Abstract
The Arabidopsis vascular system is composed of xylem and phloem, which form a well-defined collateral pattern in vascular bundles. Xylary element and fibers develop secondary cell walls (SCWs) that provide mechanical strength to support plant growth and to transport water and minerals to all above ground organs. SCWs also constitute the majority of terrestrial biomass for biofuel production. The biosynthesis of secondary cell walls are known to be under transcriptional regulation. Transcription factors, such as NAC (NAM, ATAF1/2 and CUC2) and MYB domain proteins, serve as master regulators in SCW development. Recent studies indicated that Class III homeodomain leucine zipper transcription factors (HD-ZIP III TFs) and microRNA 165/166 (miR165/166) may play important roles in SCW formation. Here we discuss the diverse functions of miR165/166 and HD-ZIPIII in vascular development and their interaction with the regulatory pathways of SCW biosynthesis.
Keywords: development, HD-ZIP III, microRNA, regulation, secondary cell wall, transcription factors, vascular system
Plant vascular tissues are composed of xylem and phloem; xylem transports water and minerals from roots to above ground organs, while phloem transports photosynthates from sources to sinks. Tracheary elements, xylary fibers and interfascicular fibers develop secondary cell walls that consist of cellulose, hemicellulose and lignin.1,2 These wall materials deposit between primary cell wall and plasma membrane soon after cells completing expansion. The accumulation of SCW gives rigidity to fiber and vessel cells facilitating their functions of mechanical support and water transport.3 In addition to the developmental importance, secondary cell walls (or better known as lignocellulosic biomass) have been used as feedstocks in biofuel industry. Therefore, understanding the regulatory mechanisms of vascular development and secondary cell wall biosynthesis has significant importance in fundamental biology as well as in biotechnological applications.
Vascular tissues are produced through proliferation and differentiation of the vascular cambium. In tree species, vascular cambium is responsible for the formation of secondary xylem or wood. Whereas, the slow growth and long life cycle impedes genetics and molecular biology studies of vascular development and wood formation in trees.4 In contrast, Arabidopsis is a fast growing model plants with notable secondary growth at the bottom part of the inflorescence stem, making Arabidopsis an excellent system to study xylem development.5 In Arabidopsis, xylem and phloem are organized in a collateral pattern, with xylem located inside and phloem outside of the vascular bundle.6,7 During secondary growth, vascular cambium and interfascicular cambium connect and form a cylindrical structure, producing secondary xylem and phloem.7 A group of HD-ZIP III TFs, whose expression is post-transcriptionally regulated by miR165/166, play key roles in the establishment of the vascular pattern.6,8-10 Mutants of HD-ZIP III TFs also exhibited obvious defects in secondary cell wall formation.11-13 In this review, we discuss the recent progress on the functions of miR165/166 and HD-ZIP III TFs in determining vascular pattern and secondary cell wall formation.
The Functions of HD-ZIP III TFs in Vascular Differentiation and Patterning
The HD-ZIP III TFs have 5 members, i.e. REVOLUTA/INTERFASCICULAR FIBERLESS1 (REV/IFL1), PHABULOSA (PHB), PHAVOLUTA (PHV), CORONA (CAN/AtHB15) and AtHB8. These proteins are characterized by an HD-ZIP domain for DNA binding and protein dimerization14 and a highly conserved lipid or steroid binding START (Steroidogenic acute regulatory protein-related lipid transfer) domain15 (Fig. 1). The HD-ZIP III TFs regulate a number of developmental processes, such as embryo patterning, meristem initiation and homeostasis, lateral organ polarity and vascular development in Arabidopsis.11-13,16-19 In this review, we mainly focus on the functions of HD-ZIP III TFs in vascular differentiation and secondary cell wall development.
Figure 1.

Domain structures of the 5 HD-ZIP III transcription factors (TFs) and the complementary sequence of miR165b. The four known functional domains (from N-terminus to C-terminus) of the HD-ZIP III TFs are: the homeodomain that is responsible for DNA binding (orange); the basic Leucine Zipper domain (b-ZIP), which is responsible for DNA binding and dimerization (yellow); the START domain, a putative lipid or steroid binding domain (blue diagonal lined). The miR165B binding site is located in the coding sequence of START domain (purple bar); the C-terminal MEKHLA domain (red). The paring of complementary sequence of AtHB15 and miR165b is shown at the bottom of the diagram.
The functions of HD-ZIP III TFs in vascular differentiation and patterning have been revealed through analyses of loss- and gain-of-function mutants. Double mutant of phb phv showed no obvious phenotypes in vascular patterning or plant growth, but aberrant amphicribral vasculature structure with xylem surrounded by phloem were observed in the triple mutant of phb phv rev.20 In contrast, gain-of-function point mutations of any of these 3 genes, PHB, PHV or REV, resulted in amphivasal vasculatures with phloem surrounded by xylem.17,21,22 These results indicate that PHB, PHV and REV function redundantly in determining the collateral vascular bundle organization. The function of AtHB15 in vascular development was determined through a comprehensive mutant analysis of the HD-ZIP III TFs.13 Triple mutant phb phv cna/athb15 developed amphivasal vasculature internally away from the stem periphery.13 The distinct vasculature phenotypes (amphicribral vs amphivasal) between phb phv rev and phb phv cna/athb15 indicates that REV and CNA/AtHB15 may have antagonistic functions in vascular patterning. This is supported by the fact that the vascular defects of rev/ifl1 were partially suppressed in the rev cna athb8 triple mutant.13,23 A dominant-negative mutation of CNA/AtHB15 further supports its function in vascular patterning as manifested by the development of ectopic amphivasal bundles in the clv3–1cna-1 double mutant.24 The fifth member of HD-ZIP III TF, AtHB8 interacts with auxin signal and functions as a positive regulator of procambium and cambium development in vascular tissues.25-29 The functions of HD-ZIP III TFs in vascular differentiation and patterning may be conserved in different plant species as shown by characterization of their orthologs in Zinnia elegans and rice.11,12,30
MiR165/166 Mediated Regulation in Vascular Development and Patterning
MicroRNAs are small (around 21 nucleotides) non-coding RNAs that post-transcriptionally regulate target genes by paring with the complementary sequences located in the target transcripts. The binding of microRNAs causes translational repression or degradation of target mRNAs.31,32 The expression of the 5 HD-ZIP III TFs is predicted to be regulated by microRNA 165/166 (miR165/166) .33 In Arabidopsis, there are 9 members in miR165/166 gene family, with 2 miR165 (miR165a and miR165b) and 7 miR166 (miR166a to miR166g). The mature microRNAs from these 2 species differ at only one single nucleotide.33
The complementary sequence of miR165/166 locates in the putative lipid/sterol-binding START domain of the HD-ZIP III family members (Fig. 1). Missense mutation in the START domain resulted in resistance to mRNA cleavage, and in turn over-accumulation of corresponding transcripts in the gain-of-function mutants phb-1d, phv-1d and avb.17,20,22,33 The common phenotype of these dominant mutants is the development of radialized amphivasal vascular bundles, despite their phenotypic difference in meristem patterning and lateral organ development. In contrast, a point mutation in the START domain in AtHB15/INCURVATA4/CORONA resulted in a higher expression of the AtHB15/ICU4 transcripts and incurvature in leaves, but vascular patterning (bilateral organization) stays normal,18,19 indicating that the function of AtHB15/ICU4/CNA is different from PHB, PHV and REV in vascular development. Taken together, interruption of the miR165/166 binding to the complementary sequence in HD-ZIP III TFs may affect vascular development.
Direct evidence of miR165/166 function in vascular development comes from the identification and analysis of the corresponding activation tagged mutants. Activation tagging of miR166a and miR166g resulted in significant reduction of the transcript levels of PHB, PHV and AtHB15, but less or no obvious effect on the expression level of REV or AtHB8.34,35 We recently reported that activation-tagging of miR165b that showed similar effect on the expression of HD-ZIP III genes, with PHB, PHV and AtHB15 as that mainly affected targets.36 The common phenotype of ectopic internalized amphivasal bundle(s) in all 3 activation-tagged lines in miR166a, miR166g and miR165b may reflect the fact that the same subset of HD-ZIP III genes were significantly repressed in these lines. In contrast, transgenic overexpression of miR165a under the control of 35S promoter caused repression on all of the 5 HD-ZIP III genes.10 The difference between overexpression (driven by 35S promoter) and activation-tagging of miR165/166 in regulating HD-ZIP III genes may be explained by the difference in their promoters. While 35S promoter causes a constitutive overexpression, activation tagging leads primarily to an enhancement of the endogenous expression pattern.37 In fact, different microRNAs showed distinct and dynamic expression patterns as characterized by Promoter:GUS analyses.38 It is also possible that the activation tagging preserved the sequence integrity around the miR165/166 loci, which may contain regulatory elements, such as small peptides.39
Transcriptional Pathways in Secondary Cell Wall Biosynthesis
NAC domain and MYB domain transcription factors (NACs and MYBs) function as master switches in secondary wall biosynthesis (Fig. 2). NACs are plant-specific proteins with a conserved NAC domain in the N-terminal region and an activation domain in the C-terminal region.40,41 NAC SECONDARY WALL THICKENING FACOTRS1 (NST1) and NST2 activate secondary cell wall thickening in anther endothecium,42 while NST1 together with SECONDARY WALL-ASSOCIATED NAC-DOMAIN PROTEIN 1 (SND1) control secondary wall biosynthesis in fiber cells.2,43 VASCULAR-RELATED NAC DOMAIN6 (VND6) and VND7 play important roles in metaxylem and protoxylem formation, respectively.44 The other VND members (VND1–5) positively regulate secondary cell wall deposition in fibers.45 Direct downstream of the NACs are the MYB master regulators, MYB46 and MYB8346,47. A number other transcription factors, including many other NACs, MYBs and KNATs, function downstream of master NACs/MYBs and form complex regulatory networks in secondary wall biosynthesis (Fig. 2).48-53
Figure 2.
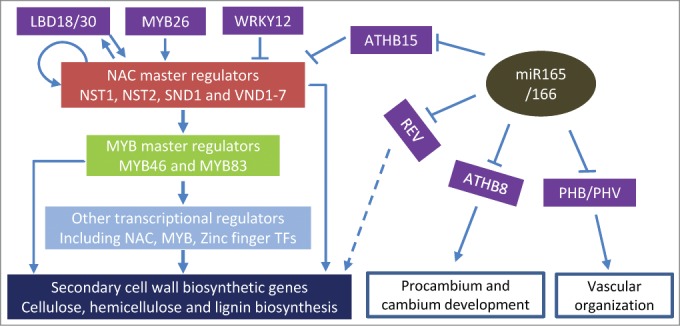
Transcriptional regulation of secondary cell wall biosynthesis and the role of miR165/166-ATHB15.
Both positive and negative regulators of the master switches have been identified in recent years. MYB26 positively regulate the expression of NST1 and NST2, and in turn activate secondary cell all thickening in anther endothecium.54,55 In dicotyledonous plants, WRKY12 functions as a negative regulator of secondary cell wall thickening by directly repressing the expression of NST2 in pith cells.1,56 Feedback regulation is also common in regulating secondary cell wall development. SND1 actually binds to its own promoter and functions as a positive regulator for its own expression.57 Another example is MYB32, a direct target of SND1, represses SND1 expression and secondary cell wall biosynthesis.57,58 Expression of LOB domain proteins LBD18/ASL20 and LBD30/ASL19 is dependent on VND6 and VND7, while ectopic expression of VND7 was detected in LBD18 overexpressing plants indicating a positive feedback regulatory mechanism.59-61
HD-ZIP III TFs and miR165/miR166 Interact with Secondary Wall Regulatory Pathways
The first line of evidence showing that HD-ZIP III TFs involve in secondary cell wall biosynthesis comes from the characterization of rev/ifl1 mutant. The null mutation of REV resulted in disruption of interfascicular fiber differentiation in stems.23,62-64 These studies indicate that REV functions as a positive regulator of the differentiation of interfascicular fibers, or secondary cell wall synthesis in general.50 AtHB8 also functions as a positive regulator of secondary wall development. Although the knock-out mutants of athb-8 showed no phenotypes in secondary cell wall development, overexpression of AtHB8 promoted xylem differentiation.16 In contrast to REV and AtHB8, AtHB15 functions as a negative regulator of secondary wall development. In Populous, synthetic miRNA knock-down of the AtHB15 ortholog, POPCORONA, resulted in an abnormal lignification in pith cells, while overexpression of a miRNA-resistant POPCORONA delayed lignification of xylem and phloem fibers during secondary growth.65 In Arabidopsis, examination of 2 athb15 knockout mutants confirmed AtHB15 function as a negative regulator of secondary wall development.36 Secondarily thickened cell walls and over-accumulation of all 3 major secondary wall components were observed in the pith cells of the athb15 mutants (Fig. 2).36
Activation tagging of miR166a resulted in meristem enlargement (the mutant was hence named as men1) and greatly expanded protoxylem and metaxylem.34 It is proposed that miR166-mediated ATHB15 mRNA cleavage is a principal mechanism for the observed vascular development phenotype.34 In both vascular and interfascicular regions, vascular cambium are formed in the periphery of the existing secondary wall bearing vessel and fiber cells. Indeed, the expanded protoxylem and metaxylem in men1 was believed to be the results of promoting the activity of fascicular cambium and interfascicular cambium. Recently, we reported a miR165b activation-tagging line, stp-2d, which showed a dominant secondary cell wall thickening phenotype. The stems of stp-2d are much thinner than the wild type indicating that the cambium activity is unlikely enhanced in the stp-2d mutant. Actually, the activation tagging of miR165b in the stp-2d mutant resulted in secondary cell wall biosynthesis in pith cells, which are located in the center of the stem. Transgenic overexpression of a microRNA resistant AtHB15 (mAtHB15) further indicate that miR165b functions through AtHB15 in regulating secondary wall development in pith.36
The miR165/166 mediated ATHB15 cleavage impact the known regulatory pathways in secondary wall development (Fig. 2). Two NAC master switches, SND1 and NST2, are upregulated in athb15 mutants confirmed that AtHB15 functions as a negative regulator of the secondary wall related regulatory pathway.36 The pith phenotypes of stp-2d and athb15 mutants are quite similar to that of previously described wrky12 mutants.56 Gene expression analysis indicated that WRKY12 and AtHB15 may not directly regulate each other.36 Although the exact relations between WRKY12 and miR165b-AtHB15 involved regulatory pathways are still unknown, these studies shed new light in our understanding of the regulation of secondary cell wall formation.
Conclusions
Plants have to invest a large amount of resources and energy to produce secondary cell walls, which once made cannot be recycled and reused by the plants. Therefore, plants have established elegant regulatory pathways to ensure only certain cell types develop secondary cell walls at certain developmental stages.66 Accumulation of secondary walls in ground tissues, such as pith, increases biomass density and may boost biofuel production.56,67 The miR165/166 and HD-ZIP III TFs control many aspects of plant growth and development. In this review, we discussed the functions of miR165/166 and HD-ZIP III TFs in regulating vascular development and secondary cell wall biosynthesis. Future studies should focus on elucidation of the functional specificity of the close members in miR165/166 family, as well as their interaction with existing regulatory pathways. The studies on miR165/166 and HD-ZIP III TFs will enhance our understanding of vascular development and facilitate genetic engineering of biomass feedstocks for biofuels production.
Funding
This study was supported, in part, by the USDA National Institute of Food and Agriculture, Hatch project number CONS00925; and by grants to HW from the Research Excellent Program.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Wang HZ, Dixon RA. On-off switches for secondary cell wall biosynthesis. Mol Plant 2012; 5:297-303; PMID:22138968; http://dx.doi.org/ 10.1093/mp/ssr098 [DOI] [PubMed] [Google Scholar]
- 2.Zhong R, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006; 18:3158-70; PMID:17114348; http://dx.doi.org/ 10.1105/tpc.106.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong R, Ye ZH. Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 2015; 56:195-214; PMID:25294860; http://dx.doi.org/ 10.1093/pcp/pcu140 [DOI] [PubMed] [Google Scholar]
- 4.Chaffey N, Cholewa E, Regan S, Sundberg B. Secondary xylem development in Arabidopsis: a model for wood formation. Physiol Plant 2002; 114:594-600; PMID:11975734; http://dx.doi.org/ 10.1034/j.1399-3054.2002.1140413.x [DOI] [PubMed] [Google Scholar]
- 5.Nieminen KM, Kauppinen L, Helariutta Y. A weed for wood? Arabidopsis as a genetic model for xylem development. Plant Physiol 2004; 135:653-9; PMID:15208411; http://dx.doi.org/ 10.1104/pp.104.040212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlsbecker A, Helariutta Y. Phloem and xylem specification: pieces of the puzzle emerge. Curr Opin Plant Biol 2005; 8:512-7; PMID:16039153; http://dx.doi.org/ 10.1016/j.pbi.2005.07.001 [DOI] [PubMed] [Google Scholar]
- 7.Jouannet V, Brackmann K, Greb T. (Pro)cambium formation and proliferation: two sides of the same coin? Curr Opin Plant Biol 2015; 23:54-60; PMID:25449727; http://dx.doi.org/ 10.1016/j.pbi.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al.. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 2010; 465:316-21; PMID:20410882; http://dx.doi.org/ 10.1038/nature08977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 2011; 138:2303-13; PMID:21558378; http://dx.doi.org/ 10.1242/dev.060491 [DOI] [PubMed] [Google Scholar]
- 10.Zhou GK, Kubo M, Zhong R, Demura T, Ye ZH. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol 2007; 48:391-404; PMID:17237362; http://dx.doi.org/ 10.1093/pcp/pcm008 [DOI] [PubMed] [Google Scholar]
- 11.Ohashi-Ito K, Fukuda H. HD-zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol 2003; 44:1350-8; PMID:14701930; http://dx.doi.org/ 10.1093/pcp/pcg164 [DOI] [PubMed] [Google Scholar]
- 12.Ohashi-Ito K, Kubo M, Demura T, Fukuda H. Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol 2005; 46:1646-56; PMID:16081527; http://dx.doi.org/ 10.1093/pcp/pci180 [DOI] [PubMed] [Google Scholar]
- 13.Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005; 17:61-76; PMID:15598805; http://dx.doi.org/ 10.1105/tpc.104.026161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sessa G, Steindler C, Morelli G, Ruberti I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol 1998; 38:609-22; PMID:9747806; http://dx.doi.org/ 10.1023/A:1006016319613 [DOI] [PubMed] [Google Scholar]
- 15.Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci 1999; 24:130-2; PMID:10322415; http://dx.doi.org/ 10.1016/S0968-0004(99)01362-6 [DOI] [PubMed] [Google Scholar]
- 16.Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G. The arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 2001; 126:643-55; PMID:11402194; http://dx.doi.org/ 10.1104/pp.126.2.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001; 411:709-13; PMID:11395776; http://dx.doi.org/ 10.1038/35079635 [DOI] [PubMed] [Google Scholar]
- 18.Ochando I, Gonzalez-Reig S, Ripoll JJ, Vera A, Martinez-Laborda A. Alteration of the shoot radial pattern in Arabidopsis thaliana by a gain-of-function allele of the class III HD-Zip gene INCURVATA4. Int J Dev Biol 2008; 52:953-61; PMID:18956325; http://dx.doi.org/ 10.1387/ijdb.072306io [DOI] [PubMed] [Google Scholar]
- 19.Ochando I, Jover-Gil S, Ripoll JJ, Candela H, Vera A, Ponce MR, Martínez-Laborda A, Micol JL. Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in arabidopsis. Plant Physiol 2006; 141:607-19; PMID:16617092; http://dx.doi.org/ 10.1104/pp.106.077149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 2003; 13:1768-74; PMID:14561401; http://dx.doi.org/ 10.1016/j.cub.2003.09.035 [DOI] [PubMed] [Google Scholar]
- 21.McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development 1998; 125:2935-42; PMID:9655815 [DOI] [PubMed] [Google Scholar]
- 22.Zhong R, Ye ZH. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol 2004; 45:369-85; PMID:15111711; http://dx.doi.org/ 10.1093/pcp/pch051 [DOI] [PubMed] [Google Scholar]
- 23.Zhong R, Ye ZH. IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 1999; 11:2139-52; PMID:10559440; http://dx.doi.org/ 10.1105/tpc.11.11.2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green KA, Prigge MJ, Katzman RB, Clark SE. CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 2005; 17:691-704; PMID:15705957; http://dx.doi.org/ 10.1105/tpc.104.026179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 1995; 121:4171-82; PMID:8575317 [DOI] [PubMed] [Google Scholar]
- 26.Donner TJ, Sherr I, Scarpella E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 2009; 136:3235-46; PMID:19710171; http://dx.doi.org/ 10.1242/dev.037028 [DOI] [PubMed] [Google Scholar]
- 27.Kang J, Dengler N. Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta 2002; 216:212-9; PMID:12447534; http://dx.doi.org/ 10.1007/s00425-002-0847-9 [DOI] [PubMed] [Google Scholar]
- 28.Mattsson J, Ckurshumova W, Berleth T. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol 2003; 131:1327-39; PMID:12644682; http://dx.doi.org/ 10.1104/pp.013623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wenzel CL, Schuetz M, Yu Q, Mattsson J. Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J 2007; 49:387-98; PMID:17217464; http://dx.doi.org/ 10.1111/j.1365-313X.2006.02977.x [DOI] [PubMed] [Google Scholar]
- 30.Itoh J, Hibara K, Sato Y, Nagato Y. Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol 2008; 147:1960-75; PMID:18567825; http://dx.doi.org/ 10.1104/pp.108.118679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 32.Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Curr Opin Plant Biol 2005; 8:38-44; PMID:15653398; http://dx.doi.org/ 10.1016/j.pbi.2004.11.008 [DOI] [PubMed] [Google Scholar]
- 33.Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region. EMBO J 2004; 23:3356-64; PMID:15282547; http://dx.doi.org/ 10.1038/sj.emboj.7600340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Narry Kim V, et al.. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J 2005; 42:84-94; PMID:15773855; http://dx.doi.org/ 10.1111/j.1365-313X.2005.02354.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 2005; 132:3657-68; PMID:16033795; http://dx.doi.org/ 10.1242/dev.01942 [DOI] [PubMed] [Google Scholar]
- 36.Du Q, Avci U, Li S, Gallego-Giraldo L, Pattathil S, Qi L, Hahn MG, Wang H. Activation of miR165b represses AtHB15 expression and induces pith secondary wall development in Arabidopsis. Plant J 2015; 83(3):388-400; PMID:26043238 [DOI] [PubMed] [Google Scholar]
- 37.Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al.. Activation tagging in Arabidopsis. Plant Physiol 2000; 122:1003-13; PMID:10759496; http://dx.doi.org/ 10.1104/pp.122.4.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung JH, Park CM. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 2007; 225:1327-38; PMID:17109148; http://dx.doi.org/ 10.1007/s00425-006-0439-1 [DOI] [PubMed] [Google Scholar]
- 39.Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Becard G, Combier JP. Primary transcripts of microRNAs encode regulatory peptides. Nature 2015; 520:90-3; PMID:25807486; http://dx.doi.org/ 10.1038/nature14346 [DOI] [PubMed] [Google Scholar]
- 40.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al.. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA 2003; 10:239-47; http://dx.doi.org/ 10.1093/dnares/10.6.239 [DOI] [PubMed] [Google Scholar]
- 41.Shen H, Yin Y, Chen F, Xu Y, Dixon R. A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. BioEnergy Res 2009; 2:217-32; http://dx.doi.org/ 10.1007/s12155-009-9047-9 [DOI] [Google Scholar]
- 42.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005; 17:2993-3006; PMID:16214898; http://dx.doi.org/ 10.1105/tpc.105.036004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007; 19:270-80; PMID:17237351; http://dx.doi.org/ 10.1105/tpc.106.047043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 2005; 19:1855-60; PMID:16103214; http://dx.doi.org/ 10.1101/gad.1331305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, Zhong R, Ye ZH. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PloS ONE 2014; 9:e105726; PMID:25148240; http://dx.doi.org/ 10.1371/journal.pone.0105726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy RL, Zhong R, Ye ZH. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 2009; 50:1950-64; PMID:19808805; http://dx.doi.org/ 10.1093/pcp/pcp139 [DOI] [PubMed] [Google Scholar]
- 47.Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007; 19:2776-92; PMID:17890373; http://dx.doi.org/ 10.1105/tpc.107.053678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhargava A, Mansfield SD, Hall HC, Douglas CJ, Ellis BE. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol 2010; 154:1428-38; PMID:20807862; http://dx.doi.org/ 10.1104/pp.110.162735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li E, Bhargava A, Qiang W, Friedmann MC, Forneris N, Savidge RA, Johnson LA, Mansfield SD, Ellis BE, Douglas CJ. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 2012; 194:102-15; PMID:22236040; http://dx.doi.org/ 10.1111/j.1469-8137.2011.04016.x [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, You S, Taylor-Teeples M, Li WL, Schuetz M, Brady SM, Douglas CJ. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 2014; 26:4843-61; PMID:25490916; http://dx.doi.org/ 10.1105/tpc.114.128322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, et al.. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015; 517:571-5; PMID:25533953; http://dx.doi.org/ 10.1038/nature14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008; 20:2763-82; PMID:18952777; http://dx.doi.org/ 10.1105/tpc.108.061325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong R, Ye ZH. Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci 2014; 229:193-207; PMID:25443846; http://dx.doi.org/ 10.1016/j.plantsci.2014.09.009 [DOI] [PubMed] [Google Scholar]
- 54.Steiner-Lange S, Unte US, Eckstein L, Yang C, Wilson ZA, Schmelzer E, Dekker K, Saedler H. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 2003; 34:519-28; PMID:12753590; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01745.x [DOI] [PubMed] [Google Scholar]
- 55.Yang C, Xu Z, Song J, Conner K, Vizcay Barrena G, Wilson ZA. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 2007; 19:534-48; PMID:17329564; http://dx.doi.org/ 10.1105/tpc.106.046391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, Avci U, Nakashima J, Hahn MG, Chen F, Dixon RA. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci USA 2010; 107:22338-43; PMID:21135241; http://dx.doi.org/ 10.1073/pnas.1016436107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H, Zhao Q, Chen F, Wang M, Dixon RA. NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J 2011; 68:1104-14; PMID:21883551; http://dx.doi.org/ 10.1111/j.1365-313X.2011.04764.x [DOI] [PubMed] [Google Scholar]
- 58.Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 2004; 40:979-95; PMID:15584962; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02280.x [DOI] [PubMed] [Google Scholar]
- 59.Endo H, Yamaguchi M, Tamura T, Nakano Y, Nishikubo N, Yoneda A, Kato K, Kubo M, Kajita S, Katayama Y, et al.. Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol 2015; 56:242-54; PMID:25265867; http://dx.doi.org/ 10.1093/pcp/pcu134 [DOI] [PubMed] [Google Scholar]
- 60.Soyano T, Thitamadee S, Machida Y, Chua NH. ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 2008; 20:3359-73; PMID:19088331; http://dx.doi.org/ 10.1105/tpc.108.061796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J 2011; 66:579-90; PMID:21284754; http://dx.doi.org/ 10.1111/j.1365-313X.2011.04514.x [DOI] [PubMed] [Google Scholar]
- 62.Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J 2001; 25:223-36; PMID:11169198; http://dx.doi.org/ 10.1046/j.1365-313x.2001.00959.x [DOI] [PubMed] [Google Scholar]
- 63.Talbert PB, Adler HT, Parks DW, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 1995; 121:2723-35; PMID:7555701 [DOI] [PubMed] [Google Scholar]
- 64.Zhong R, Taylor JJ, Ye ZH. Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 1997; 9:2159-70; PMID:9437861; http://dx.doi.org/ 10.1105/tpc.9.12.2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du J, Miura E, Robischon M, Martinez C, Groover A. The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PloS ONE 2011; 6:e17458; PMID:21386988; http://dx.doi.org/ 10.1371/journal.pone.0017458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 1993; 3:1-30; PMID:8401598; http://dx.doi.org/ 10.1111/j.1365-313X.1993.tb00007.x [DOI] [PubMed] [Google Scholar]
- 67.Hines JP. Boostign Biofuels. Science 2011; 311:11; http://dx.doi.org/ 10.1126/science.331.6013.11-a [DOI] [Google Scholar]


