ABSTRACT
Nitrate and ammonium are the 2 most common forms of inorganic nitrogen available to plants in the soil. We previously identified a group of class III glutaredoxin genes whose expression is strongly upregulated by nitrate, but not ammonium, in Arabidopsis thaliana shoots and roots. A reverse genetics approach was used to functionally characterize a subset of these nitrate-regulated glutaredoxins, and we found that the AtGRXS3,4,5, and 8 genes function as negative regulators of primary root growth. AtGRXS3/4/5/8 are arranged in a tandem array on Arabidopsis chromosome 4, and these genes show very high levels of sequence similarity. Interestingly, there is one additional glutaredoxin, AtGRXS7, in this same gene cluster, but this gene was not identified as nitrate-responsive in our previous studies. We show here that AtGRXS7 is upregulated by nitrate and shows strong co-expression with the other glutaredoxins in this gene cluster. Further, AtGRXS7 was effectively silenced by the RNAi construct used to target AtGRXS3/4/5/8 for previous functional analyses. Overall, it appears that the 5 genes in the AtGRX3/4/5/7/8 cluster share virtually identical sequences, regulatory patterns, and functions, collectively acting to regulate primary root growth in response to soil nitrate.
KEYWORDS: Arabidopsis thaliana, AtGRXS7, glutaredoxins, nitrate, plant development, primary root growth
Abbreviations
- RNAi
RNA interference
- RNA-seq
RNA sequencing
Nitrogen is a critical macronutrient for plant growth, and lack of nitrogen can be a major limitation to agricultural productivity. Most plants are dependent on inorganic sources of nitrogen in the soil, typically in the form of nitrate (NO3−) or ammonium (NH4+). Although both nitrate and ammonium can be efficiently assimilated by most plants, these 2 nitrogen sources can have differential effects on plant physiology and development.1,2 Accordingly, plant transcriptomes and proteomes are uniquely modified under different nitrogen sources.2,3
In our recent study of global transcriptional responses to nitrate and ammonium, we identified a group of 7 class III glutaredoxins which were specifically upregulated by nitrate, but not ammonium, in Arabidopsis thaliana shoots and roots.4 We functionally analyzed a subset of these nitrate-regulated glutaredoxins: AtGRXS3, AtGRXS4, AtGRXS5, and AtGRXS8. RNA silencing of these 4 glutaredoxins revealed that they act as negative regulators of primary root growth, potentially acting to regulate root system architecture in response to nitrate availability in the soil.4 Further study demonstrated that the regulation of AtGRXS3/4/5/8 is dependent upon cytokinin signaling that is activated by nitrate.4
AtGRXS3/4/5/8 are found in a tandem array on Arabidopsis chromosome 4. One additional glutaredoxin is also found in this cluster: AtGRXS7 (Fig. 1A). All of the glutaredoxins in this cluster, including AtGRXS7, show very high levels of nucleotide and amino acid sequence identity (Fig. 1B). In our initial studies, AtGRXS7 was not identified as a nitrate-regulated gene. We now know that this was because the AtGRXS7 sequence was not included on the ATH1 Affymetrix Gene Chip that was used for transcriptional profiling in these studies. Due to the absence of AtGRXS7 on the ATH1 Gene Chip, substantially less is known about the regulation of this gene compared to other glutaredoxins.5 However, recent RNA-seq analyses suggest that all of the genes in the glutaredoxin cluster, including AtGRXS7, form a co-expression network (Table 1).6 Real time RT-PCR analysis demonstrated that AtGRXS7 is strongly upregulated by nitrate (Fig. 2A). Further, nitrate-mediated induction of AtGRXS7 gene expression is dependent on the cytokinin response regulators ARR1, 10, and 12 (Fig. 2A). This mirrors the expression profiles of AtGRXS3, 4, 5 and 8, the other glutaredoxin genes in the cluster.4 Thus, it appears that the entire AtGRXS3/4/5/7/8 cluster is coordinately regulated by nitrate and cytokinin.
Figure 1.

The AtGRX3/4/5/7/8 gene cluster. (A) Location of AtGRXS3, AtGRXS4, AtGRXS5, AtGRXS7 and AtGRXS8 on chromosome 4 of Arabidopsis thaliana (image adapted from TAIR 10). (B) Nucleotide and protein sequence identity of AtGRX3/4/5/7/8.
Table 1.
Genes coexpressed with the glutaredoxin AtGRXS3 (Adapted from ATTED-II [ver. 8.0] – Aoki et al.6).
| Microarray data |
RNAseq data |
||||
|---|---|---|---|---|---|
| Correlation coefficient | Gene | Function | Correlation coefficient | Locus | Function |
| 0.85 | AtGRXS5 | Thioredoxin superfamily protein | 0.74 | AtGRXS7 | Thioredoxin superfamily protein |
| 0.75 | AtGRXS4 | Thioredoxin superfamily protein | 0.72 | AtGRXS8 | Thioredoxin superfamily protein |
| 0.71 | AtGRXS8 | Thioredoxin superfamily protein | 0.71 | AtGRXS4 | Thioredoxin superfamily protein |
| 0.55 | At5g18600 | Thioredoxin superfamily protein | 0.51 | At2g30540 | Thioredoxin superfamily protein |
| 0.47 | At3g62930 | Thioredoxin superfamily protein | 0.56 | At3g62950 | Thioredoxin superfamily protein |
| 0.37 | At2g15680 | Calcium-binding EF-hand family protein | 0.44 | At1g03020 | Thioredoxin superfamily protein |
Figure 2.
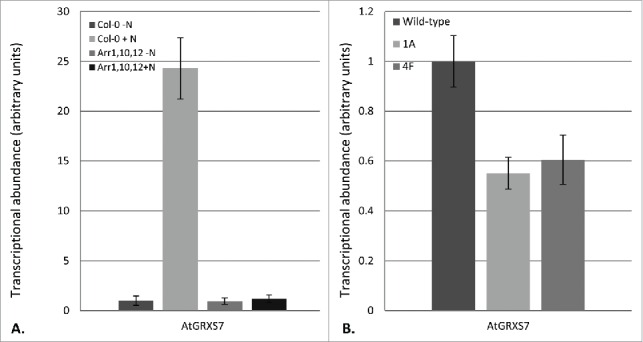
AtGRXS7 gene expression in response to nitrate and in glutaredoxin-silenced plant lines. (A) Nitrate induction of AtGRXS7 gene expression was quantified by real-time RT-PCR in hydroponically-grown wild-type (Col-0) and ARR1,10,12 mutant plants which were nitrogen-starved for 26 hr and then resupplied with 1 mM KNO3 (+N) or 0.5 mM K2SO4 (−N) for 8 h. Data points represent means ± SEM (n = 3). Transcript abundance data for wild-type plants supplied with K2SO4 was arbitrarily assigned a value of 1.0. (B) Relative transcript abundance of AtGRXS7 in wild-type and glutaredoxin-silenced lines (1A, 4F). Data points represent means ± SEM (n = 6). Transcript abundance data for wild-type plants was arbitrarily assigned a value of 1.0
We previously analyzed the functional role of nitrate-regulated glutaredoxins using transgenic lines expressing an RNAi vector based on the AtGRXS3 sequence. We showed that this vector effectively silenced AtGRXS3 as well as AtGRXS4, 5, and 8 due to their high level of sequence similarity.4 Given that AtGRXS7 is >90 % identical to AtGRXS3, it is unsurprising that the expression of AtGRXS7 was also reduced in the glutaredoxin-silenced transgenic lines (Fig. 2B). Collectively, these results suggest that AtGRXS3, 4, 5, 7, and 8 are virtually identical in sequence, chromosomal location, and gene expression. This gene cluster is likely the result of multiple relatively recent gene duplication events, and the individual genes are almost certainly functionally redundant. Thus, we consider this cluster of genes to act as a single genetic locus which plays a critical role in modulating root system architecture in response to nitrate availability and distribution in the soil.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Walch-Liu P, Filleur S, Gan YB, Forde BG. Signaling mechanisms integrating root and shoot responses to changes in the nitrogen supply. Photosynth Res 2005; 83:239-50; PMID:16143854; http://dx.doi.org/ 10.1007/s11120-004-2080-9 [DOI] [PubMed] [Google Scholar]
- 2.Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA. Distinct signaling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ 2010; 33:1486-501; PMID:20444219; http://dx.doi.org/10.1111/j.1365-3040.2010.02158.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moller AL, Pedas P, Andersen B, Svensson B, Schjoerring JK, Finnie C. Responses of barley root and shoot proteomes to long-term nitrogen deficiency, short-term nitrogen starvation and ammonium. Plant Cell Environ 2011; 34:2024-37; PMID:21736591; http://dx.doi.org/ 10.1111/j.1365-3040.2011.02396.x [DOI] [PubMed] [Google Scholar]
- 4.Patterson K, Walters LA, Cooper AM, Olvera JG, Rosas MA, Rasmusson AG, Escobar MA. Nitrate-regulated glutaredoxins control Arabidopsis thaliana primary root growth. Plant Physiol 2016; 170:989-99; PMID:26662603; http://dx.doi.org/ 10.1104/pp.15.01776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belin C, Bashandy T, Cela J, Delorme-Hinoux V, Riondet C, Reichheld JP. A comprehensive study of thiol reduction gene expression under stress conditions in Arabidopsis thaliana. Plant Cell Environ 2015; 38:299-314; PMID:24428628; http://dx.doi.org/ 10.1111/pce.12276 [DOI] [PubMed] [Google Scholar]
- 6.Aoki Y, Okamura Y, Tadaka S, Kinoshita K, Obayashi T.ATTED-II in 2016: a plant coexpression database towards lineage-specific coexpression.Plant Cell Physiol 2016; 57:e5; PMID:26546318; http://dx.doi.org/ 10.1093/pcp/pcv165 [DOI] [PMC free article] [PubMed] [Google Scholar]


