Abstract
To cope with low phosphate (Pi) availability, plants have to adjust its gene expression profile to facilitate Pi acquisition and remobilization. Sensing the levels of Pi is essential for reprogramming the gene expression profile to adapt to the fluctuating Pi environment. AtPHR1 in Arabidopsis and OsPHR2 in rice are central regulators of Pi signaling, which regulates the expression of phosphate starvation-induced (PSI) genes by binding to the P1BS elements in the promoter of PSI genes. However, how the Pi level affects the central regulator to regulate the PSI genes have puzzled us for a decade. Recent progress in SPX proteins indicated that the SPX proteins play important role in regulating the activity of central regulator AtPHR1/OsPHR2 in a Pi dependent manner at different subcellular levels.
Keywords: Phosphate starvation, PHR, Pi homeostasis, Pi-signaling, SPX-domain protein
Phosphorus (P) is an important plant nutrient. To cope with low phosphate (Pi) availability, plants have evolved sophisticated developmental strategies to facilitate Pi acquisition and remobilization. Sensing the levels of external and internal Pi is essential for reprogramming the gene expression profile and adapting to the fluctuating Pi environment. As we known, a large proportion of Pi starvation-induced (PSI) genes are under the control of PHR1 (PHOSPHATE STARVATION RESPONSE REGULATOR 1) in Arabidopsis (AtPHR1).1,2 Just like AtPHR1 is central regulator in Arabidopsis, its homolog, OsPHR2 is a central regulator of phosphate signaling in rice, which enhanced the expression of phosphate starvation-induced (PSI) genes and resulted in the enhancement of Pi acquisition under Pi deficiency condition. This occurred via AtPHR1/OsPHR2 binding to a cis-element named the PHR1 binding sequences (P1BS).3,4 However, the transcription level of AtPHR1/OsPHR2 was not responsive to Pi starvation, raising the question as to how plants sense changes in cellular Pi levels to activate the central regulator and then initiate the reprograming of gene expression.
Recent works in our lab published in Proc Natl Acad Sci USA (PNAS), Plant Cell and J. Exp. Bot. and the work in Arabidopsis published in Prof. Javier Paz-Ares' lab indicate that SPX proteins play very important role in sensing the levels of external and internal Pi level and negatively regulating Pi homeostasis in plants.5-8 SPX domain is a conservative domain named after SYG1 (suppressor of yeast gpa1), Pho81 (CDK inhibitor in yeast PHO pathway), and XPR1 (xenotropic and polytropic retrovirus receptor).9 In plants, proteins harboring the SPX domain are classified into 4 families based on the presence of additional domains in their structure, namely the SPX, SPX-EXS, SPX-MFS and SPX-RING families.10
In Arabidopsis, 20 SPX domain-containing proteins have been identified.9 Sixteen members contain SPX domain and another domain, while the other 4 proteins contain only SPX domain.11 There are 15 SPX domain-containing proteins identified in rice genome. Nine of them contain SPX domain and additional domain, while the other 6 proteins contain only SPX domain.10 SPX proteins are referenced as proteins exclusively harboring the SPX domain.10 Four SPX proteins in Arabidopsis (named AtSPX1-AtSPX4) and 6 SPX proteins in rice (named OsSPX1-OsSPX6) have been identified.11,12 Phylogenetic analysis showed that the SPX proteins can be grouped into 3 subclades. Among which OsSPX1 and OsSPX2 in rice and AtSPX1 and AtSPX2 in Arabidopsis were grouped in the same subclade, OsSPX4 and AtSPX4 were grouped in a second subclade, while OsSPX3, OsSPX5, OsSPX6 and AtSPX3 were grouped into the third subclade.10,12
SPXs Proteins Act as Negative Regulator of Pi Signaling
In plants, SPX proteins were first noticed as Pi starvation induced genes. All the SPX genes were Pi starvation induced except OsSPX4 in rice and AtSPX4 in Arabidopsis.11,12 SPX proteins were found to be negative regulator of Pi signaling in plants. It was found that overexpression of OsSPX1, OsSPX2, OsSPX3, OsSPX4, OsSPX5 can counteract the phenotype of PHR2 overexpression. In which Pi accumulated in the leaf tips and also necrosis in the leaf tips with Pi starvation induced genes such as IPS1 and PT2 were highly induced in the OsPHR2 overexpression transgenic lines. While the double mutants showed normal Pi level and expression level of PSI genes comparable to that of wild type.5-7 On the contrary, knockout lines of the OsSPX1, OsSPX2, OsSPX4 showed significant necrosis in leaf tips and Pi accumulation in shoot (5, 7). Though single mutant of OsSPX3, OsSPX5 doesn't show significant necrosis in the leaf tips, significant necrosis and Pi accumulation were found in the OsSPX3, OsSPX5 double mutant. And the Pi starvation induced genes (e.g. IPS1, miR399, PT2, miR827,PAP10, SQD2) also significantly upregulated in the double mutant.6 Similar results for AtSPX1 and AtSPX2 in Pi signaling were got from Arabidopsis which just like OsPSX1 and OsSPX2 in rice Pi signaling.8 All the results indicate that SPX proteins act redundantly as negative regulators of Pi signaling.
SPX Proteins Localized in Different Subcellular Organelles
The SPX proteins were localized in different subcellular localization. OsSPX1 and OsSPX2 and AtSPX1 and AtSPX2 were localized exclusively to the nucleus as showed by protoplast and onion epidermal cells and plants.7,8,11,12 While AtSPX4 and OsSPX4 were localized in both cytoplasm and nucleus.5,11 The third subclade of SPX porteins AtSPX3 were localized in cytoplasm in some speckles and the OsSPX3 and OsSPX5 were localized in both nucleus and cytoplasm as visualized using the fusions of SPX3-GFP and SPX5-GFP in rice protoplast.6,11,12
SPXs Regulate PHR2 Activity in Different Subcellular Levels to Regulate Pi Signaling
Arabidopsis AtPHR1 and its rice homolog OsPHR2 are known to be central transcription factors in Pi homeostasis. Our recent data showed that OsSPX1, OsSPX2, OsSPX4 and probably other SPX proteins were negative regulator of Pi signaling though affect OsPHR2 function in a Pi dependent way in rice.5-7 Similarly, AtSPX1 and AtSPX2 were also negative regulator of AtPHR1 in a Pi dependent way in Arabidopsis.8 OsSPX1 and OsSPX2 in rice and AtSPX1 and AtSPX2 in Arabidopsis function in a Pi dependent way to inhibit OsPHR2/AtPHR1 function at high cellular Pi status.7,8 And the SPX domain of SPX1 and SPX2 is critical for repressing PHR2 binding to cis-elements by protein interaction.7 While OsSPX4 also function in a Pi dependent way to inhibit PHR2 function at high cellular Pi status.5 Under Pi starvation condition, OsSPX4 would be degraded.
All the results demonstrate that SPX proteins function as components in the Pi-sensing mechanism to control the activity of OsPHR2/AtPHR1, the central regulators of Pi starvation responses, as depicted in our working model (Fig. 1). Under high cellular Pi conditions, SPX4 and maybe other SPX proteins interact with OsPHR2/AtPHR1 in the cytoplasm which inhibit OsPHR2/AtPHR1 from moving into nuclear, while in nucleus, SPX1, SPX2 and also other SPXs will further interact with OsPHR2/AtPHR1, therefore OsPHR2/AtPHR1 can't bind to the promoter of PSI genes, so PSI genes would express in a basal level. However, under low cellular Pi conditions, SPX4 will be degraded, so OsPHR2/AtPHR1 can move into nuclear, while in nuclear, interaction of SPX1 and SPX2 and possibly other SPXs with OsPHR2/AtPHR1 was weakened and OsPHR2/AtPHR1 will preferentially bind to the P1BS motif of PSI gene promoters, allowing OsPHR2/AtPHR1 to up-regulate the expression of PSI genes, including OsSPX1 and OsSPX2, OsSPX3, OsSPX5, and OsSPX6. Which make sure the plant cell to adjust itself to the changed Pi environment. The accumulation of SPXs under Pi-deficient conditions also allows plants to shut down the OsPHR2/AtPHR1 dependent Pi starvation response rapidly after Pi repletion. Altogether, all the data suggesting that SPXs possibly involved in sensing the Pi. Whether SPX proteins function as Pi sensor is a promising question to be investigated in the near future.
Figure 1.
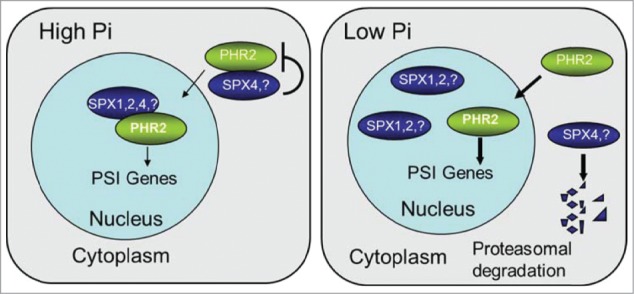
A model of SPX proteins to regulate PHR2 (or AtPHR1) activity for Pi-starvation induced transcription in response to the cellular Pi concentration. Negative and positive regulatory effects are indicated by flat-ended lines and arrows. The thick arrowed lines represent enhancement.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Ministry of Agriculture of China (2014ZX08009-003-005), the National Science Foundation of China (30971742), and the Program for New Century Excellent Talents in University and the Fundamental Research Funds for the Central Universities.
References
- 1.Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 2001; 15:2122-33; PMID:11511543; http://dx.doi.org/ 10.1101/gad.204401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Perez-Perez J, Solano R, Leyva A, Paz-Ares J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 2010; 6: e1001102; PMID:20838596; http://dx.doi.org/ 10.1371/journal.pgen.1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 2008; 146:1673-86; PMID:18263782; http://dx.doi.org/ 10.1104/pp.107.111443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu P, Shou H, Xu G, Lian X. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr Opin Plant Biol 2013; 16: 205-12; PMID:23566853; http://dx.doi.org/ 10.1016/j.pbi.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 5.Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, Wu P. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 2014; 26:1586-97; PMID:24692424; http://dx.doi.org/ 10.1105/tpc.114.123208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J, Hu H, Zhang K, Zhang W, Yu Y, Wu Z, Wu P. The paralogous SPX3 and SPX5 genes redundantly modulate Pi homeostasis in rice. J Exp Bot 2014; 65:859-70; PMID:24368504; http://dx.doi.org/ 10.1093/jxb/ert424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, Shou H, Mo X, Mao C, Wu P. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci USA 2014; 111:14953-8; PMID:25271318; http://dx.doi.org/ 10.1073/pnas.1404680111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodriguez J, Leyva A, Rubio V, Sommer H, Paz-Ares J. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc Natl Acad Sci USA 2014; 111:14947-52; PMID:25271326; http://dx.doi.org/ 10.1073/pnas.1404654111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Ribot C, Rezzonico E, Poirier Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol 2004; 135:400-11; PMID:15122012; http://dx.doi.org/ 10.1104/pp.103.037945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 2011; 193:842-51; http://dx.doi.org/ 10.1111/j.1469-8137.2011.04002.x [DOI] [PubMed] [Google Scholar]
- 11.Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 2008; 54:965-75; PMID:18315545; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03460.x [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J Integr Plant Biol 2009; 51:663-74; PMID:19566645; http://dx.doi.org/ 10.1111/j.1744-7909.2009.00834.x [DOI] [PubMed] [Google Scholar]


