ABSTRACT
Aluminum (Al) and proton (H+) are 2 coexisting rhizotoxicities limiting plant growth in acid soils. Sensitive to Proton Rhizotoxicity (STOP) 1-like zinc finger transcription factors play important roles in regulating expression of downstream genes involved in tolerance mechanism of either stress. In this mini-review, we summarized recent advances in characterizing STOP1-like proteins with respect to plant Al and H+ tolerance. The possible involvement of structure-function of STOP1-like proteins in differential regulation of Al and H+ tolerance are discussed. In addition, we also direct research in this area to protein phosphorylation.
KEYWORDS: Aluminum toxicity, protein phosphorylation, proton toxicity, sensitive to proton rhizotoxicity (STOP) 1, structure-function, transcriptional regulation
Accounting for around 50% of potential arable land and limiting plant growth and productivity, soil acidity is a serious problem in agriculture worldwide.1 When grow in acid soils, plants suffer from acid soil syndrome that includes excess of aluminum (Al3+), protons (H+), manganese (Mn2+) and iron (Fe3+), and deficiency in phosphorus (P), calcium (Ca) and magnesium (Mg).2 Among these nutritional stresses, Al and H+ have been considered as the 2 most important rhizotoxicities that severely impair root apex and stunt root development, consequently resulting in susceptible to other stresses.3-6 On the other hand, plants have evolved sophisticated mechanisms to cope with either Al or H+ stress. A number of genes that are responsible for Al tolerance have been isolated from various plant species. Genes encoding malate and citrate transporters were identified as key genes in the Al-exclusion mechanism, such as Al-activated malate transporter (ALMT) and multidrug and toxic compound extrusion (MATE) protein, and for genes encoding natural resistance-associated macrophage protein (Nramp) and ATP-binding cassette transporter, such as Nramp aluminum transporter 1 (Nrat1) and ALUMINUM SENSITIVE 1 (ALS1) are involved in the sequestration process of Al.7-10 Although less information is available on H+ tolerance mechanism, those genes involved inγ-aminobutyric acid (GABA) shunt pathway and pH-regulating metabolic pathways, such as carbon and nitrogen metabolism, are assumed to be critical for H+ tolerance.11-13 However, how Al and H+ stress signals activate and regulate expression of downstream genes that detoxify rhizotoxicities on acid soils is still limited.
Do STOP1-like proteins play differential roles in Al and H+ tolerance?
Sensitive to proton rhizotoxicity (STOP) 1, a member of C2H2-type transcription factor, was identified via map-based cloning of a low-pH-sensitive Arabidopsis (Arabidopsis thaliana) mutant that is also hypersensitive to Al toxicity.14AtSTOP1 is involved in the positive regulation of 3 main Al resistance genes AtALMT1, AtMATE, and AtALS3 in Arabidopsis.13,15 In addition to regulate Al tolerance genes, AtSTOP1 also regulates a series of potential H+ tolerance genes, including genes encoding CBL-interaction protein kinase 23, Polygalacturonase inhibiting protein 1, Glutamate dehydrogenase 1, AtSTOP2 and so on.13,16 Thus, the identification of AtSTOP1 first links Al tolerance and H+ tolerance together at molecular level. An AtSTOP1 homolog in rice, OsART1, was identified using a mutant-screening approach. Unlike AtSTOP1, OsART1 is, however, only involved in Al tolerance.17 Recently, we isolated and characterized a STOP1-like gene VuSTOP1 from rice bean (Vigna umbellata), a legume originated in tropical and subtropical acid soil regions. In planta complementation of the Atstop1 mutant assay showed that VuSTOP1 is involved mainly in H+ tolerance.18 To date, at least 10 STOP1-like proteins have been functionally characterized in a number of plant species with respect to Al and H+. At first glance, it appears that the role of STOP1-like proteins in tolerance to either stress differs among different plant species. However, the finding that STOP1-like proteins displayed preferential role to one stress tolerance is mainly based on the in planta complementation analysis in Atstop1 mutant (Table 1). The conclusion is more convincing that STOP1-like protein plays an important role in both Al tolerance and H+ tolerance based on knock-out or knock-down mutant analysis. For example, knock-down of STOP1 resulted in clear Al sensitivity, and slight but significant proton sensitivity in tobacco (RNAi) and eucalyptus (knock-down hairy root).19,20 Nonetheless, AtSTOP2-RNAi lines did not display altered Al- and proton-sensitivity, but showed significant proton tolerance in Atstop1 complemented lines.16 One possible reason is functional redundancy between AtSTOP2 and AtSTOP1, since AtSTOP2 functions in downstream of AtSTOP1. There is a similar case in rice that loss-of-function mutant of OsART1 did not show any sensitivity to proton. However, rice has more copies of STOP1-like proteins than Arabidopsis, and OsART1 is not the closest homolog of AtSTOP1 in rice (Fig. 1). Thus, whether some plant species have evolved specific STOP1-like protein to specific stress tolerance or functional redundancy masks the function of a certain STOP1-like protein with regard to Al or H+ tolerance has to be investigated yet.
Table 1.
Summary of STOP1-like genes involved in H+ and/or Al3+ tolerance.
| Species | Gene | Expression pattern | Transformation host | Transformation method | H+ sensitivity or tolerance | Al3+ sensitivity or tolerance | Reference |
|---|---|---|---|---|---|---|---|
| Abrabidopsis thaliana | AtSTOP1 | Constitutive | A.thaliana | Mut/comp | √ | √ | Iuchi et al 2007 |
| AtSTOP2 | Constitutive | A.thaliana | RNAi | × | × | Kobayashi et al., 2014 | |
| Atstop1 | Comp | √ | Slightly | ||||
| Oryza sativa | OsARTl | Constitutive | O.sativa | Mut/comp | × | √ | Yamaji et al., 2009 |
| Nicotiana tabacum | NtSTOP1 | ? | N.tabacum | RNAi | √ | √ | Ohyama et al., 2013 |
| Atstop1 | Comp | √ | Slightly | ||||
| Physcomitrella patens | PpSTOP1 | ? | P.patens | RNAi | × | √ | |
| Atstop1 | Comp | √ | √ | ||||
| Lotus japonicus | LjSTOP1 | ? | Atstop1 | Comp | √ | Slightly | |
| Camellia sinensis | CsSTOP1 | ? | Atstop1 | Comp | √ | Slightly | |
| Populus nigra | PnSTOP1 | ? | Atstop1 | Comp | √ | Slightly | |
| Eucalyptus | EguSTOP1 | ? | Eucalyptus | KD-hairy root | √ | √ | Sawaki et al., 2014 |
| Atstop1 | Comp | √ | Slightly | ||||
| Vigna umbellata | VuSTOP1 | Induced by H+ and Al3+ | Atstop1 | Comp | √ | Slightly | Fan et al., 2015 |
| TaSTOP1-A | Slightly induced by Al3+ | ? | ? | ? | ? | ||
| Triticum aestivum | TaSTOP1-B | Constitutive | ? | ? | ? | ? | Garcia-Oliveira et al., 2013 |
| TaSTOP1-C | Slightly inducedby H+ | ? | ? | ? | ? |
Figure 1.
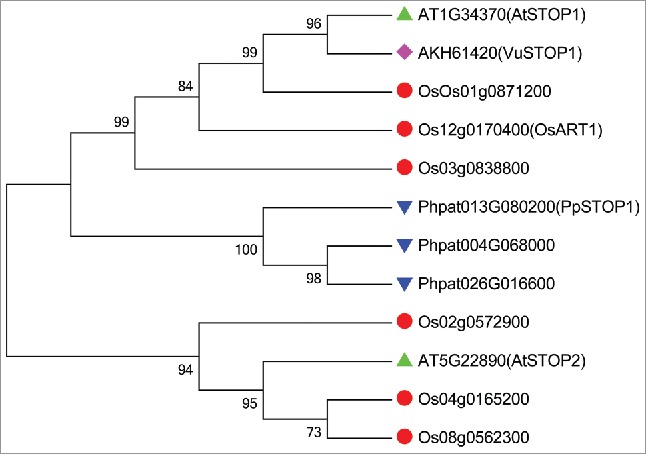
Phylogenic analysis of STOP1-like proteins among different plant species.
Transcriptional regulation versus activation
The finding that the transcription abundance of rice bean VuSTOP1 was induced by both H+ and Al3+ stresses gives rise to the complexity of regulation of STOP1-like proteins. 18,21 Either AtSTOP1 and AtSTOP2 in Arabidopsis or OsART1 in rice is constitutively expressed,14,16,17 thus Al or H+ regulates them posttranslationally, e.g. direct activation of protein activity. On the contrary, the increase in expression level of VuSTOP1 indicates that other transcription factors are involved. The requirement of protein degradation for Al-dependent VuSTOP1expression induction suggests that a transcriptional repressor may be involved in the transcriptional repression of VuSTOP1 in the absence of Al stress.18 Interestingly, 3 transcripts of TaSTOP1 displayed differential expression patterns with respect to different stresses.22 However, the expression patterns of STOP1-like genes in other plant species are still unknown. It is possible that different regulation mechanisms are related to different functions of STOP1-like proteins with respect to specific stress tolerance.
Structure-function of STOP1-like proteins
A common feature of STOP1-like proteins that could not restore Al tolerance phenotype of Atstop1 mutant is that they could not restore AtALMT1 expression and the subsequent malate secretion. It is well known that the malate secretion mediated by AtALMT1 in Arabidopsis can explain as much as 70% of Al tolerance.15 Therefore, possibility remains whether the ineffectiveness of these STOP1-like proteins in restoring the mutant phenotype is due directly to the poorly transcriptional activation of AtALTM1 expression by interaction between STOP1-like proteins and cis-acting element in the promoter of AtALMT1. Although STOP1-like proteins of all species have 4 highly conserved C2H2 zinc finger domains, there is a highly variable amino acid sequence structure in the N and C termini. Recent study has indicated that these C2H2 zinc finger domains are only responsible for directly binding to the cis-acting element GTGCCCAA in the promoter of AtALMT1, but not responsible for transcriptional activation of target genes.23 We have examined the transcriptional activity of VuSTOP1 via a yeast expression system. Interestingly, we found that both N and C regions are responsible for transactivation.18 Considering the greater variation in both N and C termini among STOP1-like proteins, one possible reason for determining specific function of STOP1-like proteins in terms of Al and H+ tolerance might be that transcriptional activation of Al-tolerance genes is more sensitive to the protein structure of STOP1 than that of H+ tolerance genes. Further research is required to examine the sequence and structural differences of N and/or C termini outside of conserved C2H2 zinc finger domains among STOP1-like proteins and the conservation of cis-acting elements in the promoters of target genes.
Protein phosphorylation
Protein phosphorylation has been implicated in Al-dependent organic acids secretion from several plant species.24-27 In both rice bean and Arabidopsis, it has been demonstrated that transcriptional regulation of AtALMT1 and VuMATE1 is responsible for protein phosphorylation-dependent secretion of organic acids, respectively.25,27 Furthermore, co-expression analysis revealed that AtSTOP1 is expressed similarly with 2 protein kinases At1g34300 and At2g02220. Thus, protein phosphorylation is likely to be involved in differential roles of STOP1-like proteins with respect to Al and H+ tolerance.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the 973 project (2014CB441002), the Natural Science Foundation of China (31501827, 31222049 and 31071849), and the Fundamental Research Funds for the Central Universities.
References
- 1.vonUexküll HR, Mutert E. Global extent, development and economic impact of acid soils. Plant Soil 1995; 171:1-15; http://dx.doi.org/ 10.1007/BF00009558 [DOI] [Google Scholar]
- 2.Ishitani M, Rao I, Wenzl P, Beebe S, Tohme J. Integration of genomics approach with traditional breeding towards improving abiotic stress adaptation: Drought and aluminum toxicity as case studies. Field Crop Res 2004; 90:35-45; http://dx.doi.org/ 10.1016/j.fcr.2004.07.004 [DOI] [Google Scholar]
- 3.Kochian LV, Hoekenga OA, Piñeros MA. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Ann Rev Plant Biol 2004; 55:459-93;PMID:15377228; http://dx.doi.org/17661006 10.1146/annurev.arplant.55.031903.141655 [DOI] [PubMed] [Google Scholar]
- 4.Koyama H, Toda T, Yotoka S, Dawair Z, Hara T. Effects of aluminum and pH on root growth and cell viability in Arabidopsis thaliana strain Landsberg in hydroponic culture. Plant Cell Physiol 1995; 36: 201-05 [Google Scholar]
- 5.Ikka T, Kobayashi Y, Iuchi S, Sakurai N, Shibata D, Kobayashi M, Koyama H. Natural variation of Arabidopsis thaliana reveals that aluminum resistance and proton resistance are controlled by different genetic factors. Theor Appl Genet 2007; 115:709-19; PMID:17661006; http://dx.doi.org/ 10.1007/s00122-007-0602-5 [DOI] [PubMed] [Google Scholar]
- 6.Yang JL, Zheng SJ, He YF, Matsumoto H. Aluminium resistance requires resistance to acid stress: a case study with spinach that exudes oxalate rapidly when exposed to Al stress. J Exp Bot 2005; 56:1197-203; PMID:15737984; http://dx.doi.org/ 10.1093/jxb/eri113 [DOI] [PubMed] [Google Scholar]
- 7.Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. A wheat gene encoding an aluminum-activated malate transporter. Plant J 2004; 37:645-53; PMID:14871306; http://dx.doi.org/ 10.1111/j.1365-313X.2003.01991.x [DOI] [PubMed] [Google Scholar]
- 8.Magalhaes JV, Liu J, Guimarães CT, Lana UG, Alves VM, Wang YH, Schaffert RE, Hoekenga OA, Piñeros MA, Shaff JE, et al.. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 2007; 39:1156-61; PMID:17721535; http://dx.doi.org/ 10.1038/ng2074 [DOI] [PubMed] [Google Scholar]
- 9.Xia J, Yamaji N, Kasai T, Ma JF. Plasma membrane-localized transporter for aluminum in rice. Proc Natl Acad Sci USA 2010; 107:18381-5; PMID:20937890; http://dx.doi.org/ 10.1073/pnas.1004949107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen PB, Cancel J, Rounds M, Ochoa V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007; 225:1447-58; PMID:17171374; http://dx.doi.org/ 10.1007/s00425-006-0452-4 [DOI] [PubMed] [Google Scholar]
- 11.Bouche N, Fromm H. GABA in plants: just ametabolite? Trends Plant Sci 2004; 9:110-5; PMID:15003233; http://dx.doi.org/ 10.1016/j.tplants.2004.01.006 [DOI] [PubMed] [Google Scholar]
- 12.Fütterer O, Agnelov A, Lieseqang H, Gottschalk G, Schleper C, Schepers B, Dock C, Antranikian G, Liebl W. Genome sequence of Picrophilu storridus and its implications for life around pH 0. Proc Natl Acad Sci USA 2004; 101:9091-96; PMID:15184674; http://dx.doi.org/ 10.1073/pnas.0401356101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawaki Y, Iuchi S, Kobayashi Y, Kobayashi Y, Ikka T, Sakurai N, Fujita M, Shinozaki K, Shibata D, Kobayashi M, et al.. STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol 2009; 150: 281-94; PMID:19321711; http://dx.doi.org/ 10.1104/pp.108.134700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iuchi S, Koyama H, Iuchi A, Kobayashi Y, Kitabayashi S, Kobayashi Y, Ikka T, Hirayama T, Shinozaki K, Kobayashi M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA 2007; 104: 9900-5; PMID:17535918; http://dx.doi.org/ 10.1073/pnas.0700117104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Magalhaes JV, Shaff J, Kochian LV. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J 2009; 57:389-99; PMID:18826429; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03696.x [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi Y, Ohyama Y, Kobayashi Y, Ito H, Iuchi S, Fujita M, Zhao CR, Tanveer T, Ganesan M, Kobayashi M, et al.. STOP2 activates transcription of several genes for Al-and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Mol Plant 2014; 7:311-22; PMID:23935008; http://dx.doi.org/ 10.1093/mp/sst116 [DOI] [PubMed] [Google Scholar]
- 17.Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009; 21:3339-49; PMID:19880795; http://dx.doi.org/ 10.1105/tpc.109.070771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan W, Lou HQ, Gong YL, Liu MY, Cao MJ, Liu Y, Yang JL, Zheng SJ. Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol 2015; 208: 456-68; PMID:25970766; http://dx.doi.org/ 10.1111/nph.13456 [DOI] [PubMed] [Google Scholar]
- 19.Ohyama Y, Ito H, Kobayashi Y, Ikka T, Morita A, Kobayashi M, Imaizumi R, Aoki T, Komatsu K, Sakata Y, et al.. Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol 2013; 162:1937-46; PMID:23749850; http://dx.doi.org/ 10.1104/pp.113.218958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawaki Y, Kobayashi Y, Kihara-Doi T, Nishikubo N, Kawazu T, Kobayashi M, Kobayashi Y, Iuchi S, Koyama H, Sato S. Identification of a STOP1-like protein in Eucalyptus that regulates transcription of Al tolerance genes. Plant Sci 2014; 223:8-15; PMID:24767110; http://dx.doi.org/ 10.1016/j.plantsci.2014.02.011 [DOI] [PubMed] [Google Scholar]
- 21.Fan W, Lou HQ, Gong YL, Liu MY, Wang ZQ, Yang JL, Zheng SJ. Identification of early Al-responsive genes in rice bean (Vigna umbellata) roots provides new clues to molecular mechanisms of Al toxicity and tolerance. Plant Cell Environ 2014; 37:1586-97; PMID:24372448; http://dx.doi.org/ 10.1111/pce.12258 [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Oliveira AL, Benito C, Prieto P, de Andrade Menezes R, Rodrigues-Pousada C, Guedes-Pinto H, Martins-Lopes P. Molecular characterization of TaSTOP1 homoeologues and their response to aluminium and in bread wheat (Triticum aestivum L.). BMC Plant Biol 2013; 13:134; PMID:24034075; http://dx.doi.org/ 10.1186/1471-2229-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tokizawa M, Kobayashi Y, Saito T, Kobayashi M, Iuchi S, Nomoto M, Tada Y, Yamamoto YY, Koyama H. STOP1, CAMTA2 and other transcription factors are involved in aluminum-inducible AtALMT1 expression. Plant Physiol 2015; 167:991-1003; PMID:25627216; http://dx.doi.org/ 10.1104/pp.114.256552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osawa H, Matsumoto H. Possible involvement of protein phosphorylation in aluminum-responsive malate efflux from wheat root apex. Plant Physiol 2001: 126:411-20; PMID:11351103; http://dx.doi.org/ 10.1104/pp.126.1.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi Y, Hoekenga OA, Itoh H, Nakashima M, Saito S, Shaff JE, Maron LG, Piñeros MA, Kochian LV, Koyama H. Characterizationof AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol 2007; 145:843-52; PMID:17885092; http://dx.doi.org/ 10.1104/pp.107.102335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ligaba A, Kochian L, Piñeros M. Phosphorylation at S384 regulates the activity of the TaALMT1 malate transporter that underlies aluminum resistance in wheat. Plant J 2009; 60:411-23; PMID:19563436; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03964.x [DOI] [PubMed] [Google Scholar]
- 27.Liu MY, Chen WW, Xu JM, Fan W, Yang JL, Zheng SJ. The role of VuMATE1 expression in aluminium-inducible citrate secretion in rice bean (Vigna umbellata) roots. J Exp Bot 2013; 64:1795-804; PMID:23408830; http://dx.doi.org/ 10.1093/jxb/ert039 [DOI] [PMC free article] [PubMed] [Google Scholar]


