Abstract
Gibberellins (GAs) are important phytohormones for plant growth and development. DELLAs are members of the plant-specific GRAS protein family and act as repressors of GA signaling. DELLAs are rapidly degraded in the presence of GAs. GA-GID1-DELLA complexes are recognized and ubiquitinated by the SCFSLY complex. The sleepy1 (sly1) F-box mutant exhibits dwarfism and low-germination phenotypes due to high accumulation of DELLAs. Overexpression of GID1 in the sly1 mutant partially rescues these phenotypes without degradation of DELLAs suggesting that proteolysis independent regulation of DELLAs exists in GA signaling. But the molecular mechanisms of non-proteolytic regulation of DELLA are largely unknown. Recently we identified a DELLA binding transcription factor, GAI-ASSOCIATED FACTOR1 (GAF1). GAF1 also interacts with co-repressor TOPLESS RELATED (TPR) in nuclei. DELLAs and TPR act as coactivator and corepressor of GAF1, respectively. GAs converts the GAF1 complex from transcriptional activator to repressor via degradation of DELLAs. The overexpression of ΔPAM, lacking of DELLAs binding region of GAF1, partially rescue dwarf phenotypes of GA deficient or GA insensitive mutant. In this study, we investigate the relationship between non-proteolytic regulation of DELLAs and GA signaling via DELLA-GAF1 complex using modified yeast two-hybrid system.
Keywords: DELLA, GAF1, gibberellin, GID1, proteolysis, signaling, SLY1, transcription factor, TOPLESS
Gibberellins (GAs) promote seed germination, root growth, stem elongation, leaf expansion, flower induction and the development of flowers, fruits, and seeds.1,2 The endogenous levels of GAs are fine-tuned by feedback control at several steps in their metabolic pathway, including GA 20-oxidase and GA 3-oxidase.2-4 GA feedback regulation has been shown to depend on GA signaling components, including the GA receptors GA-INSENSITIVE DWARF1 (GID1), the F-box proteins SLEEPY1 (SLY1) and DELLAs. Bioactive GAs are recognized by GID1, and the GA-GID1 complex binds to DELLAs. The SCF E3 ubiquitin ligase complex including SLY1 interacts with the GID1-GA-DELLA complex and promotes degradation of DELLA through the ubiquitin-proteasome pathway.5 DELLAs are major negative regulators in GA signaling. Several DELLA interaction proteins have been identified.5
For example, DELLAs regulate hypocotyl elongation by interacting with transcription factors, PHYTOCHROME INTERCTING FACTORS (PIFs)6,7 and BRASSINOZOLE RESISTANT1 (BZR1).8,9 The interaction between DELLAs and the transcription factors inhibits hypocotyl elongation by blocking their DNA binding activities. GAs trigger the degradation of DELLAs, which release PIFs and BZR1 to activate the target genes, thus promoting hypocotyl elongation. The titration of transcriptional activators by DELLAs partly explains GA-dependent transcriptional activation and how plants integrate environmental stimuli and GA signals to optimize growth and development. However, genes encoding GA 20-oxidase and GA 3-oxidase are down regulated by GA via feedback regulation. Genome-wide analysis revealed that the effect of GA on gene expression is predominantly through repression.10,11 These observations cannot be explained by the conventional titration model. Thus other molecular mechanisms underlying GA-dependent transcriptional regulation must exist.
Recently we identified a DELLAs binding transcription factor, designated GAI-ASSOCIATED FACTOR1 (GAF1) and revealed a new role of DELLAs as transcriptional coactivator. GAF1 also interacts with corepressor TOPLESS RELATED (TPR). DELLAs and TPR act as coactivator and corepressor of GAF1, respectively (Fig. 1).12 The DELLAs turn on or off two sets of GA-regulated genes by titration and co-activation, providing a novel mechanism for integrative regulation of plant growth and GA homeostasis. To investigate the biological role of DELLAs binding to GAF1, we generated transgenic plants expressing a mutant version of GAF1 that cannot bind DELLA (ΔPAM) under the control of the CaMV 35S promoter in the ga1–3, a GA deficient mutant or gai-1, a GA insensitive mutant. The overexpression of ΔPAM in ga1–3 or gai-1 partially rescue dwarf and low-germination phenotypes without affecting DELLA levels.12 ΔPAM protein acts as a constitutive repressor with TPR and promotes GA signaling (Fig. 1).
Figure 1.
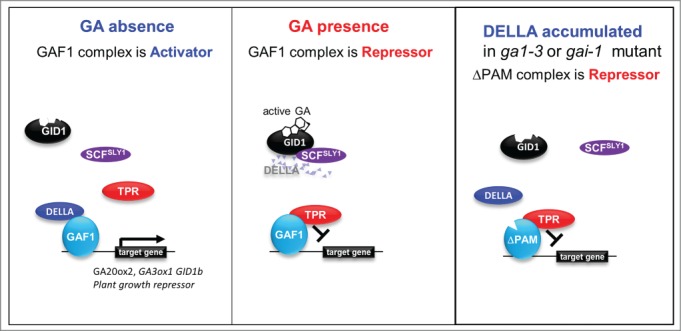
A co-activator model for GA signaling. Under GA deficient conditions, DELLAs are stable and localized in nuclei. DELLAs interact with GAF1, and exhibiting high transcriptional activity with GAF1. In the presence of GA, DELLAs are degraded via the 26S proteasome pathway. The GAF1-TPR complex exhibits transcriptional repression activity. GA-induced functional conversion of GAF1 complex in plants depends on the degradation of coactivator DELLAs. The ga1–3 or gai-1 mutant accumulate DELLA proteins at a higher level. ΔPAM cannot bind DELLA but can interact with TPR. ΔPAM protein acts as a constitutive repressor with TPR and promotes GA signaling.
The sly1 mutant exhibited seed dormant, dwarf and infertile phenotypes associated with high level accumulation of DELLA due to lack of DELLA proteolysis.13,14 Although the amounts of accumulated DELLAs in sly1 mutant are higher than those of GA deficient mutant ga1–3 or GA insensitive gid1a gid1b gid1c triple mutant, sly1 exhibited less severe dwarf and low-germination phenotypes than ga1–3 or gid1a gid1b gid1c. Moreover GID1 overexpression can partially rescue the dwarf and low-germination phenotypes of sly1 mutant in a GA-dependent manner. This observation suggests the existence of non-proteolytic repression of DELLAs, which is hidden in the wild-type genetic background because DELLAs are rapidly degraded in response to GAs. How do GAs promote GA signaling without degradation of DELLAs in sly1? One possible mechanism for the non-proteolytic repression of DELLA is that GAs promote dissociation of the complex between DELLAs and transcription factors, including GAF1. The dissociated GAF1 from DELLAs could form a repressor complex with TPR and promote GA signaling.
To examine the effect of GA-GID1 binding to DELLA for the interaction between DELLA and GAF1, we carried out modified yeast two-hybrid assay. As expected, the binding of GID1 to GAL4AD-DELLA reduced the interaction between GAL4AD-DELLA and GAL4BD-GAF1 in a GA-dependent manner (Fig. 2). Every AtGID1-DELLAs interaction was detected in a GA-dependent manner, although the interaction of AtGID1b with some DELLAs was weakly detected without GA in yeast.15,16 We could not detect the reduction of β-gal activity in the absence of GA3 (Fig. 2). It indicated that the dissociation of GAF1 from DELLAs by the binding of GID1 is GA dependently. Both DELLA/TVHYNP and SAW motif of DELLAs are necessary for the binding of GID1 to DELLAs.17 Because the binding of GAF1 to DELLAs also required SAW motif of DELLAs,12 the bindings of GA-GID1 to DELLA might be competitive with that of GAF1. Thus, these observations suggest that GA-GID1 promote GA signaling through the dissociation of GAF1 from DELLAs without degradation of DELLAs in sly1 genetic background (Fig. 3). This hypothesis is consistent with the previous observation that the overexpression of GID1 suppresses the phenotypes of sly1 in a GA dependent manner.14,18 The SAW domain of DELLAs is also necessary for the interaction with BZR1.8 Further analysis of the effect of GA-GID1 binding for the interaction between DELLAs and other interaction proteins provides new insight for the mechanism of non-proteolytic regulation of DELLA.
Figure 2.
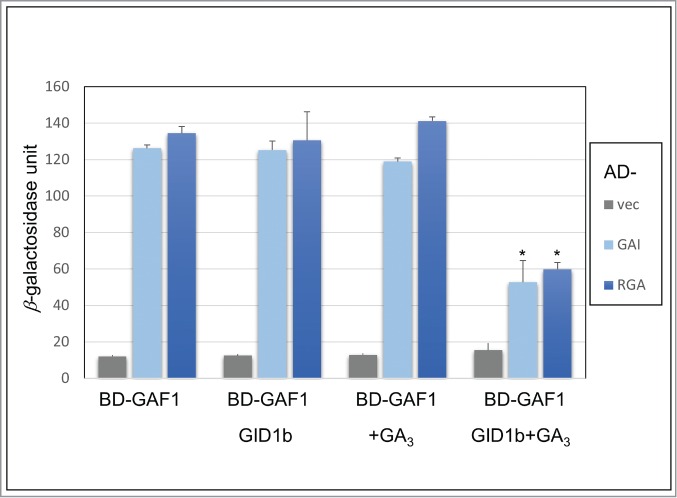
GID1 promote the dissociation of GAF1 from DELLA in GA-dependent manner. GAF1 fused with the GAL4 DNA-binding domain, GAI and RGA fused with the GAL4 activation domain and resulting β-galactosidase activity was detected in the presence or absence of GID1 and with or without 10−3 M GA3. To avoid the suppression of transactivation activity of DELLAs by the masking of GA-GID1 to DELLA motif in yeast, we use GAL4 activation domain fused to DELLAs as prey protein. GAF1 interact with DELLAs, GAI and RGA, in yeast. The binding of GID1 promote the dissociation of GAF1 from DELLA in presence GA condition. Data are means±SD, n = 3 “vec” indicates empty vector using as negative control. Asterisks represent Student's t test significance compared with BD-GAF1 (*P < 0.05).
Figure 3.
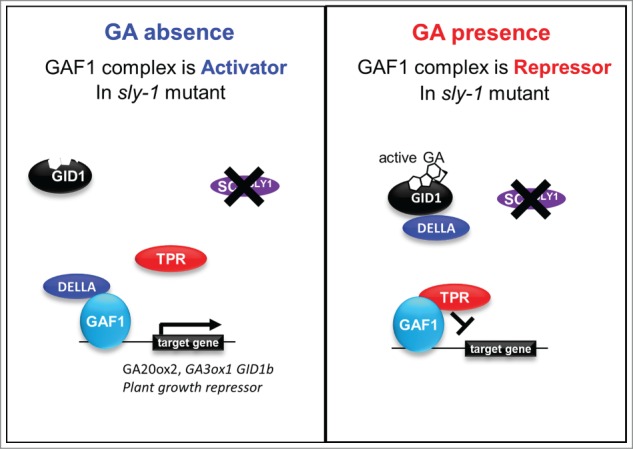
Non-proteolytic regulation model for GA signaling. The sly-1 mutants accumulate DELLA proteins at a higher level. But the binding of GA-GID1 to DELLA promotes the dissociation of GAF1 from DELLA. The dissociated GAF1 from DELLAs could form a repressor complex with TPR and promote GA signaling.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported in part by grants from the Japan Society for the Promotion of Science (JSPS) to J.F. (26440148) and Y.T. (15H04392), and by a grant from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to J.F. (24116525) and Y.T. (24118004).
References
- 1.Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Ann Rev Plant Biol 2004; 55:197-223. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi S. Gibberellin metabolism and its regulation. Ann Rev Plant Biol 2008; 59:225-51. [DOI] [PubMed] [Google Scholar]
- 3.Fukazawa J, Nakata M, Ito T, Yamaguchi S, Takahashi Y. The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J 2010; 62:1035-45; PMID:20345601 [DOI] [PubMed] [Google Scholar]
- 4.Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y. Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell 2010; 22:1592-604; PMID:20442373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 2011; 21:R338-45; PMID:21549956 [DOI] [PubMed] [Google Scholar]
- 6.de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008; 451:480-4; PMID:18216857 [DOI] [PubMed] [Google Scholar]
- 7.Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al.. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008; 451:475-9; PMID:18216856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 2012; 14:810-7; PMID:22820377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallego-Bartolome J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadi D, Blazquez MA. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 2012; 109:13446-51; PMID:22847438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, Sun TP. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007; 19:3037-57; PMID:17933900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirano K, Kouketu E, Katoh H, Aya K, Ueguchi-Tanaka M, Matsuoka M. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J 2012; 71:443-53; PMID:22429711 [DOI] [PubMed] [Google Scholar]
- 12.Fukazawa J, Teramura H, Murakoshi S, Nasuno K, Nishida N, Ito T, Yoshida M, Kamiya Y, Yamaguchi S, Takahashi Y. DELLAs function as co-activators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 2014; 26:2920-38; PMID:25035403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steber CM, Cooney SE, McCourt P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 1998; 149:509-21; PMID:9611170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ariizumi T, Murase K, Sun TP, Steber CM. Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 2008; 20:2447-59; PMID:18827182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al.. Identification and characterization of Arabidopsis gibberellin receptors. Plant J 2006; 46:880-9; PMID:16709201 [DOI] [PubMed] [Google Scholar]
- 16.Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al.. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. The Plant cell 2006; 18:3399-414; PMID:17194763; http://dx.doi.org/ 10.1105/tpc.106.047415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 2010; 22:2680-96; PMID:20716699; http://dx.doi.org/ 10.1105/tpc.110.075549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ariizumi T, Hauvermale AL, Nelson SK, Hanada A, Yamaguchi S, Steber CM. Lifting DELLA repression of Arabidopsis seed germination by nonproteolytic gibberellin signaling. Plant Physiol 2013; 162:2125-39; PMID:23818171; http://dx.doi.org/ 10.1104/pp.113.219451 [DOI] [PMC free article] [PubMed] [Google Scholar]


