ABSTRACT
The shoot apical meristem is the central organizer of plant aerial organogenesis. The molecular bases of its functions involve several cross-talks between transcription factors, hormones and microRNAs. We recently showed that the expression of the homeobox transcription factor STM is induced by mechanical perturbations, adding another layer of complexity to this regulation. Here we provide additional evidence that mechanical perturbations impact the promoter activity of CUC3, an important regulator of boundary formation at the shoot meristem. Interestingly, we did not detect such an effect for CUC1. This suggests that the robustness of expression patterns and developmental programs is controlled via a combined action of molecular factors as well as mechanical cues in the shoot apical meristem.
KEYWORDS: CUC genes, mechanical stress, Meristem, micro-RNAs
Throughout their lifetime, plants are exposed to a number of external mechanical perturbations, such as wind or touch. This leads to changes in mRNA level of many genes, and long-term developmental responses such as stem thickening and flowering delay 5 In addition to these external factors, plants are also constantly affected by intrinsic tensile stresses, notably because plant cells are under high turgor pressure. There is now accumulating evidence that these internal stresses are affecting many aspects of the cells, thus channeling growth in the long term.14
The shoot apical meristem (SAM) is the central organizer of plant aerial organogenesis. Initiation of new organs and maintenance of SAM is achieved through the concomitant action of multiple regulatory pathways. Genetic screens have identified many key players, including transcription factors, hormones, and microRNAs.38,48,53,55 In addition to these biochemical factors, mechanical forces are also present within the meristem. In particular, the boundary domain that separates newly emerging organ from the meristem is under highly anisotropic tensile stresses.4,15,32 A key question for the future is the analysis of the interplay between mechanical forces and the molecular regulators of meristem function (Fig. 1A).
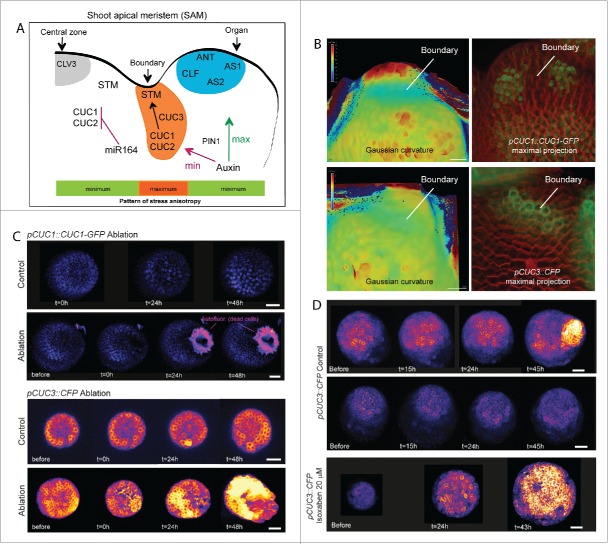
Figure 1.
Interplay between molecular and mechanical regulators at the shoot meristem boundary. (A) Shape changes at the meristem is accompanied by the expression of specific genes.52,46 Among those are ANT, CLF, AS1, AS2 that are expressed mainly in the growing organ and, CUC1, 2 and 3, the expression of which is restricted to the boundary domain.43,46,52,54 The pattern of stress anisotropy is represented by color bar with green corresponding to the regions with lower stress anisotropy (central zone and organ) and orange in the boundary domain where stress anisotropy is maximal. This stress and gene expression pattern correlates with the distribution of the plant hormone auxin that has its local maximum where new organ initiates and its local minimum at the organ boundary.8,17,38 Acting in a PIN1-dependent manner, auxin may promote organ emergence, notably through the repression of KNOX genes,11,16,41 whereas in the boundary, it may negatively regulate the axillary meristem formation.50 (B) CUC1 and CUC3 expression pattern at the boundary in meristems from greenhouse-grown plants: Signal intensity (right panels) and Gaussian curvature (left panels) of pCUC1::CUC1-GFP and pCUC3::CFP in representative SAM. Membranes were labeled with FM4–64. Gaussian curvature is extracted using the level set method and MorphoGraphX. Representative images highlighting the differences between pCUC1::CUC1-GFP and pCUC3::CFP signal intensities in young boundaries, as revealed by the curvature map. Scale bars: 10 μm. (C) Expression of pCUC1::CUC1-GFP (upper panels) and pCUC3::CFP (lower panels) in meristems from in vitro grown plants, before and after ablation. Expression is shown using the Fire lookup table in ImageJ, with the threshold of 5 in the Fire representation. No significant induction of signal is observed for pCUC1::CUC1-GFP; only an increase of autofluorescence in dead cells can be detected 24 hour after ablation. In contrast, pCUC3::CFP expression (lower panels) is induced 24 hours after ablation, when compared to control. Scale bar, 20 μm. (D) Expression of pCUC3::CFP in meristems from in vitro grown plants, before and after treatment with 20 µM isoxaben. The two upper panels display representative untreated pCUC3::CFP meristems with induction only when an organ emerges (upper panel). In contrast, after the isoxaben treatment the intensity of pCUC3::CFP signal is induced everywhere in the meristem, when compared to control. Scale bar, 20 μm.
In a recent article,26 we showed that a master regulator of meristem maintenance, the homeodomain protein SHOOT MERISTEMLESS (STM) is expressed at a higher level in the boundary domain, and that this local increase in promoter activity can be related to mechanical stress: mechanical perturbations are sufficient to induce STM expression in the meristem. Interestingly, mechanical perturbations do not affect all boundary-expressed genes in the same way. For instance, the promoter activity of the PINOID gene, which is also increased in the boundary domain, is not affected by mechanical perturbations.26
Here we focus on the most canonical genetic markers of boundary identity, the CUP SHAPED COTYLEDON (CUC) genes. CUC1, CUC2 and CUC3, belong to a group of NAC domain transcription factors and show a high level of functional redundancy. They play an essential role in shoot meristem initiation through the regulation of STM expression.1,9,19,29,34,45,49 Recently it has been reported that CUC1 and CUC2 are also required for formation and stable positioning of the carpel margin meristems.23 Depletion of these genes leads to defects in cotyledon separation, organ fusions and cup-shaped cotyledons.19,49 Although functionally connected, the expression of CUC1, CUC2 and CUC3 genes is regulated through different pathways. This is also reflected by their expression profiles, which are not identical in the SAM,25,34,45 Fig. 1B
In addition to transcriptional control, CUC1 and CUC2 are subjected to post-transcriptional regulation through the microRNA pathway.2,27,37,39,42 In plants, miRNAs are produced from larger RNA precursor transcripts that contain a self-complementary structure allowing the formation of a hairpin. After transcription, the miRNA precursor is being recognized by a protein complex that induces its cleavage and further maturation of an active miRNA. miRNAs are short (20 - 22 nt) single stranded molecules, that are predominantly associated with AGO1 and target mRNAs to cleaveage or translational repression in a sequence-dependent manner.3,20,22,28,30,51 Based on sequence similarity of the mature miRNA and the target specificity, the miRNAs are grouped to several (over 90) families (http://www.mirbase.org/; 6,7,24,36,39 The mRNA of CUC1 and CUC2 is targeted for cleavage by the miRNAs of the miR164 family, comprising 3 isoforms - miR164a, miR164b and miR164c.2,27,31,39,42,43 Plants, with the miR164-resistant versions of CUC1 or CUC2 have been shown to have severe defects in the organ boundary formation and organ separation during both vegetative and reproductive development.2,27,31,33,35 The elimination of miR164 activity leads to enlargement of the CUC1 and CUC2 expression domains in the inflorescence meristem, supporting a scenario in which miR164 acts in spatio-temporal regulation of expression of these genes, preventing the fluctuations and contributing to the robustness of developmental programs.35,43 Interestingly, the CUC3 mRNA does not contain the microRNA targeted site, yet it displays a robust boundary specific expression,12,46,52 Fig. 1B.
We recently showed that mechanical perturbations in the form of ablation in the SAM is sufficient to induce CUC3 expression in the SAM, while CUC1 expression profile remains largely unaffected in the same conditions.26 Here we further confirm this result: No significant induction of signal or change in signal patterning was detected in the pCUC1::CUC1-GFP line after ablation (Fig. 1C upper panels). This suggests that CUC1 expression at the boundary is unlikely to be controlled through mechanical cues. In contrast, the pCUC3::CFP line exhibited a strong induction of the signal in meristem (here represented at 24 hours after ablation (Fig. 1C lower panels).
To further investigate the effect of mechanical perturbation of CUC3 expression, the pCUC3::CFP reporter line was treated with isoxaben and the impact on CFP signal intensity was analyzed over time. Isoxaben is a well-known inhibitor of cellulose synthesis; such a treatment is supposed to weaken and increase tensions in cell walls. We detected an increase of the CFP signal intensity in the regions of tissue folding in the treated meristems (Fig. 1D).
This rather supports a scenario in which mechanical stress may impact CUC3 expression, and channel its expression in the boundary domain of the meristem, while CUC1 would rely on miRNA activity to achieve such specificity. Needless to say that other factors, and notably auxin depletion at the boundary, may very well add another layer of regulation to these expression patterns.
Altogether, this work illustrates how the members of a small gene family can be regulated by different cues, despite having redundant functions. It also shows how mechanical cues and miRNA activity can differentially channel molecular inputs into specific outputs. As the molecular bases of meristem functions are now well described, elucidating further such interplays represents a major challenge for the future of plant development.
Material and methods
Plant lines and growth conditions
The pCUC1::CUC1-GFP and pCUC3::CFP lines have recently been described.12
“Greenhouse-grown plants” were initially grown in short-day conditions (8 hr/16 hr light/dark period) for one month and then transferred to long-day conditions (16 hr/8 hr light/dark period). Stems were cut and the SAM was dissected when the inflorescence meristem was visible, i.e. between the appearance of the first flower to the appearance of first silique (stages 13 to 17 44 and transferred on a half MS medium with vitamins and 0.125 µg/µL of BAP for imaging as already described.10
“In vitro grown plants” were grown in a phytotron in long day conditions on Arabidopsis medium (Duchefa) supplemented with 10 µM NPA to inhibit flower initiation and generate naked meristems. NPA-treated in vitro grown plants were transferred to a medium without NPA as soon as naked meristems were formed as already described.13 Meristems were then imaged from 24h to 48h after transfer on the NPA-free medium.
Confocal laser scanning microscopy and image analysis
Dissected meristems and plants grown in vitro were imaged in water using a SP8 confocal microscope (Leica, Germany) or a LSM780 microscope (Zeiss, Germany) to generate stack of optical sections with an interval of 0.25, 1 or 2 µm between slices. In some cases, membranes were stained with FM4-64.
The maps of meristem curvature, pCUC1::CUC1-GFP and pCUC3::CFP signals at cellular levels were obtained using the MorphographX software (www.morphographix.org). The curvature maps were generated by plotting the mean Gaussian curvature on non-segmented meshes with a neighboring of 15 µm.
Ablations and isoxaben treatment
Each experiment was performed on at least 2 independent sets of plants, and with at least 4 independent plants in each set. In all experiments, the t = Xh time point corresponds to X hours after the beginning of treatment. Controls and assays were analyzed in parallel (same growth conditions, same imaging conditions). The ablations and isoxaben treatments that were carried out on WT plants were performed on plants previously grown in vitro NPA and transferred in a medium without NPA 0 to 24h before the beginning of the experiment.
The ablations were performed with a needle as already described.15,47
The isoxaben treatments were conducted by immersing the plants in aqueous solutions of 20 µM of isoxaben overnight (for 12 to 14 hours,18,47). Controls were obtained by water immersion with an equivalent volume of Dimethyl Sulfoxide (DMSO). The presence of isoxaben in the meristem could be confirmed by its impact on meristem and cell size.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the European Research Council ERC grant 615739 “MechanoDevo.” We thank Platim (UMS 3444 Biosciences Gerland-Lyon Sud) and the Sainsbury lab in Cambridge for help with imaging.
References
- 1.Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Company Of Biologists Development 1999; 126(8):1563-70; PMID:1007921915723790 [DOI] [PubMed] [Google Scholar]
- 2.Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 Mutant Uncovers a Role for MicroRNA miR164c in Regulating Petal Number in Arabidopsis. Curr Biol 2005; 15(4), pp.303-15; PMID:15723790; http://dx.doi.org/ 10.1016/j.cub.2005.02.017 [DOI] [PubMed] [Google Scholar]
- 3.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci 2005; 102(33), pp.11928-33; PMID:16081530; http://dx.doi.org/ 10.1073/pnas.0505461102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boudaoud A. An introduction to the mechanics of morphogenesis for plant biologists. Trends Plant Sci 2010; 15(6):353-60; PMID:20427223; http://dx.doi.org/10.1016/j.tplants.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 5.Braam J, Davis RW. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 1990; 60(3), pp.357-64; PMID:2302732; http://dx.doi.org/ 10.1016/0092-8674(90)90587-5 [DOI] [PubMed] [Google Scholar]
- 6.Bülow L, Bolívar JC, Ruhe J, Brill Y, Hehl R. MicroRNA Targets,” a new AthaMap web-tool for genome-wide identification of miRNA targets in Arabidopsis thaliana. BioData Min 2012; 5(1):7; PMID:22800758; http://doi.org/ 10.1186/1756-0381-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chien CH, Chiang-Hsieh YF, Chen YA, Chow CN, Wu NY, Hou PF, Chang WC. AtmiRNET: a web-based resource for reconstructing regulatory networks of Arabidopsis microRNAs. Database 2015; 2015: bav042; PMID:25972521; http://dx.doi.org/ 10.1093/database/bav042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Reuille PB, Bohn-Courseau I. Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc Natl Acad Sci U S A 2006; 103(5):1627-32; PMID:16432202; http://doi.org/ 10.1073/pnas.0510130103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst HA, Olsen AN, Larsen S, Lo Leggio L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Reports 2004; 5(3), pp.297-303; PMID:15083810; http://dx.doi.org/ 10.1038/sj.embor.7400093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez R, Das P, Mirabet V, Moscardi E, Traas J, Verdeil JL, Malandain G, Godin C. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nature Publishing Group 2010; 7:547-53; PMID:20543845; http://doi.org/15371311 10.1038/nmeth.1472 [DOI] [PubMed] [Google Scholar]
- 11.Furutani M. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 2004; 131(20), pp.5021-30; PMID:15371311; http://dx.doi.org/ 10.1242/dev.01388 [DOI] [PubMed] [Google Scholar]
- 12.Gonçalves B, Hasson A, Belcram K, Cortizo M, Morin H, Nikovics K, Vialette-Guiraud A, Takeda S, Aida M, Laufs P, et al.. A conserved role for CUP-SHAPED COTYLEDON genes during ovule development. Plant J 2015; 83(4), pp.732-42; PMID: 26119568; http://dx.doi.org/14671026 10.1111/tpj.12923. [DOI] [PubMed] [Google Scholar]
- 13.Grandjean O, Vernoux T, Laufs P, Belcram K, Mizukami Y, Traas J. In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 2004; 16:74-87; PMID:14671026; http://dx.doi.org/ 10.1105/tpc.017962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamant O. Widespread mechanosensing controls the structure behind the architecture in plants. Curr Opin Plant Biol 2013; 16(5), pp.654-60; PMID:23830994; http://dx.doi.org/ 10.1016/j.pbi.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 15.Hamant O, Heisler MG, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al.. Developmental patterning by mechanical signals in Arabidopsis. Science 2008; 322(5908), pp.1650-5; PMID:19074340; http://dx.doi.org/ 10.1126/science.1165594 [DOI] [PubMed] [Google Scholar]
- 16.Hay A. Asymmetric leaves1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 2006; 133(20), pp.3955-61; PMID:16971475 [DOI] [PubMed] [Google Scholar]
- 17.Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 2005; 15(21):1899-911; PMID:16271866; http://dx.doi.org/ 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- 18.Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jönsson H, et al.. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. Plos Biol 2010; 8:e1000516; PMID:20976043; http://dx.doi.org/ 10.1371/journal.pbio.1000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibara KI, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 2006; 18(11):pp.2946-57; PMID:17122068; http://doi.org/ 10.1105/tpc.106.045716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu W, Coller J. What comes first: translational repression or mRNA degradation? The deepening mystery of microRNA function. Cell Res 2012; 22(9):pp.1322-4; PMID:22613951; http://doi.org/ 10.1038/cr.2012.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA Directs mRNA Cleavage of the Transcription Factor NAC1 to Downregulate Auxin Signals for Arabidopsis Lateral Root Development. Plant Cell Online 2005; 17(5):pp.1376-86; PMID:15829603; http://doi.org/18073770 10.1105/tpc.105.030841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008; 9(1), pp.22-32; PMID:18073770; http://doi.org/ 10.1038/nrm2321 [DOI] [PubMed] [Google Scholar]
- 23.Kamiuchi Y, Yamamoto K, Furutani M, Tasaka M, Aida M. The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Frontiers Plant Sci 2014; 5:165; http://doi.org/ 10.3389/fpls.2014.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2013; 42:D68-73; PMID:24275495; http://doi.org/ 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D. A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 2006; 133(16):3223-30; PMID:16854978; http://doi.org/ 10.1242/dev.02508 [DOI] [PubMed] [Google Scholar]
- 26.Landrein B, Kiss A, Sassi M, Chauvet A, Das P, Cortizo M, Laufs P, Takeda S, Aida M, Traas J, et al.. Mechanical stress contributes to the expression of the STM homeobox gene in Arabidopsis shoot meristems. Elife 2015; 4:e07811 In Press; PMID:26623515; http://doi.org/ 10.7554/eLife.07811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 2004; 131(17):pp.4311-22; PMID:15294871; http://doi.org/ 10.1242/dev.01320 [DOI] [PubMed] [Google Scholar]
- 28.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA Targets Directed by a Class of Arabidopsis miRNA. Science 2002; 297(5589):2053-6; PMID:12242443 [DOI] [PubMed] [Google Scholar]
- 29.Long JA, Moan EI, Medford JI, Barton MK. A member of the knotted class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996; 379(6560):66-9; PMID:8538741; http://doi.org/ 10.1038/379066a0 [DOI] [PubMed] [Google Scholar]
- 30.Mallory A, Vaucheret H. Form, Function, and Regulation of ARGONAUTE Proteins. Plant Cell 2010; 22(12):3879-89; PMID:21183704; http://dx.doi.org/ 10.1105/tpc.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 2004; 23(16):3356-64; PMID:15282547; http://dx.doi.org/ 10.1038/sj.emboj.7600340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirabet V, Das P, Boudaoud A, Hamant O. The role of mechanical forces in plant morphogenesis. Annual Rev Plant Biol 2011; 62:365-85; http://dx.doi.org/ 10.1146/annurev-arplant-042110-103852 [DOI] [PubMed] [Google Scholar]
- 33.Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant cell 2006; 18(11):2929-45; PMID:17098808; http://dx.doi.org/ 10.1105/tpc.106.045617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen AN. Ernst HA, Leggio LL, Skriver K.. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 2005; 10(2):79-87; PMID:15708345; http://dx.doi.org/ 10.1016/j.tplants.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 35.Peaucelle A, Morin H, Traas J, Laufs P. Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 2007; 134(6), pp.1045-50; PMID:17251269; http://dx.doi.org/ 10.1242/dev.02774 [DOI] [PubMed] [Google Scholar]
- 36.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 2006; 20(24):3407-25; PMID:17182867; http://dx.doi.org/ 10.1101/gad.1476406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K. Interplay of miR164, Cup-Shaped Cotyledon genes and lateral suppressor controls axillary meristem formation in Arabidopsis thaliana. Plant J 2008; 55(1):65-76; PMID:18346190; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03483.x [DOI] [PubMed] [Google Scholar]
- 38.Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nat Cell Biol 2003; 426(6964), pp.255-60; PMID:14628043; http://doi.org/12101121 10.1038/nature02081 [DOI] [PubMed] [Google Scholar]
- 39.Reinhart BJ. MicroRNAs in plants. Genes Development 2002; 16(13):1616-26; PMID:12101121; http://dx.doi.org/ 10.1101/gad.1004402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of Plant MicroRNA Targets. Cell 2002; PMID:12202040; http://dx.doi.org/ 10.1016/S0092-8674(02)00863-2 [DOI] [PubMed] [Google Scholar]
- 41.Scanlon MJ. The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol 2003; 133(2), pp.597-605; PMID:14500790; http://dx.doi.org/ 10.1104/pp.103.026880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific Effects of MicroRNAs on the Plant Transcriptome. Dev Cell 2005; 8(4), pp.517-27; PMID:15809034; http://dx.doi.org/ 10.1016/j.devcel.2005.01.018 [DOI] [PubMed] [Google Scholar]
- 43.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 2007; 134(6):1051-60; PMID:17287247; http://dx.doi.org/ 10.1242/dev.02817 [DOI] [PubMed] [Google Scholar]
- 44.Smyth DR, Bowman JL, Meyerowitz EM. Early Flower Development in Arabidopsis. Plant Cell 1990; 2(8):755-67; PMID:2152125; http://dx.doi.org/ 10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spinelli SV, Martin AP, Viola IL, Gonzalez DH, Palatnik JF. A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol 2011; 156(4):1894-904; PMID:21685178; http://doi.org/ 10.1104/pp.111.177709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian C, Zhang X, He J, Yu H, Wang Y, Shi B, Han Y, Wang G, Feng X, Zhang C, et al.. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol Sys Biol 2014; 10(10), pp.755-5; http://dx.doi.org/ 10.15252/msb.20145470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uyttewaal M, Burian A, Alim K, Landrein B, Borowska-Wykręt D, Dedieu A, Peaucelle A, Ludynia M, Traas J, Boudaoud A, et al.. Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 2012; 149:439-51; PMID:22500806; http://dx.doi.org/ 10.1016/j.cell.2012.02.048 [DOI] [PubMed] [Google Scholar]
- 48.Veit B. Hormone mediated regulation of the shoot apical meristem. Plant Mol Biol 2008; 69:397-408; PMID:18797999; http://dx.doi.org/ 10.1007/s11103-008-9396-3 [DOI] [PubMed] [Google Scholar]
- 49.Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 2003; 15(7):1563-77; PMID:12837947; http://dx.doi.org/ 10.1105/tpc.012203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Kohlen W, Rossmann S, Vernoux T, Theres K. Auxin Depletion from the Leaf Axil Conditions Competence for Axillary Meristem Formation in Arabidopsis and Tomato. Plant Cell 2014; 26(5):2068-79; PMID: 24850851; http://doi.org/8252622 10.1105/tpc.114.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wightman B, Ha I, Ruvkun G. Posttranscriptional Regulation of the Heterochronic Gene Lin-14 by Lin-4 Mediates Temporal Pattern-Formation in C-Elegans. Cell 1993; 75(5):855-62; PMID:8252622; http://dx.doi.org/ 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- 52.Yadav RK, Tavakkoli M, Xie M, Girke T, Reddy GV. A high-resolution gene expression map of the Arabidopsis shoot meristem stem cell niche. Development 2014; 141(13):2735-44; PMID:24961803; http://doi.org/ 10.1242/dev.106104 [DOI] [PubMed] [Google Scholar]
- 53.Yruela I. Plant development regulation: Overview and perspectives. J Plant Physiol 2015; 182:62-78; PMID:26056993; http://dx.doi.org/ 10.1016/j.jplph.2015.05.006 [DOI] [PubMed] [Google Scholar]
- 54.Zadnikova P, Simon R. How boundaries control plant development. Curr Opin Plant Biol 2014; 17:116-25; PMID:24507503; http://dx.doi.org/ 10.1016/j.pbi.2013.11.013 [DOI] [PubMed] [Google Scholar]
- 55.Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X. Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to Regulate Shoot Apical Meristem Development. Cell 2011; 145(2):242-56; PMID: 21496644; http://doi.org/ 10.1016/j.cell.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]