Abstract
Correct positioning of the division plane is a prerequisite for plant morphogenesis. The preprophase band (PPB) is a key intracellular structure of division site determination. PPB forms in G2 phase as a broad band of microtubules (MTs) that narrows in prophase and specializes few-micrometer-wide cortical belt region, named the cortical division zone (CDZ), in late prophase. The PPB comprises several molecules, some of which act as MT band organization and others remain in the CDZ marking the correct insertion of the cell plate in telophase. Ran GTPase-activating protein (RanGAP) is accumulated in the CDZ and forms a RanGAP band in prophase. However, little is known about when and how RanGAPs gather in the CDZ, and especially with regard to their relationships to MT band formation. Here, we examined the spatial and temporal distribution of RanGAPs and MTs in the preprophase of onion root tip cells using confocal laser scanning microscopy and showed that the RanGAP band appeared in mid-prophase as the width of MT band was reduced to nearly 7 µm. Treatments with cytoskeletal inhibitors for 15 min caused thinning or broadening of the MT band but had little effects on RanGAP band in mid-prophase and most of late prophase cells. Detailed image analyses of the spatial distribution of RanGAP band and MT band showed that the RanGAP band positioned slightly beneath the MT band in mid-prophase. These results raise a possibility that RanGAP behaves differently from MTs during their band formation.
Keywords: confocal laser scanning microscope, cortical division zone, microtubule, onion root meristems, preprophase band, RanGAP
Abbreviations
- CBB
coomassie brilliant blue
- CDS
cortical division site
- CDZ
cortical division zone
- DMSO
dimethyl sulfoxide
- LatB
latrunculin B
- LRR_RI
leucine rich repeat of ribonuclease inhibitor
- MT
microtubule
- PPB
preprophase band
- PVDF
polyvinylidene difluoride
- RanGAP
Ran GTPase activating protein
- 5-AU
5-aminouracil.
Introduction
Control of cell division pattern during plant development is one of the essential questions in plant morphogenesis.1 The division site, the place where division plane will be inserted, is predetermined before nuclear division and the preprophase band (PPB) is involved in the process.2,3 The PPB appears as a broad microtubule (MT) band in G2, and the band gradually narrows in prophase.4-6 The width of the MT band reaches to a few micrometer by late prophase, and then the MT band disassembles when cells enter prometaphase.4,5 As the MT band narrows down, the PPB demarcates the few-micrometer-wide cortical belt region. The cell cortex of the belt region is specifically modified and imprinted as the cortical division zone (CDZ), where molecules involved in cell plate insertion act at the end of cytokinesis. Some molecules, recruited to the CDZ during PPB formation to form a molecular band, remains in the CDZ after the disassembly of the MT band and their band gradually narrows to specify the position where the cell plate attaches in cytokinesis. This cortical position is referred to as the cortical division site (CDS).7,8
A number of proteins involved in PPB formation and the CDZ maintenance have been identified.9 TONNEAU1 (TON1) and a protein phosphatase 2A regulatory subunit, TON2/FASS/DCD1/ADD1, are 2 major proteins that are involved in the PPB formation and the loss-of-function mutants of these proteins show phenotypes of the misoriented division plane and lack of PPBs.10-15 Several types of molecules, such as actins, MT-associated proteins (MAPs), kinesins, cdk/cyclins and other proteins have also been reported in PPBs.16-18 Actins are most well-known components of PPB,19-21 and the drug studies have suggested their involvement in the narrowing of the MT band.22 Several MT associated proteins (MAPs) are also located in the PPB18 and studies of the loss-of-function mutants have shown that the MICROTUBULE ORGANIZATION 1 (MOR1) and CLIP-associated proteins (CLASP) are involved in the MT band organization.23-25
Many proteins known to be in PPBs disappear from the CDZ when MTs in PPBs disassembles at prometaphase. In contrast, TANGLED (TAN) and Ran GTPase-activating protein (RanGAP), are proteins that accumulate in the CDZ and remain there even after the disassembly of the MT band.26,27 They are thought to be positive memories of the division site. TAN was originally found in the maize tan mutant that shows a phenotype of disordered pattern of leaf epidermal cell arrangement.28,29 TAN is a highly basic protein that can directly bind to MTs in vitro.30 Pharmacological studies have shown that the initial recruitment of AtTAN::YFP (an Arabidosis TAN protein fused to a yellow fluorescent protein [YFP]) into the PPB depends on MTs; however, once the TAN band is formed, MTs are not required for the retention of the band.26 The maintenance of TAN band in the CDZ is abolished in a mutant lacking a pair of kinesin-12 class proteins PHRAGMOPLAST-ORIENTING KINESINS1 (POK1) and POK2. TAN has been shown to interact with C-terminal domain of POK131 and the POK may function to maintain TAN band in the CDZ after the MT band disassembly.32
The small GTPase Ran is involved in the nucleocytoplasmic transport, spindle MT assembly, nuclear envelope re-assembly and cell cycle control in vertebrates. RanGAP1 and its associated factor RanBP1 promote the conversion of RanGTP into RanGDP, and the guanine nucleotide exchange factor RCC1 promotes the exchange of RanGDP for RanGTP. The gradient in the concentration of RanGTP is maintained by a strict compartmentalization of these factors. In vertebrates, RanGAP is located in the nuclear pore to help nucleocytoplasmic transport.33,34 Plant RanGAP proteins are located on the nuclear envelope, mitotic spindles, phragmoplast and PPBs.27,35-37 Arabidopsis has 2 RanGAP proteins, AtRanGAP1 and AtRanGAP2.35 The plant RanGAPs have a unique N-terminal domain called WPP domain,38 which is considered to be a target domain necessary for anchoring AtRanGAP1 to the nuclear envelope.37,39 Plant RanGAPs extracted from cells in mitotic stages co-assemble with MTs in vitro, whereas those extracted from interphase cells do not co-assemble.35 Xu et al.27 showed that Arabidopsis RanGAP1 is associated with PPB and that it remains after the MT band disassembly. Accumulation of RanGAP in the CDZ requires the presence of the WPP domain. Xu et al.27 also showed that RanGAP1 recruitment to the PPB site is lost in a fass/ton2 mutant whereas the RanGAP band is retained in the PPB when MTs are depolymerized by oryzalin. POK1 and POK2 are also involved in the maintenance of RanGAP1 in the CDZ after disassembly of the MT band.
Although experiments by Xu et al.27 explain several key molecular networks of the accumulation and the maintenance of RanGAPs in the CDZ, the time when the RanGAP band appears in preprophase remains unsolved. Xu et al.27 observed the RanGAP band in only 45% of dividing cells and they speculated that the low frequency of the RanGAP band is 1) caused by the detection limit of the assay system or 2) because half of the cells enter cell division lacking RanGAP band. In order to answer this question, more precise observations of RanGAP and MT distribution during preprophase is necessary.
Studies on the molecular mechanisms of RanGAP and TAN in the CDZ have been carried out using Arabidopsis seedlings and tobacco suspension cultures. However, it is not easy to analyze when and how RanGAP accumulates in the CDZ during preprophase in these systems. Onion cells have large chromosomes and onion root tip cells have been used to study the cell division,40,41 including the PPB development. Several developmental stages have been distinguished during the cell cycle progress in terms of the rate of the nuclear condensation and the width of the MT band.4-6,20,42-51 In the present study, we have employed onion root tip cells to examine relationships among RanGAPs, MTs and nuclear stages to answer when and how RanGAPs gather in the CDZ. Our observation clearly showed that the RanGAP band starts appearing when the width of the MT band reaches about 7 µm and the RanGAP band is not fully coupled with MTs during RanGAP band formation stage.
Material and Methods
Plant material, culture conditions and partial synchronization of PPB formation
Onion (Alliumu cepa L. cv. Highgold Nigou, Sakata Seed Co.) seeds were placed on a filter paper moistened with distilled water and kept in the dark at 25°C. Root tips of 4-day-old seedlings were used in the experiments. For experiments with cytoskeletal drugs, onion seedlings grown for 3 d were transferred on a filter paper soaked with 79 µM 5-aminouracil (5-AU, Sigma-Aldrich Co.) and incubated in the dark at 25°C for 17 h. These seedlings were then washed twice with distilled water and kept on a filter paper moistened with distilled water. After the incubation in the dark at 25°C for 6.75 h, the seedlings were transferred onto a filter paper with 10 µM latrunculin B (LatB, WakoPure Chemical Industries, Ltd.) or 20 µM oryzalin (Riedel-de Haën AG) and kept in the dark at 25°C for 15 min.
Cloning and sequencing of onion RanGAP cDNA
Total RNA and genomic DNA were extracted using the cetyltrimethylammonium bromide method52 from roots of 4-day-old onion seedlings that were frozen in liquid nitrogen and powdered with a mortar and pestle. Partial fragments of the onion RanGAP gene were amplified by PCR from the genomic DNA with a degenerated primer set RanGAP1_1 and a nested primer set RanGAP1_2 (Fig. S1A) designed from the several conserved amino acid sequences in the 5 plant RanGAPs (Fig. S1C). The amplified fragments were inserted into a vector pBluescriptII SK(−) and their sequences were determined. To obtain cDNA for the onion RanGAP, cDNA was synthesized from poly A+ RNA purified from the total RNA by an oligo(dT)-cellulose column (GE Healthcare). To obtain the partial cDNA for onion RanGAP, degenerated primers (Fig. S1A) were designed from the several conserved amino acid sequences in the 5 plant RanGAPs (Fig. S1C). For cloning of the entire cDNA, 5′- and 3′-RACEs products were obtained using a GeneRacer Kit (Invitrogen, Thermo Fisher Scientific Inc.) with the primer sets described in Fig. S1A. DNA fragments of 5′- and 3′-RACEs were inserted in a pBluescriptII SK(−), and those fragments were sequenced using the M13 primers set. Finally, end-to-end PCR was performed with onion cDNA using an Ex Taq DNA polymerase (Takara Bio Inc.) and a primer set ACRanGAP1_1 (Fig. S1A). In addition, a fragment of the onion RanGAP gene was amplified from the genomic DNA using a PrimeStar DNA polymerase (Takara Bio Inc.) and a primer set ACRanGAP1_2 (Fig. S1A). These amplified DNA fragments were sequenced using a 3130 Genetic Analyzer (Applied Biosystems Inc.). The nucleotide sequences of onion RanGAP cDNA and genomic DNA are available from the DNA Data Bank of Japan database (accession numbers, LC050995 for cDNA and LC050996 for genomic DNA).
Antibody production and western blotting
The DNA fragment of the onion RanGAP (1–497 amino-acid residues) was amplified by PCR using a PrimeStar DNA polymerase and a primer set ACRanGAP1_3 (Fig. S1A) to add the restriction enzyme sites KpnI and BamHI at the 5′- and 3′-end, respectively and was inserted in a pBluescriptII SK(−). The DNA fragment was then isolated from this plasmid using KpnI and BamHI restriction enzymes, and ligated into an expression vector pET-32b (Novagen, EMD Bioscience, Inc.) cut by KpnI and BamHI using DNA Ligation Kit, Mighty Mix (Takara Bio Inc.). The obtained plasmid was transformed into Escherichia coli BL21 (DE3) strain. The expressed polypeptides that were insoluble in the E. coli cells were solubilized with urea and then purified and dialyzed to remove urea using a His-Bind purification kit (Novagen, EMD Bioscience, Inc.) according to the manufacturer's instructions. Purified polypeptide (400 μg) mixed with a Freund's Complete Adjuvant (FCA, Cosmo Bio Co.) was injected into a rabbit (New Zealand white). After 2 weeks and 4 weeks, the rabbit was inoculated with the polypeptide (200 μg) mixed with Freund's Incomplete Adjuvant (FIA, Cosmo Bio Co.). Two weeks later, we prepared a serum from the blood collected from the immunized rabbit. To obtain an antibody immunoreactive to the onion RanGAP, the purified polypeptide (5 μg) was separated on a 10% SDS-PAGE gel and transferred to a polyvinylidene difluoride (PVDF) membrane. After the transfer, the membrane was immersed in a solution of 5% skim milk and TBS, consisting of 50 mM Tris-HCl (pH 7.5) and 0.15 M NaCl and a part (about 1 cm wide) of the membrane corresponding to the size of the polypeptide was cut out with clean scissors. The strip of the membrane was reacted with 5-fold dilution of the serum obtained from the immunized rabbit in a phosphate-buffered saline (PBS; 1.5 mM KH2PO4, 137 mM NaCl, 10 mM Na2HPO4, pH 7.4) with 0.05% Tween 20 at room temperature for 1 h. After washing the strip with PBS in 0.05% Tween 20 four times, the stripe was incubated in 0.1 M glycine-HCl pH 2.5 at room temperature for 7 min to elute the antibody specific to the onion RanGAP. The eluate was collected and neutralized with ice-chilled 1 M Tris. Finally, NaN3 was added to the eluate and the eluate was stored at 4°C. For immunostaining, we used mouse monoclonal anti ß-tubulin (Sigma-Aldlich Co.), sheep polyclonal anti-tubulin (Cytoskeleton, Inc.), mouse monoclonal antibody against the 65 kDa MAP53 (renamed as anti-BY2MAP65) and the anti-onion RanGAP antibody as described above. To prepare onion crude protein, root tips of 4-day-old seedlings were crushed in a liquid nitrogen with a mortar and pestle and homogenized in a Protein Extraction Buffer (GE Healthcare). Then the homogenate was centrifuged at 20,000 × g for 30 s at 4°C and the obtained supernatant was collected as the onion crude protein. The onion crude protein was separated on a 10% SDS-PAGE gel and transferred to a PVDF membrane, which was then incubated in 5% skim milk in TBS. Immunoblotting analyses were performed with the primary antibodies described above, and immuno-reactive bands were visualized with a peroxidase labeled goat anti-rabbit IgG (H + L) (Kirkegaard & Perry Laboratories) or VECTASTAIN Universal Elite ABC Kit (Vector Laboratories Ltd.) using a Pierce Western Blotting substrate (Thermo Fisher Scientific, Inc.). Negative control experiments were carried out using a preimmune serum or a mixture of purified antibody with antigen that was preincubated overnight at 4°C.
Sample preparation for immunofluorescence microscopy
Onion root tips were fixed in 3% paraformaldehyde (Electron Microscopy Sciences) and 0.05% glutaraldehyde (TAAB Laboratories Equipment Ltd.) in 50 mM PIPES buffer, pH 6.8 containing 5 mM ethylene glycol tetraacetic acid (EGTA), 1 mM MgSO4, 2% dimethyl sulfoxide (DMSO), and 1% glycerin. Fixed root tips were washed with the PIPES buffer and treated with 1% cellulase Onozuka RS and 1% macerozyme R-10 (Yakult Pharmaceutical Industry Co., Ltd.) dissolved in 0.1% Nonidet P-40, 1% cold fish gelatin, 0.4 M mannitol and 0.02 tablet/L protease inhibitors (Roche Diagnostics GmbH) at room temperature for 15 min. After washing with PBS, approximately 10 root tips were squashed on a slide glass with a cover glass and dried at room temperature for 1 h. The root tip cells were treated with PBS containing 0.05% TritonX-100, 0.1% glycine, and 1% cold fish gelatin for 10 min and washed with PBS, immersed in methanol at −20°C for 10 min. After washing with PBS, root tip cells were treated with the primary antibodies at 37°C for 90 min. Following a 10-min rinse in PBS, cells were reacted with the secondary antibodies at 37°C for 90 min and then rinsed again. Alexa Fluor 488 F (ab') 2 fragment of goat anti-mouse IgG (H+L), Alexa Fluor 594 goat anti-mouse IgG (H+L), Alexa Fluor 488 goat anti-rabbit IgG (H+L), Alexa Fluor 594 goat anti-rabbit IgG (H+L) or Alexa Fluor 594 donkey anti-sheep IgG (H+L) (Molecular probes, Thermo Fisher Scientific Inc.) was used as a secondary antibody depending on the experimental objective. After immersion in a mixture of 50 mM Tris-HCl buffer, pH 9.0, and 50% glycerin at room temperature for 3 min, the cells were treated with a mixture consisting of 50 mM Tris-HCl buffer, pH 9.0, 50% glycerin, 1 mg/mL p-phenylenediamine and 10 mg/L Hoechst 33258 and were sealed on a cover glass.
Observation and quantitative analysis by microscopy
The specimens were examined using a microscope equipped with an epifluorescence illumination (X2 + N1-EFT3, Nikon Co.) and the nuclear stages of cells with a PPB were determined by fluorescence imaging of Hoechst 33258 staining. The nuclear stages in cells with a PPB are classified into 3 groups, including interphase, mid-prophase, and late prophase. The width of MT and RanGAP bands in these cells was determined using a confocal laser scanning microscope system (C1si VBGT Dapi + ECLIPSE TE2000-E, Nikon Co.). Some images were obtained using a confocal laser scanning microscope system of TCS SP8 (Leica Microsystems GmbH). The width of the MT and RanGAP bands in a cell was determined from the mid-longitudinal optical section of the Nikon confocal laser scanning microscope images. As two cross-sectional images of a band were seen in a mid-longitudinal section of the cell, the width of the shorter MT band was used for the measurement. The intensity profile analysis was performed using ImageJ software54 from images acquired using the C1si Nikon confocal microscope. The correlations between the width of MT bands and that of RanGAP bands were evaluated by the Spearman's rank correlation.
Results
Characterization of onion RanGAP antibody
The determined cDNA sequence of onion RanGAP (AcRanGAP) and its predicted amino acid sequence are shown in Fig. S1B and C. Our results showed that the AcRanGAP was encoded by a gene encompassing 1,635 bp, and a resulting protein consisted of 544 amino acids with the predicted molecular mass of about 60 kDa (Fig. S1B and C). The amino acid sequence of AcRanGAP is about 60% identical to the equivalent Arabidopsis RanGAP1 sequence and it contains WPP, leucine-rich repeat ribonuclease inhibitor (LRR_RI)-like domain, and acidic domain that are common in plant RanGAPs39 (Fig. 1A). We obtained an anti-AcRanGAP antibody (497 amino acids in the N-terminal region, Fig. 1A). To test the specificity of the anti-AcRanGAP antibody, crude extracts of onion root tips were electrophoretically separated, blotted on a PVDF membrane, and probed with the antibody. The purified anti-AcRanGAP antibody reacted with a single band of predicted RanGAP size (Fig. 1B lane 3). As negative controls, the primary antibody was replaced by the respective preimmune serum, or by a mixture of purified anti-AcRanGAP antibody with the antigen. We could not detect any bands of the predicted RanGAP size in the negative controls (Fig. 1B lanes 2 and 4).
Figure 1.
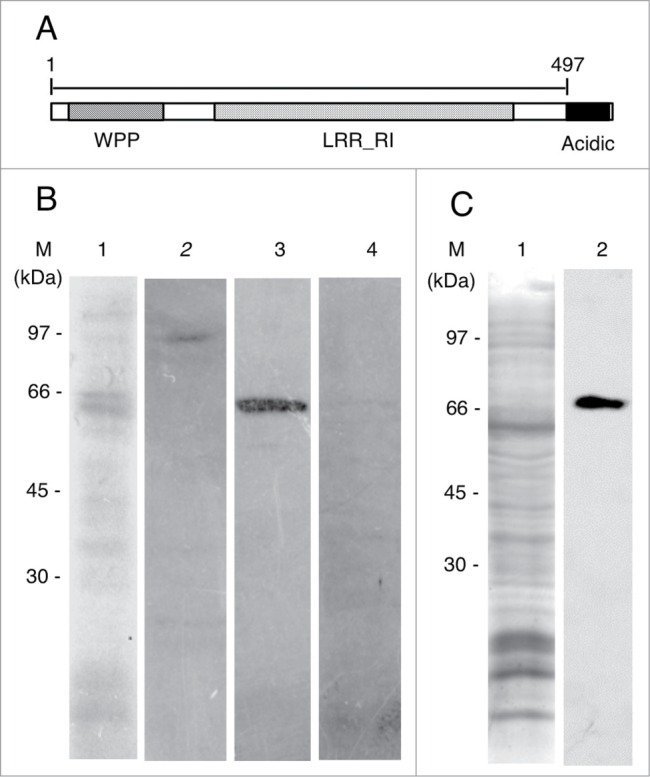
Characterizations of antibodies against RanGAP and MAP65. (A) Schematic diagram of the domain structure of an onion RanGAP, AcRanGAP. AcRanGAP contains tryptophan-proline-proline domain (WPP), leucine-rich repeat ribonuclease inhibitor (LRR_RI)-like domain, and acidic domain (Acidic). Numbers represent amino acid positions, starting at the N-terminus. A peptide of AcRanGAP fragment shown as a bar (amino acid 1 to 497) in the figure was used as an antigen to produce antibody against AcRanGAP. (B) Immunoblot analysis of an antibody against AcRanGAP fragment in crude extract from onion root tips. Lane 1, Coomassie brilliant blue staining; lane 2, control immunoblot with a preimmune rabbit serum; lane 3, immunoblot with an affinity purified antibody against AcRanGAP; lane 4, control immunoblot with a mixture of a purified antibody against AcRanGAP preincubated with an antigen of AcRanGAP fragment. (C) Immunoblot analysis of antibody against tobacco BY2MAP65 in crude extract from onion root tips. Lane 1, Coomassie brilliant blue staining of a crude extract from onion root tips; lane 2, immunoblot with a mouse monoclonal antibody against tobacco BY2MAP65. (M) indicates the position of molecular mass markers and their molecular masses (kDa) in (B) and (C).
RanGAP band appears when the width of the MT band reduces to below 7 µm
To test the immunolocalization of AcRanGAP in meristematic cells of 4-day-old onion root tips, triple labeling of chromosomes, RanGAPs, and MTs was performed using the Hoechst 33258 stain, anti-AcRanGAP antibody, and anti-tubulin antibody, respectively. Figure 2 shows a gallery of RanGAP distribution at different stages of cell division. The cell division stages were determined by reference to chromosome configurations. The strong association of AcRanGAP with the nuclear membrane was detected throughout the cell cycle, including newly formed daughter nuclei (Figs. 2B, E, H, K, Q). In mitotic stages, AcRanGAPs were concentrated in the area of spindles, especially near the kinetochore (arrowhead in Fig. 2N). The strong fluorescence signals of AcRanGAP was also detected at the edge of the phragmoplast (Fig. 2Q).
Figure 2.
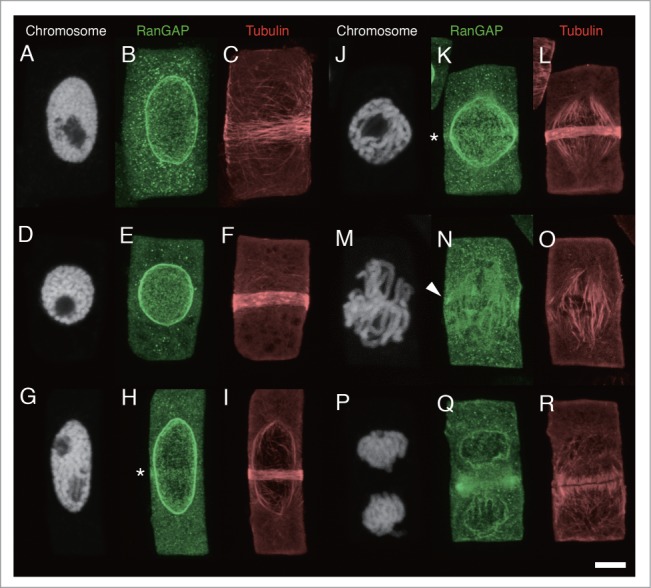
Localization of RanGAP and tubulin in onion root tip cells. Fluorescence images of chromosomes, RanGAPs and tubulins in onion root tip cells of interphase (A-C), mid-prophase (D-I), late prophase (J-L), prometaphase/metaphase transition (M-O), and telophase (P-R). A-C, D-F, G-I, J-L, M-O, and P-R are images of the same cells, respectively. Chromosomes are shown as white (A, D, G, J, M, P), RanGAP images are shown as green (B, E, H, K, N, Q) and tubulin as red (C, F, I, L, O, R). Asterisks indicates RanGAP band. Arrowhead shows the concentration of RanGAP around kinetochore. Bar: 5 μm.
We detected a RanGAP band in the PPB region of prophase cells with a relatively narrow MT band (Figs. 2H and K), but it was absent in the early stages of PPB formation (Figs. 2B and E). We also could not detect the clear RanGAP band after the prometaphase (Figs. 2N and Q). In order to determine when the RanGAP band appears during the PPB formation, we categorized the preprophase stage into 3 groups: interphase (Figs. 2A-C), mid-prophase (Figs. 2D-I), and late prophase (Figs. 2J-L), relative to the chromosome condensation. Since the boundary between the early prophase nuclei (slightly condensed chromosomes) and interphase nuclei is not always well defined, we considered the stage of interphase and early prophase as the “interphase” stage (Fig. 2A). We dealt with the “late prophase” stage as a separate from the “mid-prophase” stage. The “late prophase” was defined as a stage where chromosomes are highly condensed and individual finger-shaped chromosomes are clearly distinguishable inside the nucleus (Fig. 2J). In the late prophase stage, PPBs was very narrow (2–3 µm wide) and composed of a packed MT band, and most of the late-prophase cells had bipolar spindles (Figs. 2J and L). Other prophase cells, with the chromosome condensation rate between that of chromosomes in early and late prophase, were defined to be in the “mid-prophase” stage (Figs. 2D-I). Figure 3 shows the histograms of the distribution of the width of the MT band in interphase, mid-prophase, and late prophase. As was expected, the MT band gradually narrowed when the nuclear stages progressed from interphase to late prophase. The histograms showed that PPB in many interphase cells did not have the RanGAP band (Fig. 3A), whereas the RanGAP band was present in most of the late prophase cells (81.7 ± 9.2%, the mean and standard error of the means of 3 independent experiments) (Fig. 3C) and in 67.9 ± 7.4% of cells in mid-prophase stage (Fig. 3B). In cells with more than 7-µm-wide, only 15.8 ± 8.9% of PPBs contained the RanGAP band (Fig. 3B). About a half of the interphase cells (48.4 ± 12.1%) contained PPB with the RanGAP band when the width of the MT band was less than 7 µm, while 7.4 ± 5.4% of cells whose MT band width was more than 7 µm had the RanGAP band (Fig. 3A). These results indicated that the RanGAP band started to appear when the width of the MT band reduced to less than 7 µm. To test the correlation between the MT band and RanGAP band, we plotted the width of the RanGAP band vs. that of the MT band (Fig. 4). This figure indicates that despite the difference in the width of the RanGAP and MT bands in some interphase cells, the RanGAP band first appeared as a slightly broad (~7 µm wide) band and it gradually narrowed down to 2~5 µm-wide.
Figure 3.

Relationship between the appearance of RanGAP band and the developmental stage of PPB MTs in onion root tip cells. Data from 108 PPBs in an experiment are shown here. Width of the MT band in interphase (A), mid-prophase (B), and late prophase (C) cells. Closed boxes show cells with a RanGAP band and open boxes show cells without RanGAP bands.
Figure 4.

Relationships between the width of MT bands and the width of RanGAP bands in root tip cells of 4-day-old onion seedlings. Nuclear stages were determined by the Hoechest 33258 staining. The straight line in the figure shows the points where the width of RanGAP band corresponds to that of MT band. •, interphase cells; ◊, mid-prophase cells; ▴, late prophase cells. Spearman's R = 0.648, p < 0.001 (mid-prophase, n = 51), Spearman's R = 0.324, p = 0.281 (late prophase, n = 13).
RanGAPs in cells with aberrant MT bands induced by cytoskeletal inhibitors
Our immunofluorescence study presented above clearly showed that the RanGAP band start to appear in mid-prophase when the width of the MT bands is reduced to less than 7 µm. To further examine the relationships between RanGAP and MT bands, we observed the behavior of RanGAPs when the configuration of the MT bands was modified by cytoskeletal inhibitors. However, in 4-day-old root tips, nearly 10% of all examined cells contained PPB (including broad PPB) and 1% of all examined cells were late prophase. We carried out partial synchronization of PPBs using the method developed by Burgess and Northcote55 for wheat root tip cells by using 5-AU. After the incubation of onion seedlings with 5-AU for 17 h, seedlings were transferred to distilled water to induce cell division. Using this method, we were able to produce cells with a PPB (including broad PPBs) in nearly 40% of root tip cells 7 h after their transfer from 5-AU to distilled water. To modify the MT band, we employed 2 different methods using cytoskeletal drugs. The application of oryzalin, an inhibitor of plant MT polymerization,56 to root tip cells may induce the depolymerization of MTs in a PPB. Hence, the number of MTs in PPB is expected to decrease or MTs are expected to disperse shortly after oryzalin application. In contrast, re-broadening of the MT band is induced by the application of inhibitors for F-actin polymerization, such as cytochalasin and latrunculin. 22,49,57 In the present study, we examined 5-AU synchronized root tip cells that were treated with oryzalin and latrunculin B (LatB). Root tip cells treated with 0.2% DMSO for 15 min were used as control.
As was predicted, the treatment of oryzalin induced the dispersion of MTs from the PPB, resulting in increased tubulin fluorescence in the cytoplasm. In oryzalin-treated cells, MT band in the majority of cells became thin (Fig. 5F). In some mid-prophase cells, the MT band dispersed completely to the cell cortex, rendering it difficult to determine the edge of the MT band; nevertheless, these cells also had a RanGAP band (Figs. 5A-C, 6B). Comparison of the width of the RanGAP band (3.31 ± 0.32 μm, n = 18, the mean ± standard error obtained from 18 mid-prophase cells) and that of the MT band (2.54 ± 0.32 μm, n = 18) showed that the number of PPBs whose RanGAP band was wider than that of MT band increased in mid-prophase (Fig. 6B). Using synchronized onion root tips, we were able to examine a sufficient number of late prophase-PPBs. In oryzalin-treated late prophase cells, almost all of the MT bands became thin. In these late prophase cells, some RanGAP band remained 2–5 µm wide, while others RanGAP bands were not detectable (Fig. 6B). Band width in late prophase cells was compared among cells that had both MT and RanGAP band. Although the width of oryzalin-treated MT band was narrower than that of DMSO-control, the width of oryzalin-treated RanGAP band remained the similar size as control RanGAP band (Table 1). This result suggested that a certain group of late prophase RanGAP band was wider than the oryzalin-treated MT band, in others, RanGAP band disappeared after the treatment of oryzalin.
Figure 5.
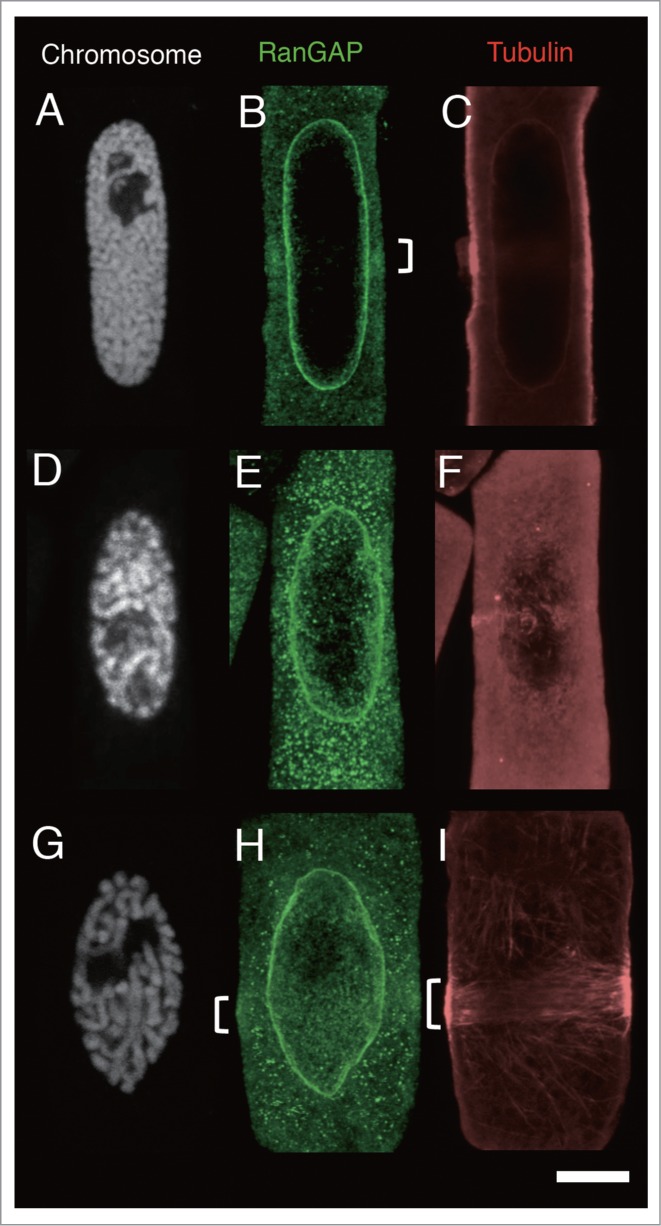
Effects of MT band modification by oryzalin and/or latrunculin B (LatB) on the localization of RanGAP and tubulin in root tip cells of 4-day-old onion seedlings 7 h after their transfer from 5-AU to distilled water. (A-C), immunofluorescence images of an onion root tip cell in mid-prophase treated with 20 µM oryzalin for 15 min. (D-F), immunofluorescence images of an onion root tip cell in late prophase treated with 20 µM oryzalin for 15 min. (G-I) immunofluorescence images of an onion root tip cell in late prophase treated with 10 µM Lat B for 15 min. A-C, and D-F are images of the same cells, respectively. Chromosomes are shown as white (A, D and G), RanGAP images are shown in green (B, E and I), and tubulin in red (C, F and I). Brackets indicate RanGAP and MT bands. Bar: 5 μm.
Figure 6.
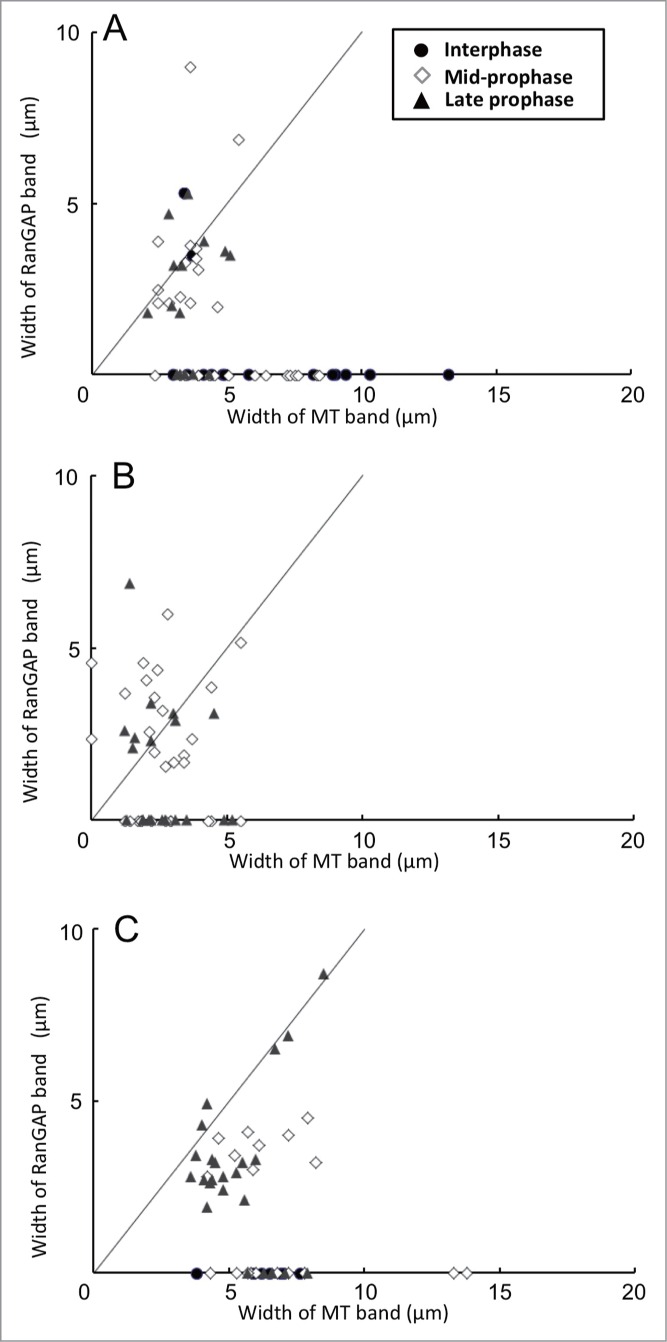
Effects of MT band modification by cytoskeletal inhibitors on the relationship between width of MT bands and that of RanGAP bands in root tip cells of 4-day-old onion seedlings 7 h after their transfer from 5- AU to distilled water. Inhibitors were applied 15 min before chemical fixation. (A) Control experiment with 0.2% DMSO. (B) Root tips treated with 20 µM oryzalin for 15 min. (C) Root tips treated with 10 µM latrunculin B (LatB) for 15 min. The straight lines in each figure shows the points where the width of RanGAP band corresponds to that of MT band. The nuclear stages were determined by the Hoechest 33258 stainings. •, interphase cells; ◊, mid-prophase cells; ▴, late prophase cells.
Table 1.
Effects of cytoskeletal inhibitors on the widths of MT and RanGAP bands in late prophase cells of 4-day-old onion root tips
| Late prophase cells with a MT band and a RanGAP band |
||
|---|---|---|
| Width of MT band | Width of RanGAP band | |
| Treatment | (µm) | (µm) |
| 0.2% DMSO | 3.51 ± 0.11a,b,x (n = 13) | 3.27 ± 0.29c,d,x (n = 13) |
| 20 µM oryzalin | 2.30 ± 0.35a*, y (n = 9) | 3.20 ± 0.49c,y (n = 9) |
| 10 µM LatB | 5.05 ± 0.29b*, z (n = 19) | 3.72 ± 0.41d,z* (n = 19) |
PPBs in root tips were synchronously induced 7 h after their transfer from 5- AU to distilled water. Seedlings were treated with drugs for 15 min before fixation. The width of MT band and that of RanGAP band in late prophase cells having both MT and RanGAP bands were compared. All values show the mean ± standard error from late prophase cells. Letter n in parenthesis indicates the number of examined late prophase cells. Student t-test was carried out between samples indicated with the same small letter (a~c and x~z).
indicates significant difference from the corresponding control value (P < 0.05).
In LatB-treated mid-prophase cells, width of MT band rewiden to 5–10 µm but the width of RanGAP band was less than 5 µm suggesting that the RanGAP behaves independently from MTs (Fig. 6C). In LatB-treated late prophase cells, the width of MT band widen 1.5-fold compared to that in control cells, and the width of the RanGAP band remained narrow (Table 1). In some LatB-treated late prophase cells, both the widths of MT and RanGAP bands became wide (7–8 µm wide) (Fig. 6C). These observations suggested that the developing RanGAP band remained in the PPB site although MT band became thinner or re-broadened by cytoskeletal inhibitors. In some late prophase cells RanGAP band dispersed by the oryzalin treatment.
Spatio-temporal differences between RanGAP band and MT band
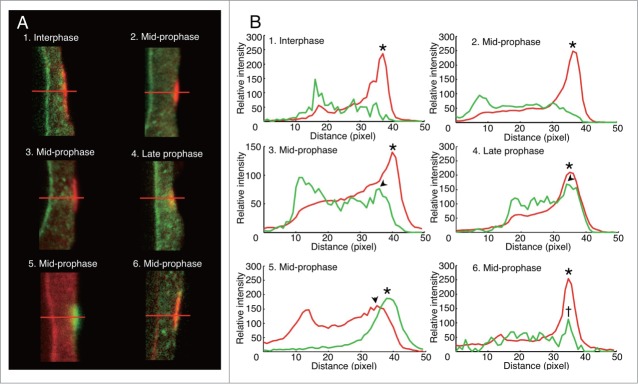
Spatial relationship between RanGAP band and MT band was observed by imaging analysis of confocal laser scanning microscope images of PPBs double stained with anti-AcRanGAP and anti-tubulin antibodies. The intensity profile of the green fluorescence signal of RanGAP and that of the red fluorescence signal of tubulin measured along the horizontal red line drawn in Fig. 7A were compared during PPB development. In interphase cells characterized by PPB with a broad MT band, the clear green-RanGAP fluorescence peak was detectable at the nuclear membrane site, but there were no peaks in the cell cortex where the MT band was positioned (Figs. 7A1 and B1). In mid-prophase cells without RanGAP band, green-RanGAP fluorescence peak was not detectable in the cortex (Figs. 7A2 and B2). In mid-prophase cells with the RanGAP band, the peak of the green-RanGAP fluorescence profile appeared in the cell cortex, and it was slightly shifted from the peak of the tubulin fluorescence toward the cytoplasmic side (Figs. 7A3 and B3). In the late prophase cells, the peak of the green-RanGAP fluorescence profile and that of the red-tubulin fluorescence profile overlapped in the cell cortex (Figs. 7A4 and B4).
Figure 7.
Spatio-temporal differences between RanGAP band and MT band in onion root tip cells. (A) Immunofluorescence images of PPB region of mid-longitudinal optical section of onion root tip cells 7 h after their transfer from 5- AU to distilled water. 1–4, RanGAP (green) and tubulin (red) immunofluorescence; 5, RanGAP (red) and tubulin (green) immunofluorescence; 6, MAP65 (green) and tubulin (red) immunofluorescence. (B) Fluorescence intensity profile along the red lines shown in (A). 1–6 in (B) correspond to 1–6 in (A), respectively. Fluorescence intensity profile colored red and green in (B)correspond to red and green fluorescence in A, respectively. Arrowheads, peaks of RanGAP intensities in PPBs; asterisks, peaks of tubulin intensities in PPB; dagger, the peak of MAP65 intensity. 1, PPB in interphase cell; 2,3,5,6, PPB in mid-prophase cells; 4, PPB in late prophase cell.
To examine whether the shift in green-RanGAP fluorescence peak from red-tubulin fluorescence peak in PPBs of mid-prophase cells was an artifact derived from the combinations of fluorescence probes, we exchanged the colors of secondary antibodies so that red-fluorescence was detected for RanGAP-signal and green fluorescence for tubulin-signal. The results showed that the RanGAP peak also shifted slightly from the tubulin peak toward the inner cytoplasmic side (Figs. 7A5 and B5). Next, we compared the location of MTs and MAP65, which is an MT-associated protein family whose isotypes have been found in the PPBs.58-62 We used anti-BY2MAP6553 which cross-reacted with a single band of the crude extract of onion root tips (Fig. 1C). The result showed that the peak of MAP65 overlapped with the peak of tubulin fluorescence in PPB of mid-prophase cells (Figs. 7A6 and B6), indicating that the RanGAP band in mid-prophase is located toward the inner cytoplasmic side from the MT band.
Discussion
The division site in cells is determined in 2 steps consisting of the formation of a broad MT band that defines the division plane orientation and the MT band narrowing that demarcates the division site. 3,63 As narrowed MT band marks a region broader than the cell plate attachment site,64 the term CDZ has been recently introduced to describe the position of PPB, whereas the CDS refers to the exact site of cell plate attachment.8 Hence, the process of division site insertion can be divided into the following steps: 1) broad PPB formation stage in which orientation of the division plane is fixed (G2 to early prophase); 2) PPB narrowing stage in which TAN and RanGAPs accumulate to establish the CDZ (mid-prophase to late prophase); 3) maintenance of the CDZ during chromosome movement (prometaphase to anaphase); and 4) CDS establishment by narrowing of the band of CDZ proteins (telophase). TON2/ FASS may be involved in the step 1 as suggested by the absence of MT band formation in ton2/fass mutants.10-15 However, once a broad MT band appears, the MT band starts to narrow. If step 2 - the CDZ formation stage - starts immediately after the appearance of the broad PPB, RanGAP should appear concomitantly with the appearance of the broad PPB. In the present study we focused on step 2 to answer the question as to when and how the RanGAP band appears and we demonstrated that the RanGAP band appears when the width of the MT band reduces to less than 7 µm (Figs. 3 and 4). In most of the cells in late prophase, PPBs have a very narrow MT band (nearly 3 µm wide) and have a RanGAP band (Figs. 2J-L, 3C and 4) of the same width as the MT band. Probably, the CDZ is established in the late prophase stage when the band is narrowest. Our results indicate that the lack of the RanGAP band in half of the preprophasic or mitotic cells observed by Xu et al.27 may be due to the detection limits of their technique and not due to the absence of the RanGAP band in some of the cells during cell division.
In the present study, we successfully examined the molecular location of RanGAP and MT bands in onion root tip cells with higher accuracy when compared to previous observations in Arabidopsis by Xu et al. 27 The process of PPB development can be divided into 3 stages (Fig. 8). In the broad MT band stage (PPB width > 7 µm), RanGAPs are not concentrated in the cortical region (Fig. 8A). When the PPB width narrows down to nearly 7 µm, RanGAPs start to gather near the cell cortex (Fig. 8B). This marks the beginning of the CDZ formation (CDZ-forming stage). RanGAPs start accumulating in the cortical region in the MT-independent manner in mid-prophase to a certain stage of late prophase. Our analysis showed that the RanGAP band is located slightly toward the inner cytoplasmic side from the MT band in mid-prophase (Fig. 7). This observation indicates that RanGAPs behave differently from MTs during CDZ formation. Our observations of the RanGAP behavior during the MT band modification by cytoskeletal inhibitors in mid-prophase cells and in some late prophase cells corroborate the results of the experiments with oryzalin carried out by Xu et al.,27 which suggest that RanGAP band is independent from MTs
Figure 8.
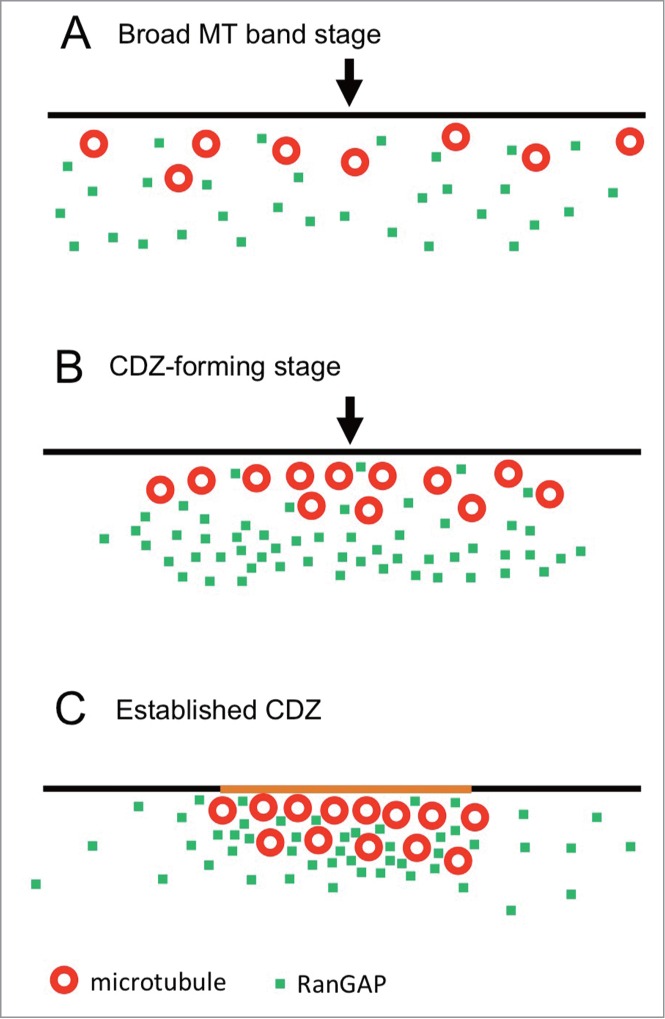
Model of MT and RanGAP band formation during PPB development. Black lines, red circles, green dots, and arrowheads show plasma membrane, MTs, RanGAP, and the center of the PPB, respectively. (A) Broad MT band forming stage. Many interphase cells and some mid-prophase cells have broad MT band and RanGAP proteins are widely diffused in the cytoplasm. (B) CDZ-forming stage. Most of mid-prophase cells and some late prophase cells have narrow MT band (less than 7 µm wide) and RanGAPs start to gather around the PPB region, but they are located slightly toward the inner cytoplasmic side from the MT band. (C) Established CDZ. A packed-narrow MT band (2–5 µm wide) and RanGAPs are concentrated into the MT band and form a CDZ in a certain time of late prophase. The plasma membrane in the CDZ is colored orange.
The CDZ is established sometime in late prophase when RanGAPs and TANs are concentrated at the CDZ (Established CDZ, Fig. 8C). In oryzalin-treated late prophase cells, half of the cells had intact RanGAP band and the remaining cells did not have RanGAP band although a remnant of the MT band still existed (Figs. 5 and 6). There is a possibility that the dependency of MTs on RanGAP maintenance may differ in its developmental stages. Possibile explanation is as follows: in the CDZ forming stage, changes in the MT configuration do not interfere with the RanGAP band formation because RanGAPs and MTs gather to the CDZ zone independently (Fig. 8B). As RanGAPs and MTs stayed very near after the establishment of CDZ, the MT deploymerization interferes with the positioning of RanGAPs. Recently, POK1 has been shown to be recruited to the CDZ in the MT-dependent and TAN-independent manner in prophase, but its functions in the maintenance of TAN in the CDZ after prometaphase suggests that POK is involved in the translation of the positional information of PPB to establish CDZ.32 These drastic changes in the molecular environment in the PPB region before and after the establishment of CDZ may influence the RanGAP band change in the dependency of MTs.
Xu et al. 27 has been presented a clear images of Arabidopsis root tip cells showing the existence of the RanGAP band after MT band disassembly. We were able to obtain clear images showing the existence of the RanGAP band in prophase cells (Figs. 2H and K) but not in mitotic stages (Fig. 2Q) of the onion root tip cells. The immunostaining method we applied is a well-established method used for molecular localization in fixed root tip cells. This method has been used for γ–tubulin65,66 and cdc242 in PPBs and existence of the actin depleted zone51. In the present experiments, the images of mitotic cells showed the presence of a possible RanGAP band only in a few mitotic cells (for example, Fig. 2N). The observed band-like signals in the cortex may have derived from the CDZ or from the spindles. One possible explanation for the inconsistency in the results between Arabidopsis and onion is that the region between the spindles and the cell cortex in onion root tip cells is narrow, causing interference between the fluorescence signal derived from the spindles and that from the RanGAP band. Further studies will be necessary to answer this problem.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was partly supported by JSPS grant 17207006 to YM.
References
- 1.Sinnott EW. Plant Morphogenesis. New York: McGraw-Hill; 1960:550p [Google Scholar]
- 2.Gunning BES. The cytokinetic apparatus: its development and spatial regulation In: The Cytoskeleton in Plant Growth and Development. Lloyd CW, ed. London: Academic Press; 1982:229-92 [Google Scholar]
- 3.Mineyuki Y. The preprophase band of microtubules: its function as a cytokinetic apparatus in higher plants. Int Rev Cytol 1999; 187:1-49 [Google Scholar]
- 4.Wick SM, Duniec J. Immunofluorescence microscopy of tubulin and microtubule arrays in plant cells. I. Preprophase band development and concomitant appearance of nuclear envelope-associated tubulin. J Cell Biol 1983; 97:235-43; PMID:6345554; http://dx.doi.org/ 10.1083/jcb.97.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wick SM, Duniec J. Immunofluorescence microscopy of tubulin and microtubule arrays in plant cells. II. Transition between the pre-prophase band and the mitotic spindle. Protoplasma 1984; 122:45-55; http://dx.doi.org/ 10.1007/BF01279436 [DOI] [Google Scholar]
- 6.Mineyuki Y, Wick SM, Gunning BES. Preprophase bands of microtubules and the cell cycle: kinetics and experimental uncoupling of their formation from the nuclear cycle in onion root-tip cells. Planta 1988; 174:518-26; PMID:24221569; http://dx.doi.org/ 10.1007/BF00634482 [DOI] [PubMed] [Google Scholar]
- 7.Van Damme D, Vanstraelen M, Geelen D. Cortical division zone establishment in plant cells. Trends Plant Sci 2007; 12:458-64; PMID:17765597; http://dx.doi.org/ 10.1016/j.tplants.2007.08.011 [DOI] [PubMed] [Google Scholar]
- 8.Van Damme D, Gadeyne A, Vanstraelen M, Inzé D, Van Montagu MCE, De Jaeger G, Russinova E, Geelem D. Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc Natl Acad Sci USA 2011; 108:615-20; PMID:21187379; http://dx.doi.org/ 10.1073/pnas.1017890108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen CG, Wright AJ, Müller S. The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J 2013; 75:258-69; PMID:23496276; http://dx.doi.org/ 10.1111/tpj.12177 [DOI] [PubMed] [Google Scholar]
- 10.Traas J, Bellini C, Nacry P, Kronenberger J, Bouchez D, Caboche M. Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 1995; 375:676-7; http://dx.doi.org/ 10.1038/375676a0 [DOI] [Google Scholar]
- 11.Azimzadeh J, Nacry P, Christodoulidou A, Drevensek S, Camilleri C, Amiour N, Parcy F, Pastuglia M, Bouchez D. Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 2008; 20:2146-59; PMID:18757558; http://dx.doi.org/ 10.1105/tpc.107.056812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spinner L, Pastuglia M, Belcram K, Pegoraro M, Goussot M, Bouchez D, Schaefer DG. The function of TONNEAU1 in moss reveals ancient mechanisms of division plane specification and cell elongation in land plants. Development 2010; 137:2733-42; PMID:20663817; http://dx.doi.org/ 10.1242/dev.043810 [DOI] [PubMed] [Google Scholar]
- 13.Camilleri C, Azimzadeh J, Pastuglia M, Bellini C, Grandjean O, Bouchez D. The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 2002; 14:833-45; PMID:11971138; http://dx.doi.org/ 10.1105/tpc.010402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torres-Ruiz RA, Jürgens G. Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 1994; 120:2967-78; PMID:10484674 [DOI] [PubMed] [Google Scholar]
- 15.Wright AJ, Gallagher K, Smith LG. Discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell 2009; 21:234-47; PMID:19168717; http://dx.doi.org/ 10.1105/tpc.108.062810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duroc Y, Bouchez D, Pastuglia M. The preprophase band and division site determination in land plants In: The Plant Cytoskeleton. Liu B, ed. Berlin: Springer; 2011:145-85 [Google Scholar]
- 17.McMichael CM, Bednarek SY. Cytoskeletal and membrane dynamics during higher plant cytokinesis. New Phytol 2013; 197:1039-57; PMID:23343343; http://dx.doi.org/ 10.1111/nph.12122 [DOI] [PubMed] [Google Scholar]
- 18.Hamada T. Microtubule organization and microtubule-associated proteins in plant cells. Int Rev Cell Mol Biol 2014; 312:1-52 [DOI] [PubMed] [Google Scholar]
- 19.Traas JA, Doonan JH, Rawlins DJ, Shaw PJ, Watts J, Lloyd CW. An actin network is present in the cytoplasm throughout the cell cycle of carrot cells and associates with the dividing nucleus. J Cell Biol 1987; 105:387-95; PMID:2440896; http://dx.doi.org/ 10.1083/jcb.105.1.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palevitz BA. Actin in the preprophase band of Allium cepa. J Cell Biol 1987; 104:1515-9; PMID:3294855; http://dx.doi.org/ 10.1083/jcb.104.6.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kakimoto T, Shibaoka H. Actin filaments and microtubules in the preprophase band and phragmoplast of tobacco cells. Protoplasma 1987; 140:151-6; http://dx.doi.org/ 10.1007/BF01273724 [DOI] [Google Scholar]
- 22.Mineyuki Y, Palvitz BA. Relationship between preprophase band organization, F-actin and the division site in Allium: fluorescence and morphometric studies on cytochalasin-treated cells. J Cell Sci 1990; 97:283-95 [Google Scholar]
- 23.Ambrose JC, Shoji T, Kotzer AM, Pighin JA, Wasteneys GO. The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 2007; 19:2763-75; PMID:17873093; http://dx.doi.org/ 10.1105/tpc.107.053777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamura E, Himmelspach R, Rashbrooke MC, Whittington AT, Gale KR, Collings DA, Wasteneys GO. Microtubule organization 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol 2006; 140:102-14; PMID:16377747; http://dx.doi.org/ 10.1104/pp.105.069989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO. MOR1 is essential for organizing cortical microtubules in plants. Nature 2001; 411:610-3; PMID:11385579; http://dx.doi.org/ 10.1038/35079128 [DOI] [PubMed] [Google Scholar]
- 26.Walker KL, Müller S, Moss D, Ehrhardt DW, Smith LG. Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Current Biol 2007; 17:1827-36; PMID:17964159; http://dx.doi.org/ 10.1016/j.cub.2007.09.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu XM, Zhao Q, Rodrigo-Peiris T, Brkljacic J, He CS, Müller S, Meier I. RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc Natl Acad Sci USA 2008; 105:18637-42; PMID:19011093; http://dx.doi.org/ 10.1073/pnas.0806157105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith LG, Hake S, Sylvester AW. The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 1996; 122:481-9; PMID:8625799 [DOI] [PubMed] [Google Scholar]
- 29.Cleary AL, Smith LG. The Tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell 1998; 10:1875-88; PMID:9811795; http://dx.doi.org/ 10.1105/tpc.10.11.1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith LG, Gerttula SM, Han S, Levy J. Tangled1 a microtubule binding protein required for the spatial control of cytokinesis in maize. J Cell Biol 2001; 152:231-6; PMID:11149933; http://dx.doi.org/ 10.1083/jcb.152.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller S, Han S, Smith LG. Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Current Biol 2006; 16:888-94; ; http://dx.doi.org/ 10.1016/j.cub.2006.03.034 [DOI] [PubMed] [Google Scholar]
- 32.Lipka E, Gadeyne A, Stöckle D, Zimmermann S, De Jaeger G, Ehrhardt DW, Kirik V, Van Damme D, Müller S. The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 2014; 26:2617-32; PMID:24972597; http://dx.doi.org/ 10.1105/tpc.114.124933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 2007; 8:761-73; PMID:17786152; http://dx.doi.org/ 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- 34.Ciciarello M, Mangiacasale R, Lavia P. Spatial control of mitosis by the GTPase Ran. Cell Mol Life Sci 2007; 64:1891-914; PMID:17483873; http://dx.doi.org/ 10.1007/s00018-007-6568-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pay A, Resch K, Frohnmeyer H, Fejes E, Nagy F, Nick P. Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J 2002; 30:699-709; PMID:12061901; http://dx.doi.org/ 10.1046/j.1365-313X.2002.01324.x [DOI] [PubMed] [Google Scholar]
- 36.Jeong SY, Rose A, Joseph J, Dasso M, Meier I. Plant-specific mitotic targeting of RanGAP requires a functional WPP domain. Plant J 2005; 42:270-82; PMID:15807788; http://dx.doi.org/ 10.1111/j.1365-313X.2005.02368.x [DOI] [PubMed] [Google Scholar]
- 37.Patel S, Rose A, Meulia T, Dixit R, Cyr RJ, Meier I. Arabidopsis WPP-domain proteins are developmentally associated with the nuclear envelope and promote cell division. Plant Cell 2004; 16:3260-73; PMID:15548735; http://dx.doi.org/ 10.1105/tpc.104.026740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meier I. A novel link between Ran signal transduction and nuclear envelope proteins in plants. Plant Physiol 2000; 124:1507-10; PMID:11115866; http://dx.doi.org/ 10.1104/pp.124.4.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose A, Meier I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA 2001; 98:15377-82; PMID:11752475; http://dx.doi.org/ 10.1073/pnas.261459698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nemec B. Ueber die karyokinetische Kerntheilung in der Wurzelspitze von Allium cepa. Jahrb Wiss Bot 1899; 33:313-36 [Google Scholar]
- 41.González-Fernández A, López-Sáez J, Moreno P, Giménez-Martin G. A model for dynamics of cell division cycle in onion roots. Protoplasma 1968; 65:263-76; PMID:5685727; http://dx.doi.org/ 10.1007/BF01682532 [DOI] [PubMed] [Google Scholar]
- 42.Mineyuki Y, Yamashita M, Nagahama Y. p34cdc2 kinase homologue in the preprophase band. Protoplasma 1991; 162:182-6; http://dx.doi.org/ 10.1007/BF02562561 [DOI] [Google Scholar]
- 43.Mineyuki Y, Iida H, Anraku Y. Loss of microtubules in the interphase cells of onion (Allium cepa L.) root tips from the cell cortex and their appearance in the cytoplasm after treatment with cycloheximide. Plant Physiol 1994; 104:281-4; PMID:12232080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mineyuki Y, Aioi H, Yamashita M, Nagahama Y. A comparative study on stainability of preprophase bands by the PSTAIR antibody. J Plant Res 1996; 109:185-92; http://dx.doi.org/ 10.1007/BF02344544 [DOI] [Google Scholar]
- 45.Nogami A, Suzaki T, Shigenaka Y, Nagahama Y, Mineyuki Y. Effects of cycloheximide on preprophase bands and prophase spindles in onion (Allium cepa L.) root tip cells. Protoplasma 1996; 192:109-21; http://dx.doi.org/ 10.1007/BF01273249 [DOI] [Google Scholar]
- 46.Nogami A, Mineyuki Y. Loosening of a preprophase band of microtubules in onion (Allium cepa L.) root tip cells by kinase inhibitors. Cell Struct Funct 1999; 24:419-24; PMID:15216900; http://dx.doi.org/ 10.1247/csf.24.419 [DOI] [PubMed] [Google Scholar]
- 47.Kubiak J, De Brabander M, De Mey J, Tarkowska JA. Origin of the mitotic spindle in onion root cells. Protoplasma 1986; 130:51-6; http://dx.doi.org/ 10.1007/BF01283330 [DOI] [Google Scholar]
- 48.Utrilla L, Giménez Abián MI, De la Torre C. Timing the phases of the microtubule cycles involved in cytoplasmic and nuclear divisions in cells of undisturbed onion root meristems. Biol Cell 1993; 78:235-41; http://dx.doi.org/ 10.1016/0248-4900(93)90135-2 [DOI] [Google Scholar]
- 49.Eleftheriou EP, Palevitz BA. The effect of cytochalasin D on preprophase band organization in root tip cells of Allium. J Cell Sci 1992; 103:989-98 [Google Scholar]
- 50.Collings DA, Harper JDI, Vaughn KC. The association of peroxisomes with the developing cell plate in dividing onion root cells depends on actin microfilaments and myosin. Planta 2003; 218:204-16; PMID:12937986; http://dx.doi.org/ 10.1007/s00425-003-1096-2 [DOI] [PubMed] [Google Scholar]
- 51.Liu B, Palevitz BA. Organization of cortical microfilaments in dividing root cells. Cell Motil Cytoskeleton 1992; 23:252-64; http://dx.doi.org/ 10.1002/cm.970230405 [DOI] [Google Scholar]
- 52.Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem 1987; 163:16-20; PMID:2441623; http://dx.doi.org/ 10.1016/0003-2697(87)90086-8 [DOI] [PubMed] [Google Scholar]
- 53.Sawano M, Shimmen T, Sonobe S. Possible involvement of 65 kDa MAP in elongation growth of azuki bean epicotyls. Plant Cell Physiol 2000; 41:968-76; PMID:11038057; http://dx.doi.org/ 10.1093/pcp/pcd022 [DOI] [PubMed] [Google Scholar]
- 54.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int 2004; 11:36-43 [Google Scholar]
- 55.Burgess J, Northcote DH. Action of colchicine and heavy water on the polymerization of microtubules in wheat root meristem. J Cell Sci 1969; 5:433-51; PMID:5362335 [DOI] [PubMed] [Google Scholar]
- 56.Morejohn LC, Bureau TE, Molè-Bajer J, Bajer AS, Fosket DE. Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta 1987; 172:252-64; PMID:24225878; http://dx.doi.org/ 10.1007/BF00394595 [DOI] [PubMed] [Google Scholar]
- 57.Granger CL, Cyr RJ. Use of abnormal preprophase bands to decipher division plane determination. J Cell Sci 2001; 114:599-607; PMID:11171328 [DOI] [PubMed] [Google Scholar]
- 58.Gaillard J, Neumann E, Van Damme D, Stoppin-Mellet V, Ebel C, Barbier E, Geelen D, Vantard M. Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol Biol Cell 2008; 19:4534-44; PMID:18667529; http://dx.doi.org/ 10.1091/mbc.E08-04-0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Zeng X, Liu Z-Q, Meng Q-T, Yuan M, Mao T-L. Arabidopsis microtubule-associated protein AtMAP65-2 acts as a microtubule stabilizer. Plant Mol Biol 2009; 69:313-24; PMID:19002591; http://dx.doi.org/ 10.1007/s11103-008-9426-1 [DOI] [PubMed] [Google Scholar]
- 60.Mao G, Chan J, Calder G, Doonan JH, Lloyd CW. Modulated targeting of GFP AtMAP65 1 to central spindle microtubules during division. Plant J 2005; 43:469-78; PMID:16098102; http://dx.doi.org/ 10.1111/j.1365-313X.2005.02464.x [DOI] [PubMed] [Google Scholar]
- 61.Smertenko AP, Chang H-Y, Wagner V, Kaloriti D, Fenyk S, Sonobe S, Lloyd C, Hauser M-T, Hussey PJ. The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell 2004; 16:2035-47; PMID:15273298; http://dx.doi.org/ 10.1105/tpc.104.023937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smertenko AP, Kaloriti D, Chang H-Y, Fiserova J, Opatrny Z, Hussey PJ. The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell 2008; 20:3346-58; PMID:19060108; http://dx.doi.org/ 10.1105/tpc.108.063362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mineyuki Y, Marc J, Palevitz BA. Development of the preprophase band from random cytoplasmic microtubules in guard mother cells of Allium cepa L. Planta 1989; 178:291-6; PMID:24212895; http://dx.doi.org/ 10.1007/BF00391856 [DOI] [PubMed] [Google Scholar]
- 64.Marcus A, Dixit R, Cyr RJ. Narrowing of the preprophase microtubule band is not required for cell division plane determination in cultured plant cells. Protoplasma 2005; 226:169-74; PMID:16333576; http://dx.doi.org/ 10.1007/s00709-005-0119-1 [DOI] [PubMed] [Google Scholar]
- 65.Liu B, Marc J, Joshi HC, Palevitz BA. A γ-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J Cell Sci 1993; 104:1217-28; PMID:7686171 [DOI] [PubMed] [Google Scholar]
- 66.Liu B, Joshi HC, Wilson TJ, Silflow CD, Palevitz BA, Snustad DP. γ-Tubulin in Arabidopsis: gene sequence, immunoblot, and immunofluorescence studies. Plant Cell 1994; 6:303-14; PMID:8148650; http://dx.doi.org/ 10.1105/tpc.6.5.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.