or mutant Tkv were treated with cycloheximide to block protein synthesis, and the level of myc-Tkv was detected to determine protein stability. (C) Statistical analysis of wild-type and mutant Tkv protein levels at different time points after protein synthesis blockade by cycloheximide (n = 3, mean ± SEM). (D) Verification of Tkv ubiquitination sites targeted by Ube3a. S2 cells were transfected with plasmids encoding Ube3a and wild-type or mutant Tkv. After 48 h transfection, the level of Tkv was examined by western blotting (upper blot). For ubiquitination assays, S2 cells over-expressing Ube3a and wild-type or mutant Tkv, together with HA-ubiquitin (+HA-Ub) and treated with MG132 (+MG132) to block ubiquitin-mediated degradation were immunoprecipitated with anti-myc antibody. Western blotting was performed to analyze the levels of ubiquitinated and un-ubiquitinated Tkv. (E) Statistical analysis of wild-type and mutant Tkv protein levels normalized to the loading control tubulin when co-expressed with Ube3a in S2 cells (n = 3, one-way ANOVA test, **P < 0.01, mean ± SEM).
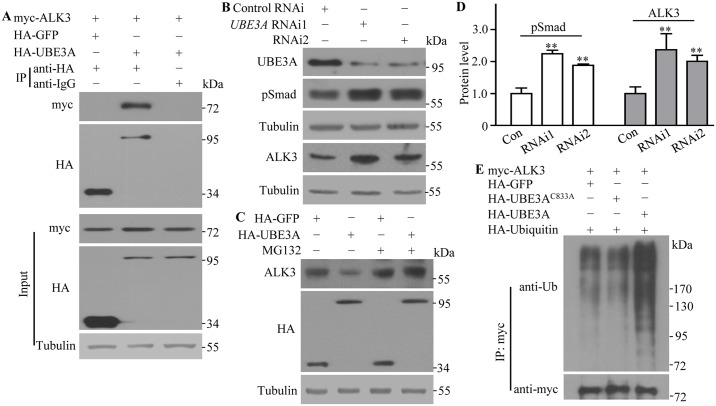
Human UBE3A binds and ubiquitinates ALK3, a mammalian homolog of Tkv
Given that Ube3a is evolutionarily conserved from Drosophila to vertebrates, we hypothesized that human UBE3A would interact with and negatively regulate ALK3, a mammalian homolog of Tkv [43]. To test this possibility, we examined the co-immunoprecipitation of ALK3 with UBE3A in lysates of human HEK293 cells. An anti-HA antibody, but not a control anti-IgG, co-precipitated myc-ALK3 with HA-UBE3A (Fig 9A). UBE3A knockdown in HEK293 cells using two different small interfering RNAs (siRNAs) produced a significantly increased level of endogenous ALK3 as well as a near two-fold increase in the level of pSmad; the mammalian counterpart of Drosophila pMad (Fig 9B and 9D). Conversely, overexpression of UBE3A in HEK293 cells led to a significant decrease in ALK3 protein level (38.1% of the control; n = 3, P < 0.001), which was blocked by the proteasome inhibitor MG132 (Fig 9C), implying that human UBE3A promotes the proteasomal degradation of ALK3. Overexpression of wild type but not the ligase dead C833A mutant UBE3A significantly increased ubiquitination of myc-ALK3, while GFP control did not (Fig 9E). Despite the fact that the corresponding K227 in ALK3 is not critical for ubiquitination in HEK293 cells (S10 Fig), consistent with the fact that some proteins have little specificity in ubiquitination sites [44], our results demonstrate that Ube3a-mediated suppression of BMP signaling is conserved in mammalian cells.
Fig 9. Human UBE3A negatively regulates the protein level of BMP receptor ALK3 in HEK293 cells.
(A) Human UBE3A interacts with the BMP receptor ALK3 in HEK293 cells. HEK293 cells were co-transfected with myc-tagged ALK3 and HA-tagged UBE3A or GFP as a control. Lysates from transfected cells were immunoprecipitated with anti-HA and anti-IgG as a control, followed by western blotting to detect the presence of HA- and myc-tagged proteins. (B) Increased levels of the endogenous pSmad protein and ALK3 were observed in HEK293 cells expressing reduced levels of UBE3A by two independent siRNAs. (C) A decreased level of the endogenous ALK3 protein was observed in UBE3A-overexpressing cells; this effect was abolished by treatment with the proteasome inhibitor MG132. (D) Statistical analysis of pMAD and ALK3 protein levels in HEK293 cells expressing reduced levels of UBE3A by two independent siRNAs (n = 4, one-way ANOVA test, mean ± SEM, **P < 0.01). (E) HA-tagged GFP, wild-type or C833A UBE3A was co-expressed with myc-ALK3 and HA-Ub in HEK293 cells as indicated. Lysates from transfected cells were immunoprecipitated with anti-myc and the levels of ubiquitination were analyzed with an anti-ubiquitin antibody.
Discussion
Ube3a regulates synapse growth and endocytosis by inhibiting BMP signaling
The synapse is a critical substrate for cognitive function, so disorders associated with intellectual and cognitive deficits, such as AS and autism, likely involve altered pathways that disrupt synaptic development and plasticity. The present study shows for the first time that ube3a regulates synapse formation and synaptic endocytosis at the Drosophila NMJ. A previous study reported normal NMJ growth in ube3a15B mutants [35]. However, we showed here that the genetic background in homozygous ube3a15B mutants may suppress expression of satellite phenotype, as trans-allelic ube3a15B/ube3a35 and hemizygous ube3a15B/Def mutants showed excess satellite boutons (S3 Fig). We noted that the satellite phenotype in ube3a mutants is mild compared to the classical endocytic endophilin and dap160/intersectin mutants [32, 45, 46]. Also, the fewer and larger SVs at active zones accompanied by an increase in amplitudes of mEJPs reported in endocytic mutants such as tweek and dap160/intersectin mutants [45–47], were not observed in ube3a mutants. However, the significantly higher number of satellite boutons, reduced dye uptake, and defective recovery of EJP amplitudes under high-frequency stimulation, implicate a regulatory role for ube3a in endocytosis at NMJs. It is worth noting that a similar endocytic function has been reported for mammalian Ube3a at inhibitory synapses in the visual cortex [14].
There appears to be a reciprocal regulation between BMP signaling and endocytosis at the Drosophila NMJ. Endocytic mutants show upregulated BMP signaling as demonstrated by increased pMad staining at NMJs [40]. Trio, a Rac GTPase-specific GEF, is positively regulated by pMad at the transcriptional level in motoneuron nuclei for proper NMJ development [48]. Conversely, upregulation of BMP signaling by mutations in Dad, an inhibitory Mad that negatively regulates BMP signaling, and in S6KL (S6 kinase like) results in apparent endocytic defects [33, 34]. In addition to the mutual regulation between the two processes, the multi-domain actin regulator Nwk functions at the interface of endocytosis and BMP signaling by physically interacting with both Tkv and endocytic proteins dynamin and Dap160/intersectin [40]. We found normal expression of endocytic proteins in ube3a mutant NMJs (S11 Fig), and endocytic endophilin A mutants have been shown previously to have a normal level of Tkv [34]. Thus, we favor a model in which upregulated BMP signaling leads to overgrown NMJs and endocytic defects in ube3a mutants, and the NMJ defects in ube3a mutants could be attributable to altered actin-cytoskeletal formation via Trio-Rac-mediated and/or Nwk-mediated pathways.
While ube3a mutation leads to an elevated level of BMP signaling, neuronal overexpression of Ube3a by elav-Gal4 or OK6-Gal4 does not result in downregulation of BMP signaling. This was demonstrated by overgrown NMJ terminals and a normal level of pMad at both NMJs and motoneuron nuclei (S12 Fig). These results indicate that overexpression of Ube3a alone is not sufficient to downregulate BMP signaling in the neuromusculature. How Ube3a overexpression leads to overgrown NMJs remains to be determined. Similar overgrown NMJ phenotypes caused by Ube3a mutation and overexpression are consistent with previous reports of defective locomotion [24] and circadian rhythms [25] upon bidirectional changes in Ube3a expression.
Ube3a ubiquitinates and promotes proteasomal degradation of the BMP receptor Tkv
Using a series of biochemical assays, we showed that Ube3a physically interacts with Tkv and facilitates its proteasome-mediated degradation (Figs 6 and 7). This function of Ube3a was conserved in mammalian cell cultures (Fig 9). We identified K227 but not K218 in the cytoplasmic region of Tkv as being critical for its ubiquitination (Fig 8). We demonstrated that Ube3a normally functions specifically to downregulate Tkv expression by proteasomal degradation, resulting in reduced presynaptic BMP signaling. In the absence of Ube3a, BMP signaling is upregulated, resulting in overgrown NMJs and endocytic defects.
The impact of Ube3a on Tkv is limited to the NMJ, as altered expression of Ube3a does not lead to corresponding changes in Tkv expression in wing discs, where the role of Tkv is well known (S2 Fig). Thus, the negative regulation of Tkv by Ube3a occurs in a tissue specific manner.
In which neuronal compartment is Tkv ubiquitinated? Given that Ube3a (S1 Fig), RING finger E3 ligase Highwire [49], E3 ligase anaphase promoting complex/cyclosome (APC/C) [50], and 20s proteasome subunit [51] are all enriched at NMJ synapses, we favor the possibility that Ube3a ubiquitinates Tkv at presynaptic terminals.
We recently identified that an evolutionarily conserved, previously uncharacterized S6KL similarly regulates NMJ growth and endocytosis by promoting proteasomal degradation of Tkv [34]. As Ube3a biochemically and genetically interacts with S6KL (S13 Fig), it remains possible that Ube3a and S6KL work synergistically in inhibiting NMJ growth by targeting Tkv for degradation
In addition to Ube3a, another HECT-domain-containing E3 ligase Smurf also inhibits BMP signaling by targeting two independent signaling components, BMP receptors and downstream effector Smad in distinct developmental processes [43, 52, 53]. However, Smurf does not regulate NMJ growth [34]. As the protein level of Mad was mildly but significantly increased in ube3a mutant brains (Fig 5C and 5D), it would be of interest to determine if Ube3a also targets Mad for proteasome-mediated degradation in NMJ development.
In addition to its role as an ubiquitin E3 ligase, UBE3A has been reported to affect nuclear hormone receptor-mediated transcription by E3 ligase-independent mechanisms [21, 54]. For instance, UBE3A was shown to inhibit the expression of the cyclin-dependent kinase inhibitor p27 at both transcriptional and post-translational levels [55]. Moreover, UBE3A inhibits estradiol-mediated transcription of the target gene Arc [21]. As with mammalian UBE3A, Drosophila Ube3a also acts as a transcriptional regulator; it activates transcription of Punch, which encodes GTP cyclohydrolase 1, a component of the monoamine biosynthesis pathway [27]. In support of a transcriptional role, Ube3a orthologs of different species are localized in the nucleus [16, 23, 27]. However, the present study reveals that Ube3a functions as an ubiquitin ligase for Tkv but not as a transcriptional regulator since the level of tkv mRNA in ube3a brains was similar to wild type (Fig 5G).
Involvement of BMP signaling upregulation in the pathophysiology of AS
Human UBE3A negatively regulated the protein levels of ALK3, and consequently the downstream effector pSmad, in HEK293 cells (Fig 9). Therefore, it is tempting to speculate that elevated BMP signaling may contribute to a subset of the abnormalities observed in patients with AS. BMP receptors are expressed at specific sites, such as the hippocampus and cerebellum, where Ube3a is also expressed [10, 56]. It remains to be determined if BMP signaling is upregulated in these brain regions that correlate with the clinical features of seizures, learning deficit and ataxia in patients with AS.
BMP signaling regulates synapse development specifically at the calyx of Held in the auditory system in a way similar to that at Drosophila NMJ, but shows no effect on synapse development in the lateral superior olive of the auditory nucleus [31]. Given that AS mice show an enhanced seizure-like response to audiogenic challenge [22], it would be improtant to determine if the development and function of the calyx of Held synapses are normal in Ube3a mutant mice. At the microscopic level, AS model mice exhibit normal dendritic elaboration but fewer and shorter dendritic spines in cerebellar Purkinje cells and pyramidal neurons in the hippocampus and cortex [16]. It is not known if BMP signaling plays a role in the formation of dendritic spines, although other targets of Ube3a such as Ephexin5 may regulate spine morphogenesis in hippocampal neurons [20].
In summary, we demonstrated that Drosophila ube3a plays an important role in regulating synapse formation and endocytosis by inhibiting BMP signaling via proteasome-mediated degradation of Tkv. The negative regulation of BMP signaling by Ube3a is conserved in mammals. These findings indicate that increased BMP signaling resulting from loss of UBE3A during development may contribute, at least in part, to the etiology of AS.
Materials and Methods
Drosophila stocks
All Drosophila stocks were fed standard cornmeal food and maintained at 25°C. w1118 was used as a wild-type control unless otherwise indicated. We generated intragenic ube3a deletions (ube3a8 and ube3a35) through P-element-mediated excisions of EP3214. Transgenic lines expressing genomic ube3a+ and ube3aC941A (containing a missense mutation in the codon for the critical catalytic cysteine residue) were from J. Fischer. UAS-Tkv-GFP was from T. B. Kornberg. UAS-DTS5 was a gift from J. Belote via K. Broadie [51]. The EMS-induced nonsense mutations ube3a549 and ube3a689 were obtained from a TILLING (targeting-induced local lesions in genomes) service (http://tilling.fhcrc.org). For tissue-specific rescue experiments, the pan-neural elav-Gal4 (from Bloomington) and muscle-specific C57-Gal4 (from V. Budnik) lines were used. RNAi lines P{GD450}v45876 and Thu3266 were obtained from the Vienna Drosophila RNAi Center (VDRC) and Tsinghua University (Beijing, China), respectively. A yellow fluorescent protein (YFP) genetrap line labeling the endogenous Tkv with YFP was obtained from the Drosophila Genome Research Center (stock number 115298; Kyoto, Japan). EP(3)3214, Df(3L)ED4470, tkv8, tkvK16713, mad12, and witB11 were obtained from the Bloomington Stock Center.
For overexpression studies, UAS-ube3a was obtained by amplifying the full-length cDNA of ube3a from a home-made cDNA pool derived from whole adults, and cloned into the vector pUAST. An UAS-ube3a insertion in the X chromosome was used in this study.
Generation of monoclonal antibodies against Ube3a
To generate specific antibodies against Ube3a, a fusion protein containing amino acids 568–973 of Drosophila Ube3a with an N-terminal 6×His tag was produced in E. coli. Mouse monoclonal antibodies produced by clone 8F7 were shown to be specific for Ube3a by western blotting and immunostaining.
Immunohistochemical analysis
Immunohistochemical analysis of larval NMJ synapses was performed as previously described [57]. The following antibodies were used: rabbit anti-Tkv (1:200; from M. B. O’Connor, University of Minnesota, Minneapolis, Minnesota), anti-CSP (1:1,000, 6D6 from Developmental Studies Hybridoma Bank (DSHB)), rabbit anti-GFP (1:100, #632460 from Clontech), rabbit anti-pMad (1:500; from P. ten Dijke, Leiden University, Leiden, Netherlands), anti-HRP conjugated with FITC or Texas red (1:50, Jackson ImmunoResearch Laboratories), and anti-Ube3a (1:30, 8F7 as described above). All primary antibodies were visualized using fluorophore-conjugated secondary antibodies, including Alexa Fluor 488 or 568-conjugated goat anti-mouse or anti-rabbit IgG (1:1,000, Molecular Probes). To quantify fluorescence intensities from immunostaining, images of the entire muscle 4 NMJ elaboration or motoneuron nuclei were taken at identical settings for all genotypes without overexposure. The intensity of pMad and Tkv-GFP staining in Fig 5 was calculated as gray values normalized to the pMad- and HRP-positive areas, respectively. The gray values were automatically calculated using ImageJ software (NIH; Bethesda, USA) and presented as arbitrary units (a.u.). All images were collected using a Leica SP5 confocal microscope and processed with Adobe Photoshop.
For statistical analysis of NMJ morphology, the number of total boutons and satellite boutons of muscle 4 NMJ in abdominal segments A2 and A3 was quantified as previously described [57].
Electrophysiological and FM1-43 uptake assays
Electrophysiological recordings were performed at 20°C essentially following a conventional procedure with minor modifications [33, 34]. For the basal transmission assay, wandering third-instar larvae were dissected in Ca2+-free HL3.1 saline and recorded in HL3.1 saline containing 0.5 mM CaCl2. Intracellular microelectrodes with a resistance greater than 5 MΩ filled with 3 M KCl were used for the assay. Excitatory junction potential (EJPs) and miniature EJPs (mEJPs) were recorded from muscle 6 in the abdominal segments A2 or A3 for 120 seconds. EJPs were elicited by low frequency (0.3 Hz) stimulation. We analyzed recordings from muscles cells with physiological resting potentials ≤ -60 mV and input resistances > 5 MΩ. For examining synaptic transmission under tetanic stimulation, synapses were stimulated at 10 Hz for 10 minutes and recorded in a modified HL3.1 saline with 0.5 mM extracellular Ca2+. For the FM1-43 uptake assay, we followed a previously published protocol [34, 58].
Cell culture
S2 cells were cultured in Schneider’s medium (Invitrogen, Carlsbad, USA) and transfected using Cellfectin II reagent (Invitrogen). For the production of double-strand RNA (dsRNA) targeting ube3a and gfp, DNA fragments were synthesized by PCR using primers containing a T7 promoter sequence (italicized): ube3a sense: 5’-TAA TAC GAC TCA CTA TAG GGA TTG CCG GAA ACC ACT GAT A-3’ and antisense: 5’-TAA TAC GAC TCA CTA TAG GGC TCC GTT CTC AAA TGG TGT G-3’, and gfp sense: 5’-TAA TAC GAC TCA CTA TAG GGA TGG TGA GCA AGG GCG AGG A-3’ and antisense: 5’-TAA TAC GAC TCA CTA TAG GGC TTG TAC AGC TCG. The dsRNAs were transcribed from both strands of the PCR-amplified DNA fragments using a Megascript T7 kit (AM1333; Invitrogen), and purified using an RNeasy kit (Qiagen).
To examine Tkv protein turnover, S2 cells were transfected with dsRNAs against ube3a or gfp (control) at an average transfection efficiency of about 46%, and treated 48 h later with 40 μg/ml cycloheximide (Sigma) for 0, 1.5, 3, 6, and 9 h. To determine whether the degradation of Tkv was proteasome-dependent, S2 cells were co-transfected with flag-Ube3a or flag-GFP plasmid together with myc-Tkv. After 48 h, cells were treated with 20 μM MG132 (Sigma) or mock treated for 6 h before being harvested for western blotting.
Human HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Invitrogen) at 37°C in a humidified incubator with 5% CO2. For efficient RNAi knockdown of human UBE3A, UBE3A siRNA1 (5’-GGG UCU ACA CCA GAU UGC UTT-3’) [59], siRNA2 (5’-CAA CUC CUG CUC UGA GAU ATT-3’) [60], and a scrambled siRNA control (5’-UUG CGG GUC UAA UCA CCG ATT-3’) were synthesized and transfected into HEK293 cells using X-tremeGENE siRNA transfection reagent (Roche). UBE3A and ALK3 (also named BMPR1A) clones were provided, respectively, by Dr. Peter M Howley and William C. Hahn at Harvard Medical School (Cambridge, USA). UBE3A and ALK3 coding sequences were subcloned to generate pCMV-Tag2B-HA-UBE3A and pCDNA3-myc-ALK3, respectively.
Immunoprecipitation (IP) and western blotting
IP assays of larval brains and ventral ganglia were carried out according to a previous report [61]. Immunoprecipitation from S2 or HEK293 cell lysates was performed following a previously described protocol [43]. Antibodies used for western blotting were: anti-myc (1:1000; 9E10 from Clontech), anti-flag (1:1,000; M2 from Sigma), anti-K48 (1:1,000; D9D5 from Cell Signaling Technologies), anti-GFP (1:1,000), anti-Ube3a (1:300), anti-HA (1:1,000; 3F10 from Roche), anti-ubiquitin (1:1000; P4D1 from Cell Signaling Technologies), anti-Wit (1:50; DSHB), anti-pSmad (1:1000; Cell Signaling Technologies), and anti-α-tubulin (1:25,000; mAb B-5-1-2 from Sigma).
To analyze protein levels in different genotypes, larval brains and ventral ganglia were homogenized in ice-cold lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% (v/v) NP-40, 0.1% (w/v) SDS, 1% (v/v) proteinase inhibitor). Western blots were probed with primary antibodies anti-Ube3a, anti-pMad (1:500, Cell Signaling Technologies), anti-Wit, anti-GFP, and anti-α-tubulin, followed by HRP-conjugated secondary antibody (1:25,000; Sigma).
In vivo and in vitro ubiquitination assay
For in vivo ubiquitination assay, S2 cells were transfected with plasmids using Cellfectin II reagent (Invitrogen). The ubiquitination assay was performed according to the protocols described previously [43]. In brief, at 48 hr posttransfection, MG132 was added into the media at the final concentration 50μM. Cells were harvested 4 hr later and lysed with a lysis buffer (50 mM Tris [pH 7.5], 120 mM NaCl, and 0.5% NP40) containing 1% (w/v) SDS to disrupt protein–protein interaction and boiled for 10 min, then diluted 10 times with cell lysis buffer.
To detect the levels of ubiquitinated Tkv in vivo, 200 larval brains of each genotype (elav-Gal4; tkv-GFP and elav-Gal4; tkv-GFP; ube3a35) were dissected and incubated with 50 μM MG132 (Sigma) in Schneider’s medium for 4 h to inhibit proteasome-mediated degradation before proceeding with IP. The brains were then homogenized in lysis buffer and the lysates were immunoprecipitated with anti-GFP. The ubiquitinated Tkv was examined by anti-ubiquitin antibody. For in vitro ubiquitination assay, the myc-tagged Tkv C-terminus (myc-TkvC) was synthesized by an in vitro translation kit containing rabbit reticulocyte lysates (Promega). Ube3a was fused to GST, then expressed and purified from E. coli. E1, E2, HA-Ub (all three from Boston Biochem), and the potential substrate myc-TkvC, together with the GST control, the E3 ligase Parkin control, or GST-Ube3a, were incubated at 30°C for 1.5 h in 40 μl ubiquitination reaction buffer (50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 50 mM NaCl, 5 mM MgCl2, 2 mM ATP). Reactions were terminated by adding SDS-PAGE sample buffer and analyzed by western blotting.
Mass spectrometry for identification of ubiquitination sites
Mass spectrometry was performed as described previously [62]. myc-Tkv expressed in S2 cells was immunoprecipitated with an anti-myc antibody. The target protein was excised, digested with trypsin, fractionated by HPLC, and analyzed by a LTQ Orbitrap Elite mass spectrometer (ThermoFisher Scientific, Waltham, MA). To further verify the sites of Tkv targeted by Ube3a, myc-Tkv mutants K218R and K227R were generated by site-directed mutagenesis.
Quantitative RT-PCR of tkv mRNA
Total RNA was extracted from brains and ventral nerve ganglia of 40 third instar larvae following the standard Trizol reagent protocol (Invitrogen). Power SYBR Green PCR Master Mix (Applied Biosystems) was used for quantitative real-time PCR. Actin mRNA level was amplified as an internal control using published primers [33], whereas tkv cDNA was amplified with primers 5’-GTG ATA GGG CAG GGC GTA GT-3’ and 5’-AGT GGG TCT CGT TCT GTG GG-3’.
Statistical analysis
Student’s t tests were used for statistical comparisons between two groups. Multiple group means were compared by one-way ANOVA. Asterisks above a column indicate comparisons between a specific genotype and wild type, whereas asterisks above a bracket denote comparisons between two specific genotypes. Data are presented as mean ± standard error of the mean (SEM). P-values < 0.05 were considered statistically significant.
Supporting Information
(A) Ube3a protein is cytoplasmic and is highly expressed in the ventral ganglion of a third instar larva. The ventral ganglion was double-labeled with a monoclonal antibody 8F7 against Ube3a (green) and propidium iodide (magenta) to visualize nuclei. (B) No specific Ube3a staining was detected in ube3a35 ventral ganglion. (C) Ube3a is expressed in muscles and axons with specific enrichment at presynaptic NMJ terminals compared with background expression of Ube3a in mutants. Larval preparations were double-stained with anti-Ube3a (green) and anti-HRP (magenta). Ube3a is cytoplasmic and expressed in the adult ovary (D) and adult testis (E).
(TIF)
(A, B) The specificity of anti-Ube3a was demonstrated by staining wing disc expressing a reduced level (en-Gal4/+; RNAi /+; A) or an upregulated level (UAS-ube3a/+; en-Gal4/+; B) of Ube3a in the posterior compartment by en-Gal4. Tkv protein level remains unchanged upon altered expressions of Ube3a. Scale bar = 40 μm. (C) The ventral ganglion of a OK6-Gal4/UAS-CD8-GFP larva was double-labeled with GFP (bright enough without staining) and anti-Ube3a. Ube3a expresses in GFP-positive motoneurons as well as GFP negative interneurons. (D) No specific Ube3a staining was detected in ube3a35 ventral ganglion. Scale bar = 40 μm.
(TIF)
(A) Muscle 4 NMJ synapses were double-stained with anti-HRP (green) and anti-CSP (magenta). OK6-Gal4-driven Thu3266 RNAi against ube3a in motoneurons led to more total boutons and satellite boutons compared with the control. Scale bar = 10 μm. (B) Quantification of total boutons and satellite boutons in control (OK6-Gal4/+) and OK6-Gal4/+; RNAi/+ animals. n ≥ 16 NMJs; ***P < 0.001 by t test; error bars represent SEM. (C) The NMJ growth appeared normal in ube3a15B homozygotes, but there were more total boutons and more satellite boutons in ube3a15B/ube3a35 and ube3a15B/Def mutants compared with ube3a15B. (D) Quantification of the total boutons and satellite boutons in the three genotypes. n ≥ 16 NMJs; ***P < 0.001 by one-way ANOVA test; error bars represent SEM.
(TIF)
(A, B) Electron micrographs of synaptic boutons from wild type (A) and ube3a35 mutants (B). Compared with wild type, ube3a35 mutants exhibited significantly more cisternae with diameters >60 nm (arrowheads in B). Arrows in A and B indicate active zone. Scale bar, 500 nm. (C, D) High magnification view of representative active zones from wild type (C) and ube3a35 mutants (D). Dashed line defines a 200 nm radius around the active zone for quantitative analysis of SVs. Scale bar, 100 nm. (E) High magnification view of a presynaptic bouton from ube3a35 mutants. Arrowheads indicate cisternae. Scale bar, 100 nm. (F, G) Quantification of the number of SVs within a 200 nm radius of the active zone (F) and the mean number of cisternae per cross-sectioned presynaptic bouton (G). n ≥ 38 boutons from more than 4 larvae analyzed, t test, mean ± s.e.m., ***p < 0.001.
(TIF)
(A, B) Representative traces of EJP and mEJP of wild type (A) and ube3a35 mutant (B) NMJs. Scale bars for EJP and mEJP are annotated.
(TIF)
(A, B) Increased levels of pMad at NMJ synapses (A) and motoneuron nuclei (B) of ube3a35 mutants were rescued by a heterozygous mutation of tkv8. Scale bars, 5 μm and 10 μm in A and B, respectively. (C) Quantification of the fluorescence intensities of pMad at NMJ and nuclei of different genotypes including wild type, ube3a35, and tkv8/+; ube3a35. ***p < 0.001 by one-way ANOVA; error bars indicate SEM. (D) NMJ4 synapses in abdominal segment A3 loaded with FM1-43 in wild type, ube3a35, and tkv8/+; ube3a35. Scale bar, 5 μm. (E) Quantification of FM1-43 intensities in NMJ boutons following high K+-stimulated endocytosis. n ≥ 14 NMJs; ***p < 0.001 by one-way ANOVA; error bars indicate SEM. (F) Percentage of average EJP amplitudes under tetanic stimulation for 10 min in wild type, ube3a35, and tkv8/+; ube3a35. n ≥ 10 animals. Representative traces are shown on the right.
(TIF)
(A) The ventral ganglion of a wild-type larva was double-labeled with anti-GFP and anti-HRP. No GFP signals were detected in the ventral ganglion. (B) Tkv-YFP under the control of the endogenous promoter expresses in the soma of various neurons including motoneurons along the midline of a ventral ganglion. Scale bar = 40 μm. (C, D) Tkv-YFP is functional as the genetrap line shows NMJ growth (D) comparable to wild-type control (C).
(TIF)
(A) Schematic representation of full-length and various truncated Ube3a used for co-IP assays. (B) The N-terminal regions of Ube3a mediate the interaction with myc-Tkv. S2 cell lysates were transfected with different combinations of constructs and immunoprecipitated with anti-flag antibody, followed by western blotting using anti-flag and anti-myc. IgG heavy chain is indicated by an asterisk. α-tubulin was used as a loading control. (C) Schematic diagram of full-length Tkv and its various truncations. Different functional domains are indicated. (D) Co-IP assays showed that the C-terminus of Tkv (TkvC), lacking the extracellular and transmembrane (TM) regions, was able to bind Ube3a. More precisely, the dual-specificity serine-threonine/tyrosine protein kinase catalytic (STYKc) domain exhibited strong binding to Ube3a; consistently, deletion of this domain greatly reduced the interaction.
(TIF)
(A) The level of pMad increased at NMJ synapses and motoneuron nuclei of dissected wild-type larvae treated with 50 μM MG132 inhibitor in Schneider’s medium for 4 h. (B) Quantification of fluorescence intensities of pMad at NMJ and nuclei of non-treated and MG1320-treated larvae. n ≥ 16; **P < 0.01, ***p < 0.001 by one-way ANOVA; error bars indicate SEM. (C) Expressing DTS5, a dominant temperature-sensitive mutation of the β6 subunit of the 26s proteasome (Speese et al., Curr Biol, 2003), by elav-Gal4 (elav-Gal4/+; DTS5/+) also led to an increase of pMad at NMJ synapses and the motoneuron nuclei. (D) Statistical analysis of fluorescence intensities of pMad at NMJs and motoneuron nuclei of elav/+ control and elav-Gal4-driven DTS5 larvae. n ≥ 18; **P < 0.01, ***p < 0.001 by one-way ANOVA; error bars represent SEM.
(TIF)
(A) Sequence alignment of Tkv and human ALK3 spanning the ubiquitination site K227. (B) Wild-type and mutant ALK3 showed a similar level of proteins in the presence of UBE3A. Tubulin was probed as a loading control.
(TIF)
Confocal images of NMJ4 from wild type (A, C, and E) and ube3a35 mutants (B, D, and F) labeled with anti-Dynamin (A and B), anti-Eps15 (C and D), and anti-Endophilin A (E and F).
(TIF)
(A) Overexpression of Ube3a in neurons by elav-Gal4 but not in muscles by C57-Gal4 leads to overgrown NMJ terminals. (B, C) Neuronal overexpression of Ube3a does not drive down BMP signaling as pMad staining appears normal at both NMJs (B) and motoneuron nuclei (C).
(TIF)
(A) Different plasmid constructs were singly or co-transfected to S2 cells. Transfected cells were harvested 48 hr later. Cell lysates were used in a two-step immunoprecipitation step by anti-myc and anti-HA sequentially. (B) Confocal images of NMJ4 labeled with anti-HRP. Loss of one copy of S6KL or ube3a had no effect on the number of satellite boutons, while the number of satellite boutons increased significantly in S6KL140 and ube3a35 trans-heterozygotes and double mutants. (C) Quantification of satellite bouton number in different genotypes. n ≥ 16 NMJs analyzed, one-way ANOVA, mean ± s.e.m., ***p < 0.001.
(TIF)
(DOC)
Acknowledgments
We thank Dr. J. Fischer for genomic ube3a+ and ube3aC941A transgenic flies, the TILLING service at Seattle for providing the nonsense mutation of ube3a flies, T. B. Kornberg for UAS-Tkv-GFP, J. Belote for UAS-DTS5, the Bloomington Stock Center (NIH P40OD018537), Kyoto Stock Center, Vienna RNAi Stock Center, and Tsinghua Stock Center for other fly stocks. We thank Dr. M. B. O’Connor for rabbit anti-Tkv and P. ten Dijke for pMad antibody, D. Chen for Tkv and HA-ubiquitin plasmids, Dr. P. M. Howley for UBE3A clone and Dr. W. C. Hahn for ALK3 and ALK6 clone. Dr. Y. Wang at the proteomics facility in the Institute of Genetics and Developmental Biology assisted with the mass spectrometry. We also thank H. Bellen, A. DiAntonio, J. Fischer, Y. Jiang, T.L. Schwarz, Q. Xie and Z. Xu, for helpful suggestions and comments.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Ministry of Science and Technology (2014CB942803) http://www.most.gov.cn/, the Strategic Priority Research Program B of the Chinese Academy of Sciences (XDB02020400) http://www.cas.cn/, the National Science Foundation of China to YQZ (31110103907 and 31490592) http://www.nsfc.gov.cn/ and the National Science Foundation of China to AY (31271121). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Clayton-Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet. 2003;40(2):87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dagli A, Buiting K, Williams CA. Molecular and Clinical Aspects of Angelman Syndrome. Mol Syndromol. 2012;2(3–5):100–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15(1):70–3. [DOI] [PubMed] [Google Scholar]
- 4.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15(1):74–7. [DOI] [PubMed] [Google Scholar]
- 5.Cook EH Jr., Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60(4):928–34. [PMC free article] [PubMed] [Google Scholar]
- 6.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459(7246):569–73. 10.1038/nature07953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith SE, Zhou YD, Zhang G, Jin Z, Stoppel DC, Anderson MP. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med. 2011;3(103):103ra97 10.1126/scitranslmed.3002627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yi JJ, Berrios J, Newbern JM, Snider WD, Philpot BD, Hahn KM, et al. An Autism-Linked Mutation Disables Phosphorylation Control of UBE3A. Cell. 2015;162(4):795–807. 10.1016/j.cell.2015.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends Neurosci. 2011;34(6):293–303. 10.1016/j.tins.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21(4):799–811. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Zhu G, Liu Y, Standley S, Ji A, Tunuguntla R, et al. UBE3A Regulates Synaptic Plasticity and Learning and Memory by Controlling SK2 Channel Endocytosis. Cell reports. 2015;12(3):449–61. 10.1016/j.celrep.2015.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, Prakash R, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12(6):777–83. 10.1038/nn.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proc Natl Acad Sci USA. 2010;107(12):5611–6. 10.1073/pnas.1001281107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace ML, Burette AC, Weinberg RJ, Philpot BD. Maternal Loss of Ube3a Produces an Excitatory/Inhibitory Imbalance through Neuron Type-Specific Synaptic Defects. Neuron. 2012;74(5):793–800. 10.1016/j.neuron.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaphzan H, Buffington SA, Jung JI, Rasband MN, Klann E. Alterations in intrinsic membrane properties and the axon initial segment in a mouse model of Angelman syndrome. J Neurosci. 2011;31(48):17637–48. 10.1523/JNEUROSCI.4162-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17(1):111–8. [DOI] [PubMed] [Google Scholar]
- 17.Miao S, Chen R, Ye J, Tan GH, Li S, Zhang J, et al. The Angelman syndrome protein Ube3a is required for polarized dendrite morphogenesis in pyramidal neurons. J Neurosci. 2013;33(1):327–33. 10.1523/JNEUROSCI.2509-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valluy J, Bicker S, Aksoy-Aksel A, Lackinger M, Sumer S, Fiore R, et al. A coding-independent function of an alternative Ube3a transcript during neuronal development. Nat Neurosci. 2015;18(5):666–73. 10.1038/nn.3996 [DOI] [PubMed] [Google Scholar]
- 19.Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140(5):704–16. 10.1016/j.cell.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margolis SS, Salogiannis J, Lipton DM, Mandel-Brehm C, Wills ZP, Mardinly AR, et al. EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell. 2010;143(3):442–55. 10.1016/j.cell.2010.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhnle S, Mothes B, Matentzoglu K, Scheffner M. Role of the ubiquitin ligase E6AP/UBE3A in controlling levels of the synaptic protein Arc. Proc Natl Acad Sci USA. 2013;110(22):8888–93. 10.1073/pnas.1302792110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandel-Brehm C, Salogiannis J, Dhamne SC, Rotenberg A, Greenberg ME. Seizure-like activity in a juvenile Angelman syndrome mouse model is attenuated by reducing Arc expression. Proc Natl Acad Sci USA. 2015;112(16):5129–34. 10.1073/pnas.1504809112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reiter LT, Seagroves TN, Bowers M, Bier E. Expression of the Rho-GEF Pbl/ECT2 is regulated by the UBE3A E3 ubiquitin ligase. Hum Mol Genet. 2006;15(18):2825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Bolduc FV, Bell K, Tully T, Fang Y, Sehgal A, et al. A Drosophila model for Angelman syndrome. Proc Natl Acad Sci USA. 2008;105(34):12399–404. 10.1073/pnas.0805291105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gossan NC, Zhang F, Guo B, Jin D, Yoshitane H, Yao A, et al. The E3 ubiquitin ligase UBE3A is an integral component of the molecular circadian clock through regulating the BMAL1 transcription factor. Nucleic Acids Res. 2014;42(9):5765–75. 10.1093/nar/gku225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y, Wang F, Li Y, Ferris J, Lee JA, Gao FB. The Drosophila homologue of the Angelman syndrome ubiquitin ligase regulates the formation of terminal dendritic branches. Hum Mol Genet. 2009;18(3):454–62. 10.1093/hmg/ddn373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferdousy F, Bodeen W, Summers K, Doherty O, Wright O, Elsisi N, et al. Drosophila Ube3a regulates monoamine synthesis by increasing GTP cyclohydrolase I activity via a non-ubiquitin ligase mechanism. Neurobiol Dis. 2011;41(3):669–77. 10.1016/j.nbd.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen L, Farook MF, Reiter LT. Proteomic profiling in Drosophila reveals potential dube3a regulation of the actin cytoskeleton and neuronal homeostasis. PLoS One. 2013;8(4):e61952 10.1371/journal.pone.0061952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu A, Niswander LA. Bone morphogenetic protein signalling and vertebrate nervous system development. Nat Rev Neurosci. 2005;6(12):945–54. [DOI] [PubMed] [Google Scholar]
- 30.Collins CA, DiAntonio A. Synaptic development: insights from Drosophila. Curr Opin Neurobiol. 2007;17(1):35–42. [DOI] [PubMed] [Google Scholar]
- 31.Xiao L, Michalski N, Kronander E, Gjoni E, Genoud C, Knott G, et al. BMP signaling specifies the development of a large and fast CNS synapse. Nat Neurosci. 2013;16(7):856–64. 10.1038/nn.3414 [DOI] [PubMed] [Google Scholar]
- 32.Dickman DK, Lu Z, Meinertzhagen IA, Schwarz TL. Altered synaptic development and active zone spacing in endocytosis mutants. Curr Biol. 2006;16(6):591–8. [DOI] [PubMed] [Google Scholar]
- 33.Shi W, Chen Y, Gan G, Wang D, Ren J, Wang Q, et al. Brain tumor regulates neuromuscular synapse growth and endocytosis in Drosophila by suppressing mad expression. J Neurosci. 2013;33(30):12352–63. 10.1523/JNEUROSCI.0386-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao G, Wu Y, Du L, Li W, Xiong Y, Yao A, et al. Drosophila S6 Kinase Like Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins. PLoS Genet. 2015;11(3):e1004984 10.1371/journal.pgen.1004984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valdez C, Scroggs R, Chassen R, Reiter LT. Variation in Dube3a expression affects neurotransmission at the Drosophila neuromuscular junction. Biology open. 2015;4(7):776–82. 10.1242/bio.20148045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuentes-Medel Y, Ashley J, Barria R, Maloney R, Freeman M, Budnik V. Integration of a retrograde signal during synapse formation by glia-secreted TGF-beta ligand. Curr Biol. 2012;22(19):1831–8. 10.1016/j.cub.2012.07.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhaes TR, Goodman CS. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33(4):545–58. [DOI] [PubMed] [Google Scholar]
- 38.McCabe BD, Marques G, Haghighi AP, Fetter RD, Crotty ML, Haerry TE, et al. The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron. 2003;39(2):241–54. [DOI] [PubMed] [Google Scholar]
- 39.Rawson JM, Lee M, Kennedy EL, Selleck SB. Drosophila neuromuscular synapse assembly and function require the TGF-beta type I receptor saxophone and the transcription factor Mad. J Neurobiol. 2003;55(2):134–50. [DOI] [PubMed] [Google Scholar]
- 40.O'Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58(4):507–18. 10.1016/j.neuron.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merino C, Penney J, Gonzalez M, Tsurudome K, Moujahidine M, O'Connor MB, et al. Nemo kinase interacts with Mad to coordinate synaptic growth at the Drosophila neuromuscular junction. J Cell Biol. 2009;185(4):713–25. 10.1083/jcb.200809127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 43.Xia L, Jia S, Huang S, Wang H, Zhu Y, Mu Y, et al. The Fused/Smurf complex controls the fate of Drosophila germline stem cells by generating a gradient BMP response. Cell. 2010;143(6):978–90. 10.1016/j.cell.2010.11.022 [DOI] [PubMed] [Google Scholar]
- 44.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–78. [DOI] [PubMed] [Google Scholar]
- 45.Koh TW, Verstreken P, Bellen HJ. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 2004;43(2):193–205. [DOI] [PubMed] [Google Scholar]
- 46.Marie B, Sweeney ST, Poskanzer KE, Roos J, Kelly RB, Davis GW. Dap160/intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron. 2004;43(2):207–19. [DOI] [PubMed] [Google Scholar]
- 47.Verstreken P, Ohyama T, Haueter C, Habets RL, Lin YQ, Swan LE, et al. Tweek, an evolutionarily conserved protein, is required for synaptic vesicle recycling. Neuron. 2009;63(2):203–15. 10.1016/j.neuron.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ball RW, Warren-Paquin M, Tsurudome K, Liao EH, Elazzouzi F, Cavanagh C, et al. Retrograde BMP signaling controls synaptic growth at the NMJ by regulating trio expression in motor neurons. Neuron. 2010;66(4):536–49. 10.1016/j.neuron.2010.04.011 [DOI] [PubMed] [Google Scholar]
- 49.Wan HI, DiAntonio A, Fetter RD, Bergstrom K, Strauss R, Goodman CS. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26(2):313–29. [DOI] [PubMed] [Google Scholar]
- 50.van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell. 2004;119(5):707–18. [DOI] [PubMed] [Google Scholar]
- 51.Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr Biol. 2003;13(11):899–910. [DOI] [PubMed] [Google Scholar]
- 52.Liang YY, Lin X, Liang M, Brunicardi FC, ten Dijke P, Chen Z, et al. dSmurf selectively degrades decapentaplegic-activated MAD, and its overexpression disrupts imaginal disc development. J Biol Chem. 2003;278(29):26307–10. [DOI] [PubMed] [Google Scholar]
- 53.Miyares RL, Stein C, Renisch B, Anderson JL, Hammerschmidt M, Farber SA. Long-chain Acyl-CoA synthetase 4A regulates Smad activity and dorsoventral patterning in the zebrafish embryo. Dev Cell. 2013;27(6):635–47. 10.1016/j.devcel.2013.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nawaz Z, Lonard DM, Smith CL, Lev-Lehman E, Tsai SY, Tsai MJ, et al. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19(2):1182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishra A, Godavarthi SK, Jana NR. UBE3A/E6-AP regulates cell proliferation by promoting proteasomal degradation of p27. Neurobiol Dis. 2009;36(1):26–34. 10.1016/j.nbd.2009.06.010 [DOI] [PubMed] [Google Scholar]
- 56.Zhang D, Mehler MF, Song Q, Kessler JA. Development of bone morphogenetic protein receptors in the nervous system and possible roles in regulating trkC expression. J Neurosci. 1998;18(9):3314–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao A, Jin S, Li X, Liu Z, Ma X, Tang J, et al. Drosophila FMRP regulates microtubule network formation and axonal transport of mitochondria. Hum Mol Genet. 2011;20(1):51–63. 10.1093/hmg/ddq431 [DOI] [PubMed] [Google Scholar]
- 58.Shi WW, Chen Y, Gan GM, Wang D, Ren JQ, Wang QF, et al. Brain Tumor Regulates Neuromuscular Synapse Growth and Endocytosis in Drosophila by Suppressing Mad Expression. J Neurosci. 2013;33(30):12352–63. 10.1523/JNEUROSCI.0386-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimoji T, Murakami K, Sugiyama Y, Matsuda M, Inubushi S, Nasu J, et al. Identification of annexin A1 as a novel substrate for E6AP-mediated ubiquitylation. J Cell Biochem. 2009;106(6):1123–35. 10.1002/jcb.22096 [DOI] [PubMed] [Google Scholar]
- 60.Kelley ML, Keiger KE, Lee CJ, Huibregtse JM. The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J Virol. 2005;79(6):3737–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian X, Li J, Valakh V, DiAntonio A, Wu C. Drosophila Rae1 controls the abundance of the ubiquitin ligase Highwire in post-mitotic neurons. Nat Neurosci. 2011;14(10):1267–75. 10.1038/nn.2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shibata S, Zhang J, Puthumana J, Stone KL, Lifton RP. Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc Natl Acad Sci U S A. 2013;110(19):7838–43. 10.1073/pnas.1304592110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Ube3a protein is cytoplasmic and is highly expressed in the ventral ganglion of a third instar larva. The ventral ganglion was double-labeled with a monoclonal antibody 8F7 against Ube3a (green) and propidium iodide (magenta) to visualize nuclei. (B) No specific Ube3a staining was detected in ube3a35 ventral ganglion. (C) Ube3a is expressed in muscles and axons with specific enrichment at presynaptic NMJ terminals compared with background expression of Ube3a in mutants. Larval preparations were double-stained with anti-Ube3a (green) and anti-HRP (magenta). Ube3a is cytoplasmic and expressed in the adult ovary (D) and adult testis (E).
(TIF)
(A, B) The specificity of anti-Ube3a was demonstrated by staining wing disc expressing a reduced level (en-Gal4/+; RNAi /+; A) or an upregulated level (UAS-ube3a/+; en-Gal4/+; B) of Ube3a in the posterior compartment by en-Gal4. Tkv protein level remains unchanged upon altered expressions of Ube3a. Scale bar = 40 μm. (C) The ventral ganglion of a OK6-Gal4/UAS-CD8-GFP larva was double-labeled with GFP (bright enough without staining) and anti-Ube3a. Ube3a expresses in GFP-positive motoneurons as well as GFP negative interneurons. (D) No specific Ube3a staining was detected in ube3a35 ventral ganglion. Scale bar = 40 μm.
(TIF)
(A) Muscle 4 NMJ synapses were double-stained with anti-HRP (green) and anti-CSP (magenta). OK6-Gal4-driven Thu3266 RNAi against ube3a in motoneurons led to more total boutons and satellite boutons compared with the control. Scale bar = 10 μm. (B) Quantification of total boutons and satellite boutons in control (OK6-Gal4/+) and OK6-Gal4/+; RNAi/+ animals. n ≥ 16 NMJs; ***P < 0.001 by t test; error bars represent SEM. (C) The NMJ growth appeared normal in ube3a15B homozygotes, but there were more total boutons and more satellite boutons in ube3a15B/ube3a35 and ube3a15B/Def mutants compared with ube3a15B. (D) Quantification of the total boutons and satellite boutons in the three genotypes. n ≥ 16 NMJs; ***P < 0.001 by one-way ANOVA test; error bars represent SEM.
(TIF)
(A, B) Electron micrographs of synaptic boutons from wild type (A) and ube3a35 mutants (B). Compared with wild type, ube3a35 mutants exhibited significantly more cisternae with diameters >60 nm (arrowheads in B). Arrows in A and B indicate active zone. Scale bar, 500 nm. (C, D) High magnification view of representative active zones from wild type (C) and ube3a35 mutants (D). Dashed line defines a 200 nm radius around the active zone for quantitative analysis of SVs. Scale bar, 100 nm. (E) High magnification view of a presynaptic bouton from ube3a35 mutants. Arrowheads indicate cisternae. Scale bar, 100 nm. (F, G) Quantification of the number of SVs within a 200 nm radius of the active zone (F) and the mean number of cisternae per cross-sectioned presynaptic bouton (G). n ≥ 38 boutons from more than 4 larvae analyzed, t test, mean ± s.e.m., ***p < 0.001.
(TIF)
(A, B) Representative traces of EJP and mEJP of wild type (A) and ube3a35 mutant (B) NMJs. Scale bars for EJP and mEJP are annotated.
(TIF)
(A, B) Increased levels of pMad at NMJ synapses (A) and motoneuron nuclei (B) of ube3a35 mutants were rescued by a heterozygous mutation of tkv8. Scale bars, 5 μm and 10 μm in A and B, respectively. (C) Quantification of the fluorescence intensities of pMad at NMJ and nuclei of different genotypes including wild type, ube3a35, and tkv8/+; ube3a35. ***p < 0.001 by one-way ANOVA; error bars indicate SEM. (D) NMJ4 synapses in abdominal segment A3 loaded with FM1-43 in wild type, ube3a35, and tkv8/+; ube3a35. Scale bar, 5 μm. (E) Quantification of FM1-43 intensities in NMJ boutons following high K+-stimulated endocytosis. n ≥ 14 NMJs; ***p < 0.001 by one-way ANOVA; error bars indicate SEM. (F) Percentage of average EJP amplitudes under tetanic stimulation for 10 min in wild type, ube3a35, and tkv8/+; ube3a35. n ≥ 10 animals. Representative traces are shown on the right.
(TIF)
(A) The ventral ganglion of a wild-type larva was double-labeled with anti-GFP and anti-HRP. No GFP signals were detected in the ventral ganglion. (B) Tkv-YFP under the control of the endogenous promoter expresses in the soma of various neurons including motoneurons along the midline of a ventral ganglion. Scale bar = 40 μm. (C, D) Tkv-YFP is functional as the genetrap line shows NMJ growth (D) comparable to wild-type control (C).
(TIF)
(A) Schematic representation of full-length and various truncated Ube3a used for co-IP assays. (B) The N-terminal regions of Ube3a mediate the interaction with myc-Tkv. S2 cell lysates were transfected with different combinations of constructs and immunoprecipitated with anti-flag antibody, followed by western blotting using anti-flag and anti-myc. IgG heavy chain is indicated by an asterisk. α-tubulin was used as a loading control. (C) Schematic diagram of full-length Tkv and its various truncations. Different functional domains are indicated. (D) Co-IP assays showed that the C-terminus of Tkv (TkvC), lacking the extracellular and transmembrane (TM) regions, was able to bind Ube3a. More precisely, the dual-specificity serine-threonine/tyrosine protein kinase catalytic (STYKc) domain exhibited strong binding to Ube3a; consistently, deletion of this domain greatly reduced the interaction.
(TIF)
(A) The level of pMad increased at NMJ synapses and motoneuron nuclei of dissected wild-type larvae treated with 50 μM MG132 inhibitor in Schneider’s medium for 4 h. (B) Quantification of fluorescence intensities of pMad at NMJ and nuclei of non-treated and MG1320-treated larvae. n ≥ 16; **P < 0.01, ***p < 0.001 by one-way ANOVA; error bars indicate SEM. (C) Expressing DTS5, a dominant temperature-sensitive mutation of the β6 subunit of the 26s proteasome (Speese et al., Curr Biol, 2003), by elav-Gal4 (elav-Gal4/+; DTS5/+) also led to an increase of pMad at NMJ synapses and the motoneuron nuclei. (D) Statistical analysis of fluorescence intensities of pMad at NMJs and motoneuron nuclei of elav/+ control and elav-Gal4-driven DTS5 larvae. n ≥ 18; **P < 0.01, ***p < 0.001 by one-way ANOVA; error bars represent SEM.
(TIF)
(A) Sequence alignment of Tkv and human ALK3 spanning the ubiquitination site K227. (B) Wild-type and mutant ALK3 showed a similar level of proteins in the presence of UBE3A. Tubulin was probed as a loading control.
(TIF)
Confocal images of NMJ4 from wild type (A, C, and E) and ube3a35 mutants (B, D, and F) labeled with anti-Dynamin (A and B), anti-Eps15 (C and D), and anti-Endophilin A (E and F).
(TIF)
(A) Overexpression of Ube3a in neurons by elav-Gal4 but not in muscles by C57-Gal4 leads to overgrown NMJ terminals. (B, C) Neuronal overexpression of Ube3a does not drive down BMP signaling as pMad staining appears normal at both NMJs (B) and motoneuron nuclei (C).
(TIF)
(A) Different plasmid constructs were singly or co-transfected to S2 cells. Transfected cells were harvested 48 hr later. Cell lysates were used in a two-step immunoprecipitation step by anti-myc and anti-HA sequentially. (B) Confocal images of NMJ4 labeled with anti-HRP. Loss of one copy of S6KL or ube3a had no effect on the number of satellite boutons, while the number of satellite boutons increased significantly in S6KL140 and ube3a35 trans-heterozygotes and double mutants. (C) Quantification of satellite bouton number in different genotypes. n ≥ 16 NMJs analyzed, one-way ANOVA, mean ± s.e.m., ***p < 0.001.
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.